Abstract
Seed germination is regulated through elaborately interacting signaling networks that integrate diverse environmental cues into hormonal signaling pathways. Roles of gibberellic acid and abscisic acid in germination have been studied extensively using Arabidopsis (Arabidopsis thaliana) mutants having alterations in seed germination. Auxin has also been implicated in seed germination. However, how auxin influences germination is largely unknown. Here, we demonstrate that auxin is linked via the IAA30 gene with a salt signaling cascade mediated by the NAM-ATAF1/2-CUC2 transcription factor NTM2/Arabidopsis NAC domain-containing protein 69 (for NAC with Transmembrane Motif1) during seed germination. Germination of the NTM2-deficient ntm2-1 mutant seeds exhibited enhanced resistance to high salinity. However, the salt resistance disappeared in the ntm2-1 mutant overexpressing the IAA30 gene, which was induced by salt in a NTM2-dependent manner. Auxin exhibited no discernible effects on germination under normal growth conditions. Under high salinity, however, whereas exogenous application of auxin further suppressed the germination of control seeds, the auxin effects were reduced in the ntm2-1 mutant. Consistent with the inhibitory effects of auxin on germination, germination of YUCCA 3-overexpressing plants containing elevated levels of active auxin was more severely influenced by salt. These observations indicate that auxin delays seed germination under high salinity through cross talk with the NTM2-mediated salt signaling in Arabidopsis.
Seed germination is a plant-specific developmental process during which an embryonic plant grows to form a seedling. It is critical for the establishment of plant growth in soil. As sessile organisms, most plant species cannot leave the place where their seeds germinate throughout their life span. Therefore, seeds possess an array of strategies by which they constantly monitor intrinsic and environmental conditions to determine when to germinate (Finkelstein et al., 2008).
The most critical external factors that influence germination include ambient temperature, water availability, oxygen, and soil salinity. Light intensity and quality also play a role in seed germination (Finch-Savage and Leubner-Metzger, 2006). These environmental signals are integrated into internal developmental programs, such as growth hormone signaling, to regulate the timing of germination (Finkelstein et al., 2008). Therefore, identification of signaling molecules and elucidation of underlying molecular mechanisms linking external cues with internal growth hormonal signals are crucial for understanding the germination process.
The roles of many signaling molecules, particularly abscisic acid (ABA) and GA, have been studied for decades using mutants with defective germination phenotypes, mostly in Arabidopsis (Arabidopsis thaliana). Whereas ABA is required for seed dormancy and thus acts as a negative regulator of germination, GA promotes seed germination. Consequently, whereas mutants having defects in ABA biosynthesis exhibit a reduction in seed dormancy, those with mutations in ABA catabolic pathways show enhanced seed dormancy (Okamoto et al., 2006). Soil salinity influences seed germination in an ABA-dependent manner at least partly by repressing GA biosynthesis (Achard et al., 2006; Kim et al., 2008). Consistent with the promotive role of GA in germination, mutants lacking GA biosynthesis or defective in GA signaling do not germinate even when external conditions are favorable (Peng and Harberd, 1997; Kim et al., 2008).
Auxin plays diverse roles in virtually all aspects of plant growth and developmental processes (Weijers and Jürgens, 2004; Quint and Gray, 2006; Abel, 2007). It also plays a role in plant responses to biotic and abiotic stresses (Navarro et al., 2006; Park et al., 2007). Notably, recent studies in several plant species support the role of auxin in seed germination (Birgit et al., 2005), although the underlying signaling schemes have not been explored at the molecular level.
Global gene expression studies in Arabidopsis support the role of auxin in seed germination. It has been observed that genes encoding auxin flux carriers and biosynthetic enzymes are up-regulated by GA in germinating seeds, suggesting that GA activity influences auxin level and transport during seed germination (Ogawa et al., 2003; Carrera et al., 2007). Investigation of auxin homeostasis and measurements of endogenous auxin contents also support its role in seed germination. Assays using the DR5-GUS reporter system have shown that GUS activity is elevated in the radicle after cold imbibition (Ni et al., 2001; Liu et al., 2007). In addition, it has been found that whereas endogenous contents of free indole-3-acetic acid (IAA) are elevated, those of conjugated forms are reduced in the germinating seeds of Scots pine (Pinus sylvestris; Ljung et al., 2001). A similar result has been obtained from germination assays of bean (Phaseolus vulgaris) seeds (Bialek and Cohen, 1989).
More direct evidence sustaining the involvement of auxin in seed germination has been inferred from germination assays using transgenic plants overexpressing miR160 or its target Auxin Response Factor10 (ARF10; Liu et al., 2007). Seed germination of the miR160-overproducing transgenic plants is hyposensitive to ABA. In contrast, that of transgenic plants overexpressing a miR160-resistant ARF10 gene is hypersensitive to ABA. Interestingly, this ABA hypersensitivity is mimicked in the germinating seeds of wild-type plants in the presence of exogenous auxin application, supporting signaling cross talk between auxin and ABA.
The NAC (for NAM-ATAF1/2-CUC2) transcription factors constitute one of the largest transcription factor families in plant genomes (Ooka et al., 2003; Olsen et al., 2005b). The Arabidopsis genome contains more than 100 NAC members. Roles of many NAC transcription factors have been demonstrated in diverse developmental processes and plant responses to biotic and abiotic stresses, such as floral development (Sablowski and Meyerowitz, 1998), apical meristem formation (Hibara et al., 2003), stress responses and signaling (Balazadeh et al., 2010; Jensen et al., 2010; Seo et al., 2010), cell cycle control (Kim et al., 2006), and germination under high salinity (Kim et al., 2008). A few NAC transcription factors, such as AtNAC2 functioning in root development (He et al., 2005), have been suggested to mediate auxin signaling in the salt stress response. However, no NAC protein has been shown to play a role in auxin-salt stress signaling cross talk during seed germination. Notably, some NAC proteins are membrane associated, and controlled proteolytic activation of the membrane-bound NAC transcription factors has been proposed to serve as an adaptive strategy that ensures rapid transcriptional responses to abrupt environmental changes (Seo et al., 2008).
In this work, we demonstrate that the plasma membrane-bound NAC transcription factor NTM2 integrates auxin and salt signals in regulating Arabidopsis seed germination. In this signaling scheme, auxin signals are incorporated via the IAA30 gene into the NTM2-mediated salt signaling pathway. Therefore, we propose that NTM2 serves as a molecular link that interconnects a developmental feedback loop of auxin signaling with a salt signal transduction pathway during seed germination.
RESULTS
NTM2 Is a Plasma Membrane-Bound NAC Transcription Factor
We have recently reported that a membrane-bound NAC transcription factor, NTM1 (for NAC with Transmembrane Motif1; At4G01540) regulates cell division by modulating cytokinin signaling (Kim et al., 2006). Notably, an adjacent locus (At4G01550) also encodes a NAC protein, designated NTM2, having a similar structural organization and a high sequence homology to the NTM1 protein (Supplemental Fig. S1). The NTM2 protein was designated Arabidopsis NAC domain-containing protein 69 (ANAC069) in the previous report (Ooka et al., 2003). Therefore, we decided to examine whether NTM2/ANAC069 is functionally similar to NTM1.
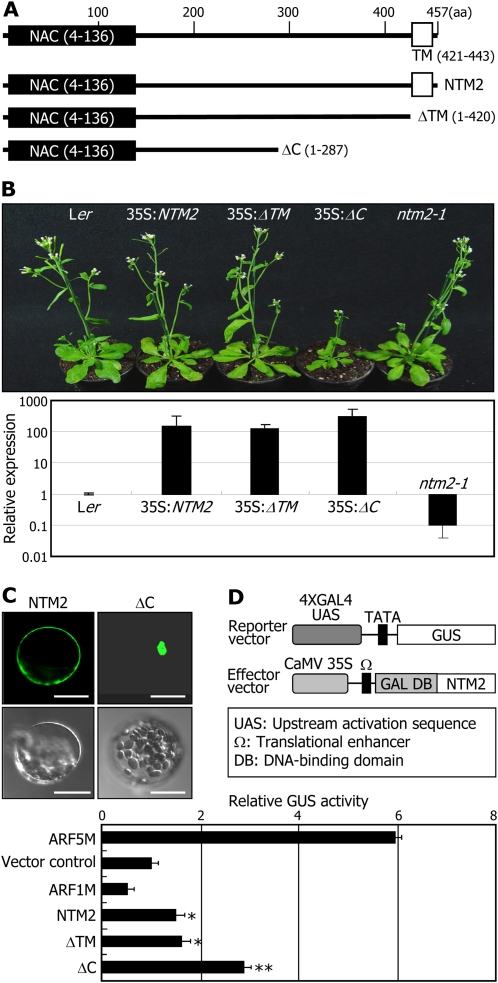
The NTM2 protein consists of 457 residues (Fig. 1A). Like the NTM1 protein, a NAC DNA-binding domain resides in the N-terminal region, and a transmembrane (TM) motif is predicted in the far C-terminal region. To investigate the physiological role of NTM2, we produced a few truncated NTM2 forms (Fig. 1A). The ΔTM construct included residues 1 to 420 and lacked the TM motif. The ΔC construct consisted of residues 1 to 287 and was similar in size to the transcriptionally active nuclear forms of NTM1 (Kim et al., 2006) and NTL8 (Kim et al., 2008) as well as to the known nuclear NAC proteins (Kim et al., 2006; Ruiming et al., 2007). The NTM2 gene constructs were transformed into Landsberg erecta (Ler) plants under the control of the cauliflower mosaic virus (CaMV) 35S promoter.
Figure 1.
NTM2 is a plasma membrane-bound NAC transcription factor. A, NTM2 constructs used. Numbers indicate amino acid (aa) residue positions. The TM was predicted using the ARAMEMNON membrane protein database (http://aramemnon.botanik.uni-koeln.de/). B, Transgenic plants overexpressing NTM2 gene constructs driven by the CaMV 35S promoter. Ler plants were used for transformation. Five-week-old plants grown in soil under long days were photographed (top panel). Transcript levels were determined by qRT-PCR using RNA samples extracted from 2-week-old whole plants grown on a MS-agar plate (bottom panel). Biological triplicates were averaged. Error bars represent se. The y axis is presented on a logarithmic scale for better comparison of fold changes. C, Subcellular localization of NTM2 and ΔC proteins. The GFP-NTM2 and GFP-ΔC fusion constructs were expressed transiently in Arabidopsis protoplasts and visualized by fluorescence microscopy (top panels) and differential interference contrast microscopy (bottom panels). Bars = 20 μm. D, Transcriptional activation activity assays in Arabidopsis protoplasts. The GAL4 transient expression assays were carried out using Arabidopsis protoplasts, as described previously (Miura et al., 2007). Vector control, Transformation with the effector vector without gene inserts; ARF5M and ARF1M, transformations with the effector vectors containing the ARF5M gene (activator control) and the ARF1M gene (repressor control), respectively (Tiwari et al., 2003). Five measurements were averaged. Error bars indicate se. Statistical significance was determined by Student’s t test (* P < 0.01, ** P < 0.005).
As previously reported with NTM1, NTL6, and NTL8 (Kim et al., 2006, 2008; Seo et al., 2010), transgenic plants overexpressing the full-size NTM2 form (35S:NTM2) and those overexpressing the ΔTM form (35S:ΔTM) were phenotypically indistinguishable from control plants (Fig. 1B). In contrast, those overexpressing the ΔC form, which is presumably analogous to a transcriptionally active NTM2 form, exhibited a dwarfed appearance with small, curled leaves, suggesting that membrane release of the NTM2 protein is essential for its activity. However, no leaf serration was observed, unlike the transgenic plants overexpressing a ΔC form of the NTM1 protein (Kim et al., 2006), suggesting that the physiological roles of the NTM1 and NTM2 proteins are at least partly distinct from each other.
To determine the subcellular localization of the NTM2 protein, a GFP-coding sequence was fused in-frame to the 5′ ends of the NTM2 gene sequences and the GFP-NTM2 gene fusions were transiently expressed in Arabidopsis protoplasts. The results showed that whereas the full-size NTM2 protein was localized at the plasma membranes, the ΔC form was detected predominantly in the nucleus (Fig. 1C).
We employed a GAL4 transient expression system in Arabidopsis protoplasts (Miura et al., 2007) to investigate the transcriptional activation activities of NTM2 protein. The NTM2 gene sequences were fused in-frame to the 3′ end of the GAL4 DNA-binding domain-coding sequence in the effector vector (Fig. 1D, top panel). The effector vectors, the reporter vector having the GUS reporter gene, and the vector containing the Renilla luciferase gene, which was included to normalize the measurements, were cotransformed into Arabidopsis protoplasts. The assays revealed that the NTM2 and ΔTM proteins possess discernible levels of transcriptional activation activities, showing that the NTM2 protein is a transcriptional activator (Fig. 1D, bottom panel). The ΔC protein exhibited the highest activity, which was much higher than that of the ΔTM protein, suggesting that membrane release itself is not adequate for the transcriptional activation activity. It is also envisioned that the C-terminal sequence of ΔTM, which is missing in ΔC, may confer an inhibitory effect on the transcriptional activation activity of NTM2, as suggested with NTL6 (Seo et al., 2010). Together, these observations indicate that the NTM2 protein is a plasma membrane-localized transcriptional activator.
The NTM2 Gene Is Induced by High Salinity
Many NAC proteins are involved in plant responses to various environmental stresses, such as high salinity, cold, and dark-induced leaf senescence (Olsen et al., 2005b; Kim et al., 2008; Seo et al., 2010). We found that the phenotypes of 35S:ΔC transgenic plants were similar to those observed in plants grown under stressful conditions (Stanton et al., 2000; Kim et al., 2007), suggesting that the NTM2 gene is related to plant stress responses.
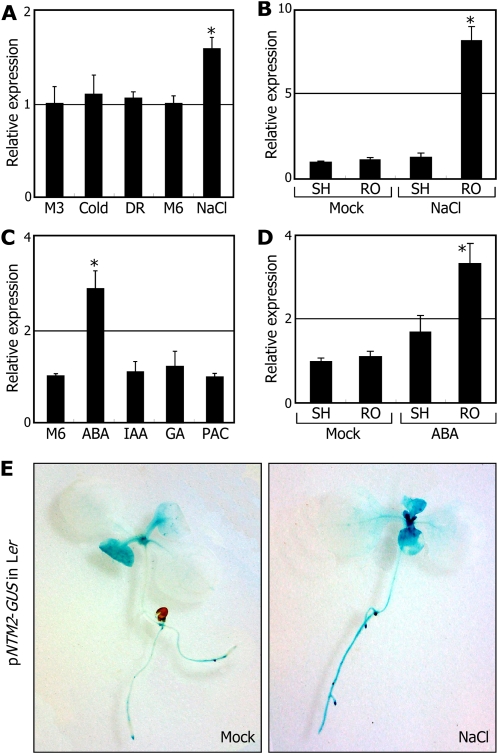
To obtain clues to the role played by NTM2, the effects of abiotic stresses and growth hormones on NTM2 gene expression were examined by quantitative real-time reverse transcription (qRT)-PCR. The NTM2 gene was slightly induced only by high salinity among the stress conditions examined when whole plants were used for total RNA extraction (Fig. 2A). Notably, expression studies using the shoot and root samples revealed that whereas the NTM2 gene was uninfluenced by high salinity in the shoot, it was induced approximately 8-fold in the roots (Fig. 2B). This result is also consistent with the strong induction of ANAC069 by NaCl in the roots (Jiang and Deyholos, 2006). The NTM2 gene was also induced by ABA (Fig. 2C), particularly in the root (Fig. 2D). These observations suggest that the NTM2 gene plays a role in salt stress responses, which is likely to be mediated by ABA.
Figure 2.
The NTM2 gene is induced by high salt. Transcript levels were determined by qRT-PCR. Biological triplicates were averaged. Error bars indicate se. Statistical significance was determined by Student’s t test (* P < 0.01). Two-week-old plants grown on MS-agar plates were used for extraction of total RNA or subsequent treatments. A, Effects of abiotic stress conditions on NTM2 gene transcription. Plants were exposed to cold (4°C, 3 h), drought (DR; 3 h), or NaCl (150 mm, 6 h). Whole plants were used for extraction of total RNA. M3 and M6, Mock treatments for 3 and 6 h, respectively. B, Effects of high salinity on NTM2 gene transcription in the shoots (SH) and roots (RO). Plants were soaked for 6 h in MS liquid cultures supplemented with 150 mm NaCl, and the shoot and root samples were harvested separately for extraction of total RNA. C, Effects of growth hormones on NTM2 gene transcription. Plants were transferred to MS liquid cultures containing appropriate concentrations of growth hormones, such as ABA (50 μm, 6 h), IAA (20 μm, 6 h), GA (50 μm, 6 h), or PAC (50 μm, 6 h). Whole plants were used for extraction of total RNA. D, Effects of ABA on NTM2 gene transcription in the shoots and roots. Plants were treated with ABA as described in C, and the shoot and root samples were harvested separately for extraction of total RNA. E, Effects of high salinity on the promoter activities of the NTM2 gene. Two-week-old transgenic plants expressing the pNTM2-GUS fusion grown on MS-agar plates were incubated for 6 h in MS liquid cultures supplemented with 150 mm NaCl and subjected to GUS staining.
We also examined the effects of high salinity on NTM2 gene expression using a promoter-GUS gene fusion, in which the GUS-coding sequence was fused to the 3′ end of the NTM2 gene promoter sequence covering an approximately 2-kb region upstream of the transcription start site. The promoter-GUS construct (pNTM2-GUS) was transformed into Ler plants. Whereas GUS activity was uninfluenced by high salinity in the leaves, it was elevated significantly in the roots (Fig. 2E), further supporting the predominant induction of the NTM2 gene in the roots under high salinity.
ntm2-1 Seedling Growth Is Less Sensitive to High Salinity
The phenotypes of 35S:ΔC transgenic plants and the effects of salt on NTM2 expression suggested that the NTM2 gene might be involved in the plant response to high salinity. To examine this hypothesis, we analyzed the phenotypes and salt responsiveness of a transposon insertional NTM2 knockout mutant (ntm2-1; CSHL-ET8732), which was obtained from the Arabidopsis Genetrap Database at Cold Spring Harbor Laboratory (Supplemental Fig. S2).
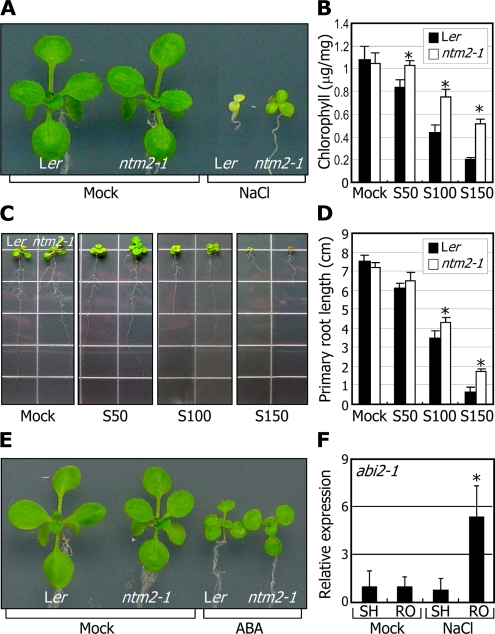
The ntm2-1 mutant did not exhibit any discernible phenotypes compared with control plants when grown under normal growth conditions (Fig. 1B). However, it responded differentially to high salinity. The ntm2-1 seedling growth was influenced to a lesser degree by salt (Fig. 3A). In particular, unlike control seedlings, leaf yellowing was not observed in the mutant seedlings under high salinity (Fig. 3B). Primary root growth of the ntm2-1 mutant was also less sensitive to high salinity (Fig. 3, C and D), supporting that the NTM2 gene is related to salt responses.
Figure 3.
The ntm2-1 mutant exhibits reduced sensitivity to high salinity. A, Effects of high salinity on ntm2-1 seedling growth. Three-day-old seedlings grown on MS-agar plates were transferred to fresh MS-agar plates supplemented with 150 mm NaCl and further grown for 2 weeks. B, Measurements of chlorophyll contents. Aerial parts of 2-week-old seedlings grown on MS-agar plates containing various concentrations of NaCl were used for extraction of chlorophylls. Ten measurements were averaged. Statistical significance was determined by Student’s t test (* P < 0.01). Error bars indicated se. S50, S100, and S150, NaCl at 50 mm, 100 mm, and 150 mm, respectively. C and D, Effects of high salinity on primary root growth. Three-day-old seedlings grown on MS-agar plates were transferred to fresh MS-agar plates supplemented with various concentrations of NaCl and further grown for 10 d (C). Primary root lengths of 20 seedlings were measured and averaged (D). Error bars represent se (t test; * P < 0.01). E, Effects of ABA on ntm2-1 seedling growth. Plants were treated as described in A, but 0.6 μm ABA was used instead of NaCl. F, Effects of high salinity on NTM2 gene transcription in the abi2-1 mutant. Two-week-old plants grown on MS-agar plates were transferred to MS liquid cultures supplemented with 150 mm NaCl and soaked for 6 h. The shoot (SH) and root (RO) samples were harvested separately for extraction of total RNA. Levels of the NTM2 transcripts were determined by qRT-PCR. Biological triplicates were averaged. Error bars indicate se (t test; * P < 0.01).
We also examined the effects of ABA on seedling growth. Notably, the effects of ABA on seedling growth were similar in control and ntm2-1 mutant plants (Fig. 3E). Furthermore, salt induction of the NTM2 gene still occurred in the roots of abi2-1, an ABA-insensitive mutant with disrupted ABA signaling (Zhu, 2002; Fig. 3F), as observed in control plants treated with salt (Fig. 2D). These observations indicate that the role of the NTM2 gene in plant salt responses does not depend on ABA.
Germination of ntm2-1 Seeds Is Resistant to High Salinity
One of the plant developmental processes affected by salt is seed germination. Soil salinity represses seed germination either by imposing osmotic stress (Perruc et al., 2007) or by reducing GA biosynthesis (Achard et al., 2006; Kim et al., 2008). We found that the NTM2 gene is induced by high salinity. Therefore, it was hypothesized that the NTM2 gene might be related to seed germination under high salinity.
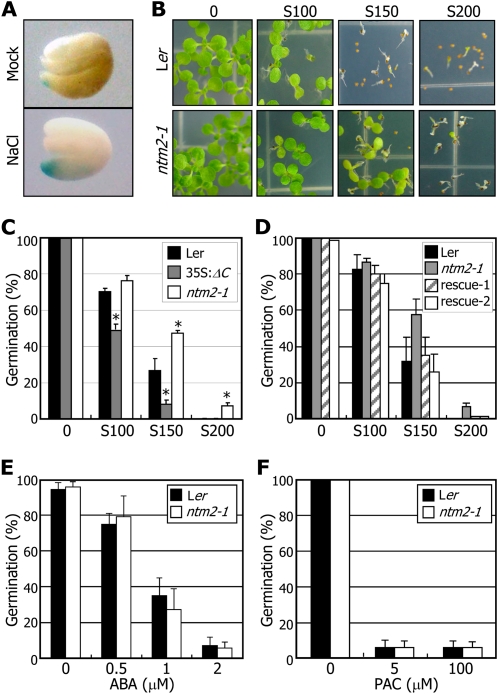
We first examined the expression pattern of the NTM2 gene in germinating seeds of transgenic plants expressing the pNTM2-GUS construct. Reporter gene expression was detected at a low level in germinating seeds under normal conditions (Fig. 4A). In contrast, it was highly induced in the emerging radicle under high salinity.
Figure 4.
Germination of the ntm2-1 seeds is less sensitive to high salinity. A, NTM2 expression in germinating seeds. Seeds of transgenic plants expressing the pNTM2-GUS reporter were cold imbibed and allowed to germinate for 2 d on MS-agar plates containing 150 mm NaCl before GUS staining. B and C, Effects of high salinity on the germination of ntm2-1 seeds. Cold-imbibed seeds were germinated on MS-agar plates containing varying concentrations of NaCl and photographed 7 d after cold imbibition (B). Germination percentages were calculated 4 d after cold imbibition (C). Radicle emergence was used as a morphological marker for germination. Three measurements, each consisting of approximately 50 to 60 seeds, were averaged for each plant group. Error bars indicate se (* P < 0.01). S100, S150, and S200, NaCl at 100 mm, 150 mm, and 200 mm, respectively. D, Complementation of the ntm2-1 mutant. The ntm2-1 mutant was transformed with the NTM2 gene with its own promoter, resulting in pNTM2:NTM2/ntm2-1 (rescue). Two independent lines were subject to germination assays as described in B. E, Effects of ABA on the germination of ntm2-1 seeds. Germination assays were carried out as described in B, but ABA was used instead of NaCl. F, Effects of PAC on the germination of ntm2-1 seeds. Germination assays were carried out as described in B, but PAC was used instead of NaCl.
We next examined the germination phenotypes of the ntm2-1 mutant seeds under high salinity. Germination of control seeds was delayed in the presence of 100 to 150 mm NaCl (Fig. 4B), as reported previously (Achard et al., 2006; Kim et al., 2008). In contrast, that of the ntm2-1 mutant seeds exhibited reduced sensitivity to high salinity. When counted 4 d after cold imbibition, whereas germination of control seeds was reduced by approximately 70% in the presence of 150 mm NaCl, that of the ntm2-1 seeds was reduced by approximately 40% (Fig. 4C). Germination of the 35S:ΔC transgenic seeds was reduced by more than 90% under identical conditions. To confirm the role of the NTM2 gene in germination under high salinity, the ntm2-1 mutant was transformed with the NTM2 gene with its own promoter. The resultant pNTM2-NTM2/ntm2-1 plants were phenotypically similar to control plants (data not shown). Furthermore, its germination phenotype was also similar to that of control seeds under high salinity (Fig. 4D), indicating that the NTM2 gene plays a role in salt regulation of seed germination.
We also examined the effects of ABA and GA on the germination phenotypes of the ntm2-1 seeds. Germination of the ntm2-1 seeds was influenced by ABA in a pattern similar to that observed in control seeds (Fig. 4E). The germination phenotypes of control and ntm2-1 seeds were also similar in the presence of the GA biosynthetic inhibitor paclobutrazol (PAC; Sankhla et al., 2006; Fig. 4F). Together, these observations indicate that the NTM2-mediated salt signaling in seed germination is not related to ABA and GA, which is in agreement with the ABA independence of the NTM2 gene in seedling growth (Fig. 3, C and D).
Auxin Modulates the Germination of ntm2-1 Seeds under High Salinity
Arabidopsis mutants containing lower levels of active auxin exhibit reduced growth and, in many cases, also show enhanced resistance to environmental stresses (Wang et al., 2001; Park et al., 2007; Shibasaki et al., 2009). Auxin has also been implicated in seed germination under stressful conditions (Liu et al., 2007). We found that plant growth was severely retarded in the 35S:ΔC transgenic plants. In addition, seedling growth and seed germination of the ntm2-1 mutant were resistant to high salinity. Therefore, we examined whether the NTM2 gene is related to auxin.
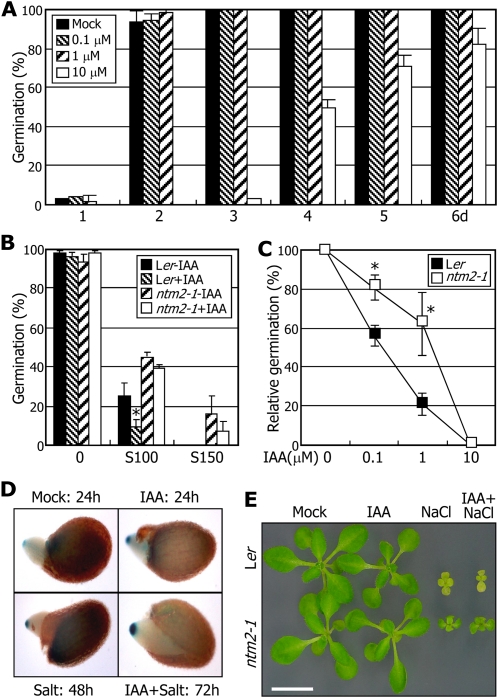
Under normal growth conditions, germination of control seeds was uninfluenced to a discernible level by 1 μm IAA (Fig. 5A). Interestingly, when counted 2 d after cold imbibition under high salinity, the repressive effects of high salinity on germination were exaggerated by IAA in control seeds (Fig. 5B). In contrast, germination of ntm2-1 seeds was influenced to a lesser degree by auxin. To further examine the role of IAA in salt regulation of seed germination, germination assays were carried out in the presence of 150 mm NaCl and varying concentrations of IAA. The results showed that germination of the ntm2-1 seeds was less sensitive to auxin (Fig. 5C), which is consistent with the reduced sensitivity of ntm2-1 seed germination. These observations indicate that whereas auxin does not contribute to seed germination under normal growth conditions, it delays seed germination under high salinity.
Figure 5.
Auxin influences the germination of the ntm2-1 seeds under high salinity. A, Effects of auxin on the germination of Ler seeds. Seeds were cold imbibed and germinated on MS-agar plates supplemented with varying concentrations of IAA. Germination assays were carried out as described in Figure 4B. B and C, Effects of auxin on the germination of ntm2-1 seeds under high salinity. Seeds were cold imbibed and germinated on MS-agar plates supplemented with 1 mm IAA and varying concentrations of NaCl (B) or on MS-agar plates supplemented with 150 mm NaCl and varying concentrations of IAA (C). Germination assays were carried out as described in Figure 4B, except that germination percentages were calculated 2 d after cold imbibition. Error bars indicate se. Statistical significance was determined by Student’s t test (* P < 0.01). S100 and S150, NaCl at 100 mm and 150 mm, respectively. In C, germination percentages were calculated relative to those on NaCl at 150 mm without auxin. D, Effects of auxin and salt on DR5-GUS expression. Cold-imbibed seeds of DR5-GUS plants were allowed to germinate for the indicated time periods on MS-agar plates supplemented with 10 μm IAA or 150 mm NaCl or both. Germinating seeds at similar germination stages were subjected to GUS staining. E, Effects of auxin on ntm2-1 seedling growth under high salinity. Seedlings were grown for 2 weeks on MS-agar plates supplemented with 10 μm IAA or 100 mm NaCl or both. Bar = 10 mm.
The relationship between auxin and NTM2-mediated salt signaling during seed germination was also explored by employing the DR5-GUS reporter system (Ni et al., 2001). A relatively high level of GUS activity was detected in the emerging radicles of germinating seeds (Fig. 5D), as has been reported previously (Ni et al., 2001; Liu et al., 2007). When seeds of the DR5-GUS plants were allowed to germinate on half-strength Murashige and Skoog-agar plates (hereafter referred to as MS-agar plates) containing 100 mm NaCl, the GUS activity was elevated in the emerging radicles, as observed on those supplemented with 1 μm auxin. Notably, the GUS activity was further elevated in the emerging radicles when both NaCl and IAA were included in the assays, showing that elevation of the GUS activity in the emerging radicles goes well with delayed seed germination.
We next examined whether the suppressive effects of auxin on germination were also applicable to other salt stress responses, such as seedling growth. We found that growth retardation and leaf yellowing still occurred in the salt-treated control plants regardless of IAA (Fig. 5E). In addition, ntm2-1 seedling growth under high salinity was also uninfluenced by auxin, indicating that auxin is not related to the NTM2-mediated salt signaling during seedling growth.
IAA30 Is Involved in the NTM2-Mediated Salt Signaling during Seed Germination
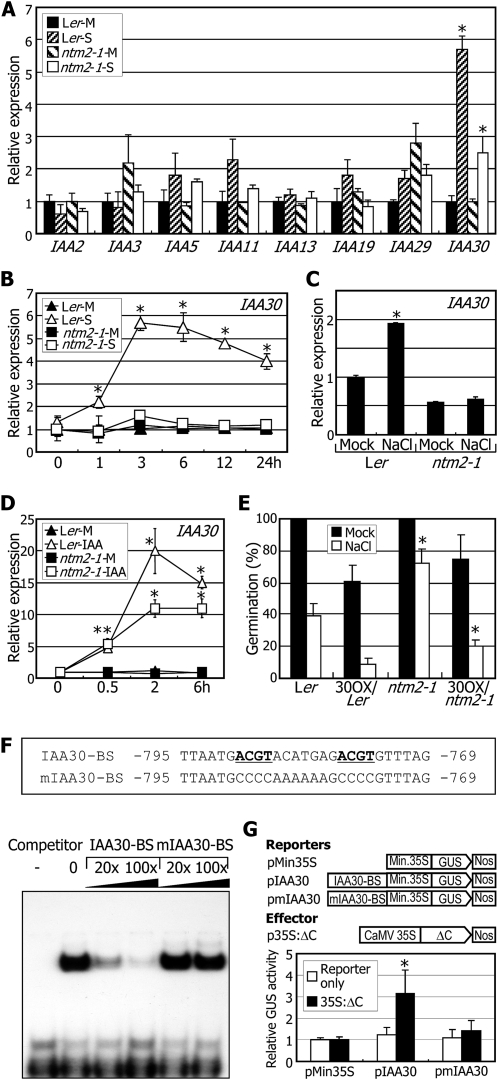
Our data indicated that auxin is linked with the NTM2 gene in seed germination under high salinity. To further investigate the salt-auxin signaling connection, we examined the expression of genes involved in auxin signaling and metabolism, such as Transport Inhibitor Response1 (TIR1) and its potential homologs, Aux/IAAs, and GH3s, in the ntm2-1 mutant. Among the genes examined, the Aux/IAA30 gene (hereafter referred to as IAA30) was most prominently influenced by high salinity (Fig. 6A; Supplemental Fig. S3). It was induced by approximately 6-fold by 150 mm NaCl, as inferred from the digital northern data available in the Genevestigator database (https://www.genevestigator.com/gv/index.jsp). Notably, the salt induction of IAA30 largely disappeared in the ntm2-1 mutant (Fig. 6A). In addition, gene expression studies using the shoot and root samples revealed that whereas the IAA30 gene was not discernibly affected by high salinity in the shoots, it was induced approximately 6-fold in the roots (Supplemental Fig. S4), like the NTM2 gene. These observations indicate that salt induction of the IAA30 gene at least in part depends on NTM2.
Figure 6.
The IAA30 gene is involved in the NTM2-mediated salt regulation of germination. In A to D, transcript levels were determined by qRT-PCR. Biological triplicates were averaged. Error bars indicate se. Statistical significance was determined by Student’s t test (* P < 0.01). Two-week-old plants grown on MS-agar plates were used for treatments with NaCl and IAA. Whole plants were used for extraction of total RNA. M, Mock; S, NaCl. A, Effects of high salinity on IAA gene transcription. Plants were transferred to MS liquid cultures supplemented with 150 mm NaCl and soaked for 3 h before harvesting plant materials. B, Kinetic expression pattern of the IAA30 gene under high salinity. Plants were soaked in MS liquid cultures supplemented with 150 mm NaCl for the indicated time periods before harvesting plant materials. C, IAA30 gene transcription in germinating seeds under high salinity. Cold-imbibed seeds were allowed to germinate for 30 h, and total RNA was extracted from the germinating seeds. D, Kinetic expression pattern of the IAA30 gene after auxin treatments. Plants were transferred to MS liquid cultures supplemented with 10 μm IAA and incubated for the indicated time periods before harvesting plant materials. E, Germination phenotypes of the ntm2-1 mutant overexpressing the IAA30 gene. The ntm2-1 mutant was transformed with the IAA30 gene driven by the CaMV 35S promoter (30OX/ntm2-1). Germination assays were carried out as described in Figure 4B. F, Electrophoretic mobility shift assays on ΔC binding to a conserved sequence in the IAA30 gene promoter. The recombinant ΔC protein was prepared as a MBP-ΔC fusion in E. coli cells. The minus (–) lane is a control without adding ΔC protein. Approximately 0.5 μg of the ΔC protein was used for each assay. The conserved nucleotides underlined were mutated in the mIAA30-BS sequence. Unlabeled DNA fragments were added as competitors. G, Transcriptional activation activity assays in Arabidopsis protoplasts. The IAA30-BS or mIAA30-BS sequence was fused upstream to a 35S minimal TATA promoter (Min35S)-GUS reporter construct (top panel). The reporter and the 35S:ΔC effector constructs were cotransformed into Arabidopsis protoplasts. Luciferase gene expression was used to normalize the GUS activity. Five measurements were averaged (bottom panel). Error bars indicate se. Statistical significance was determined by Student’s t test (* P < 0.01). Nos, Terminator in the pCAMBIA 1305.1 vector.
In addition to the IAA30 gene, other Aux/IAA and GH3 genes, such as IAA11, IAA19, and GH3.4, were also induced by high salinity, but not in the ntm2-1 mutant (Fig. 6A; Supplemental Figs. S3 and S5), suggesting that the NTM2-mediated salt signaling is linked to multiple auxin signaling pathways.
The NTM2-dependent salt induction of IAA30 was further examined by histochemical staining using the pIAA30-GUS fusion construct, in which a GUS-coding sequence was transcriptionally fused to the IAA30 gene promoter sequence, consisting of approximately 1.9 kb upstream of the transcriptional start site. GUS staining assays of transgenic plants expressing the pIAA30-GUS fusion construct revealed that GUS activity was elevated after salt treatments in the roots (Supplemental Fig. S6). In contrast, no elevation of GUS activity was observed in the ntm2-1 mutant background.
Kinetic measurements of the IAA30 transcript revealed that its induction was initiated within 1 h after salt treatments, reaching the plateau at a time point of 3 h (Fig. 6B). In the ntm2-1 mutant, the inductive effects of salt on IAA30 expression were significantly reduced, as observed in histochemical staining assays (Supplemental Fig. S5), showing that the salt induction of the IAA30 gene depends on the NTM2 gene.
We next asked whether the NTM2 dependence of the IAA30 gene expression occurred in the germinating seeds. Cold-imbibed seeds were allowed to germinate for 30 h under high salinity, and the IAA30 transcript levels were measured in the germinating seeds. The results showed that whereas the transcript level was elevated in the salt-treated control seeds, no induction of the IAA30 gene was observed in the germinating ntm2-1 mutant (Fig. 6C), supporting that the IAA30 gene is a component of NTM2-mediated salt signaling in seed germination. Meanwhile, we observed that the inductive effects of auxin on the IAA30 gene decreased by approximately 50% in the ntm2-1 mutant (Fig. 6D), suggesting that the NTM2 gene contributes to the auxin induction of IAA30 expression.
To further examine the relationship between auxin and NTM2-mediated salt signaling in seed germination, the IAA30 gene was overexpressed driven by the CaMV 35S promoter in the ntm2-1 mutant and the germination phenotypes of the resultant transgenic seeds (30OX/ntm2-1) were analyzed. Under high salinity, the germination percentage of the transgenic seeds was lower than that of the ntm2-1 seeds but was comparable to that of control seeds (Fig. 6E), demonstrating that the effects of auxin on seed germination under high salinity are mediated at least in part by the IAA30 gene.
NTM2 Binds to the IAA30 Gene Promoter
We found that the IAA30 gene is induced in the 35S:ΔC transgenic plants. In addition, salt induction of the IAA30 gene requires NTM2. Therefore, we asked whether NTM2 directly regulates the IAA30 gene.
Nucleotide sequence analysis revealed that the IAA30 gene promoter contained a conserved sequence region (Fig. 6F), which is similar to the known NAC-binding sequence (Olsen et al., 2005a). We carried out electrophoretic mobility shift assays to examine whether NTM2 binds to the conserved sequence (IAA30-BS). The recombinant ΔC protein was prepared as a maltose-binding protein (MBP)-ΔC in an Escherichia coli expression system. The MBP-ΔC protein bound to the IAA30-BS sequence (Fig. 6F). In addition, its binding was reduced significantly in the presence of excess amounts of unlabeled competitor DNA. However, this interaction was unaffected by the addition of the mutated competitor DNA (mIAA30-BS), showing that NTM2 binds specifically to the IAA30 gene promoter.
To examine the transcriptional activation activity of NTM2 in planta, we carried out transient coexpression assays in Arabidopsis protoplasts using a series of GUS reporter constructs and the p35S:ΔC effector plasmid (Fig. 6G, top panel). Cotransformation with the pIAA30-BS reporter increased GUS activity approximately 3-fold (Fig. 6G, bottom panel). In contrast, cotransformation with the pmIAA30 reporter did not influence GUS activity, indicating that the NTM2 protein functions as a transcriptional activator of the IAA30 gene by binding directly to the gene promoter.
The NTM2 protein is associated with the plasma membranes. Our data indicate that it activates the IAA30 gene under high salinity, necessitating that it is released from the plasma membranes upon exposure to high salinity. To examine this, the GFP-NTM2 gene fusion was expressed transiently in Arabidopsis protoplasts, and they were exposed to high salinity. We observed that whereas GFP signals were localized predominantly in the plasma membranes (Fig. 1C), a large portion of GFP signals were detected in the nucleus after salt treatments (Supplemental Fig. S7), indicating that NTM2 processing is induced by high salinity.
Germination Is Delayed by Auxin under High Salinity
We found that germination was suppressed by auxin under high salinity. Therefore, it was suspected that the effects of salt on germination would be exaggerated in plants containing higher levels of active auxin.
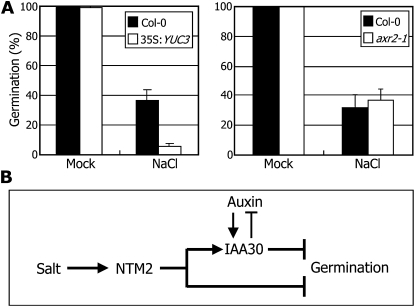
We first examined the germination phenotype of transgenic plants overexpressing the YUCCA3 (YUC3) gene, which encodes an auxin biosynthetic enzyme (Zhao et al., 2001). As inferred from assays on the effects of auxin on germination under high salinity, germination of the 35S:YUC3 transgenic seeds was more severely suppressed by high salt than that of control seeds (Fig. 7A). In contrast, seed germination of the axr2-1 mutant, which has a gain-of-function mutation in the IAA7 gene (Nagpal et al., 2000), was uninfluenced to a discernible level by high salt. In addition, the IAA7 gene was uninfluenced by salt (data not shown), suggesting that the effect of auxin on seed germination under high salinity is not related to the IAA7 gene.
Figure 7.
Auxin interacts with NTM2-mediated salt stress signals during seed germination. A, Germination phenotypes of auxin-related mutants under high salinity. Seeds of axr2-1 mutants and transgenic plants overexpressing the YUC3 gene (At1G04610) driven by the CaMV 35S promoter were cold imbibed and germinated on MS-agar plates supplemented with 150 mm NaCl. Germination assays were carried out as described in Figure 4B. Error bars indicate se. Statistical significance was determined by Student’s t test (* P < 0.01). Col-0, Ecotype Columbia. B, Schematic working model of NTM2 during seed germination under high salinity. A part of the NTM2-mediated salt signals is mediated by IAA30, which functions as a negative regulator of auxin signaling.
Altogether, our observations demonstrate that auxin signals are incorporated into the NTM2-mediated salt signal transduction pathway during seed germination (Fig. 7B). In this signaling cross talk, the IAA30 gene incorporates auxin and salt signals into the germination process.
DISCUSSION
NTM2-Mediated Salt Signaling in Seed Germination
Soil salinity influences a broad spectrum of physiological and developmental events in plants, from seed germination to flowering initiation (Achard et al., 2006; Jamil et al., 2006). High salinity imposes ionic and osmotic stresses on plants (Zhu, 2002; Munns and Tester, 2008). In earlier stages of plant responses, a rapid osmotic phase is caused by the osmotic pressure of high salinity in the soil. After salts accumulate above a threshold level within the plant, a slower ionic phase occurs because of the toxic effects of salt ions.
Molecular genetic and physiological studies have identified numerous genes and molecular mechanisms underlying resistance responses to salt stress. In particular, roles of ABA have been extensively studied in salt regulation of seed germination (Zhu, 2002). Furthermore, it has been shown that the ABA-mediated salt signaling during the germination process is also interconnected with other growth hormone signaling, such as GA and ethylene (Kucera et al., 2005; Holdsworth et al., 2008).
In this work, we found that a NAC transcription factor, NTM2, constitutes a salt signaling pathway functioning in seed germination. Although the NTM2 gene was induced by ABA to some degree, its role in seed germination and seedling growth under high salinity was independent of ABA. Instead, the NTM2-mediated salt signaling was linked to auxin signaling. The NTM2 gene was induced by high salinity primarily in the roots, a major plant organ that perceives soil salinity. Germination of the ntm2-1 mutant seeds was less sensitive to high salinity, and the salt resistance response disappeared in the ntm2-1 mutant complemented with a wild-type NTM2 gene, confirming the role of NTM2 in the salt regulation of seed germination.
Concerning the role of NTM2 in salt signaling, an additional factor that should be considered is its association with the plasma membranes. Whereas the full-size NTM2 protein having a putative TM motif in the C-terminal region was localized at the plasma membranes, the ΔC form lacking the TM motif was found mostly in the nucleus. Transgenic plants overexpressing the full-size NTM2 protein exhibited no discernible phenotypes, but those overexpressing the ΔC form showed distinct phenotypes, as observed with other membrane-bound NAC proteins (Kim et al., 2006, 2008; Seo et al., 2010). Therefore, it is possible that proteolytic release of the NTM2 protein from the plasma membranes would be an additional step regulated by high salinity in addition to the salt effects on NTM2 gene transcription.
The NTM2 protein is released from the plasma membranes under high salinity. It is known that membrane-bound transcription factors are released proteolytically from the membranes by either intramembrane proteases or ubiquitin/proteasome-dependent processing (Seo et al., 2008). All the NAC membrane-bound transcription factors characterized so far, such as NTM1, NTL6, and NTL8, are processed by calpain- or metalloprotease-like activities (Kim et al., 2006, 2008; Seo et al., 2010). It will be interesting to examine whether NTM2 processing is also mediated by specific proteases or by ubiquitin-mediated processing.
Auxin and Seed Germination
Our data demonstrate that auxin signals are interconnected with NTM2-mediated salt signaling during seed germination, in which the IAA30 gene plays a role (Fig. 7B). Whereas auxin does not influence the germination process under normal growth conditions, it acts as a negative regulator of seed germination under high salinity. Consistent with this notion, seed germination of the YUC3-overexpressing plants was more sensitive to high salinity. Histochemical assays using the DR5-GUS reporter system also showed that elevation of GUS activity in the radicles is correlated with delayed seed germination. We observed that GUS activity was elevated by high salinity as well as by auxin in the emerging radicle. It was further elevated when the seeds were allowed to germinate in the presence of both auxin and salt, the combination of which significantly delays seed germination.
However, the relationship between auxin and salt signaling during seed germination does not seem to be simple. The germination process is completed with the protrusion of the radicle through the seed coat (Finch-Savage and Leubner-Metzger, 2006), suggesting that growth potential of the emerging radicle is essential for the completion of the germination process. Therefore, a question to be answered is how the positive regulatory role of auxin on the emergence of the radicles is compromised under high salinity.
The Aux/IAA proteins are short-lived transcription factors that act as repressors of early auxin response genes by forming heterodimers with ARFs that act as positive regulators (Reed, 2001). Auxin enhances the interaction of the Aux/IAA proteins with TIR1, a component of the SCF E3 ligase complex, and directs degradation of the Aux/IAA proteins, resulting in activation of the ARF-regulated genes (Tan et al., 2007). Canonical Aux/IAA proteins are distinguished by having four conserved sequence motifs, designated domains I, II, III, and IV. It is notable that a few Aux/IAA members, including IAA30, IAA31, and IAA20, lack domain II (Dreher et al., 2006), which mediates the interaction with TIR1 (Woodward and Bartel, 2005). Accordingly, the noncanonical AUX/IAA proteins have a longer half-life (Remington et al., 2004). These noncanonical, long-lived Aux/IAA members have been suggested to play distinct roles in auxin response and signaling (Remington et al., 2004; Dreher et al., 2006).
The IAA30 gene is auxin inducible, unlike IAA20 and IAA31 (Remington et al., 2004; Dreher et al., 2006), and expressed mainly in the root apical meristem. The IAA30-overexpressing plants exhibit dwarfed growth with stunted primary root growth because of disturbed root apical meristem activity (Sato and Yamamoto, 2008). We found that it is also induced by high salt. However, the inductive effect of salt disappeared in the ntm2-1 mutant, indicating that salt induction of the IAA30 gene depends on NTM2. Notably, the inductive effect of auxin on IAA30 expression was reduced in the ntm2-1 mutant, further supporting the correlation between the IAA30-mediated auxin signal and the NTM2-mediated salt signal. Previous data and our own data suggest that high salinity, when applied together with auxin, elevates the level of active auxin and thus induces the IAA30 gene to a level that confers an inhibitory effect in the radicle. As a result, the growth potential of the radicle would be reduced, resulting in repression of the germination process.
It has been reported that endogenous auxin contents are elevated in the radicle after cold imbibition in several plant species (Bialek and Cohen, 1989; Ni et al., 2001; Liu et al., 2007), which is necessary for the growth potential of the radicle. However, further elevation of endogenous auxin contents by salt stress in the radicle would also activate negative regulators of auxin signaling, such as IAA30 (Remington et al., 2004). Therefore, it is envisioned that the IAA30-mediated cross talk between salt stress and auxin signals is an adaptation strategy that ensures the germination of seeds only under favorable growth conditions.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) lines used were in the Ler background. The transposon insertion mutant ntm2-1 (CSHL-ET8732), obtained from the Arabidopsis Genetrap Database at Cold Spring Harbor Laboratory, was in the Ler ecotype. Plants were grown in a controlled culture room set at 22°C with a relative humidity of 60% under long days (16 h of light and 8 h of dark) with white light illumination (120 μmol photons m−2 s−1) provided by fluorescent FLR40D/A tubes (Osram). The absence of gene expression in the ntm2-1 mutant was verified by RT-PCR before use.
Arabidopsis Transformation
To produce transgenic plants overexpressing the NTM2, ΔTM, and ΔC genes, individual cDNAs were amplified by RT-PCR using SuperScript II reverse transcriptase (Invitrogen) and the Pfu Turbo DNA polymerase (Stratagene). The PCR products were subcloned into the pB2GW7 expression vector under the control of the CaMV 35S promoter using the Gateway vector system (Invitrogen; Karimi et al., 2002). Agrobacterium tumefaciens-mediated Arabidopsis transformation was performed according to a modified floral dip method (Clough and Bent, 1998). Homozygotic transgenic lines were obtained by selection for three or more consecutive generations.
Analysis of Transcript Levels
Total RNAs were extracted from appropriate plant materials using the RNeasy Plant Mini Kit (Qiagen) or from germinating seeds according to the procedure described previously (Suzuki et al., 2004). They were pretreated with an RNase-free DNase I to eliminate contaminating genomic DNA and cleaned up using the Qiagen Plant Total RNA Isolation Kit.
Transcript levels were determined by qRT-PCR carried out in 96-well blocks using the Applied Biosystems 7500 Real-Time PCR System and the SYBR Green I Master Mix in a volume of 20 μL. The PCR primers were designed using the Primer Express software installed in the system and listed in Supplemental Table S1. The two-step thermal cycling profile used was 15 s at 94°C and 1 min at 68°C. Reactions were carried out in biological triplicates using plant samples harvested separately for each run. The comparative ΔΔCT method was used to evaluate relative quantities of each amplified product. The threshold cycle (CT) was automatically determined for each reaction by the system set with default parameters. PCR specificity was determined by melt curve analysis of amplified products using the standard method installed in the system.
Histochemical Staining
Transgenic plants overexpressing the DR5-GUS reporter, in which GUS gene expression is driven by an artificial DR5 auxin-responsive cis-element (Ni et al., 2001), were used. To examine the effects of IAA and high salt on GUS expression in germinating seeds, the transgenic seeds were germinated on MS-agar plates supplemented with 10 μm IAA or 150 mm NaCl or both.
Transgenic plants overexpressing the pNTM2-GUS and pIAA30-GUS fusions, in which GUS gene expression is driven by the NTM2 and IAA30 gene promoters, respectively, were subject to salt treatments. Plants grown for 2 weeks on MS-agar plates were soaked for 6 h in MS liquid cultures supplemented with 150 mm NaCl.
For histochemical detection of GUS activities, plant materials were incubated in 90% acetone for 15 min on ice, washed twice with rinsing solution [50 mm sodium phosphate, pH 7.2, 0.5 mm K3Fe(CN)6, and 0.5 mm K4Fe(CN)6], and subsequently incubated at 37°C for 1 to 3 h in fresh rinsing solution containing 2 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Duchefa). They were subsequently dipped in ethanol and visualized using a DIMIS-M digital camera (JMTECH).
Germination Assays
Routinely, 2-week-old, air-dried seeds were used for germination assays. Seeds were imbibed on MS-agar plates at 4°C for 3 d in complete darkness and allowed to germinate at 22°C under long days. Emergence of a visible radicle was used as a morphological marker for germination. To examine the effects of IAA, PAC, ABA, and NaCl, MS-agar plates supplemented with 0.1 to 10 μm IAA, 5 to 100 μm PAC, 0.5 to 2 μm ABA, or 50 to 150 mm NaCl were used for the germination assays. Approximately 50 to 60 seeds were routinely counted for each measurement, and three independent measurements were averaged.
Transient Expression Assays in Arabidopsis Protoplasts
For transcriptional activation activity assays, we employed a GAL4 transient expression system using Arabidopsis protoplasts (Miura et al., 2007). The NTM2 gene constructs were fused in-frame to the 3′ end of the GAL4 DNA-binding domain-coding sequence in the effector plasmid. The effector plasmid, the reporter plasmid containing the GUS reporter gene, and the plasmid containing the Renilla luciferase gene, which is used to normalize the measurements, were cotransformed into Arabidopsis protoplasts by a polyethylene glycol (PEG)-mediated transformation method (Yoo et al., 2007). GUS activities were assayed by the fluorometric method as described previously (Jefferson et al., 1987). Five measurements were averaged and statistically treated for individual NTM2 gene constructs.
To examine the transcriptional activation activity of the ΔC protein on the IAA30 gene, several reporter and effector plasmids were prepared. The pMin35S reporter plasmid contains a minimal CaMV 35S promoter (the −56 to −8 sequence region, including the TATA box) and the GUS reporter gene. In the pIAA30 reporter plasmid, a putative NAC-binding sequence (IAA30-BS) identified from the IAA30 gene promoter was fused to the 35S minimal promoter. The IAA30-BS sequence was mutated, resulting in mIAA30-BS, to verify specific binding of the ΔC protein. To construct the p35S:ΔC effector plasmid, the ΔC gene sequence was subcloned into a plant expression vector containing the CaMV 35S promoter. The reporter and effector plasmids were cotransformed into Arabidopsis protoplasts by a PEG-mediated transformation method.
Subcellular Localization of NTM2 Proteins
A GFP-coding sequence was fused in-frame to the 5′ end of the NTM2 gene sequences. The gene fusions were subcloned into the p2FGW7 expression vector (Invitrogen) and transformed into Arabidopsis protoplasts by a PEG-mediated method. The NTM2 proteins were visualized by differential interference contrast microscopy and fluorescence microscopy.
Electrophoretic Mobility Shift Assays
The ΔC gene sequence was subcloned into the pMAL-c2X Escherichia coli expression vector (NEB) containing a MBP-coding sequence. The MBP-ΔC fusion protein was purified according to the manufacturer’s instructions using the pMAL Protein Fusion and Purification System (no. E8000S). A 27-bp IAA30-BS DNA fragment was end labeled with [32P]γdATP using T4 polynucleotide kinase. Labeled probes were incubated for 30 min at 25°C with 0.5 μg of the recombinant MBP-ΔC protein in binding buffer (10 mm Tris-HCl, pH 7.6, 50 mm NaCl, 1 mm EDTA, 5 mm dithiothreitol, and 5% glycerol) supplemented with 100 ng of poly(dI-dC) in the presence or absence of competitor DNAs. The reaction mixtures were electrophoresed on 6% native polyacrylamide gels. The gels were dried on Whatman 3MM paper and exposed to x-ray films.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers IAA30 (At3g62100) and NTM2 (At4g01550).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of NTM1 and NTM2 proteins.
Supplemental Figure S2. Mapping of the transposon insertion site and absence of NTM2 gene expression in the ntm2-1 mutant.
Supplemental Figure S3. Expression patterns of TIR-related and GH3 genes under high salinity.
Supplemental Figure S4. Effects of high salinity on IAA30 gene expression in the shoots (SH) and roots (RO).
Supplemental Figure S5. Kinetic expression patterns of IAA11, IAA19, and GH3.4 genes under high salinity.
Supplemental Figure S6. Effects of high salinity on the promoter activities of the IAA30 gene.
Supplemental Figure S7. High salinity-induced nuclear localization of NTM2.
Supplemental Table S1. List of PCR primers used in this work.
Supplementary Material
Acknowledgments
We thank Dr. Pil Joon Seo for technical support and scientific discussion on seed germination assays.
References
- Abel S. (2007) Auxin is surfacing. ACS Chem Biol 2: 380–384 [DOI] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B. (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Bialek K, Cohen JD. (1989) Free and conjugated indole-3-acetic acid in developing bean seeds. Plant Physiol 91: 775–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgit K, Marc AC, Gerhard LM. (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15: 281–307 [Google Scholar]
- Carrera E, Holman T, Medhurst A, Peer W, Schmuths H, Footitt S, Theodoulou FL, Holdsworth MJ. (2007) Gene expression profiling reveals defined functions of the ATP-binding cassette transporter COMATOSE late in phase II of germination. Plant Physiol 143: 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY. (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44: 903–916 [DOI] [PubMed] [Google Scholar]
- Hibara K, Takada S, Tasaka M. (2003) CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J 36: 687–696 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ. (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Jamil M, Lee DB, Jung KY, Ashraf M, Lee SC, Rhal ES. (2006) Effect of salt (NaCl) stress on germination and early seedling growth of four vegetables species. Cent Eur Agric 7: 273–282 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K. (2010) The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim SG, Lee AK, Yoon HK, Park C-M. (2008) A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J 55: 77–88 [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, Park C-M. (2007) Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res 35: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park C-M. (2006) A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18: 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera B, Cohn MA, Leubner-Metzger G. (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15: 281–307 [Google Scholar]
- Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52: 133–146 [DOI] [PubMed] [Google Scholar]
- Ljung K, Ostin A, Lioussanne L, Sandberg G. (2001) Developmental regulation of indole-3-acetic acid turnover in Scots pine seedlings. Plant Physiol 125: 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Ni DA, Wang LJ, Ding CH, Xu ZH. (2001) Auxin distribution and transport during embryogenesis and seed germination of Arabidopsis. Cell Res 11: 273–278 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. (2005a) DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci 169: 785–797 [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. (2005b) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park C-M. (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282: 10036–10046 [DOI] [PubMed] [Google Scholar]
- Peng J, Harberd NP. (1997) Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol 113: 1051–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E, Kinoshita N, Lopez-Molina L. (2007) The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J 52: 927–936 [DOI] [PubMed] [Google Scholar]
- Quint M, Gray WM. (2006) Auxin signaling. Curr Opin Plant Biol 9: 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135: 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiming L, Wensheng Z, Xiangbing M, Min W, Youliang P. (2007) Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci 172: 120–130 [Google Scholar]
- Sablowski RW, Meyerowitz EM. (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Sankhla D, Davis TD, Sankhla N. (2006) Effect of gibberellin biosynthesis inhibitors on shoot regeneration from hypocotyl explants of Albizzia julibrissin. Plant Cell Rep 13: 115–118 [DOI] [PubMed] [Google Scholar]
- Sato A, Yamamoto KT. (2008) Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant 133: 397–405 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park C-M. (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim SG, Park C-M. (2008) Membrane-bound transcription factors in plants. Trends Plant Sci 13: 550–556 [DOI] [PubMed] [Google Scholar]
- Shibasaki K, Uemura M, Tsurumi S, Rahman A. (2009) Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell 21: 3823–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton ML, Roy BA, Thiede DA. (2000) Evolution in stressful environments. I. Phenotypic variability, phenotypic selection, and response to selection in five distinct environmental stresses. Evolution 54: 93–111 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kawazu T, Koyama H. (2004) RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37: 542–544 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mopper S, Hasenstein KH. (2001) Effects of salinity on endogenous ABA, IAA, JA, and SA in Iris hexagona. J Chem Ecol 27: 327–342 [DOI] [PubMed] [Google Scholar]
- Weijers D, Jürgens G. (2004) Funneling auxin action: specificity in signal transduction. Curr Opin Plant Biol 7: 687–693 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.