Abstract
Starch-branching enzyme (SBE), a glucosyl transferase, is required for the highly regular pattern of α-1,6 bonds in the amylopectin component of starch. In the absence of SBEIIa, as shown previously in the sbe2a mutant of maize (Zea mays), leaf starch has drastically reduced branching and the leaves exhibit a severe senescence-like phenotype. Detailed characterization of the maize sbe2a mutant revealed that SBEIIa is the primary active branching enzyme in the leaf and that in its absence plant growth is affected. Both seedling and mature sbe2a mutant leaves do not properly degrade starch during the night, resulting in hyperaccumulation. In mature sbe2a leaves, starch hyperaccumulation is greatest in visibly senescing regions but also observed in green tissue and is correlated to a drastic reduction in photosynthesis within the leaf. Starch granules from sbe2a leaves observed via scanning electron microscopy and transmission electron microscopy analyses are larger, irregular, and amorphous as compared with the highly regular, discoid starch granules observed in wild-type leaves. This appears to trigger premature senescence, as shown by an increased expression of genes encoding proteins known to be involved in senescence and programmed cell death processes. Together, these results indicate that SBEIIa is required for the proper diurnal cycling of transitory starch within the leaf and suggest that SBEIIa is necessary in producing an amylopectin structure amenable to degradation by starch metabolism enzymes.
In contrast to starch storage organs (e.g. maize [Zea mays] endosperm), which function as long-term storage sinks for starch, leaves undergo starch synthesis and degradation in accordance with the diurnal cycle. During the light phase, photosynthesis in maize leaves produces both Suc for export and starch, which is stored within bundle sheath chloroplasts. During the dark phase, or when photosynthetic rates are quite low, this transitory starch is mobilized for use in leaf metabolism as well as for export to nonphotosynthetic sink organs (for review, see Zeeman et al., 2007).
Both transitory and storage starches are composed of both the essentially linear polymer, amylose, and the branched polymer, amylopectin. Amylose contains α-1,4-linked Glc monomers, whereas amylopectin contains 5% α-1,6-linked Glc in addition to linear regions of α-1,4-linked Glc (for review, see Smith et al., 1997; Shannon et al., 1998). Amylopectin is the major constituent of starch, representing approximately 70% of maize storage starch (Swinkels, 1985) and equaling or exceeding 85% of transitory starch in rice (Oryza sativa; Taira et al., 1991), tobacco (Nicotiana tabacum; Matheson and Wheatley, 1962), pea (Pisum sativum; Tomlinson et al., 1997), and maize (Blauth et al., 2001) leaves.
The synthesis of amylopectin requires the action of starch-branching enzyme (SBE; EC 2.4.1.18), a glucosyl transferase that catalyzes the formation of the α-1,6 linkages in starch. During the branching process, first a maltooligosaccharide chain is cut from a growing glucan polymer by cleavage of an internal α-1,4 bond, which is followed by its transfer to the C-6 position of the same or an adjacent chain, forming an α-1,6 branch point on a linear chain. The addition of each branch adds a new nonreducing end, from which starch synthesis can continue. Therefore, the branching pattern in amylopectin not only determines the structure but can also influence the amount of starch synthesized. The characterization of glucan substrate specificities for purified SBE isoforms (Boyer and Preiss, 1978; Dang and Boyer, 1988; Guan and Preiss, 1993; Mu-Forster et al., 1996; Guan et al., 1997; Seo et al., 2002) and studies of maize mutants with altered SBE activity (Moore and Creech, 1972; Boyer and Preiss, 1981; Blauth et al., 2001, 2002; Yao et al., 2004; Li et al., 2007) clearly indicate that SBEs play a significant role in determining the fine structure of starch molecules.
Three SBE isoforms have been identified in the maize kernel: SBEI, SBEIIa, and SBEIIb. The class II (or A) enzymes (SBEIIa and SBEIIb) share similar biochemical properties, including similar Km values for amylopectin and a preference to branch amylopectin rather than amylose (Guan and Preiss, 1993), and they preferentially branch shorter chains (Takeda et al., 1993) than the class I (or B) enzyme, SBEI. In amylose-extender (ae) mutants, which lack functional SBEIIb enzyme, the kernel is viable but exhibits a slightly collapsed crown (Vineyard and Bear, 1952; Moore and Creech, 1972; Stinard et al., 1993) due to an approximately 20% reduction in endosperm starch quantity and a severe decrease in the amount of branching within amylopectin (Garwood et al., 1976). Whereas the ae mutant phenotype is observed only in the endosperm, the sbe2a mutant lacking functional SBEIIa appears to confer normal endosperm starch and a mutant phenotype is observed only in the leaf (Blauth et al., 2001). Transitory starch within the sbe2a mutant leaf exhibits a reduction in branching more extreme than in ae endosperm, and the leaves undergo premature and severe senescence (Blauth et al., 2001).
Unlike in endosperm, where the functions of the SBEs have been well characterized and increasingly understood, the role that SBEIIa plays in the synthesis and possibly in the diurnal cycling of transitory starch in the leaf is less clear. To further understand the importance of branching to transitory starch production and metabolism, we have characterized in detail maize leaves lacking functional SBEIIa (sbe2a mutants).
Our results demonstrate that SBEIIa is required in leaves for the formation of uniform starch granules that can be degraded during the dark phase of the diurnal cycle. In the absence of proper branching of amylopectin, irregular starch granules are formed that cannot be properly degraded and the accumulation of these granules within the chloroplast triggers senescence, a form of programmed cell death (PCD). Thus, in the absence of SBEIIa, the leaf sequesters large amounts of carbon as starch rather than fully mobilizing starch to provide carbon for metabolism and growth at night.
RESULTS
SBEIIa Is the Primary Starch-Branching Enzyme in the Leaf and Is Required for Plant Growth
Starch-branching activity in leaves has previously been attributed to the presence of both SBEIIa and SBEI, because fractionation of leaf extracts yields a peak of SBE activity in a similar position to that of SBEI from maize endosperm (Dang and Boyer, 1988). However, SBEI protein has not been detectable in western analysis of soluble leaf extracts (Blauth et al., 2001, 2002; Fig. 1). Whereas ae transcripts have not been detected in leaves via northern analyses (Stinard et al., 1993; Gao et al., 1996), both sbe1 (Blauth et al., 2002) and ae (Blauth et al., 2001) transcripts have been detected by reverse transcription (RT)-PCR. RT-PCR analysis of RNA from wild-type and sbe2a mutant leaves used in this study also shows expression of sbe1 but not ae transcripts (data not shown). The apparent presence of ae transcripts observed by Blauth et al. (2001) could be due to the amplification of genomic DNA present in the RNA samples, because the primers, designed within a single exon, would not distinguish between transcript and genomic templates.
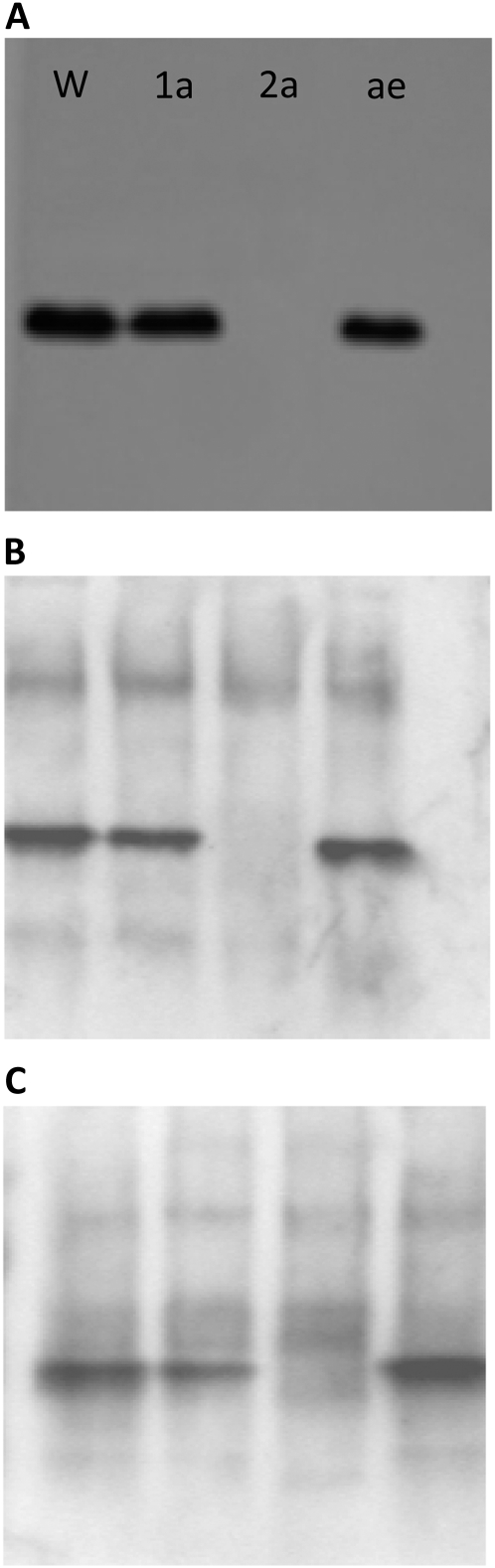
Figure 1.
SBEIIa is responsible for starch-branching activity in the leaf. Crude protein extracts from W64A (W), sbe1a (1a), sbe2a (2a), and ae (ae) mutants were electrophoresed on companion native PAGE gels and were either incubated with phosphorylase a and Glc-1-P and stained with iodine to visualize a glucan product at the site of branching activity (A) or western blotted and reacted with SBEIIa antibody (B) or SBEI antibody (C), which cross-reacts with SBEIIa and SBEIIb.
Crude protein extracts from wild-type, sbe1, sbe2a, and ae leaves were subjected to qualitative and semiquantitative starch-branching activity assays. SBE activity, as quantified by in vitro phosphorylase a stimulation assay of protein extracts from leaves isolated midway through the light phase, was approximately 7-fold higher in wild-type leaves as compared with sbe2a mutant leaves (5.24 ± 1.87 versus 0.70 ± 0.12 nmol inorganic phosphate μg−1 crude extract h−1; n = 3). When crude soluble protein extracts were separated on native PAGE gels, a single band of branching activity (Fig. 1A) was observed in wild-type W64A, sbe1, and ae extracts but not in extracts from the sbe2a mutant. This activity precisely comigrated with a protein reactive to an anti-SBEIIa antibody (Fig. 1B). In addition, SBEI was not detectable via western analysis with a nonspecific SBE antibody that cross-reacts to all three isoforms of maize SBE (Fig. 1C). Since a low level of SBE activity is detectable in leaves of the sbe2a mutant, and Sbe1 transcript was also observed by RT-PCR, we cannot rule out the possibility that low levels of SBEI protein might be expressed, even though we cannot detect the protein on western blots. Regardless, our data demonstrate that at a minimum, SBEIIa is the primary starch-branching enzyme detectable by multiple methods in the leaf and is responsible for most soluble starch-branching activity observed in leaves.
Progeny from selfed Sbe1/sbe1; Sbe2a/sbe2a plants were analyzed to assess whether the lack of SBEIIa affects aspects of plant growth. Genotyped plants were shown to segregate 9:3:3:1 Sbe1; Sbe2a:sbe1; Sbe2a:Sbe1; sbe2a:sbe1; sbe2a, as expected (data not shown), demonstrating that there are no adverse effects on allele transmission in these sbe mutant combinations. Growth of sbe2a homozygous mutants was reduced, as evidenced by shorter height (Fig. 2A) and fewer leaves (Fig. 2B) as well as by an observed reduction in ear size and seed set (data not shown) in sbe2a as compared with Sbe2a genotypes. Consistent with the observation that only SBEIIa accumulates to significant amounts in leaf tissue, the sbe1 mutation had no effect on plant height (Fig. 2) as well as no observed reduction in ear size or seed set (data not shown).
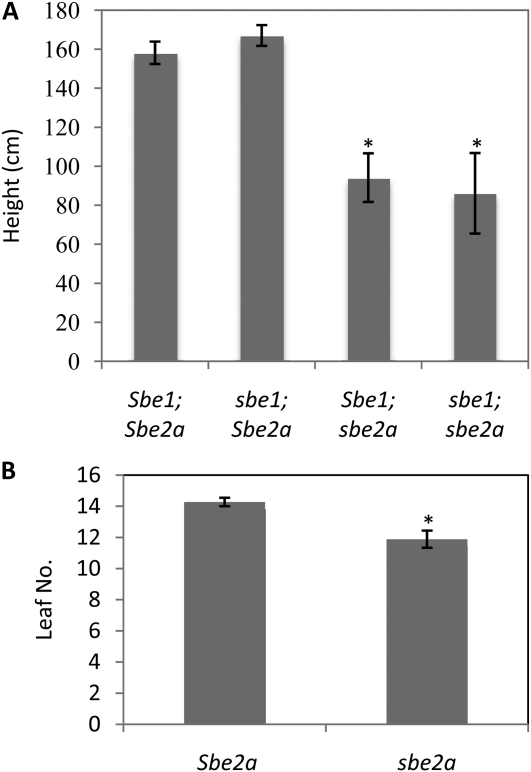
Figure 2.
sbe2a plants have reduced height and leaf number. A, Plant height was measured after anthesis in Sbe2a and sbe2a plants grown in 2006 (n = 51) and 2007 (n = 162). Because statistical differences were not seen between years for each genotype, the average over 2 years is shown. Asterisks denote significant differences in height in the sbe2a mutant as compared with the Sbe2a classes (P < 10−3). B, Leaf number was counted at maturity for Sbe2a (n = 30) and sbe2a (n = 16) plants. The asterisk denotes that the value significantly differs from the wild type (P < 0.01). Error bars represent se.
Starch Degradation in the Dark Is Reduced in Plants Lacking SBEIIa
To understand how starch accumulation is affected in the sbe2a mutants, leaves from 3-week-old seedlings harvested at the end of both the dark and light phases were harvested and either decolorized and stained with iodine to visualize starch content (Fig. 3, A–D) or used for starch extraction and quantification (Fig. 3E). In Sbe2a leaves, starch is present at the end of the light phase (Fig. 3A) and is reduced by 80% at the end of the dark phase (Fig. 3, C and E; P < 0.001). The amount of starch at the end of the light phase did not significantly differ between Sbe2a and sbe2a leaves (Fig. 3E; P = 0.149). At the end of the dark phase, however, sbe2a mutant leaves, which have not started to visibly senesce, still contained starch (Fig. 3D). Only approximately 40% of starch was degraded during the dark phase (Fig. 3E), which significantly differs from the amount of starch degraded during the dark phase in wild-type plants (P < 0.05). Suc levels were unaffected in the sbe2a mutant (data not shown). Both the qualitative and quantitative assays suggest that starch in sbe2a leaves is not being degraded at the same rate as in Sbe2a leaves.
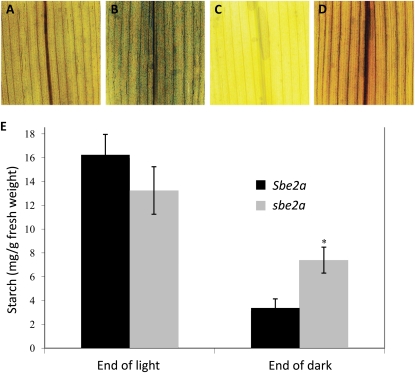
Figure 3.
sbe2a leaves have reduced starch degradation during the dark phase. A to D, Leaves from Sbe2a/Sbe2a (A and C) and sbe2a/sbe2a (B and D) 3-week-old seedlings were harvested at the end of the 16-h-light (A and B) and 8-h-dark (C and D) phases, decolorized in boiling ethanol, and stained with iodine to visualize starch levels. E, Starch in replicate leaves was quantified at the end of light and dark phases (n = 4). Starch was partially purified and quantified. For sbe2a/sbe2a seedlings, only nonsenescing leaves were sampled. The asterisk denotes that the value significantly differs from Sbe2a at the end of the dark phase (P < 0.05). Error bars represent se.
Starch Content and Photosynthetic Rates Are Inversely Related in Senescing Regions of sbe2a Leaves
Differential starch accumulation between visibly senescing and “green” portions of leaves from sbe2a were compared with leaves of wild-type Sbe2a plants. Leaves harvested in early morning from wild-type Sbe2a and mutant sbe2a 2.5-month-old field-grown plants (Fig. 4, A and C) were decolorized and stained with iodine to visualize starch (Fig. 4, B and D). Whereas little starch was present in Sbe2a leaves at the beginning of the light phase, the entire sbe2a leaf stained blue, indicating the presence of starch. Interestingly, regions that were visibly senescing stained more intensely, suggesting a greater accumulation of starch in senescing regions. Starch extracted from both genotypes was quantified, and approximately 6-fold more starch was observed in visibly senescing portions of sbe2a leaves as compared with Sbe2a leaves (Fig. 4E). Consistent with increased starch content in 3-week-old seedling leaves that had not yet begun to visibly senesce, green portions of 2.5-month-old sbe2a leaves also exhibited increased starch content, 4-fold higher than in the wild type.
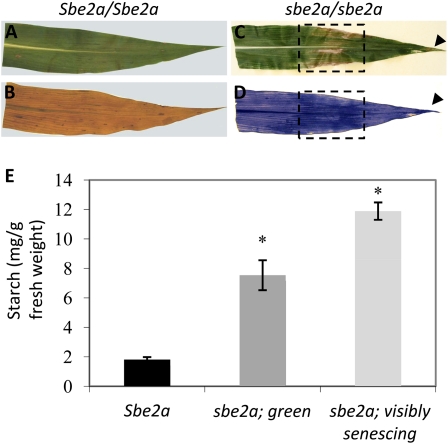
Figure 4.
Starch accumulates within visibly senescing regions of sbe2a leaves. A to D, Wild-type Sbe2a (A) and homozygous sbe2a mutant (C) leaves exhibiting visibly senescing regions (hatched boxes and arrowheads) were harvested at 8 am, decolorized in boiling ethanol, and stained with iodine to visualize increased starch levels (B and D). E, Starch was extracted and quantified from leaves similar to A. Because senescing regions contain less moisture than nonsenescing leaves, starch values were normalized to the dry weights of the samples. Green and senescing leaf samples from sbe2a plants had significantly higher starch contents as compared with the wild type (denoted by the asterisk; P < 0.001). Error bars represent se of six biological replicates.
Photosynthetic rates were measured in Sbe2a leaves and in both green and visibly senescing regions of sbe2a leaves (Fig. 4E) from 2.5-month-old plants. Visibly senescing leaves exhibited no photosynthesis, whereas green leaves from sbe2a plants exhibited a photosynthetic rate 18% lower than wild-type leaves (Table I). For chlorophyll content, there is a 13% reduction in green sections and a 50% reduction in senescing portions of sbe2a leaves (Table II). Together, these data suggest that the accumulation of starch or associated metabolic changes within the sbe2a leaf down-regulates photosynthesis and that in the chlorotic, visibly senescing regions photosynthesis is essentially eliminated. This suggests that starch hyperaccumulates prior to and may be the cause of premature senescence in sbe2a leaves.
Table I. Photosynthetic rate and stomatal conductance in wild-type and sbe2a leaves.
Data represent means from 18 replicates ± se.
| Genotype | Leaf Tissue | Photosynthetic Rate | Percentage of the Wild Type | Stomatal Conductance | Percentage of the Wild Type |
| μmol CO2 fixed m−2 s−1 | μmol | ||||
| Sbe2a/Sbe2a | Green | 30.8 ± 0.3 | 100 | 0.238 ± 0.007 | 100 |
| sbe2a/sbe2a | Green | 25.3 ± 0.8a | 82.1 | 0.183 ± 0.007a | 76.9 |
| Senescing | −3.4 ± 0.6a | −11.0 | 0.032 ± 0.004a | 13.5 |
Amount significantly differs from the wild type (P < 0.0001; ANOVA).
Table II. Chlorophyll content.
Data represent means from 18 replicates ± se.
| Genotype | Leaf Tissue | Chlorophyll Contenta | Percentage of the Wild Type |
| Sbe2a/Sbe2a | Green | 52.4 ± 0.5 | 100 |
| sbe2a/ sbe2a | Green | 45.7 ± 0.6b | 87.2 |
| Senescing | 24.3 ± 0.9b | 46.4 |
Values represent relative units that are linearly correlated to extractable chlorophyll.
Amount significantly differs from the wild type (P < 0.0001; ANOVA).
Starch Granules in sbe2a Mutants Are Abnormal and Irregular
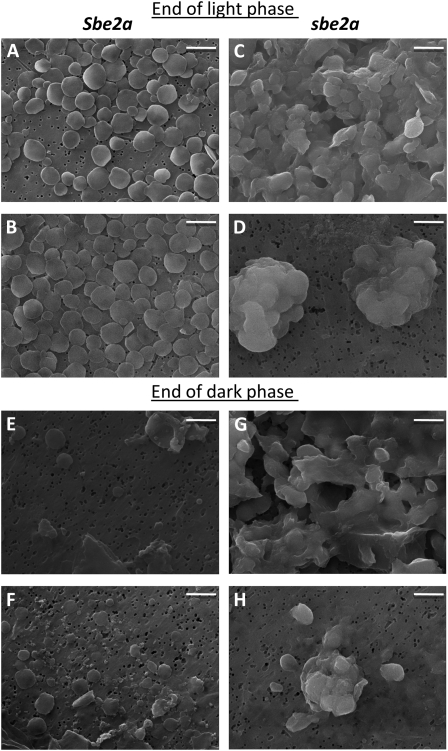
We hypothesize that the deficiency in SBEIIa in mutant leaves results in an alteration of granule and/or starch molecular structure, which could impact the enzymatic degradation of starch. Starch purified from wild-type and sbe2a mutant (not visibly senescing) leaves was harvested at the end of the light and dark phases, and granules were examined via scanning electron microscopy (SEM). The majority of Sbe2a granules at the end of the light phase were uniform discs of similar size (1.27 ± 0.01 μm; Fig. 5, A and B). At the end of the dark phase, discoid granules (Fig. 5, E and F) were much fewer in number (per gram fresh weight) and had a smaller diameter (1.14 ± 0.05 μm), which is significantly different from granules at the end of the light phase (P = 0.01). In addition, irregularly shaped granules and small particles were also recovered (Fig. 5, E and F); very small granules may not have been recoverable by the method used to extract the starch. Analysis of Sbe2a starch by flow cytometry, which allows for relative comparisons of granule size and complexity, suggested that Sbe2a granules are uniform in size and smaller at the end of the dark as compared with the light phase (data not shown). These observations are consistent with the fact that starch granules remaining at the end of the dark phase might be smaller or irregularly shaped due to incomplete degradation.
Figure 5.
Starch granules from sbe2a leaves are abnormal and irregular. SEM of starch granules is shown at the end of the light (A–D) and dark (E–H) phases of the diurnal cycle. Starch was extracted from Sbe2a wild-type (A, B, E, and F) and sbe2a mutant (C, D, G, and H) leaves harvested from 1-month-old plants at the end of the light and dark phases. Bars = 2 μm.
The structure and complexity of sbe2a starch at the end of both the light and dark phases differ greatly from the wild type. Whereas Sbe2a starch granules at the end of the light phase are highly regular in shape and size (Fig. 5, A and B), only some of the starch granules isolated from sbe2a leaves are discoid (Fig. 5, C, G, and H). Predominantly, sbe2a granules are abnormal and appear to be lobular (Fig. 5, C, D, G, and H) and fused (Fig. 5, C and G). Consistent with SEM, flow cytometry of sbe2a granules yielded a broader particle size distribution, with a portion of sbe2a being larger than Sbe2a granules (data not shown). Although precise changes in starch structure cannot be inferred from flow cytometry or microscopy, the data clearly demonstrate significant differences in sbe2a versus wild-type starch granular structure. Starch from sbe2a mutants, which lack proper branching, lose granular uniformity and fail to be properly degraded during the dark phase. This suggests that in the sbe2a mutant, the leaf becomes an organ of abnormal starch accumulation.
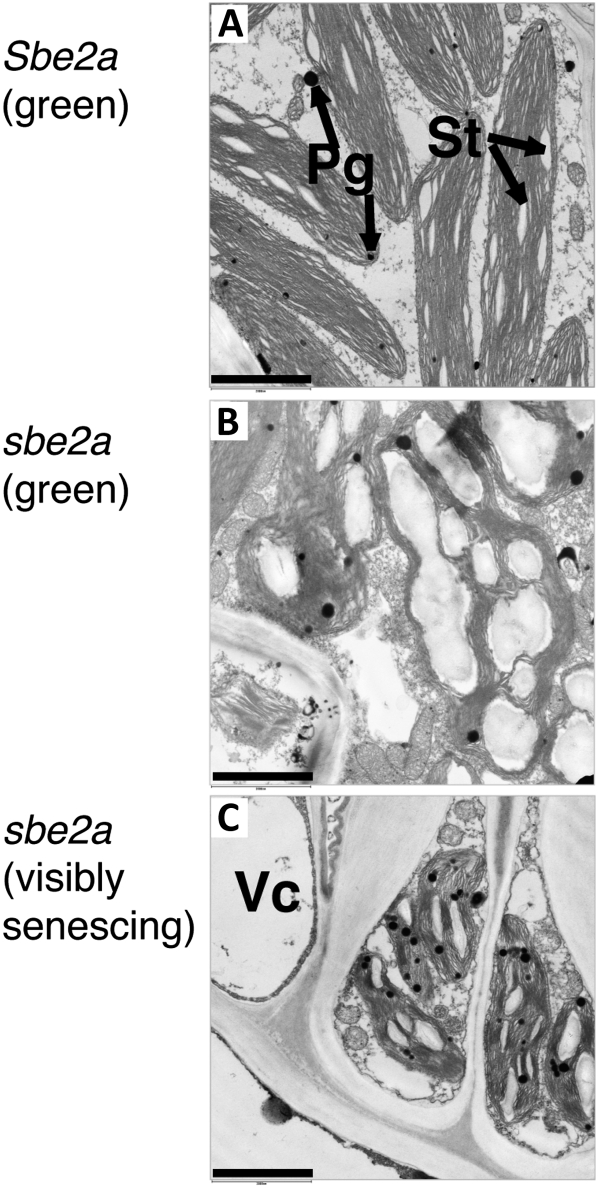
sbe2a Chloroplasts Are Distended and Contain Large Numbers of Irregular Starch Granules
To ascertain how the abnormal starch granules in sbe2a leaves affect cellular ultrastructure, green and visibly senescing cells harvested from sbe2a mutants during the middle of the light phase were examined using transmission electron microscopy (TEM). In mature wild-type leaves, bundle sheath chloroplasts are oblong and contain few grana and several crystalline starch granules that are relatively uniform in shape and size (Fig. 6A; Kirchanski, 1975). In contrast, bundle sheath chloroplasts in green portions of sbe2a mutant leaves are misshapen and contain larger, irregular, and sometimes fused granules (Fig. 6B) similar to sbe2a starch granules analyzed by SEM (Fig. 5, C, G, and H). Cellular ultrastructure is further affected in visibly senescing portions of sbe2a leaves (Fig. 6C). In addition to the persistence of starch granules first identified in green tissue of sbe2a leaves, cells in visibly senescing parts of the leaf exhibit a hallmark of senescence, an increased number of plastoglobules (i.e. lipoprotein bodies) necessary for the mobilization of lipids (for review, see Bréhélin and Kessler, 2008). In addition, chloroplasts are compacted, membranes appear larger, and starch granules appear to be present both within and sometimes outside of chloroplasts. These data suggest that the persistence of aberrant starch granules in sbe2a chloroplasts triggers changes in cellular ultrastructure and may lead to programmed senescence or PCD.
Figure 6.
Chloroplasts within sbe2a leaves are misshapen and contain large, irregular starch granules. TEM micrographs of chloroplasts are shown within bundle sheath cells from Sbe2a wild-type leaves (A), green tissue from sbe2a mutant leaves (B) and visibly senescing tissue from sbe2a mutant leaves (C). Starch granules (St), plastoglobules (Pg), and vacuoles (Vc) are indicated with arrows. Bars = 2 μm.
The Accumulation of Aberrant Starch Granules in sbe2a Leaves Directly or Indirectly Induces Genes Involved in PCD
The presence of abnormally large starch granules (Fig. 6) or associated metabolic changes could potentially trigger the expression of genes involved in starch degradation or other genes responding to the abnormal physiological and physical conditions.
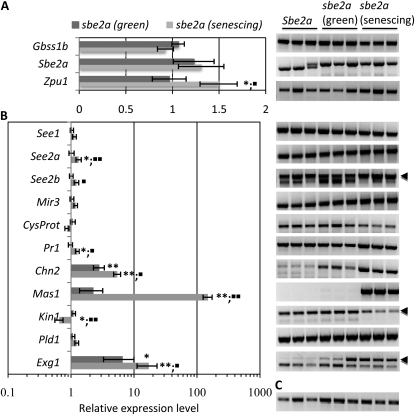
By semiquantitative RT-PCR analysis, the relative expression levels of Sbe2a and the gene encoding granule-bound starch synthase (GBSS), responsible for amylose synthesis, are unaffected in green and visibly senescing leaves of sbe2a plants (Fig. 7A). However, transcript levels of Zea mays pullulanase-type debranching enzyme (Zpu1), the pullulanase-type debranching enzyme involved in amylopectin degradation (Dinges et al., 2003), increase by approximately 50% in visibly senescing sbe2a leaves (Fig. 7A). Increased expression of this debranching enzyme in a plant in which starch lacks significant branching is perhaps surprising, but expression could be induced by metabolic changes stemming from altered starch turnover.
Figure 7.
A number of genes involved in starch degradation and PCD are differentially regulated in sbe2a leaves. A and B, Total RNA isolated from green leaves of Sbe2a/Sbe2a and green or visibly senescing leaves of sbe2a/sbe2a plants grown for approximately 2.5 months in the field were examined for the expression of starch synthesis and metabolism genes (A) and genes involved in senescence/PCD (B). C, Semiquantitative RT-PCR results were standardized to tubulin. Results are expressed as relative expression as compared with Sbe2a leaves. Aberrant sbe2a transcripts are found in sbe2a leaves (see “Materials and Methods”). Significantly different expression as compared with the wild-type is denoted by asterisks (* P < 0.05 or ** P < 0.01) and as compared with green sbe2a leaves is denoted by one black square (P < 0.05) or two black squares (P < 0.01). Error bars indicate se of three biological replicates. For genes that yielded multiple bands via RT-PCR, arrowheads denote the product that was of appropriate size and was used for quantification. Gbss1b, granule-bound starch synthase1b; Sbe2a, starch-branching enzyme2a; Zpu1, pullulanase-type starch-debranching enzyme1; See1, senescence-enhanced1; See2a, senescence-enhanced2a; See2b, senescence-enhanced2b; Mir3, maize insect resistance3; CysProt, Cys protease1; Pr1, pathogenesis-related1; Chn2, chitinase2; Mas1, malate synthase1; Kin1, knotted1-induced1; Pld1, phospholipase D1; Exg1, exoglucanase1.
To ascertain whether PCD/senescence is prematurely triggered in leaves lacking SBEIIa, transcript levels of several genes known to be up-regulated during this process (Buchanan-Wollaston et al., 2003; Gepstein et al., 2003; Table III) were also assessed (Fig. 7B). Genes involved in PCD-related processes such as protein degradation (i.e. senescence-enhanced2a [See2a] and See2b), defense (i.e. pathogenesis-related1 [Pr-1] and chitinase2 [Chn2]), lipid mobilization (i.e. malate synthase1 [Mas1] and phospholipase D1 [Pld1]), and cell wall degradation (i.e. exoglucanase1 [Exg1]) were up-regulated in visibly senescing leaf tissue from sbe2a mutants. In the case of Chn2, significantly enhanced gene expression was observed in sbe2a mutant leaf tissue that was green and not visibly showing signs of senescence. Remarkably, the Exg1 gene was induced approximately 10-fold over the wild type and the Mas1 gene was induced over 100-fold in the senescing tissue. Together, these data suggest that the accumulation of aberrant starch caused by the lack of SBE activity and the concomitant negative impact on starch turnover triggers a senescence-type PCD response in the sbe2a mutant leaf.
Table III. Genes included in transcriptome analysis.
| Gene | Protein | Protein Class | GenBank Accession No. | Forward Primer | Sequencea | Reverse Primer | Sequencea |
| See2a | Legumain-like protease | Protease | AJ131718 | See2A-Fb | GTCCTGGTGTCCTTGGAATG | See2A-R | TTGAGAACGATGGCAGTGAG |
| See2b | Legumain-like Cys protease | Protease | AJ131719 | See2b-Fb | AGCAGCAATACAACTTGGTCA | See2b-R | ACCTTCGACACTGCTTCAGG |
| See1 | Cys protease | Protease | X99936 | See1F | GCTCGAGTCCACGGTCTTC | See1Rb | AGTTCATCCTCAGCACCCAG |
| Mir3 | Cys protease | Protease | AF019147 | Mir3Fb | GATGTCAACAGGAAAAACGC | Mir3R | GTTCGCTTCGTAGCCTTCAC |
| Cys protease | Protease | EU970148 | CysProt-Fc | GTCACCGGCGTCAAGGAC | CysProt-Rc | CCCAGGAGTTCTTCATCAGC | |
| PR-1d | Pathogenesis-related protein 1 | Defense | U82200 | PR1F | AGAACTCGCCGCAGGACTAC | PR1R | CGCCTGCATGGTTTTATTG |
| Chn2d | Acidic class I chitinase | Defense | L00973 | ChiF | AACGGCTTCTACACCTACGC | ChiR | TTGGAGATGTTGGTGACGAC |
| Mas1 | Malate synthase | Carbon mobilization | L35914 | Mas1Fb | CCAAGGTCTTCATGGCTGAC | Mas1R | CCTTGATCGGGATCTGAGC |
| Kin1 | S-like RNase | U66241 | Kin1Fb | ACCAGCTGGCTCTTATGTGG | Kin1R | CACTTCTTGAAGGGGTGGAA | |
| Pld1 | Phospholipase D | Lipid degradation | D73410 | PldFb | GACTGGAACCGGACGCTG | PldR | AACCTTACACCCCTGCCTCT |
| Exg1 | Exoglucanase I | Cell wall glucan degradation | AF064707 | ExhFb | GCCGGAAAGGACAAGGTC | ExhR | GGCTTGCCGTTCTTGAGTAG |
| Sbe2a | Starch-branching enzyme IIa | Starch synthesis | U65948 | sbe2a-Fb | CTTTCTCTCGCACCGTTCTG | sbe2a-R | GCATTTGGGTTCCAGTTGTT |
| Zpu1 | Pullulanase | Starch synthesis/degradation | AF080567 | Zpu1-Fb | TGGACTTCCATCCAGTGTGA | Zpu1-R | AATCAAGCCTTGGCTTTTCA |
| Gbss1b | Granule-bound starch synthase | Starch synthesis | EU970982 | GBSSIb-Fb | CTTTTGTGCCTTGCTGCTTT | GBSSIb-R | TGAGAGCCCTTGCTTCAGTT |
| Tub6 | Tubulin | Housekeeping | L10633.1 | Tub6-Fb | ACTGCTTGCAAGGATTCCA | Tub6-R | GGCACACATCATGTTCTTGG |
Sequences are listed 5′ to 3′.
Primer spans intron/exon boundary to prevent the amplification of genomic DNA.
As compared with maize genome sequence (http://www.maizesequence.org/); gene does not contain introns.
No genomic sequence available, as compared with the first draft of the maize genome sequence.
DISCUSSION
Functional SBEIIa Is Required for the Transitory Property of Leaf Starch
SBEIIa is required for proper degradation of transitory starch. In absolute terms, wild-type leaves degraded more than twice as much starch as mutant leaves. Numerous enzymes involved in starch degradation, when mutated, lead to a “starch-excess” phenotype similar to that observed in sbe2a leaves. These enzymes include phosphoglucan phosphatase (SEX4), BAM3 β-amylase, noncatalytic BAM4 β-amylase, α-glucan, water dikinase (Lorberth et al., 1998; Zeeman et al., 1998; Zeeman and ap Rees, 1999; Yu et al., 2001; Ritte et al., 2002; Scheidig et al., 2002; Kötting et al., 2005, 2009; Niittylä et al., 2006; Fulton et al., 2008), and, in maize, pullulanase-type debranching enzyme (Dinges et al., 2003). While it is expected that mutation of genes involved in starch degradation will lead to an accumulation of starch, reasons for starch accumulation in mutants deficient in enzymes of starch synthesis are less obvious. Arabidopsis (Arabidopsis thaliana) mutants deficient in starch synthases or SBEs do not exhibit this phenomenon, with the exception of the starch synthase3 mutant (lacking starch synthase III), which has elevated levels of starch at the end of the day but not the end of the night (Zhang et al., 2005), and the ss4 mutant (lacking starch synthase IV), which has reduced levels of starch at the end of the day and elevated levels at the end of the night (Roldán et al., 2007). The ss4 mutant has a strongly reduced number of starch granules, and it has been proposed that degradation at night is limited by the granule surface area available to the degradative enzymes (Roldán et al., 2007). This explanation is unlikely to apply to the elevated starch levels in the sbe2a mutant of maize.
The starch-excess phenotype of the sbe2a mutant suggests an altered starch structure that is not easily degraded. Consistent with this idea, in vitro studies showed that whereas wild-type leaf starch was completely amenable to digestion, approximately 50% of sbe2a leaf starch, which exhibits aberrant granule structure (Fig. 5), was resistant to digestion with porcine α-amylase (Xia, 2009). This radically altered granular structure could be caused by the change in molecular structure of the starch polymers. First, the relative ratio of high Mr amylopectin to lower Mr amylose and/or altered amylopectin is greatly reduced in sbe2a leaves (Fig. 5A in Blauth et al., 2001; Fig. 4 in Dinges et al., 2003). Second, the branching pattern of the starch polymers is significantly altered. Starch from sbe2a leaves has fewer short (degree of polymerization 10–20) and more long (greater than degree of polymerization 30) chains than wild-type leaves (Dinges et al., 2003), resulting in an overall reduction in branching. Only SBEIIa protein and associated SBE activity were detected in soluble extracts from the wild type, and a small amount of SBE activity was detected from sbe2a soluble extracts. It is possible that SBEI, for which transcripts were present but protein was not identified in soluble extracts, is bound to the starch granule and contributes a low level of branching. SBEI, SBEIIa, and SBEIIb have been shown to be associated with starch granules in maize endosperm but are also present in the soluble fraction (Mu-Forster et al., 1996; Grimaud et al., 2008).
Alterations in storage granule morphology have been observed in plants with altered complements of starch synthases or SBEs. For example, the starch synthase II mutant of pea (Craig et al., 1998) and transgenic potatoes (Solanum tuberosum) with reduced activities of starch synthases II and III (Edwards et al., 1999) or SBEI and II (Schwall et al., 2000; Blennow et al., 2003) all have abnormal storage starch granules with twisted and lobed shapes. There are few reports of such alterations in the shape of transitory starch granules in plant leaves. However, the sheet-like morphology of sbe2a starch is strongly reminiscent of starch from Chlamydomonas after 24 h of incubation in vitro with ADPglucose. The normally flattened, discoid granules become highly distorted and fused into a network, which is attributed to the elongation of amylopectin outer chains within the granule by the amylose-synthesizing isoform of starch synthase, GBSS (Wattebled et al., 2002). GBSS activity on outer chains of amylopectin has similar disruptive effects on granule morphology of tubers of transgenic potatoes with reduced levels of the amylopectin-synthesizing starch synthases, SS2 and SS3 (Fulton et al., 2002). It seems possible that the low levels of glucan branching in sbe2a maize leaves severely restrict chain elongation by soluble starch synthases, which would lead to a rise in levels of the starch synthase substrate ADPglucose, for which GBSS has a relatively high Km (Denyer et al., 2001). Increased activity of GBSS inside the granule and very limited addition of new material at the periphery could thus account for the distorted, sheet-like morphology of the starch in sbe2a leaves.
The Effects of Loss of Class II SBEs Differ between Endosperm and Leaf
Endosperm starch extracted from ae mutants lacking SBEIIb exhibits a similar, albeit less severe, branching defect to that of sbe2a leaf starch (Klucinec and Thompson, 1998; Blauth et al., 2001). Although ae endosperm produces aberrant starch granules (Wang et al., 1993), the granules do not exhibit the extreme fused granule phenotype seen in sbe2a leaf starch, and there is no affect on the kernel’s viability, ability to germinate, etc. (Stinard et al., 1993), suggesting that this starch is, to at least a certain extent, degradable in vivo. The difference between leaf and endosperm in the nature and severity of effect of the loss of class II SBEs could have several causes. First, SBEI and/or SBEIIa could partially compensate for the lack of SBEIIb in endosperm. Second, ae endosperm starch may be more amenable to degradation than sbe2a leaf starch due either to its higher level of branching or to the presence of a different complement of degradation enzymes. Third, the different functions of endosperm (storage) versus leaf (transitory) starch could impact the effects of SBE mutations on these organs.
The Lack of SBEIIa in Leaves Alters Carbon Partitioning and Triggers Premature Leaf Senescence
The loss of SBEIIa and consequent hyperaccumulation of starch in sbe2a leaves have profound effects on leaf metabolism and plant growth as a whole. It seems likely that reduced plant growth is due to either or both the strongly reduced rate of photosynthesis and the reduced availability of carbohydrate in the leaf at night. However, the nature of the link between starch accumulation and the reduction in photosynthesis is less obvious. The link could be direct, through physical disruption of chloroplast function by the fused and irregular starch structures, or alternatively, the link may be indirect.
The sbe2a leaf appears to undergo programmed senescence (Lu and Zhang, 1998; for review, see Hörtensteiner, 2006; Lim et al., 2007; Bréhélin and Kessler, 2008) as starch accumulates. In addition to the decline in photosynthesis, plastoglobules accumulate in chloroplasts of visibly senescing tissue (Fig. 6). Plastoglobules are generally larger and more prevalent during senescence, as they accumulate triacylglycerol by-products of thylakoid membrane degradation (for review, see Bréhélin and Kessler, 2008), an intermediate step in the conversion of fatty acids from the thylakoid membranes into Suc, for export to other parts of the plant (Kaup et al., 2002). Mas1, which encodes the gluconeogenic enzyme malate synthase, facilitates the conversion of products of fatty acid β-oxidation via the glyoxylate cycle into Suc for export and has been shown to be triggered by the metabolic status of senescing leaves, including lipid breakdown products and reduced Suc levels due to reduced photosynthesis (Graham et al., 1992). The Mas1 gene is up-regulated more than 100-fold in visibly senescing portions of sbe2a leaves, suggesting a role of senescence in the reallocation of valuable resources in the sbe2a mutant. We also observed up-regulation of several other genes known to be induced during senescence, further implicating programmed senescence in the sbe2a phenotype.
Transitory Starch Synthesis and/or Degradation Differ between Maize, Arabidopsis, and Pea
In maize, Arabidopsis, and pea, SBEs are required for the normal synthesis and function of transitory starch. Whereas SBEIIa functions in maize leaves, in Arabidopsis, three genes encoding SBEs (i.e. BE1, BE2, and BE3) are active in the leaf (Fisher et al., 1996a; Khoshnoodi et al., 1998; Dumez et al., 2006). BE2 and BE3, class IIa SBEs with similarity to maize SBEIIa, contribute significantly to the synthesis of transitory starch. BE1 belongs to a novel SBEIII class (Han et al., 2007), for which a putative homolog has recently been identified in the maize genome (Yan et al., 2009). BE1 is apparently not required for transitory starch synthesis in Arabidopsis (Dumez et al., 2006). In pea leaves, approximately 90% of the SBE activity is accounted for by a single class II SBE, SBE A, and the remaining 10% by a class I enzyme, SBE B (Tomlinson et al., 1997).
The phenotypic effects of removing functional class II SBE(s) from leaves of maize, Arabidopsis, and pea differ substantially. Starch extracted from be2 and be3 single Arabidopsis mutants exhibits only minor changes in amylopectin structure. However, in stark contrast to the starch-excess phenotype in leaves of sbe2a maize mutants, removal of the class IIa SBEs from Arabidopsis (i.e. be2; be3 double mutant) completely abolishes starch synthesis in the leaf and results in the accumulation of maltose (Dumez et al., 2006). Dumez et al. (2006) propose that in the absence of BE2 and BE3, starch synthases produce unbranched glucan chains that are more readily digestible by amylases, even during the light phase, such that maltose accumulates in place of starch. The impact of the loss of class II SBE in pea is in some respects similar to that of the loss of SBEIIa in maize: leaf starch contains very little amylopectin and an increased amount of glucan of the same molecular mass range as amylose (Tomlinson et al., 1997). In contrast to the maize sbe2a mutant, however, there is no starch accumulation or premature leaf senescence.
Differences in the phenotypes of maize, Arabidopsis, and pea mutants lacking class II SBEs may reflect differences among the leaves of these species in other aspects of the pathways of starch synthesis and degradation. The pathway of starch degradation has been elucidated in detail only in Arabidopsis (Zeeman et al., 2010). Although maize leaves appear to contain at least some of the same components of these pathways as Arabidopsis (Friso et al., 2010), the relative contributions of these components to flux have not been systematically explored. In addition, the consequences for starch turnover due to C4 anatomy and biochemistry are not understood. In a NADP-malic enzyme C4 leaf, starch is synthesized in the bundle sheath cell, whereas Suc is synthesized in the mesophyll cell. A significant proportion of the triose phosphate that forms the substrate for starch synthesis is imported into the bundle sheath cell chloroplast from the mesophyll. This is a very different situation from a C3 leaf, in which triose phosphate is partitioned between Suc synthesis, starch synthesis, and the regenerative phase of the Calvin cycle within the same cell. Thus, lesions in starch synthesis may have different consequences for the operation of the Calvin cycle and Suc synthesis, and hence leaf physiology as a whole, in C3 and C4 leaves. Taken together, while Arabidopsis has proven to be a good model system to study transitory starch synthesis and metabolism, maize is a much more relevant system in which to study starch synthesis in the C4 grasses.
MATERIALS AND METHODS
Plant Growth, Morphometric Analysis, and Sample Collection
The maize (Zea mays) sbe2a-Mu mutant line (herein referred to as sbe2a) contains a Mu insertion in exon 2 of Sbe2a and has been backcrossed into W64A three times. sbe2a homozygous mutant leaves exhibit an early-senescence phenotype (Blauth et al., 2001).
Plants used for height and leaf number analyses were grown at the Pennsylvania State University Horticulture Research Farm in Rock Springs in 2006 and 2007 from progeny seed of a selfed Sbe2a/sbe2a heterozygote. Plants were sampled 3 weeks after germination, DNA was isolated (Dietrich et al., 2002), and plants were PCR genotyped using primers described previously (Blauth et al., 2001, 2002). The height of mature plants was measured from the soil surface to the tip of the central spike of the tassel.
Seedlings used in the diurnal studies were grown in a walk-in high-light (650 μmol m−2 s−1) Conviron growth chamber (BDW120) at 35% humidity under a diurnal cycle of 16 h of light and 8 h of dark at 28°C and 25°C, respectively. Plants used for analysis of starch content were grown for approximately 3 weeks, and the fourth vegetative leaf was sampled, flash frozen in liquid nitrogen, and stored at −80°C. Sampling of both mutant and wild-type leaves was done 3 cm from the leaf tip to exclude the senescent portion of sbe2a leaves. For qualitative starch analyses, leaves were sampled and decolorized in boiling 80% ethanol to remove chlorophyll and subsequently stained with 0.1% (w/v) iodine and 1.0% (w/v) potassium iodide.
Senescing and healthy leaf tissues used in starch quantification, RNA transcript, and photosynthesis studies were harvested at midsummer in early morning from approximately 2.5-month-old wild-type and sbe2a field-grown plants (Rock Springs, PA). Samples were taken midway along fully expanded leaves on wild-type and sbe2a mutant plants. From sbe2a plants, adjacent green and visibly senescing tissue samples (i.e. chlorotic tissue exhibiting pink sectors, most likely due to anthocyanin accumulation) were taken from each plant. Each sample was split down the midrib, and the two pieces were used for starch and transcript analyses. These same plants were used for photosynthesis and chlorophyll measurements.
Photosynthesis and Chlorophyll Measurements
Photosynthetic rate and stomatal conductance were measured using the LI-6400 Portable Photosynthesis System (LICOR Biosciences). Measurements were taken between 12 noon and 3 pm. Programmed light and humidity conditions were as described (Braun et al., 2006) under ambient temperature. Total chlorophyll was quantified in the same plants using the SPAD-502 Chlorophyll Meter (Konica Minolta). A total of 36 wild-type and sbe2a plants (green versus visibly senescing tissue) were tested, and each sector tested was assayed three times.
Crude Protein Extracts from Leaves
Total protein was extracted from frozen leaves with a chilled mortar and pestle in 600 μL of extraction buffer (100 mm HEPES, pH 7.4, 20% [v/v] glycerol, 1 mm Na fluoride, 1 mm Na orthovanadate, 5 μg mL−1 leupeptin, 10 μL mL−1 phosphatase inhibitor cocktail I [P2850; Sigma], and 1 mm phenylmethylsulfonyl fluoride) per 100 mg of tissue. Extracts were centrifuged at 6,000g for 10 min at 4°C. Supernatants were used immediately for native PAGE.
Native PAGE and Western Analyses
Crude protein extracts were separated on native polyacrylamide gels according to Dinges et al. (2001) using a mini-Protean II cell (Bio-Rad Laboratories). For western analysis, gels were blotted as described by Blauth et al. (2001) and immunodetection was performed using an enhanced chemifluorescence western-blotting detection kit according to the manufacturer’s instructions (Amersham Biosciences). Anti-SBEI and anti-SBEIIa antibodies (Blauth et al., 2001, 2002) were used at a 1:2,000 dilution.
SBE Activity Assays
In-gel SBE activity was assayed in proteins separated via native PAGE as described by Yamanouchi and Nakamura (1992). Activity was visualized by staining with 0.1% (w/v) iodine and 1.0% (w/v) potassium iodide, and gels were immediately photographed. Relative band intensities were compared on digital images using ImageJ version 1.37 (Abramoff et al., 2004).
In vitro SBE activity was measured with the phosphorylase a stimulation assay (Boyer and Preiss, 1978), modified to be nonradioactive by Fisher et al. (1996b). Assay reactions (200 μL volume) were prepared in 96-well format using 1 μg of crude extract per reaction and were incubated for 1 h at 30°C and then heat killed at 99°C for 90 s. Reactions were diluted with water, and inorganic phosphate released during the reaction was quantified with a colorimetric assay (Lanzetta et al., 1979). Measured values from control reactions containing no phosphorylase a were subtracted from values of corresponding reactions containing the enzyme.
Starch Purification and Quantification
Crude starch was purified from seedling leaves by homogenization in 0.7 m perchloric acid and 10 mg of sand. From centrifuged extracts, pelleted starch was repeatedly washed with 80% (v/v) ethanol, dried, and stored at −20°C. Dimethyl sulfoxide was added to crude starch pellets and incubated in a boiling water bath for 5 to 20 min to disperse starch. Starch was then enzymatically digested, and the resulting Glc was quantified using the Total Starch Assay Kit (Megazyme) per the manufacturer’s instructions.
Leaf starch isolated for microscopy was extracted from 5 g of leaf tissue as described by Dinges et al. (2003). The resulting starch pellet was washed repeatedly with 80% ethanol and stored in water supplemented with 0.1% (w/v) sodium azide.
Microscopy
Starch granules purified from leaf tissue were gold coated and viewed by SEM using a JSM 5400 (JEOL). Leaf punches for TEM were collected in the middle of the light phase from 6-week-old greenhouse-grown plants and fixed in 2.5% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.4, for 24 h at 4°C. Samples were rinsed in 0.1 m cacodylate buffer, fixed with 1% osmium tetroxide, and dehydrated in an ethanol series. Samples were imaged on a JEM1200 EXII (JEOL).
RNA Isolation and Transcript Analysis
Primers for transcript analysis (Table III) were designed from mRNA sequences such that one primer in the pair spanned an intron/exon boundary and prevented the amplification of genomic DNA.
RNA was extracted from wild-type (green tissue) and sbe2a leaves (visibly senescing versus green tissue) using the RNeasy Plant Mini Kit (Qiagen) per the manufacturer’s instructions. Total RNA (2 μg) was reverse transcribed by oligo(dT) priming using Moloney murine leukemia virus reverse transcriptase (New England Biolabs), and first-strand cDNA (0.5 μL) was used as a template for semiquantitative PCR using the primers listed in Table III. The PCR program consisted of 94°C for 2 min, 35 cycles of 94°C for 30 s, 4°C below primer melting temperature for 30 s, and 72°C for 1 min, and a final extension at 72°C for 5 min. Cycle numbers were titrated to determine the linear range of the amplification kinetics. PCR products were electrophoresed on 1% agarose gels containing ethidium bromide. Band intensities were quantified with ImageQuant software (GE Healthcare Life Sciences) using rectangle mode/local background correction/volume integration. Resulting values were normalized to the band intensity of tubulin.
Transcript analysis of sbe2a mutants with primers sbe2a-F and sbe2a-R (Table III) yielded an unexpected product. Whereas no product was amplified in a previous study (Blauth et al., 2001), RT-PCR on sbe2a template RNA yielded a product containing a 140-bp insertion as compared with the wild-type. Sequencing of the RT-PCR product demonstrated that the insertion was derived from a truncation of the original Mu element inserted into the sbe2a gene; the insertion contained only a portion of a single terminal inverted repeat, whereas genomic DNA contained an intact Mu element. This unexpected product is likely due to aberrant splicing within the Mu insertion during RNA processing, as has previously been documented in maize at an Adh1-Mu1 allele (Ortiz and Strommer, 1990) and at transposon insertions in Drosophila (Horowitz and Berg, 1995). The aberrant transcript present in sbe2a leaves does not produce a protein product recognizable by the SBEIIa antibody (Fig. 1B) and therefore does not produce active SBEIIa.
Statistical Analyses
Height, leaf number, and starch amounts were compared among genotypes using paired t tests assuming unequal variance. ANOVA was used to analyze photosynthesis, stomatal conductance, and RNA expression data. se values are reported for all measurements.
Acknowledgments
We thank undergraduate students Jennifer Blackman and Timothy Silberg and graduate student Yufan Zhang for technical assistance and Tom Slewinski for helpful discussions and assistance with photosynthetic measurements. We acknowledge the Electron Microscopy Facility, University Park (Huck Institutes of the Life Sciences, Pennsylvania State University), for performing electron microscopy imaging.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Blauth SL, Kim KN, Klucinec J, Shannon JC, Thompson D, Guiltinan M. (2002) Identification of Mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol Biol 48: 287–297 [DOI] [PubMed] [Google Scholar]
- Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guilitinan MJ. (2001) Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol 125: 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow A, Hansen M, Schulz A, Jørgensen K, Donald AM, Sanderson J. (2003) The molecular deposition of transgenically modified starch in the starch granule as imaged by functional microscopy. J Struct Biol 143: 229–241 [DOI] [PubMed] [Google Scholar]
- Boyer CD, Preiss J. (1978) Multiple forms of starch branching enzyme of maize: evidence for independent genetic control. Biochem Biophys Res Commun 80: 169–175 [DOI] [PubMed] [Google Scholar]
- Boyer CD, Preiss J. (1981) Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. Plant Physiol 67: 1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Ma Y, Inada N, Muszynski MG, Baker RF. (2006) tie-dyed1 regulates carbohydrate accumulation in maize leaves. Plant Physiol 142: 1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréhélin C, Kessler F. (2008) The plastoglobule: a bag full of lipid biochemistry tricks. Photochem Photobiol 84: 1388–1394 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. (2003) The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnol J 1: 3–22 [DOI] [PubMed] [Google Scholar]
- Craig J, Lloyd JR, Tomlinson K, Barber L, Edwards A, Wang TL, Martin C, Hedley CL, Smith AM. (1998) Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell 10: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang PL, Boyer CD. (1988) Maize leaf and kernel starch synthases and branching enzymes. Phytochemistry 27: 1255–1259 [PubMed] [Google Scholar]
- Denyer K, Johnson P, Zeeman S, Smith AM. (2001) The control of amylose synthesis. J Plant Physiol 158: 479–487 [Google Scholar]
- Dietrich CR, Cui F, Packila ML, Li J, Ashlock DA, Nikolau BJ, Schnable PS. (2002) Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160: 697–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, James MG, Myers AM. (2003) Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell 15: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, Myers AM, James MG. (2001) Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol 125: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumez S, Wattebled F, Dauvillee D, Delvalle D, Planchot V, Ball SG, D’Hulst C. (2006) Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. Plant Cell 18: 2694–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A, Fulton DC, Hylton CM, Jobling SA, Gidley M, Rossner U, Martin C, Smith AM. (1999) A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J 17: 251–261 [Google Scholar]
- Fisher DK, Gao M, Kim K-N, Boyer CD, Guiltinan MJ. (1996a) Two closely related cDNAs encoding starch branching enzyme from Arabidopsis thaliana. Plant Mol Biol 30: 97–108 [DOI] [PubMed] [Google Scholar]
- Fisher DK, Gao M, Kim KN, Boyer CD, Guiltinan MJ. (1996b) Allelic analysis of the maize amylose-extender locus suggests that independent genes encode starch-branching enzymes IIa and IIb. Plant Physiol 110: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. (2010) Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Edwards A, Pilling E, Robinson HL, Fahy B, Seale R, Kato L, Donald AM, Geigenberger P, Martin C. (2002) Role of granule-bound starch synthase in determination of amylopectin structure and starch granule morphology in potato. J Biol Chem 277: 10834–10841 [DOI] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G, et al. (2008) β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Fisher DK, Kim K-N, Shannon JC, Guiltinan MJ. (1996) Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol Biol 30: 1223–1232 [DOI] [PubMed] [Google Scholar]
- Garwood DL, Shannon JC, Creech RG. (1976) Starches of endosperms possessing different alleles at the amylose-extender locus in Zea mays L. Cereal Chem 53: 355–364 [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M. (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Graham IA, Leaver CJ, Smith SM. (1992) lnduction of malate synthase gene expression in senescent and detached organs of cucumber. Plant Cell 4: 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud F, Rogniaux H, James MG, Myers AM, Planchot V. (2008) Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J Exp Bot 59: 3395–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan HP, Li P, Imparl-Radosevich J, Preiss J, Keeling P. (1997) Comparing the properties of Escherichia coli branching enzyme and maize branching enzyme. Arch Biochem Biophys 342: 92–98 [DOI] [PubMed] [Google Scholar]
- Guan HP, Preiss J. (1993) Differentiation of the properties of the branching isozymes from maize (Zea mays). Plant Physiol 102: 1269–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Sun FJ, Rosales-Mendoza S, Korban SS. (2007) Three orthologs in rice, Arabidopsis, and Populus encoding starch branching enzymes (SBEs) are different from other SBE gene families in plants. Gene 401: 123–130 [DOI] [PubMed] [Google Scholar]
- Horowitz H, Berg CA. (1995) Aberrant splicing and transcription termination caused by P element insertion into the intron of a Drosophila gene. Genetics 139: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Kaup MT, Froese CD, Thompson JE. (2002) A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol 129: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnoodi J, Larsson CT, Larsson H, Rask L. (1998) Differential accumulation of Arabidopsis thaliana Sbe2.1 and Sbe2.2 transcripts in response to light. Plant Sci 135: 183–193 [Google Scholar]
- Kirchanski SJ. (1975) Ultrastructural development of dimorphic plastids of Zea mays L. Am J Bot 62: 695–705 [Google Scholar]
- Klucinec JD, Thompson DB. (1998) Fractionation of high-amylose maize starches by differential alcohol precipitation and chromatography of the fractions. Cereal Chem 75: 887–896 [Google Scholar]
- Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves: the phosphoglucan, water dikinase. Plant Physiol 137: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M. (2009) STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem 100: 95–97 [DOI] [PubMed] [Google Scholar]
- Li JH, Guiltinan MJ, Thompson DB. (2007) Mutation of the maize sbe1a and ae genes alters morphology and physical behavior of wx-type endosperm starch granules. Carbohydr Res 342: 2619–2627 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lorberth R, Ritte G, Willmitzer L, Kossmann J. (1998) Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotechnol 16: 473–477 [DOI] [PubMed] [Google Scholar]
- Lu CM, Zhang JH. (1998) Modifications in photosystem II photochemistry in senescent leaves of maize plants. J Exp Bot 49: 1671–1679 [Google Scholar]
- Matheson NK, Wheatley JM. (1962) Starch changes in developing and senescing tobacco leaves. Aust J Biol Sci 15: 445–458 [Google Scholar]
- Moore CW, Creech RG. (1972) Genetic fine structure analysis of the amylose-extender locus in Zea mays L. Genetics 70: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu-Forster C, Huang R, Powers JR, Harriman RW, Knight M, Singletary GW, Keeling PL, Wasserman BP. (1996) Physical association of starch biosynthetic enzymes with starch granules of maize endosperm: granule-associated forms of starch synthase I and starch branching enzyme II. Plant Physiol 111: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen JC. (2006) Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem 281: 11815–11818 [DOI] [PubMed] [Google Scholar]
- Ortiz DF, Strommer JN. (1990) The Mu1 maize transposable element induces tissue-specific aberrant splicing and polyadenylation in two Adh1 mutants. Mol Cell Biol 10: 2090–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. (2002) The starch-related R1 protein is an α-glucan, water dikinase. Proc Natl Acad Sci USA 99: 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán I, Wattebled F, Mercedes Lucas M, Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D’Hulst C, Mérida A. (2007) The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J 49: 492–504 [DOI] [PubMed] [Google Scholar]
- Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J. (2002) Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. Plant J 30: 581–591 [DOI] [PubMed] [Google Scholar]
- Schwall GP, Safford R, Westcott RJ, Jeffcoat R, Tayal A, Shi YC, Gidley MJ, Jobling SA. (2000) Production of very-high-amylose potato starch by inhibition of SBE A and B. Nat Biotechnol 18: 551–554 [DOI] [PubMed] [Google Scholar]
- Seo BS, Kim S, Scott MP, Singletary GW, Wong KS, James MG, Myers AM. (2002) Functional interactions between heterologously expressed starch-branching enzymes of maize and the glycogen synthases of Brewer’s yeast. Plant Physiol 128: 1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Garwood DL, Boyer CD. (1998) Genetics and physiology of starch development. BeMiller JN, Whistler RL, , Starch: Chemistry and Technology, Ed 3 Academic Press, New York, pp 23–82 [Google Scholar]
- Smith AM, Denyer K, Martin C. (1997) The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol 48: 67–87 [DOI] [PubMed] [Google Scholar]
- Stinard PS, Robertson DS, Schnable PS. (1993) Genetic isolation, cloning, and analysis of a Mutator-induced, dominant antimorph of the maize amylose extender1 locus. Plant Cell 5: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels JJM. (1985) Composition and properties of commercial native starches. Starch/Starke 37: 1–5 [Google Scholar]
- Taira T, Uematsu M, Nakano Y, Morikawa T. (1991) Molecular identification and comparison of the starch synthase bound to starch granules between endosperm and leaf blades in rice plants. Biochem Genet 29: 301–311 [DOI] [PubMed] [Google Scholar]
- Takeda Y, Guan HP, Preiss J. (1993) Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydr Res 240: 253–263 [Google Scholar]
- Tomlinson KL, Lloyd JR, Smith AM. (1997) Importance of isoforms of starch-branching enzyme in determining the structure of starch in pea leaves. Plant J 11: 31–43 [Google Scholar]
- Vineyard ML, Bear RP. (1952) Amylose content. Maize Genet Coop News Lett 26: 5 [Google Scholar]
- Wang Y-J, White P, Pollack L, Jane J. (1993) Characterization of starch structures of 17 maize endosperm mutant genotypes with Oh43 inbred line background. Cereal Chem 70: 171–179 [Google Scholar]
- Wattebled F, Buléon A, Bouchet B, Ral JP, Liénard L, Delvallé D, Binderup K, Dauvillée D, Ball S, D’Hulst C. (2002) Granule-bound starch synthase I: a major enzyme involved in the biogenesis of B-crystallites in starch granules. Eur J Biochem 269: 3810–3820 [DOI] [PubMed] [Google Scholar]
- Xia H. (2009) Structure and function of endosperm starch from maize mutants deficient in one or more starch-branching enzyme isoform activities. PhD thesis Pennsylvania State University, State College [Google Scholar]
- Yamanouchi H, Nakamura Y. (1992) Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant Cell Physiol 33: 985–991 [Google Scholar]
- Yan HB, Pan XX, Jiang HW, Wu GJ. (2009) Comparison of the starch synthesis genes between maize and rice: copies, chromosome location and expression divergence. Theor Appl Genet 119: 815–825 [DOI] [PubMed] [Google Scholar]
- Yao Y, Thompson DB, Guiltinan MJ. (2004) Maize starch-branching enzyme isoforms and amylopectin structure: in the absence of starch-branching enzyme IIb, the further absence of starch-branching enzyme Ia leads to increased branching. Plant Physiol 136: 3515–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Häusler RE, Hille D, Flügge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al. (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, ap Rees T. (1999) Changes in carbohydrate metabolism and assimilate export in starch-excess mutants of Arabidopsis. Plant Cell Environ 22: 1445–1453 [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM. (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61: 209–234 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, Rees T. (1998) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J 15: 357–365 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. (2007) The diurnal metabolism of leaf starch. Biochem J 401: 13–28 [DOI] [PubMed] [Google Scholar]
- Zhang X, Myers AM, James MG. (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol 138: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]