Abstract
We have previously shown that the Arabidopsis (Arabidopsis thaliana) RING-H2 E3 ligase RHA2a positively regulates abscisic acid (ABA) signaling during seed germination and postgerminative growth. Here, we report that RHA2b, the closest homolog of RHA2a, is also an active E3 ligase and plays an important role in ABA signaling. We show that RHA2b expression is induced by ABA and that overexpression of RHA2b leads to ABA-associated phenotypes such as ABA hypersensitivity in seed germination and seedling growth, enhanced stomatal closure, reduced water loss, and, therefore, increased drought tolerance. On the contrary, the rha2b-1 mutant shows ABA-insensitive phenotypes and reduced drought tolerance. We provide evidence showing that a rha2a rha2b-1 double mutant generally enhances ABA insensitivity of rha2b-1 in seed germination, seedling growth, and stomatal closure, suggesting that RHA2b and RHA2a act redundantly in regulating ABA responses. Genetic analyses support that, like RHA2a, the RHA2b action in ABA signaling is downstream of a protein phosphatase 2C, ABA-INSENSITIVE2 (ABI2), and in parallel with that of the ABI transcription factors ABI3/4/5. We speculate that RHA2b and RHA2a may have redundant yet distinguishable functions in the regulation of ABA responses.
The phytohormone abscisic acid (ABA) regulates many processes of plant growth and development, such as seed maturation, germination, and seedling growth. ABA is also a key hormone mediating plant adaptation to various environmental challenges, including drought, salt, cold, and other abiotic stresses (Koornneef et al., 1989; Leung and Giraudat, 1998; Finkelstein et al., 2002). For example, ABA is synthesized in response to water deficit and then induces stomatal closure via the efflux of K+ and anions from guard cells (Schroeder et al., 2001; Pandey et al., 2007). The ABA-controlled stomatal movement is vital for plant survival, and ABA-deficient and ABA-responsive mutants are susceptible to water stress (Kang et al., 2002).
Through characterization of a series of ABA-insensitive mutants that are resistant to ABA-mediated inhibition of seed germination and/or postgerminative growth, several components regulating ABA signaling in seeds and/or guard cells have been identified in the model system of Arabidopsis (Arabidopsis thaliana; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Merlot et al., 2001). Among them, the phosphatase 2C proteins ABA-INSENSITIVE1 (ABI1) and ABI2 are negative regulators of ABA signaling. It was recently shown that ABI1/2 physically interact with and inhibit downstream target proteins such as SNF1-related kinases. Stress-induced ABA promotes the recently identified ABA receptors PYR (pyrabactin resistance)/PYL (PYR1-like)/RCARs (regulatory components of ABA receptors) to interact with these protein phosphatase 2C (PP2C) proteins, and this ABA-dependent interaction down-regulates PP2C activity and therefore relieves the inhibition of their downstream target protein kinases (Fujii et al., 2009; Ma et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Park et al., 2009; Santiago et al., 2009). In contrast to ABI1/2, the ABI transcription factors ABI3, ABI4, and ABI5 are positive regulators of ABA signaling that share overlapping functions during seed germination and early seedling development (Giraudat et al., 1992; Parcy et al., 1994; Parcy and Giraudat, 1997; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001, 2002).
Ubiquitination and proteolysis by the 26S proteasome pathway is the dominant selective protein turnover system in plants (Moon et al., 2004; Schwechheimer and Schwager, 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007). Three enzymes, E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase), act sequentially to catalyze the covalent attachment of a 76-amino acid protein called ubiquitin to the target protein. The specificity of ubiquitination is largely determined by E3, which recruits appropriate substrates (Smalle and Vierstra, 2004). A growing body of evidence indicates that RING class E3 enzymes play important roles in regulating ABA signaling and related abiotic stress responses (Lee et al., 2001; Lopez-Molina et al., 2003; Zhang et al., 2005, 2007; Stone et al., 2006). For example, the E3 ligase AIP2 (for ABI3 interaction protein 2), which serves as a negative regulator of ABA signaling, is believed to target ABI3 for proteolysis (Zhang et al., 2005). ABI5 was reported to be regulated by the ubiquitin-dependent proteolysis (Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001, 2003; Smalle et al., 2003; Liu and Stone, 2010). The RING E3 ligase KEG probably targets ABI5 for degradation (Stone et al., 2006). The RING-H2 finger protein XERICO confers drought tolerance through increased ABA biosynthesis and interacts with E2 ubiquitin-conjugating enzyme (AtUBC8) and ASK1-interacting F-box protein (AtTLP9), which is involved in the ABA signaling pathway (Ko et al., 2006). SALT- AND DROUGHT-INDUCED RING FINGER1 (SDIR1) is an active E3 ligase and involved in ABA-related stress signal transduction. Overexpression of SDIR1 leads to ABA hypersensitivity and ABA-associated phenotypes, such as salt hypersensitivity in germination, enhanced ABA-induced stomatal closing, and enhanced drought tolerance (Zhang et al., 2007). These results suggest a link between protein ubiquitination and ABA-mediated stress responses in plants.
We have previously shown that the Arabidopsis RING-H2 E3 ligase RHA2a positively regulates ABA signaling during seed germination and postgerminative growth (Bu et al., 2009). Here, we report that RHA2b, the closest homolog of RHA2a, is also an active E3 ligase and plays an important role in ABA signaling. We provide evidence showing that RHA2b and RHA2a have redundant yet distinguishable functions in regulating ABA signaling and drought response.
RESULTS
RHA2b Exhibits E3 Ligase Activity
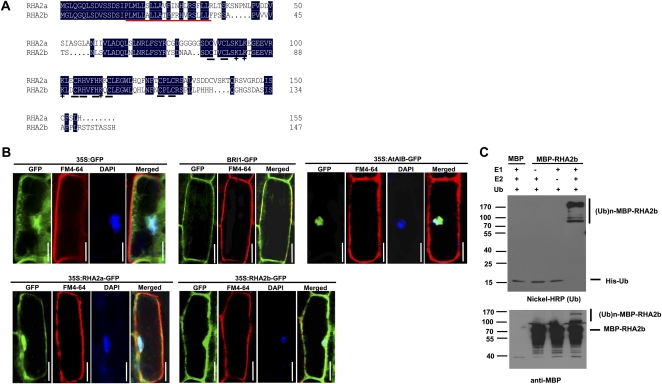
RHA2b, which is the most closely related protein to RHA2a (Lechner et al., 2002), shares 65% amino acid sequence identity with RHA2a (Fig. 1A). Both RHA2b and RHA2a contain a putative transmembrane domain and noncanonical nuclear localization sequence (Fig. 1A). To examine the subcellular localization of RHA2b and RHA2a, a GFP reporter gene was fused in frame to their coding region under the control of the 35S promoter to produce 35S:RHA2b-GFP and 35S:RHA2a-GFP translational fusions. Stable transgenic plants containing the 35S:RHA2b-GFP or 35S:RHA2a-GFP constructs showed increased sensitivity to ABA over the wild type (Supplemental Fig. S1A). When crossed into the rha2b-1 or rha2a mutant background, 35S:RHA2b-GFP and 35S:RHA2a-GFP can rescue the ABA insensitivity of rha2b-1 and rha2a (Supplemental Fig. S1B). These results demonstrated that these translational fusions are functional. Laser confocal microscopic analyses revealed that, in the presence of MG132, GFP fluorescence was detected both in the plasma membrane and the nucleus of root epidermal cells of transgenic seedlings (Fig. 1B; Supplemental Fig. S1C). As controls, our parallel experiments indicated that the previously described 35S:AtAIB-GFP fusion (Li et al., 2007) is mainly expressed in the nucleus, the BRI1-GFP fusion (Friedrichsen et al., 2000) is mainly expressed in the plasma membrane, whereas the 35S:GFP fusion is ubiquitously expressed in the membrane, the cytosol, and the nucleus (Fig. 1B). These results suggest that RHA2b and RHA2a are likely localized both in the plasma membrane and the nucleus.
Figure 1.
RHA2b exhibits E3 ligase activity. A, Alignment of deduced amino acid sequences of RHA2b and RHA2a. Conserved residues are highlighted in black, and Cys and His residues in the RING finger domain are underlined in black. The putative transmembrane domain is underlined in red, and basic amino acids in the RING finger domain are marked by cross stars. B, Subcellular localization of RHA2b and RHA2a. For 35S:GFP, 35S:AtAIB-GFP, 35S:RHA2a-GFP, or 35S:RHA2b-GFP, roots of 1-week-old seedlings were immersed in 2 μg mL−1 DAPI solution for 10 to 15 min and then stained with 5 μg mL−1 cold FM4-64 solution for 1 min. Before staining, the whole seedlings of 35S:RHA2a-GFP or 35S:RHA2b-GFP were pretreated with 50 μm MG132 for 6 h under dim-light conditions; for BRI1-GFP, roots of 1-week-old seedlings were stained with 5 μg mL−1 cold FM4-64 solution for 1 min. All seedlings were observed with fluorescence microscopy. Bars = 30 μm. C, MBP-RHA2b fusion protein was assayed for E3 activity in the presence of E1 (from wheat [Triticum aestivum]), E2 (UBCh5b), and 6× His tag ubiquitin (Ub). The numbers at left denote the molecular masses of marker proteins in kD. MBP itself was used as a negative control. Samples were resolved by 8% SDS-PAGE. Nickel-horseradish peroxidase (HRP) was used to detect His tag ubiquitin (top panel), and the anti-MBP antibody was used for maltose fusion proteins (bottom panel).
Like RHA2a, RHA2b contains a typical C3H2C3 domain in the C terminus (Fig. 1A). To test that RHA2b may exhibit E3 ligase activity, we produced a maltose-binding protein (MBP)-RHA2b fusion protein and conducted an in vitro E3 ligase activity assay. In the presence of E1 and E2 (UbcH5B), purified MBP-RHA2b was able to execute autoubiquitination, as evidenced by the formation of high-molecular-mass proteins detected by immunoblot analyses using anti-His (Fig. 1C, top panel) or anti-MBP (Fig. 1C, bottom panel) antibodies. As negative controls, when E1 or E2 was omitted from the reaction, no polyubiquitination of MBP-RHA2b was detected (Fig. 1C, second and third lanes from left). These results indicate that, like RHA2a, RHA2b also has E3 ligase activity (Bu et al., 2009).
RHA2b Shows a Distinct Expression Pattern from RHA2a
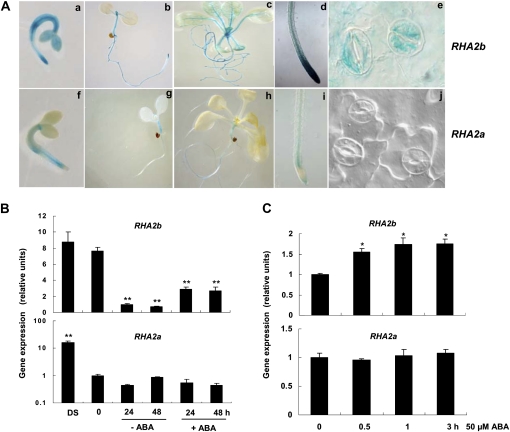
To examine the expression pattern of RHA2b, we generated a translational fusion of the RHA2b coding sequence with the GUS coding region from the uidA gene under the control of the RHA2b promoter. The resulting pRHA2b:RHA2b-GUS construct was introduced into the rha2b-1 mutant. ABA response assays showed that this construct restored the ABA-insensitive phenotype of the rha2b-1 mutant, indicating that the fusion protein is functional (Supplemental Fig. S2A). Similarly, we generated a functional pRHA2a:RHA2a-GUS construct, which restored the rha2a mutant phenotype (Supplemental Fig. S2A). These transgenic plants were subjected to histochemical staining to compare the expression pattern of RHA2b and RHA2a. As shown in Figure 2A, in 2-, 7-, and 14-d-old seedlings, pRHA2b:RHA2b-GUS was ubiquitously expressed in both root and shoot tissues, with a rich expression in shoot and root meristems and vascular tissues (Fig. 2A, a–d). Compared with pRHA2b:RHA2b-GUS, pRHA2a:RHA2a-GUS expression was generally weak, and low GUS expression can be observed in hypocotyls and roots (Fig. 2A, f–i). Significantly, in 14-d-old seedlings, RHA2b was found to be richly expressed in stomata, but RHA2a was not (Fig. 2A, e and j).
Figure 2.
Comparison of RHA2b and RHA2a expression. A, pRHA2b:RHA2b-GUS and pRHA2a:RHA2a-GUS seedling staining. a and f, Two-day-old germinating seedlings. b and g, Seven-day-old seedlings. c and h, Fourteen-day-old seedlings. d and i, Primary roots of 7-d-old seedlings. e and j, Guard cells in leaves of 14-d-old seedlings. B, RHA2b and RHA2a expression pattern in dry seeds (DS) and during seed imbibition revealed by qRT-PCR. Imbibed seeds were kept in darkness at 4°C for 72 h and then transferred to medium with or without ABA (5 μm) in constant light at 22°C for germination. Total RNA was extracted at the indicated times (0 indicates the time immediately before transfer). Transcript levels were quantified by qRT-PCR against ACTIN2. Data shown are means ± sd of three independent biological determinations. C, ABA-induced RHA2b and RHA2a expression revealed by qRT-PCR. Two-week-old wild-type seedlings were treated with 50 μm ABA for 0, 0.5, 1, and 3 h. ACTIN2 primers were used as an internal control. Data shown are means ± sd of three independent biological determinations. Asterisks in B and C indicate significant differences from the corresponding control values determined by Student’s t test (* 0.01 ≤ P < 0.05, ** P < 0.01).
We further examined the stratification-induced expression of RHA2b and RHA2a using quantitative reverse transcription (qRT)-PCR assays. As previously reported, RHA2a transcripts were abundant in dry seeds and significantly reduced when seeds were stratified at 4°C for 72 h (Bu et al., 2009). Stratification-induced reduction of RHA2b transcripts was slower than that of RHA2a, and an obvious decrease of RHA2b expression occurred 24 h after the stratification (Fig. 2B). When the stratified seeds were transferred to medium for germination, the expression of RHA2b was higher in medium containing 5 μm ABA than that containing no ABA. In contrast, parallel experiments did not show any obvious induction of RHA2a transcription by ABA (Fig. 2B). These results indicate that the phytohormone ABA induces the expression of RHA2b during early stages of seed germination.
Next, we compared ABA- and stress-induced expression patterns of RHA2b with RHA2a using 2-week-old seedlings (see “Materials and Methods”). As shown in Figure 2C and Supplemental Figure S2B, RHA2b expression was up-regulated by ABA and drought treatment, but RHA2a expression was not induced by these treatments.
RHA2b Acts Additively with RHA2a in Regulating ABA-Mediated Seed Germination and Postgerminative Growth
To explore the biological function of RHA2b in ABA signaling, we obtained a T-DNA insertion mutant, SALK_014943 (named rha2b-1), from the Arabidopsis Biological Resource Center (ABRC) that contains a T-DNA insertion at 168 bp upstream of the translational start codon of RHA2b (Supplemental Fig. S3A). qRT-PCR assay revealed that the rha2b-1 mutation reduced the expression of RHA2b by approximately 50% (Supplemental Fig. S3B). At the same time, we generated RHA2b-overexpressing plants containing the 35S promoter fused to the RHA2b coding region. RHA2b expression in these lines was assessed by qRT-PCR (Supplemental Fig. S4A). The RHA2b-overexpressing lines tested showed increased sensitivity to ABA. In our seed germination and cotyledon greening assays (Supplemental Fig. S4, C–F), line 9 was more sensitive to ABA than line 1. Given that RHA2b expression levels in line 9 were higher that those in line 1 (Supplemental Fig. S4A), our results suggest that the ABA hypersensitivity in these RHA2b-overexpressing lines is correlated with their RHA2b expression levels. Line 9 was named 35S:RHA2b and used for further studies.
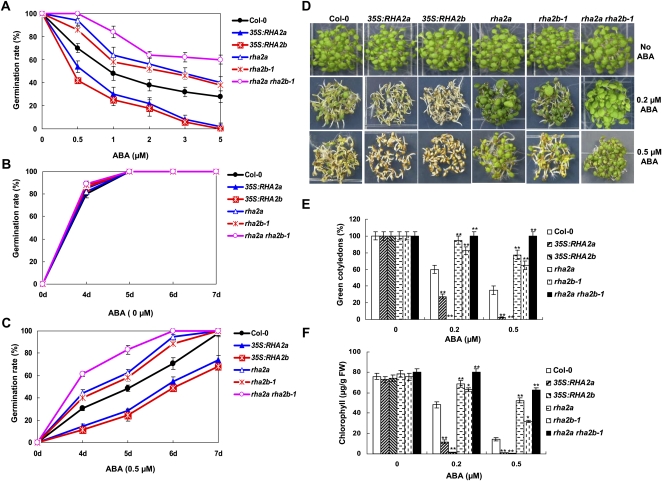
rha2b-1 and 35S:RHA2b were compared with the wild type for their responses to ABA during seed germination. In an ABA dose-response assay, seeds were sown on medium containing different concentrations of ABA and seed germination (obvious radicle emergence) percentage was scored at 3 d after the end of stratification. As shown in Figure 3A, in the presence of different concentrations of ABA, rha2b-1 plants showed higher seed germination percentage than the wild type, whereas 35S:RHA2b plants exhibited significantly reduced seed germination percentage. In the absence of ABA, seed germination of different genotypes was similar (Fig. 3B). When germinated on medium containing 0.5 μm ABA, germination of rha2b-1 seeds was earlier than in the wild type, whereas germination of 35S:RHA2b seeds was much later than in the wild type (Fig. 3C).
Figure 3.
ABA responses of Col-0, 35S:RHA2a, rha2a, 35S:RHA2b, rha2b-1, and rha2a rha2b-1 plants in seed germination and postgerminative growth. A, Seed germination percentage of the indicated genotypes grown on different concentrations of ABA was recorded at 3 d after the end of stratification. Data shown are means ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. B and C, Seed germination time course of the six genotypes grown on medium without ABA (B) or containing 0.5 μm ABA (C). Data shown are means ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. D, Photographs of young seedlings at 5 d after the end of stratification. Seeds were germinated and allowed to grow on horizontal agar medium containing 0, 0.2, and 0.5 μm ABA. E, Cotyledon greening percentage of the seedlings described in D. Values represent means ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. F, Quantification of chlorophyll content. Seedlings described in D were collected for chlorophyll a/b extraction and measurement. Values represent means ± sd of three replicates. FW, Fresh weight of whole seedlings. Asterisks in E and F indicate significant differences from the corresponding wild-type values determined by Student’s t test (* 0.01 ≤ P < 0.05, ** P < 0.01). At least three independent experiments were conducted, and similar results were obtained.
rha2b-1 and 35S:RHA2b plants were also assessed for their responses to ABA during the postgerminative growth stage. For these experiments, seeds were grown on medium containing different concentrations of ABA for 7 d (including 2 d of stratification) before cotyledon greening percentage and cotyledon chlorophyll content were examined. In the absence of ABA, cotyledon greening of the three genotypes was similar (Fig. 3, D–F). In the presence of 0.2 or 0.5 μm ABA, cotyledon greening of rha2b-1 seedlings, as measured by cotyledon greening percentage and chlorophyll content, was higher than that of the wild type. In contrast, cotyledon greening of 35S:RHA2b was lower than that of the wild type (Fig. 3, D–F). Together, these results demonstrate that whereas rha2b-1 is less sensitive than the wild type to ABA during seed germination and postgerminative growth, 35S:RHA2b is more sensitive than the wild type to ABA during the same stage, suggesting that RHA2b acts as a positive regulator of ABA signaling during seed germination and postgerminative growth.
To test the possible genetic interaction of RHA2b with RHA2a, which has been shown to exhibit similar action as RHA2b in ABA-mediated seed germination and postgerminative growth (Bu et al., 2009), we generated a rha2a rha2b-1 double mutant line and compared its ABA response with that of the single mutants. The rha2a rha2b-1 double mutant was significantly more insensitive than rha2a or rha2b-1 to ABA in seed germination (Fig. 3, A–C) and postgerminative growth (Fig. 3, D–F). These results support that RHA2b acts additively with RHA2a in regulating ABA-mediated inhibition of seed germination and postgerminative growth.
RHA2b Acts Additively with RHA2a in Regulating ABA-Mediated Inhibition of Seedling Growth
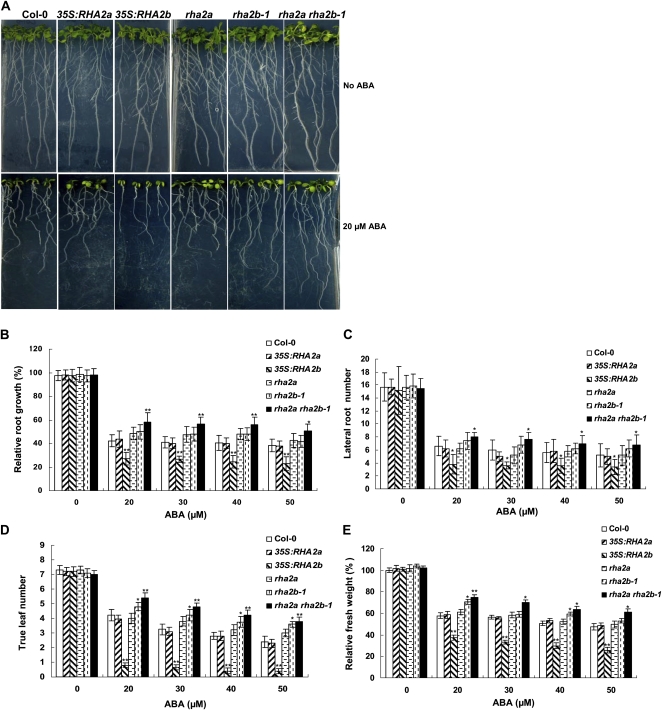
In addition to inhibiting seed germination and early seedling establishment, high concentrations of ABA also show a suppression effect on shoot and root growth of seedlings (Fujii et al., 2007). To test the possible role of RHA2b in ABA-mediated inhibition of seedling growth, 4-d-old seedlings grown on Murashige and Skoog (MS) medium were transferred to medium containing different concentrations of ABA (0, 20, 30, 40, and 50 μm) and grown for another 11 d before root and shoot growth were assessed (see “Materials and Methods”). In the absence of ABA, seedling growth of different genotypes was largely similar (Fig. 4). In the presence of different concentrations of ABA, root and shoot growth of the 35S:RHA2b plants were more severely inhibited than in the wild type, whereas shoot growth of rha2b-1 was slightly better than in the wild type. For root growth, 35S:RHA2b plants had shorter taproot length and fewer lateral roots than the wild type (Fig. 4, B and C); however, the differences between rha2b-1 and the wild type in taproot length and lateral root number were not significant. For shoot growth, 35S:RHA2b seedlings generally showed poor true leaf establishment and reduced whole seedling fresh weight as compared with the wild type, while rha2b-1 seedlings were slightly better than the wild type (Fig. 4, D and E). These results indicated that RHA2b is important for ABA-mediated inhibition of seedling growth. Interestingly, our parallel experiments indicated that, in ABA-containing medium, shoot and root growth of 35S:RHA2a or rha2a seedlings showed little, if any, difference from those of the wild type (Fig. 4). The rha2a rha2b-1 double mutant, however, grew much better than any of the single mutants and the wild type in ABA-containing medium, as assessed by root growth (Fig. 4B), lateral root number (Fig. 4C), true leaf number (Fig. 4D), and seedling fresh weight (Fig. 4E). These results suggest that RHA2b acts redundantly with RHA2a in regulating ABA-mediated inhibition of seedling growth.
Figure 4.
ABA responses of Col-0, 35S:RHA2a, rha2a, 35S:RHA2b, rha2b-1, and rha2a rha2b-1 plants in seedling growth. A, Sensitivity of seedlings to ABA. Four-day-old seedlings grown without ABA were transferred to a plate containing 20 μm ABA. The photographs were taken 11 d after the transfer. B, Measurement of taproot length. Taproot lengths of the seedlings in medium containing the indicated concentrations of ABA were measured (see “Materials and Methods”). Relative root growth compared with that on ABA-free medium is indicated. Data shown are means ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. C, Quantification of lateral root number. D, Quantification of true leaf number. E, Measurement of seedling fresh weight. Seedlings were collected for measurement of fresh weight. Relative fresh weight compared with that on ABA-free medium is indicated. Asterisks in B to E indicate significant differences from the corresponding wild-type values determined by Student’s t test (* 0.01 ≤ P < 0.05, ** P < 0.01). At least three independent experiments were conducted, and similar results were obtained. [See online article for color version of this figure.]
Genetic Analysis of RHA2b Action in ABA Signaling
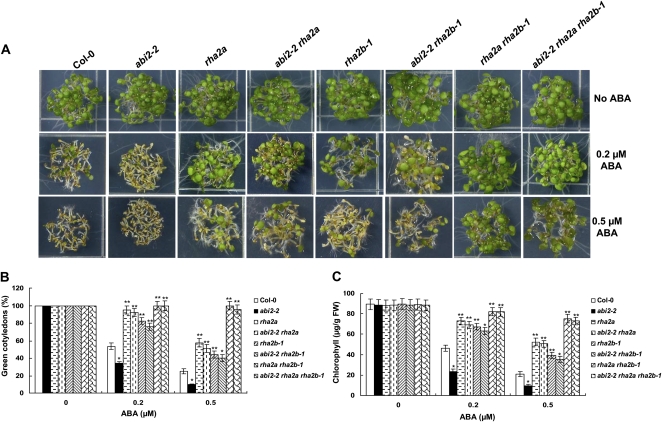
Based on a recent model of ABA signaling, the ABI2 protein, which is a PP2C that negatively regulates ABA response, interacts with the ABA receptors PYR/PYL/RCARs in the presence of ABA (Ma et al., 2009; Park et al., 2009). The abi2-2 mutant (SALK_015166), which contains a T-DNA that disrupts ABI2 expression, showed increased sensitivity to ABA in our postgerminative growth assays (Fig. 5; Supplemental Fig. S5). In the abi2-2 rha2a or abi2-2 rha2b-1 double mutants as well as the abi2-2 rha2a rha2b-1 triple mutant, the ABA hypersensitivity of abi2-2 was relieved (Fig. 5), suggesting that RHA2a and RHA2b act downstream of ABI2 in ABA signaling.
Figure 5.
ABA responses of double mutant plants abi2-2 rha2a and abi2-2 rha2b-1 and triple mutant plants abi2-2 rha2a rha2b-1 in early seedling growth. A, Photographs of young seedlings grown horizontally on medium containing 0, 0.2, or 0.5 μm ABA for 5 d after the end of stratification. B, Cotyledon greening percentage of the seedlings described in A. Values represent means ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. C, Quantification of chlorophyll content. Seedlings described in A were collected for chlorophyll a/b extraction and measurement. Values represent means ± sd of three replicates. FW, Fresh weight of whole seedlings. Asterisks in B and C indicate significant differences from the corresponding wild-type values determined by Student’s t test (* 0.01 ≤ P < 0.05, ** P < 0.01). At least three independent experiments were conducted, and similar results were obtained.
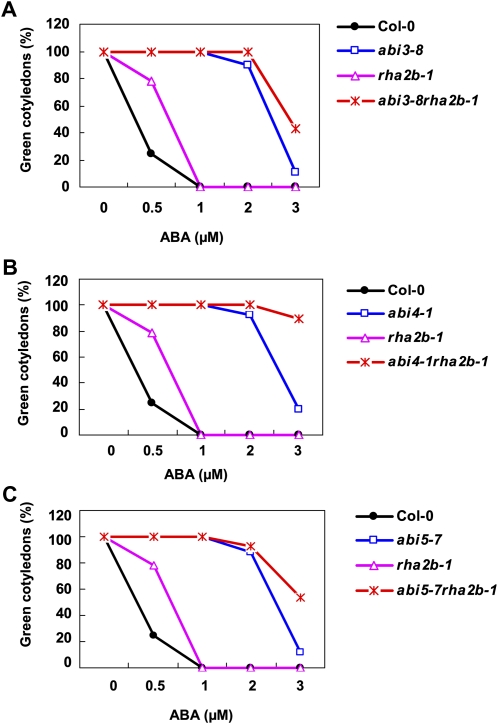
We also examined the genetic relationship of RHA2b with ABI3, ABI4, and ABI5, which are positive regulators of ABA-mediated gene transcription. Cotyledon greening assays indicated that double mutant lines rha2b-1 abi3-8 (Fig. 6A), rha2b-1 abi4-1 (Fig. 6B), and rha2b-1 abi5-7 (Fig. 6C) are more insensitive to ABA than any of the single mutants, suggesting that the action of RHA2b in ABA signaling is independent of that of the ABI transcription factor genes, including ABI3, ABI4, and ABI5.
Figure 6.
Double mutant analysis between rha2b-1 and abi3-8, abi4-1, or abi5-7. Cotyledon greening percentage of the indicated genotypes grown on medium containing different concentrations of ABA was recorded at 5 d after the end of stratification. Data shown are means ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. [See online article for color version of this figure.]
RHA2b Affects Plant Drought Tolerance
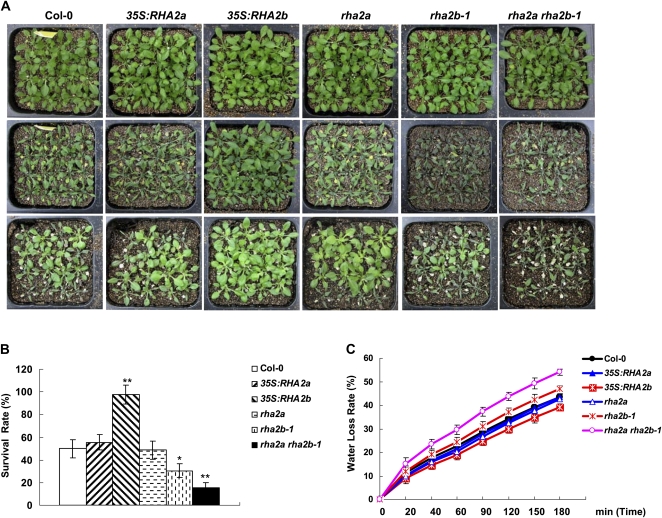
To test that RHA2b may play a role in regulating plant response to drought, 1-week-old 35S:RHA2b, rha2b-1, and wild-type plants were transplanted to soil for drought tolerance assays (see “Materials and Methods”). As shown in Figure 7A, most of the 35S:RHA2b plants survived the water stress and exhibited continued growth, but water stress led to most of the wild-type and rha2b-1 plants withering. After rewatering, 35S:RHA2b plants showed a high survival rate (97%), whereas the corresponding survival rates were 52% for wild-type plants and 33% for rha2b-1 mutants (Fig. 7B). Even though the drought tolerance of 35S:RHA2a and rha2a did not exhibit significant differences compared with the wild type, the rha2a rha2b-1 double mutants were more susceptible to water stress than the single mutants (Fig. 7, A and B), suggesting an additive interaction between RHA2b and RHA2a in regulating plant drought tolerance. In line with their drought-tolerant performance, 35S:RHA2b detached leaves exhibited slower water loss than wild-type plants and rha2b-1 detached leaves exhibited quicker water loss than wild-type plants (Fig. 7C). Consistent with our previous results (Bu et al., 2009), RHA2a itself showed little effect on leaf water loss, but the rha2a rha2b-1 double mutant showed increased leaf water loss over the single mutants (Fig. 7C).
Figure 7.
Drought responses of Col-0, 35S:RHA2a, rha2a, 35S:RHA2b, rha2b-1, and rha2a rha2b-1 plants. A, Drought tolerance assay. Seven-day-old seedlings were transferred to soil for another 2 weeks (top row), subjected to progressive drought by withholding water for specified times (middle row), and then rewatered for 2 d (bottom row). B, Survival rate of the plants from A after rewatering; sd (error bars) was calculated from results of three independent experiments (n > 30 for each experiment). Asterisks indicate significant differences from the corresponding wild-type values determined by Student’s t test (* 0.01 ≤ P < 0.05, ** P < 0.01). C, Water loss rate. Leaves of the same developmental stage were excised and weighed at various time points after detachment. Values are means ± sd of three individual plants per genotype. Experiments were repeated at least three times with similar results.
RHA2b Regulates ABA-Mediated Stomatal Closure
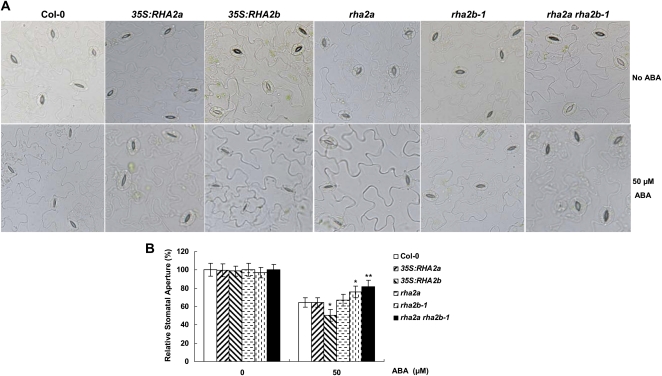
RHA2b is expressed in stomata (Fig. 2Ae) and 35S:RHA2b plants show increased drought tolerance (Fig. 7), which prompted us to investigate whether RHA2b regulates ABA-mediated stomatal closure, one of the ABA-controlled processes that determine the rate of transpiration under water deficit conditions. For these experiments, plants were exposed to high humidity for 12 h to open stomata fully, and epidermal peels from these plants were used to analyze stomatal responses to ABA. As shown in Figure 8, treatment of wild-type plants with 50 μm ABA for 3 h led to stomatal apertures being reduced to 64% of the control level (from 4.55 ± 0.74 μm to 2.94 ± 0.56 μm), whereas the same treatment led to stomatal apertures of 35S:RHA2b plants being reduced to 50% of the control level (from 4.50 ± 0.55 μm to 2.47 ± 0.62 μm) and those of rha2b-1 plants being reduced to 76% of the control level (from 4.42 ± 0.56 μm to 3.46 ± 0.65 μm), indicating that whereas 35S:RHA2b plants were more sensitive than the wild type to ABA in stomatal closure, rha2b-1 plants were less sensitive. Parallel experiments indicated that overexpression or mutation of RHA2a showed negligible effects on ABA-induced stomatal closure but the rha2a rha2b-1 double mutant was more insensitive than any of the single mutants to ABA in stomatal closure, suggesting that RHA2b acts additively with RHA2a in regulating ABA-mediated stomatal closure (Fig. 8).
Figure 8.
ABA-induced stomatal aperture of Col-0, 35S:RHA2a, rha2a, 35S:RHA2b, rha2b-1, and rha2a rha2b-1. A, Comparison of stomatal aperture in response to ABA. Epidermal peels from different genotype plants were kept for 12 h in the dark, incubated under light in stomata-opening solution for 1.5 h, and then treated with 0 and 50 μm ABA for 3 h before being observed by scanning electron microscopy. B, Measurement of stomatal aperture in response to ABA. The stomata width of epidermal peels from A was measured. Relative stomatal aperture compared with that on ABA-free medium is indicated. Data shown are means ± sd of three independent experiments (n = 40–50). Asterisks indicate significant differences from the corresponding wild-type values determined by Student’s t test (* 0.01 ≤ P < 0.05, ** P < 0.01).
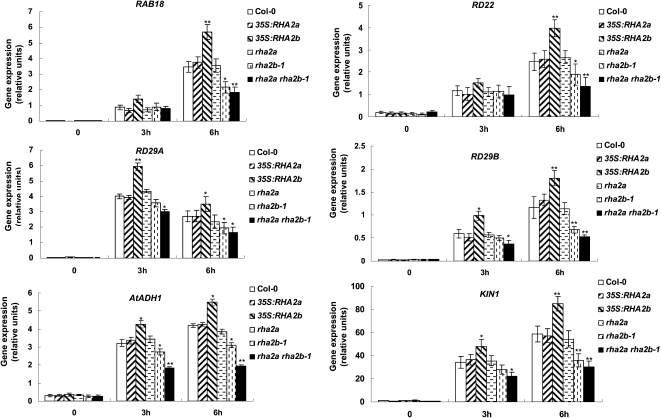
RHA2b Affects the Expression of ABA-Induced Stress-Responsive Genes
qRT-PCR assays indicated that ABA treatment led to increased expression of several ABA-inducible marker genes, including RAB18, RD29A, KIN1, AtADH1, RD22, and RD29B (Kurkela and Borg-Franck, 1992; Lång and Palva, 1992; Abe et al., 2003) in wild-type plants (Fig. 9). In line with the drought-tolerant performance of 35S:RHA2b or rha2b-1 plants, ABA-induced expression of these marker genes was generally intensified in 35S:RHA2b plants but attenuated in rha2b-1 plants (Fig. 9). Overexpression or mutation of RHA2a showed little effect on ABA-induced marker gene expression, but the attenuation of ABA-induced marker gene expression was more severe in rha2a rha2b-1 than in rha2b-1, suggesting that RHA2b acts redundantly with RHA2a in regulating the expression of ABA-induced stress-responsive genes (Fig. 9).
Figure 9.
Expression of ABA- and stress-responsive genes. Expression of ABA- and stress-responsive genes was assayed by qRT-PCR in seedlings of Col-0, 35S:RHA2a, rha2a, 35S:RHA2b, rha2b-1, and rha2a rha2b-1. Two-week-old seedlings were treated with 100 μm ABA for 0, 3, and 6 h. Data shown are means ± sd of three independent experiments. Asterisks indicate significant differences from the corresponding wild-type values determined by Student’s t test (* 0.01 ≤ P < 0.05, ** P < 0.01).
DISCUSSION
We report here the role of the RHA2b gene, which encodes an active E3 ubiquitin ligase (Fig. 1, A and C) and plays a positive role in regulating ABA signaling and stress responses. Overexpression of RHA2b leads to ABA-associated phenotypes such as ABA hypersensitivity in seed germination (Fig. 3, A–C), seedling growth (Figs. 3, D–F, and 4), enhanced stomatal closure (Fig. 8), reduced water loss (Fig. 7C), and, therefore, increased drought tolerance (Fig. 7, A and B). On the contrary, the rha2b-1 mutant shows ABA-insensitive phenotypes and reduced drought tolerance (Figs. 3, 4, 7, and 8). These data support that RHA2b is a positive regulator of the ABA signal transduction pathway. It is worthy of note that the RHA2b gene shows high sequence similarity to RHA2a, which is also a positive regulator of ABA signaling (Jensen et al., 1998; Lechner et al., 2002; Bu et al., 2009). We provide here several lines of evidence showing that the expression of the two homologous genes is distinguishable in several respects. First, compared with RHA2b, RHA2a expression shows a relatively higher abundance in dry seeds and a faster down-regulation upon stratification (Fig. 2B). Second, compared with RHA2a, RHA2b expression keeps high in later growth stages, especially in the vascular tissue and meristematic regions of shoots and roots of 14-d-old seedlings (Lechner et al., 2002; Fig. 2A). Third, RHA2b expression is induced by ABA and drought stress, whereas RHA2a expression is not (Fig. 2C; Supplemental Fig. S2B). These results suggest that RHA2a and RHA2b could have distinct physiological functions. Indeed, we show that 35S:RHA2a and rha2a exhibit opposite ABA sensitivity during seed germination and postgerminative growth (Fig. 3), suggesting that RHA2a mainly acts in these stages. We also show that, in addition to controlling ABA-mediated seed germination and postgerminative growth like RHA2a, RHA2b is also important in developmentally more advanced plants by controlling several aspects of ABA-regulated processes, including seedling growth (Fig. 4), stomatal closure (Fig. 8), and drought tolerance (Fig. 7). Importantly, the rha2a rha2b-1 double mutant is more insensitive to ABA (Figs. 3, 4, and 8) and more susceptible to drought stress (Fig. 7) than any of the single mutants, suggesting that RHA2a acts redundantly with RHA2b in ABA-signaled stress responses. Taken together, our analyses reveal that, correlating with their expression patterns, RHA2a and RHA2b show redundant yet distinguishable physiological functions in ABA signaling and drought response.
Our genetic analyses suggest that RHA2a and RHA2b could act downstream of ABI2 and in parallel with the ABI3/4/5 transcription factor genes. These two functional E3 ligases could mediate the polyubiquitination and degradation of substrates that are negative regulators of ABA signaling. Alternatively, RHA2a/RHA2b could mediate the stabilization and activation of substrates that are positive regulators of ABA signaling. Future identification of RHA2a and RHA2b substrates will further our understanding of the ABA signaling pathway.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild type in this study. Seeds of each genotype were surface sterilized with 10% bleach for 13 min and washed three times with sterile water. Sterilized seeds were then suspended in 0.1% agarose and plated on MS medium. Plants were stratified at 4°C in darkness for 2 d and then transferred to a photoperiod set at 22°C with a 16-h-light/8-h-dark photoperiod (light intensity of 120 μmol m−2 s−1). After 2 to 3 weeks, seedlings were also potted in soil and placed in a growth room at 22°C with a 16-h-light/8-h-dark cycle (light intensity of 120 μmol m−2 s−1).
Verification of the Single and Double Mutants
T-DNA insertion mutants rha2b-1 (SALK_014943) and abi2-2 (SALK_015166) were obtained from the ABRC. Seeds were harvested separately from individual plants. Subsequently, to confirm the mutant line as homozygous, PCR was performed with the genomic DNA using gene-specific primers (for rha2b-1, LP, 5′-TTTCCATGTTGGGAAATTCTG-3′, and RP, 5′-CGTGTCTGCAATCTAGCTTCC-3′; for abi2-2, LP, 5′-AAACTGTTGGGTCTACCTCGG-3′, and RP, 5′-ACCATCCCATATTCTGGTTGG-3′) and one specific primer (Lbal, 5′-TGGTTCACGTAGTGGGCCATCG-3′).
Mutants abi2-2, rha2b-1, rha2a (Bu et al., 2009), abi5-7 (Nambara et al., 2002; Tamura et al., 2006), abi4-1 (Finkelstein et al., 1998), and abi3-8 (Nambara et al., 2002; Tamura et al., 2006) were used to generate double mutants. The double mutant lines rha2a rha2b-1, abi2-2 rha2b-1, and abi2-2 rha2a were constructed by crossing between the two single mutants, and their genotypes were confirmed by PCR-based genotyping. Triple mutant abi2-2 rha2a rha2b-1 was constructed by crossing abi2-2 with rha2a rha2b-1. For double mutant lines rha2b-1 abi3-8, rha2b-1 abi4-1, and rha2b-1 abi5-7, which were also constructed by crossing, putative double mutant plants were screened from the resulting F2 progeny by their insensitivity to ABA (2 μm) during seed germination and early seedling development. Then, each double mutant line was identified by PCR-based genotyping of the RHA2b locus and sequence confirmation of the ABI3, ABI4, and ABI5 locus, respectively.
Transformation Vectors and Construction of Transgenic Plants
For overexpression of RHA2b, the RHA2b coding sequence was PCR amplified with the primers 5′-GGGGATCCATGGGACTACAAGGTCAGCTCT-3′ and 5′-GGGAGCTCTCAATGAGATGATGCAGTAGAGA-3′ using Col-0 cDNA as template. The resulting PCR product was cloned into the BamHI and SacI sites of the binary vector pCanG-HA under the control of the cauliflower mosaic virus 35S promoter.
To construct 35S:RHA2a-GFP or 35S:RHA2b-GFP, the coding sequence of RHA2a or RHA2b cDNA was amplified by PCR, digested by XbaI and KpnI, and cloned into intermediate vector pGFP-2 to form p35S:RHA2a-GFP or p35S:RHA2b-GFP. To get transgenic plants, the fused coding sequence of 35S:GFP, 35S:RHA2a-GFP, or 35S:RHA2b-GFP was released by EcoRI and PstI and then ligated into the binary vector pCAMBIA1300. The primers used were as follows: for RHA2b, P1 (5′-GGTCTAGAATGGGACTACAAGGTCAGCTC-3′) and P2 (5′-GGGGTACCATGAGATGATGCAGTAGAGGT-3′); for RHA2a, P1 (5′-GGTCTAGAATGGGGCTACAAGGTCAGCTA-3′) and P2 (5′-GGGGTACCGTGGAGAGAGAAACACGAGAT-3′).
To construct pRHA2b:RHA2b-GUS or pRHA2a:RHA2a-GUS, a 2-kb promoter sequence and coding sequence region of RHA2b or RHA2a, respectively, was amplified from genomic DNA by PCR and verified by sequencing. The PCR fragment was cloned into the PstI and EcoRI sites of binary vector pCAMBIA1391Z or HindIII and BamHI sites of PBI121 to obtain the constructs containing RHA2b or RHA2a native promoter and coding sequence fused with the GUS coding region from the uidA gene. The primers were as follows: for RHA2b, P1 (5′-GGCTGCAGCCGAGGAATCAACGGATGTCT-3′) and P2 (5′-GGGAATTCATGAGATGATGCAGTAGAGGTAG-3′); for RHA2a, P1 (5′-GGAAGCTTTCCAACTGTCATATATAACCCC-3′) and P2 (5′-GGGGATCCGTGGAGAGAGAAACACGAGATC-3′).
Transformation of Arabidopsis plants was performed by the floral dip infiltration method (Bechtold and Pelletier, 1998) using Agrobacterium tumefaciens strain GV3101 (EHA105). T2 seeds from each of the selected transgenic plants were plated on germination medium containing 50 μg mL−1 kanamycin (for pCanG-HA and PBI121) or 30 μg mL−1 hygromycin (for pCAMBIA1300 and pCAMBIA1391Z) as selection antibiotics, and the homozygous lines were selected. The constructs of 35S:RHA2a have been reported (Bu et al., 2009).
Phenotype Analysis
For seed germination, plants of different genotypes were grown in the same conditions and seeds were collected at the same time. For each comparison, seeds were planted on the same plate containing MS medium (0.5× MS salts, 1% Suc, and 0.8% agar) with different concentrations of ABA as indicated. Plates were chilled at 4°C in the dark for 3 d (stratified) and moved to 22°C with a 16-h-light/8-h-dark cycle. The percentage of seed germination was scored at the indicated times. Germination was defined as an obvious emergence of the radicle through the seed coat.
To study the inhibition effect of ABA in postgerminative growth, seeds were sown on medium containing different concentrations of ABA as indicated. The percentage of cotyledon greening was recorded at 5 d after the end of 2 d of stratification. Cotyledon greening is defined as obvious cotyledon expansion and turning green. The length of primary roots was also measured using a ruler.
The effect of ABA on cotyledon greening was also quantified by measurement of chlorophyll content of the seedlings as follows: after recording cotyledon greening percentage, the seedlings were harvested, weighed, and ground into fine powder in liquid nitrogen for chlorophyll extraction (Lichtenthaler, 1987). Chlorophyll a/b contents were determined according to a described method (Lichtenthaler, 1987).
To study the effect of ABA on seedling growth, seeds were sown on ABA-free medium for 4 d after stratification, and then the seedlings were transferred to medium containing different concentrations of ABA. After growing for 11 d on the treatment medium vertically, seedlings were photographed with a digital camera and the length of taproot growth, the number of lateral roots, the number of true leaves, and seedling fresh weight were measured.
RT-PCR Amplification
The expression of RHA2b, RHA2a, and ABI2 in mutants was examined by RT-PCR. RNA was isolated from 200-mg tissue samples using Trizol solution (Invitrogen), and first-strand cDNA synthesis was performed using 5 μg of total RNA and Moloney murine leukemia virus reverse transcriptase (Promega). The yield of cDNA was measured according to the PCR signal generated from the internal standard, the housekeeping gene ACTIN2, amplified from 26 to 28 cycles starting with 0.5 μL of the cDNA solution. Cycling conditions were as follows: 5 min at 94°C; and 25 or 27 cycles of 30 s at 94°C, 30 s at 52°C, and 20 s at 72°C. The resulting cDNAs were subjected to PCR with the following primers: for RHA2a, 5′-ACTGGATCCAAGATGGGGCTACAAGGTCAG-3′ and 5′-ACTGAGCTCTACTCAGTGGAGAGAGAAAC-3′; for RHA2b, 5′-GCTACCTCATCATCATCAGGG-3′ and 5′-TCAATGAGATGATGCAGTAGAGG-3′; for ABI2, 5′-ATGGACGAAGTTTCTCCTGCA-3′ and 5′-CATCCCAAAGACCATCACTTGC-3′; for ACTIN2, 5′-TTGACTACGAGCAGGAGATGG-3′ and 5′-ACAAACGAGGGCTGGAACAAG-3′. The expression of ACTIN2 was used as an internal control. The RT-PCR product was analyzed by electrophoresis on a 1.0% agarose gel. PCR was performed in triplicate.
Real-Time PCR Analysis
To examine the expression levels of RHA2b and RHA2a genes, quantitative real-time PCR analysis was performed. RNA extraction and RT were performed as described above for RT-PCR. PCR amplification was performed with primers of specific genes as follows: for RHA2b, 5′-GCTACCTCATCATCATCAGGG-3′ and 5′-TCAATGAGATGATGCAGTAGAGG-3′; for RHA2a, 5′-TCTCTCTCCTCGCCGTCTTC-3′ and 5′-TCCAGCTTCCTCACCTCTTCA-3′; for ACTIN2, 5′-TTGACTACGAGCAGGAGATGG-3′ and 5′-ACAAACGAGGGCTGGAACAAG-3′. Amplification of the ACTIN2 gene was used as an internal control. The cDNA was amplified with the RealMasterMix kit (SYBR Green; Tiangen) using the DNA engine Opticon 3 thermal cycler (Bio-Rad) in a 20-μL volume. PCR was performed on 96-well optical reaction plates heated for 5 min at 95°C to activate hot-start Taq DNA polymerase, followed by 40 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 58°C, and extension for 30 s at 68°C. The amplification of the target genes was monitored every cycle by SYBR Green fluorescence. The data are presented after normalizing to the reference gene, and three technical replicates were performed for each experiment.
To assay the expression of ABA-responsive genes, real-time PCR analysis was also performed with the RNA samples isolated from 2-week-old seedlings harvested at the indicated times after treatment with 100 μm ABA. Total RNA isolation and RT were performed as described above. PCR amplification was performed with primers specific for various ABA-responsive genes as follows: for RD29A (At5g52310), 5′-CAGAGGAACCACCACTCAACACA-3′ and 5′-CTCTAGGTTTACCTGTTACGCCTG-3′; for RD29B (At5g52300), 5′-ATGGAGTCACAGTTGACACGTCCT-3′ and 5′-CTTCTGGGTCTTGCTCGTCATACT-3′; for RAB18 (At5g66400), 5′-ATGGCGTCTTACCAGAACCGTCCA-3′ and 5′-ACCACCACTTTCCTTGTGGAGTTG-3′; for KIN1 (At5g15960), 5′-ATGTCAGAGACCAACAAGAATGCC-3′ and 5′-CTACTTGTTCAGGCCGGTCTTG-3′; for RD22 (At5g25610), 5′-ATGGCGATTCGTCTTCCTCTGATC-3′ and 5′-ACTCCGCCTTTACCTACTTGGACG-3′; and for AtADH1 (At1g77120), 5′-ATGTCTACCACCGGACAGATT-3′ and 5′-CGAGTGGCAATGACGACACTC-3′.
Amplification of ACTIN2 genes was used as an internal control, and quantitative real-time PCR experimental procedures were performed as described above. Three technical replicates were performed for each experiment.
Drought Treatment and Measurement of Transpiration Rate
For gene expression analysis, 2-week-old seedlings from the agar plate were transferred onto a filter paper in a covered petri dish and subjected to drought treatment. The treatment was conducted in an environment of 70% relative humidity. For the soil-grown plant drought-tolerance test, 1-week-old seedlings were transplanted to the soil for another 2 weeks under standard growth conditions, and then plants were subjected to progressive drought by withholding water for specified times and then rewatered for 2 d. To minimize experimental variations, the same number of plants were grown on the same tray. The entire test was repeated a minimum of three times. To measure the transpiration rate, detached fresh leaves were placed abaxial side up on open petri dishes and weighed at different time intervals at room temperature. Leaves of similar developmental stages (third to fifth true rosette leaves) from 4-week-old soil-grown plants were used.
Stomatal Aperture Analysis
Stomatal bioassay experiments were performed as described (Hugouvieux et al., 2001; Zhang et al., 2001, 2007; Song et al., 2005) with slight modifications. To study the promotion of stomatal closure by ABA, leaves from 4- to 5-week-old plants grown in the same conditions described above were harvested in darkness at the end of the night. Paradermal sections of abaxial epidermis obtained were incubated in solution containing 50 mm KCl, 10 mm CaCl2, and 10 mm MES-KOH, pH 6.15, at 22°C to 25°C and exposed to light for 1.5 h. Subsequently, 50 μm ABA was added to the solution to assay for stomatal closing. After treatment for 3 h, stomatal apertures were measured as described with a confocal microscope (DM5000B; Leica) fitted with a camera lucida and a digitizing table (SPOT; Houston Instrument) linked to a personal computer. The apertures of usually 50 to 60 stomata were measured in three independent experiments.
GUS Bioassays
Young seedlings at different developmental stages, and different parts from transgenic plants, were collected and used for histochemical detection of GUS expression. For general detection, materials were stained at 37°C overnight in 100 mm sodium phosphate, pH 7.0, 0.1 mm EDTA, 0.5 mm ferricyanide, 0.5 mm ferrocyanide, 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 0.1% Triton X-100. Chlorophyll was cleared from the plant tissues by immersing them in 70% ethanol.
Subcellular Localization
For 35S:GFP, 35S:AtAIB-GFP, 35S:RHA2a-GFP, and 35S:RHA2b-GFP, roots of 1-week-old seedlings were immersed in 2 μg mL−1 4′,6-diamidino-2-phenylindole (DAPI) solution for 10 to 15 min and then stained with 5 μg mL−1 ice-cold FM4-64 solution for 1 min. DAPI is extensively used for nuclei labeling, and FM4-64 is widely used for membrane labeling. Before staining, whole seedlings of 35S:RHA2a-GFP or 35S:RHA2b-GFP were pretreated with 50 μm MG132 for 6 h under dim-light conditions; for BRI1-GFP, roots of 1-week-old seedlings were stained with 5 μg mL−1 ice-cold FM4-64 solution for 1 min. All seedlings were observed by fluorescence microscopy.
Expression of MBP-RHA2b Fusion Protein and Ubiquitination Assay
To generate MBP-RHA2b, the coding sequence of RHA2b was amplified with the primers 5′-GGTCTAGAATGGGACTACAAGGTCAGCTC-3′ and 5′-GGGGATCCTCAATGAGATGATGCAGTAGAGA-3′ and cloned into the EcoRI and BamHI sites of pMal-c2 (New England Biolabs). MBP-RHA2b fusion proteins were prepared following the manufacturer’s instructions. Ubiquitination assay of RHA2b was conducted following our recently described method (Bu et al., 2009).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: RHA2b, At2g01150; RHA2a, At1g15100; ABI2, At5g57050; RD29A, At5g52310; RD29B, At5g52300; RAB18, At5g66400; KIN1, At5g15960; RD22, At5g25610; AtADH1, At1g77120.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization of RHA2b and RHA2a.
Supplemental Figure S2. Comparison of RHA2b and RHA2a expression.
Supplemental Figure S3. Molecular analysis of RHA2b/RHA2a mutants.
Supplemental Figure S4. ABA responses of RHA2b OE 1 and RHA2b OE 9 plants in seed germination and postgerminative growth.
Supplemental Figure S5. Expression of ABI2 in the abi2-2 (SALK_015166) mutant.
Supplementary Material
Acknowledgments
We thank the ABRC and the Nottingham Arabidopsis Stock Centre for providing T-DNA insertion mutants. We thank Dr. Eiji Nambara (RIKEN Plant Science Center) for providing the abi5-7 and abi3-8 mutants and Dr. Xuelu Wang (Fudan University) for providing seeds of BRI1-GFP plants.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bu Q, Li H, Zhao Q, Jiang H, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Wang D. (2009) The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol 150: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI. (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Jensen KL, Jespersen HM, Skriver K. (1998) Widespread occurrence of a highly conserved RING-H2 zinc finger motif in the model plant Arabidopsis thaliana. FEBS Lett 436: 283–287 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH. (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47: 343–355 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Hilhorst HW, Karssen CM. (1989) In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol 90: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Borg-Franck M. (1992) Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol Biol 19: 689–692 [DOI] [PubMed] [Google Scholar]
- Lång V, Palva ET. (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Lechner E, Goloubinoff P, Genschik P, Shen WH. (2002) A gene trap Dissociation insertion line, associated with a RING-H2 finger gene, shows tissue specific and developmental regulated expression of the gene in Arabidopsis. Gene 290: 63–71 [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J. (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49: 199–222 [DOI] [PubMed] [Google Scholar]
- Li H, Sun J, Xu Y, Jiang H, Wu X, Li C. (2007) The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol Biol 65: 655–665 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Liu H, Stone SL. (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH. (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P. (2002) A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Zhang W, Assmann SM. (2007) Roles of ion channels and transporters in guard cell signal transduction. FEBS Lett 581: 2325–2336 [DOI] [PubMed] [Google Scholar]
- Parcy F, Giraudat J. (1997) Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J 11: 693–702 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Schwager K. (2004) Regulated proteolysis and plant development. Plant Cell Rep 23: 353–364 [DOI] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD. (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N, Yoshida T, Tanaka A, Sasaki R, Bando A, Toh S, Lepiniec L, Kawakami N. (2006) Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol 47: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH. (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, Song CP. (2001) Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol 126: 1438–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.