Abstract
Fruit provide essential nutrients and vitamins for the human diet. Not only is the lipid-rich fleshy mesocarp tissue of the oil palm (Elaeis guineensis) fruit the main source of edible oil for the world, but it is also the richest dietary source of provitamin A. This study examines the transcriptional basis of these two outstanding metabolic characters in the oil palm mesocarp. Morphological, cellular, biochemical, and hormonal features defined key phases of mesocarp development. A 454 pyrosequencing-derived transcriptome was then assembled for the developmental phases preceding and during maturation and ripening, when high rates of lipid and carotenoid biosynthesis occur. A total of 2,629 contigs with differential representation revealed coordination of metabolic and regulatory components. Further analysis focused on the fatty acid and triacylglycerol assembly pathways and during carotenogenesis. Notably, a contig similar to the Arabidopsis (Arabidopsis thaliana) seed oil transcription factor WRINKLED1 was identified with a transcript profile coordinated with those of several fatty acid biosynthetic genes and the high rates of lipid accumulation, suggesting some common regulatory features between seeds and fruits. We also focused on transcriptional regulatory networks of the fruit, in particular those related to ethylene transcriptional and GLOBOSA/PISTILLATA-like proteins in the mesocarp and a central role for ethylene-coordinated transcriptional regulation of type VII ethylene response factors during ripening. Our results suggest that divergence has occurred in the regulatory components in this monocot fruit compared with those identified in the dicot tomato (Solanum lycopersicum) fleshy fruit model.
Fruit development, maturation, and ripening are complex biological processes unique to plants. The monocotyledonous oil palm (Elaeis guineensis) fruit is a drupe whose thick fleshy mesocarp is exceptionally rich in oil (80% dry mass), making this species the highest oil-yielding crop in the world (Murphy, 2009). The mesocarp is also especially abundant in carotenoids, and crude palm oil is the richest dietary source of provitamin A (Sambanthamurthi et al., 2000; Solomons and Orozco, 2003). Surprisingly, the molecular basis of oil palm fruit development, maturation, and ripening has received very little attention. In contrast, the fleshy berries such as tomato (Solanum lycopersicum) and grape (Vitis vinifera) are considered models due to the wealth of genome resources and genetic transformability. In these species, hormones play key roles. For example, ripening appears to be controlled by both ethylene-dependent and independent pathways, shown by the characterization of the tomato ripening-related mutants ripening-inhibitor (rin), Colorless non-ripening (cnr), Never-ripe, and Green-ripe (Wilkinson et al., 1995; Vrebalov et al., 2002; Barry and Giovannoni, 2006; Manning et al., 2006; Giovannoni, 2007). Abscisic acid (ABA) can also control tomato ripening through the activation of ethylene biosynthesis (Zhang et al., 2009a). In grape, whereas ripening was previously thought to be ethylene independent, ABA appears to control ripening along with auxin and ethylene (Coombe and Hale, 1973; Davies et al., 1997; Chervin et al., 2004, 2008).
Another common regulatory feature of fruit has emerged from functional analyses with the MADS box family of transcription factors (TFs), including RIN and TOMATO AGAMOUS-LIKE1 (TAGL1) from tomato, PLENA from peach (Prunus persica), and FaMADS9 from strawberry (Fragaria × ananassa), which have provided evidence for roles during ripening (Vrebalov et al., 2002, 2009; Tadiello et al., 2009; Seymour et al., 2011). The recent identification of diverse MADS expressed during banana (Musa acuminata) ripening raises the questions of whether the function of some MADS box proteins is common to monocots and whether divergence occurred after the separation of dicots and monocots (Liu et al., 2009; Elitzur et al., 2010). It is clear that tomato and grape, with their different modes of ripening, provide an excellent framework to compare the maturation and ripening regulatory mechanisms in diverse eudicot species. However, due to the lack of data, it is not clear whether similar or diverse regulatory mechanisms function during the maturation and ripening in monocot fruit species in general and oil-accumulating fruits tissues such as the oil palm mesocarp in particular. Indeed, the low content or lack of triacylglycerols (TAGs) and provitamin A in these eudicot model species makes them inadequate to understand the regulatory mechanisms that function during the maturation and ripening in the oil-rich mesocarp.
Current knowledge about oil synthesis is derived mainly from research on seeds in which TAGs accumulate during maturation (Bates et al., 2009; Baud and Lepiniec, 2010). In particular, the use of genomics and the model species Arabidopsis (Arabidopsis thaliana) has allowed new insights into seed lipid synthesis (Lu et al., 2009; Zhang et al., 2009b; Li-Beisson et al., 2010) and its regulation (Baud and Lepiniec, 2009). Although genes and enzymes involved in some steps of fatty acid (FA) and TAG synthesis still have to be identified or functionally validated, the knowledge is advanced enough to enable oil synthesis pathway reconstruction from transcriptome data in nonmodel species (Joët et al., 2009). In contrast, far less is known about the molecular basis of lipid metabolism in the fleshy fruits that accumulate high amounts of TAG in their mesocarp, such as avocado (Persea americana), olive (Olea europea), and oil palm. Furthermore, very few lipid-related genes have been cloned in oil palm (Othman et al., 2000; Nakkaew et al., 2008), and most of the studies on oil metabolism in this species deal with metabolic control analysis experiments using callus cultures (Ramli et al., 2009). To our knowledge, lipid synthesis in the developing oil palm fruit has never been investigated through transcriptome analysis. In contrast, transcriptomic studies of developing seeds have revealed many key features of oil synthesis. The main processes leading to the accumulation of TAGs in the seed begin in the plastid with the de novo formation of acyl chains (up to 18 carbons), followed by desaturation at the Δ9 position and FA release, then continues in the endoplasmic reticulum (ER) with the sequential acylation on the three positions of the glycerol backbone, additional FA desaturation, esterification to phosphatidylcholine (PC) and acyl editing, and finally storage within specialized organelles called oil bodies. The identification of a very limited number of expression patterns for the numerous genes involved in FA and TAG synthesis first suggested that the transcriptional network that controls seed lipid metabolism is remarkably coordinated (Ruuska et al., 2002; Baud and Lepiniec, 2009). In the Arabidopsis embryo, almost all genes involved in de novo FA synthesis show the same timing and pattern of expression (i.e. a bell-shaped pattern whose peak coincides with the onset of lipid accumulation). In contrast, the transcription of genes required for TAG assembly in the ER rises later and remains high during the maturation process. These patterns have also been observed in the copious persistent endosperm of coffee (Coffea arabica), another seed tissue that accumulates oil (Joët et al., 2009). The analysis of Arabidopsis seed microarray data also revealed that the fold increase of most genes involved in TAG assembly and storage was significantly higher than that of genes of the core FA biosynthetic machinery (Baud and Lepiniec, 2009). Whether these important characteristics of seed oil synthesis are conserved and function in nonseed tissues such as the mesocarp of the oil palm fruit remains unknown.
The fact that most genes governing de novo FA synthesis in the plastid share the same temporal transcription pattern strongly suggests that they are coregulated and probably share common cis- and trans-regulatory elements. The isolation of the Arabidopsis wrinkled1 (wri1) mutant constituted a key milestone in the validation of this hypothesis. This mutant is specifically impaired in seed TAG accumulation and shows lower transcript levels for key enzymes involved in lipid and carbohydrate metabolism (Focks and Benning, 1998; Ruuska et al., 2002). Furthermore, overexpression of WRI1, which encodes an APETALA2 (AP2)/ethylene-responsive element binding protein family TF, led to increased seed oil content and up-regulation of certain FA biosynthesis genes (Cernac and Benning, 2004; Maeo et al., 2009). Indeed, WRI1 interacts directly with an AW box sequence [CnTnG(n)7CG] in the upstream regions of several FA biosynthesis genes (Maeo et al., 2009). In addition, WRI1 expression appears to be under the control of LEAFY COTYLEDON1 (LEC1) and LEC2, two master regulators of seed maturation (Baud et al., 2007; Mu et al., 2008). While an analogous regulatory system appears to function in maize (Zea mays) through the ZmWRI1 and ZmLEC1 orthologs (Shen et al., 2010), a maize ortholog of LEC2 has not been identified, suggesting differences in this key regulatory network between monocots and dicots. Finally, whether similar regulatory systems function in the lipid-rich mesocarp during fruit ripening compared with that during seed maturation is unknown.
The oil palm mesocarp not only accumulates the largest amount of TAGs compared with all other crop species but also large amounts of carotenoids (500 μg g−1 mesocarp dry mass), predominantly α- and β-carotene (Ikemefuna and Adamson, 1984). Carotenes are precursors of vitamin A, an essential nutrient for human beings. Carotenoids and their oxygenated derivatives xanthophylls give fruits their pigmentation and play an important role as visible signals to attract animals for seed dispersal (Tanaka et al., 2008). The cleavage of carotenoids also generates apocarotenoids, including the plant hormone ABA, with emerging roles during fruit ripening (Davies et al., 1997; Deluc et al., 2007; Gambetta et al., 2010). In addition, volatile apocarotenoid compounds such as β-ionone and geranylacetone have animal-attracting characteristics with very low odor thresholds and play crucial roles in fruit dispersion (Auldridge et al., 2006a, 2006b).
Despite the attractive quantitative and qualitative carotenoid composition of the oil palm mesocarp, very few studies have focused on the transcriptional mechanisms underlying carotenoid gene metabolism or performed metabolite profiling during mesocarp development (Ikemefuna and Adamson, 1984; Khemvong and Suvachittanont, 2005; Rasid et al., 2008). Only two genes, PHYTOENE SYNTHASE (PSY) and 1-DEOXY-XYLULOSE 5-PHOSPHATE SYNTHASE (DXS), have been investigated to date in oil palm. PSY and DXS are involved in the first committed step of carotenoid biosynthesis and the upstream 2-C-methyl-d-erythritol 4-phosphate pathway, respectively. By contrast, virtually all carotenogenic genes have been identified in model plants (Hirschberg, 2001; Howitt and Pogson, 2006; Chen et al., 2010), and this genomic knowledge should facilitate the identification of gene orthologs in the oil palm mesocarp. During the ripening of the fleshy tissue of tomato, chloroplasts redifferentiate to chromoplasts that accumulate large amounts of lycopene, a compound with strong antioxidant properties but no provitamin A activity (Arango and Heise, 1998; Josse et al., 2000). While tomato is a reference for carotenogenesis in fleshy fruit, it does not constitute an optimal model to study the transcriptional regulation of downstream pathways of carotene synthesis.
Our objective is to provide a basis to understand the molecular regulation and coordination of TAG and carotenoid biosynthesis during oil palm mesocarp maturation and ripening compared with those found in seeds and nonoily fruit. We used morphological, histological, and biochemical analyses to define phases of oil palm fruit development, maturation, and ripening, and in parallel, we generated and analyzed the mesocarp transcriptome during maturation and ripening using 454 pyrosequencing. By studying the accumulation of oil and carotenoids and hormonal profiling during oil palm fruit development and analyzing the differentially expressed transcripts, we have characterized key regulatory steps of the specific pathways and corresponding genes that are likely to function during fruit maturation and ripening. We describe the unique characters of lipid and carotenoid metabolism in this species and provide insight into the transcriptional coordination and hormonal regulation in the mesocarp tissue compared with those in developing seeds and fleshy fruits. An overview of the metabolic networks involved in lipid and carotenoid accumulation and transcriptional networks in the oil palm mesocarp will allow comparison with model species and help identify candidate genes and molecular markers associated with these important chemical traits for breeding programs.
RESULTS
Phases of Oil Palm Fruit Development, Maturation, and Ripening Defined by Morphological, Cellular, Biochemical, and Hormonal Parameters of the Mesocarp
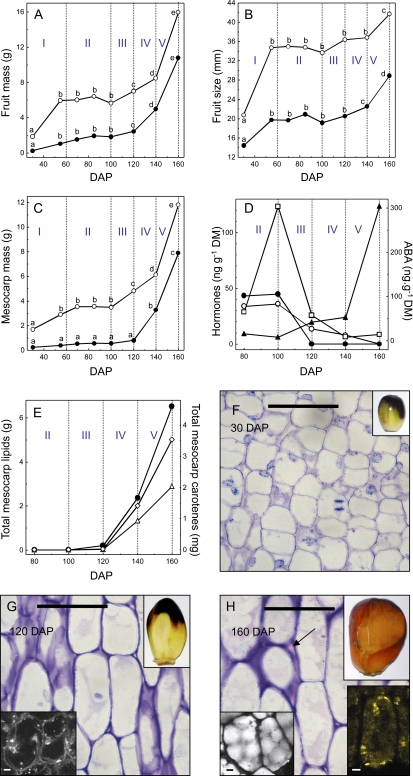
Under the field conditions and with the genetic material used, the fruit of oil palm completed their development, maturation, and ripening in approximately 160 d. Similar to what occurs in other drupes, a biphasic growth curve was observed, with an initial increase in fruit mass and size measured between 30 and 60 d after pollination (DAP; Fig. 1, A and B). After a 40-d lag period (60–100 DAP), a further increase in fruit mass was observed between 100 and 160 DAP, in particular between 140 and 160 DAP, accompanied by an increase in fruit size (Fig. 1, A and B). A similar pattern was observed with the mesocarp fresh mass, which represents approximately 75% of that of the ripe fruit. Notably, a large increase in dry mass between 120 and 160 DAP reflects lipid accumulation in this tissue (Fig. 1, C and E). Further analysis of selected histological, biochemical, and hormonal parameters allowed us to define five distinct phases of mesocarp development. Phase I, between 30 and 60 DAP, is defined by anticlinal cell divisions and expansion along with the initial increase in fruit mass and size (Fig. 1, A–C and F). Phase II, between 60 and 100 DAP, is a transition period characterized by a lag in the accumulation of fresh mass and also by peak amounts of indole-3-acetic acid (IAA) and IAA conjugates (Fig. 1D; Supplemental Fig. S1). Phase III, between 100 and 120 DAP, is the end of the transition period, during which decreases in auxin, gibberellic acid (GA), and cytokinin metabolites are observed (Fig. 1D; Supplemental Fig. S1). Phase IV is concomitant with the beginning of maturation, characterized by an increase in mesocarp fresh mass and the beginning of lipid accumulation detected by 120 DAP (Fig. 1, C, E, and G). Phase IV, between 120 and 140 DAP, is characterized by lipid (more than 2 g per fruit) and carotenoid accumulation in addition to the low amounts of all hormone metabolites examined, including auxin, GA and cytokinin, ABA, and ethylene (Fig. 1, D and E; Supplemental Fig. S1). Finally, during the ripening phase V, there is a large increase in the hormones ABA and ethylene, and cell wall detachment related to ripening processes in the mesocarp are visualized (Fig. 1, D and H; Supplemental Fig. S1). During this phase, dry and fresh fruit mass increase massively, concurrently with lipid and carotenoid accumulation in the mesocarp (Fig. 1, A, C, and E). As observed at 160 DAP, lipids accumulate within subcellular spherical organelles (10–15 μm in diameter, six to 12 per mesocarp cell) that occupy the volume of the cells (Fig. 1H). In addition, as observed in ripe fruits, mesocarp cells contain distinct regions, presumably chromoplasts, with high carotenoid concentrations (Fig. 1H).
Figure 1.
Morphological, biochemical, and histological analysis of oil palm fruit development from 30 to 160 DAP. A, Evolution of fresh (white circles) and dry (black circles) mass of whole fruits. B, Evolution of fruit length (white circles) and diameter (black circles). C, Evolution of fresh (white circles) and dry (black circles) mass of mesocarp. D, Quantification of auxin (IAA; white squares), cytokinin (trans-zeatin-O-glucoside; white circles), GA3 (black circles), and ABA (black triangles). DM, Dry mass. E, Lipid (black circles), α-carotene (white triangles), and β-carotene (white diamonds) content. F to H, Cellular characteristics of 30-DAP (F), 70-DAP (G), and 160-DAP (H) mesocarp. Bottom left insets in G and H show confocal microscopy of mesocarp lipids detected by Nile red staining. Bottom right inset in H shows confocal microscopy of mesocarp β-carotene localization. Phases of development are I, II, III, IV, and V. Lipid and carotenoid content values are expressed as total g of mesocarp lipid or mg of carotenoid on a per fruit basis. The arrow indicates cell separation and enlarged intercellular spaces. Lowercase letters above data points indicate statistically significant differences. Bars = 50 μm; inset bars = 5 μm.
Statistical Analysis and Gene Ontology Annotations of Contigs Differentially Represented during Mesocarp Development
A total of 29,034 contigs were obtained after clustering and assemblage of good-quality sequence reads (Supplemental Figs. S2 and S3). Read amounts associated with each of the 29,034 contigs were then analyzed using Audic-Claverie and false discovery rate (FDR) statistics, leading to the identification of 2,629 differentially expressed contigs, which are henceforth referred to as contig group I, while the remaining 26,405 contigs with either nondifferential representation or low read abundance are referred to as contig group II. Hierarchical cluster analysis of group I contigs allowed the identification of four major clusters, termed A, B, C, and D, which contain 695, 418, 1,167, and 349 contigs, divided into five, four, five, and three subclusters, respectively (Supplemental Fig. S4). In general, contigs that exhibited a transcription peak at 100, 120, 140, or 160 DAP were grouped in cluster A, B, C, or D, respectively. Gene Ontology (GO) accessions were assigned to the 2,629 group I contigs using the Blast2Go platform (Götz et al., 2008). A total of 2,074 contigs (77%) were assigned GO accessions, 488 in cluster A (70%), 377 in cluster B (90%), 908 in cluster C (78%), and 301 in cluster D (86%). To identify differences in the GO annotations of the four clusters that reflect underlying molecular processes, an enrichment analysis of the annotations of each individual cluster compared with the annotations of the 2,074 contigs was performed using the GOSSIP platform (Blüthgen et al., 2005). Cluster B had the most overrepresented GO annotations (131 terms), followed by cluster D (22) and cluster C (four), while cluster A had only underrepresented GO annotations (Supplemental Table S1). Cluster C had the most underrepresented annotations (51 terms), followed by cluster D (43), cluster B (32), and cluster A (17). The most significant underrepresented GO terms in cluster A were related to carbohydrate metabolic processes, including glycolysis, while the most significantly overrepresented terms in cluster B were FA biosynthetic process, cellular carbohydrate metabolic process, and lipid biosynthetic process. The results suggest that in the 120-DAP mesocarp, at the beginning of phase IV, significant changes to the transcriptome coincide with the onset of the observed lipid accumulation (Fig. 1E). By contrast, terms related to nucleic acid binding, nucleus, DNA binding, and lipid catabolic process are overrepresented in cluster C, which suggests that major transcriptional regulatory changes are initiated by the end of phase IV at the onset of ripening. Finally, the overrepresented terms in cluster D relate to carbohydrate metabolic processes, including O-glycosyl compound hydrolysis, glycosyl bond hydrolysis, glucan endo-1,3-β-d-glucosidase activity, chitinase activity, and cellular aromatic compound metabolic processes, which suggests that ripening processes such as cell wall modifications are taking place by 160 DAP. Overall, this analysis indicates that certain changes in the transcriptome are coordinated and reflect the biochemical and physiological processes observed during the maturation and ripening phases of the mesocarp (Fig. 1).
Reconstructing FA and TAG Biosynthetic Pathways in the Oil Palm Mesocarp Using a Pyrosequencing-Based Transcriptome
Based on our current knowledge of oil synthesis in seeds, the pyrosequencing-based transcriptome approach enabled us to reconstruct the FA and TAG synthesis pathways in the oil palm mesocarp (Fig. 2). Indeed, for almost all lipogenic steps, contigs that exhibited high similarity to genes from Arabidopsis and other oleaginous species were found (Supplemental Table S2).
Figure 2.
FA synthesis and TAG assembly in the oil palm mesocarp as reconstructed from the 454 pyrosequencing transcriptome. Contig names followed by asterisks indicate contigs showing significant variations during development according to Audic-Claverie and FDR statistics (contig group I). Gene expression levels at 100, 120, 140, and 160 DAP are indicated with colored bars. For the developmental stage displaying maximal expression level, the normalized transcript abundance, expressed as the number of transcripts per 200,000 transcripts, is given. For the other stages, expression levels are indicated as percentages of the maximal normalized transcript abundance of the gene, as described in the color code from 0% (white) to 100% (dark blue). ACC BC, Biotin carboxylase subunit of heteromeric acetyl-CoA carboxylase (ACCase); ACC Ctα, carboxyltransferase α-subunit of heteromeric ACCase; ACC Ctβ, carboxyltransferase β-subunit of heteromeric ACCase; CPT, diacylglycerol cholinephosphotransferase; DGAT, acyl-CoA:diacylglycerol acyltransferase; EAR, enoyl-acyl carrier protein (ACP) reductase; ENOp, enolase; FAD2, oleate desaturase; FAD3, linoleate desaturase; FATA, acyl-ACP thioesterase A; FATB, acyl-ACP thioesterase B; GPAT, glycerol-3-phosphate acyltransferase; HAD, hydroxyacyl-ACP dehydrase; KAR, ketoacyl-ACP reductase; KAS I, ketoacyl-ACP synthase I; KAS II, ketoacyl-ACP synthase II; KAS III, ketoacyl-ACP synthase III; LACS, long-chain acyl-CoA synthetase; LPAAT, 1-acylglycerol-3-phosphate acyltransferase; LPCAT, 1-acylglycerol-3-phosphocholine acyltransferase; MAT, malonyl-CoA:ACP malonyltransferase; PAP, phosphatidate phosphatase; PDAT, phospholipid:diacylglycerol acyltransferase; PDH, dihydrolipoamide acetyltransferase, E2 component of pyruvate dehydrogenase complex; PK, pyruvate kinase; SAD, stearoyl-ACP desaturase.
Three enzymes appear to participate in the acylation of diacylglycerol (DAG) to TAG in the oil palm mesocarp. First, the presence of two contigs (CL875Contig1 and CL1Contig9221) similar to type 1 and type 2 diacylglycerol acyltransferases (DGAT), respectively, suggests that both classes of DGAT play a role in TAG synthesis in this tissue. Moreover, the route involving PC as the acyl donor may make a similar contribution to the final acylation step of DAG, given that a contig (CL1451Contig2) highly similar to an Arabidopsis phospholipid:diacylglycerol acyltransferase (AtPDAT1; At5g13640) had comparable total read amounts to that of DGAT1- and DGAT2-like contigs (about 40 reads per 200,000). Furthermore, no contig was found for Arachis hypogea cytosolic DGAT or the Arabidopsis bifunctional wax ester synthase/DGATs, nor did we identify contigs for an acyltransferase with membrane-bound O-acyltransferase domains that displayed read levels higher than the DGAT- and PDAT-like contigs described above. Overall, these results suggest that the three DGAT1-, DGAT2-, and PDAT-like contigs identified indeed encode the enzymes responsible for final TAG assembly in the oil palm mesocarp.
Because the reaction catalyzed by PDAT also produces lysophosphatidylcholine, a lysophospholipid acyltransferase is necessary to cooperate with PDAT to regenerate PC from lysophosphatidylcholine. Interestingly, a contig (CL212Contig1) highly similar to AtLPLAT1 and AtLPLAT2, two lysophospholipid acyltransferases recently characterized in Arabidopsis (Ståhl et al., 2008), had read amounts similar to the PDAT- and DGAT-like contigs. It is also possible that this lysophospholipid acyltransferase-like contig participates in acyl editing (Bates et al., 2009), although the requirement of this mechanism in a tissue producing an oil poor in polyunsaturated FAs is unknown. In this regard, no contig with high similarity to Arabidopsis phospholipase A2 was found.
Finally, no contig was identified for the recently characterized phosphatidylcholine:DAG cholinephosphotransferase, which is encoded by ROD1 and contributes to PC-DAG interconversion in developing Arabidopsis seeds together with CDP-choline:DAG cholinephosphotransferase (Lu et al., 2009). Furthermore, the absence of any oleosin transcript in our libraries may provide an explanation for the considerable size of the oil droplets that accumulate in the oil palm mesocarp (10–15 μm; Fig. 1H). This size is similar to that of the large lipid bodies in the seeds of Arabidopsis lines in which oleosins were suppressed or severely attenuated (Siloto et al., 2006).
FA Synthesis in the Plastid and TAG Assembly in the ER Are Governed by Two Different Transcriptional Programs
The reconstruction of the FA and TAG biosynthetic pathways revealed that de novo formation of acyl chains in the plastid and TAG assembly in the ER follows two distinct transcriptional programs. Indeed, almost all genes involved in glycolysis and FA synthesis in the plastid were from group I, which suggests strong developmental transcriptional regulation of this pathway in the mesocarp. In addition, almost all contigs of the core FA biosynthetic machinery showed maximal transcription at 120 DAP and grouped in cluster B, as determined by hierarchical clustering analysis (Supplemental Fig. S4), and displayed high read levels (average of 424 per 200,000 reads, up to 1,993 reads for enoyl-ACP reductase). Furthermore, the transcription peak of the core FA biosynthetic machinery coincided with the onset of oil accumulation in the mesocarp at the beginning of the maturation phase IV (Figs. 1E and 2). By contrast, for the TAG assembly pathway, only contigs for oleate and linoleate desaturases (FAD2 and FAD3) were found within group I. For the enzymes involved in the sequential acylation on the three positions of the glycerol backbone, PC-DAG interconversion, and acyl editing, contigs were identified in group II only, suggesting very little or no developmental control of transcription of genes involved in TAG assembly (Fig. 2). Moreover, contigs related to TAG assembly displayed no clear peak but rather showed low (81 reads per 200,000) and constant read amounts. This unambiguous transcriptional dissimilarity between the pathways in the two compartments and the high homogeneity of patterns observed within each block suggest that FA synthesis in the plastid and TAG assembly in the ER are controlled by two different transcriptional programs.
Because transcripts encoding enzymes involved in de novo FA synthesis were grouped in cluster B, which is characterized by profiles that peak at 120 DAP, and in particular in subcluster B1 (Supplemental Fig. S4), a careful examination of the most highly represented contigs of this subcluster, which contains only 148 contigs, was carried out in order to identify potential regulatory elements of FA synthesis. This analysis revealed that the most abundant contig (CL1Contig558, renamed here EgLIP1) of this subcluster is highly similar to a TAG lipase class 3 family protein, which could account for the intensive TAG hydrolysis in the oil palm mesocarp found previously (Ngando Ebongue et al., 2006). The second most abundant contig within subcluster B1 (CL1Contig8373) has high similarity to the transcriptional regulator WRI1 (Supplemental Table S2), demonstrating that the pyrosequencing-based approach chosen in this study together with appropriate statistics constitute a reliable and straightforward method to examine the coregulation of genes involved in a given pathway and to identify candidates involved in their regulation.
A Transcript Encoding a TF Similar to WRI1 Is Highly Expressed in the Mesocarp during FA Biosynthesis
CL1Contig8373 belongs to the WRI clade (Supplemental Fig. S5), which contains WRI1 (with 68% identity) and two other WRI1-like sequences from Brassica napus (BnWRI1, ABD72476; Liu et al., 2010) and maize (ZmWRI1, AY103852; Shen et al., 2010), for which similar WRI function was shown in the seed. The contig CL1Contig8373, renamed EgAP2-2, peaks at 120 DAP, which corresponds to the onset of lipid accumulation and the transcript profile peak for many FA biosynthesis genes (Figs. 1E and 2). From the 19 transcripts expressed in the mesocarp encoding FA and TAG biosynthetic enzymes that contained a proximal upstream region long enough (more than 150 bp), canonical AW box elements were found in six transcripts (Supplemental Fig. S5). All of the transcripts encode enzymes similar to those that are active in the plastid and involved in glycolysis (ENOp and PK) and FA synthesis (ACCase CTα, KASIII, SAD, and FATB). The 15-bp element described by Baud et al. (2009) was not found in the 5′ untranslated region of oil palm sequences available.
WRI1 is known to be a target of LEC2 and LEC1, which are both master regulators of the seed maturation process and also involved in FA biosynthesis regulation (Baud et al., 2007; Mu et al., 2008; Shen et al., 2010). The analysis revealed an absence of any sequences with significant similarity to transcripts encoding ABSCISIC ACID INSENSITIVE3/VIVIPAROUS1, FUSCA3, and LEC2, all of which belong to the B3 superfamily. Similarly, no contig with HPA3 subunits of the CCAAT-binding factor (IPR003956) was found among group I contigs.
The Massive Carotene Accumulation Precedes ABA Biosynthesis
During mesocarp development, there is a spectacular increase in α- and β-carotene contents (expressed as total mesocarp carotenoid per fruit) during the maturation and ripening phases IV and V (Fig. 1E). While prior to 120 DAP only minimal amounts of the xanthophylls lutein and violaxanthin were detected, the total carotene content (α- and β-carotenes) reached very high amounts (718 μg g−1 dry mass) by 160 DAP. Only trace to low quantities of ABA were observed from 80 to 140 DAP, whereas a 6-fold increase in the ABA content was observed between 140 and 160 DAP (approximately 300 ng g−1 dry mass). The amount of the conjugated storage form ABA Glc ester also increased 3-fold between 140 and 160 DAP (Fig. 3).
Figure 3.
Carotenoid and ABA biosynthetic pathways with related metabolites and transcripts present during mesocarp development. Metabolite profiles are presented in graph insets, and relative expression is as in Figure 2. ABA-GE, ABA Glc ester; ABAGT, ABA glucosyltransferase; ABAO, abscisic-aldehyde oxidase; ABA8ox, ABA 8'-hydroxylase; CCD, carotenoid cleavage dioxygenase; CRTISO, carotenoid isomerase; CYP97A, P450 β-ring carotene hydroxylase; CYP97C, P450 -ring carotene hydroxylase; DPA, dihydrophaseic acid; DXR, 1-deoxy-xylulose 5-phosphate reductoisomerase; FPS, farnesyl pyrophosphate synthase; GGPPS, geranyl geranyl pyrophosphate synthase; GPPS, geranyl diphosphate synthase; HYD, nonheme di-iron β-carotene hydroxylase; LCY-B, lycopene β-cyclase; LCY-E, lycopene -cyclase; MEP, methyl-erythritol phosphate; NCED, 9-cis-epoxycarotenoid dioxygenase; NSY, neoxanthin synthase; NXS, neoxanthin synthase; Or, plastid-associated protein containing a DNAJ Cys-rich domain; PA, phaseic acid; PDS, phytoene desaturase; PHY, phytochrome; PIF, phytochrome-interacting factor; SET, histone methyltransferase (with SET domain); XO, xanthosin dehydrogenase; ZDS, ζ-carotene desaturase; ZEP, zeaxanthin epoxidase; Z-ISO, 15-cis-ζ-carotene isomerase; #, no contig.
As with FA and TAG synthesis pathways, the 454 sequencing data set enabled the identification of at least one contig for almost each step of the carotene, xanthophyll, and ABA biosynthetic pathways (Fig. 3; Supplemental Table S2). However, only three contigs involved in carotenogenesis were found within group I. These three genes, PSY, PDS, and PLASTID TERMINAL OXIDASE (PTOX) all encode for enzymes involved in the first two committed steps of carotenogenesis: condensation of two molecules of geranylgeranyl diphosphate (PSY), desaturation of the resulting phytoene (PDS), and recycling of the plastoquinol pool used as an electron acceptor for phytoene desaturation (PTOX). Moreover, these three contigs displayed maximal expression levels at 140 to 160 DAP, during which α- and β-carotene accumulated massively (Figs. 1E and 3). Therefore, these findings suggest that the first two committed steps of the carotenoid pathway, accomplished by PSY and PDS, are the major transcriptional control points for mesocarp carotenoid metabolism. Furthermore, despite the fact that the other carotenogenic genes were found only within group II, many displayed similar expression profiles. For example, two contigs similar to 15-CIS-ζ-CAROTENE ISOMERASE and ζ-CAROTENE DESATURASE, also involved in the desaturation steps, and fibrillins that are associated with carotenoid storage, all have higher read amounts at 160 DAP. Overall, these results suggest a coordinated transcriptional regulation of carotenogenesis in the oil palm mesocarp.
By contrast, genes involved in the upstream pathway, which is common with the biosynthesis of other isoprenoids, appeared to be expressed at lower levels, although there are putative paralogs for both DXS and FARNESYL PYROPHOSPHATE SYNTHASE expressed during the carotenogenic phase. Similarly, genes acting downstream of the desaturation steps, in particular those encoding LCY-E and LCY-B, which direct lycopene toward α- or β-carotene, were expressed rather weakly. However, considering that lycopene was not detected and that the mesocarp accumulates exceptional amounts of carotenes, LCY-E and LCY-B were obviously not rate-limiting steps. Therefore, as described previously for the TAG assembly pathway, this work provides evidence that a given metabolic pathway may be highly active without requiring that all biosynthetic steps display high and regulated transcript abundance.
Coordinated Transcriptional Activation of ABA Metabolism at Ripening
Genes encoding neoxanthin synthase, xanthosin dehydrogenase, and ABA glucosyl transferase all show a significant (group I) increase in transcript abundance between 140 and 160 DAP (Fig. 3). In addition, genes encoding for carotenoid cleavage dioxygenases and ABA 8'-HYDROXYLASE (ABA8OX) contigs displayed a similar transcription pattern, even though they were found in group II. Furthermore, this marked up-regulation of ABA biosynthetic genes during mesocarp ripening matched perfectly the dramatic increase in ABA content at this stage (Figs. 1D and 3). The metabolism of ABA, therefore, clearly displayed a coordinated transcriptional activation at ripening. These results also suggest local transcriptional control of ABA biosynthesis rather than transport from other tissues. Finally, the delay between carotenoid accumulation and subsequent ABA biosynthesis is consistent with the well-established control of ABA synthesis through the cleavage of 9-cis-xanthophylls rather than through the activation of de novo xanthophyll synthesis (Thompson et al., 2000).
Chloroplasts and Chromoplasts Share a Common Repertoire of Carotenogenesis Regulators
In contrast to our extensive knowledge of the enzymatic steps required for carotenoid synthesis in plants, relatively little is known about the regulatory mechanisms of this pathway (Lu and Li, 2008). Carotenoid content in the chromoplast has been related to the transcriptional control of genes involved in light signaling (PHY-A; UV-DAMAGED DNA-BINDING PROTEIN1 [DDB1]) and plastid morphogenesis (Or; Liu et al., 2004; Lu et al., 2006; Cazzonelli and Pogson, 2010), while for chloroplasts, genes involved in ethylene signaling (RAP2.2), chromatin modification (SDG8), and light signaling (DEETIOLATED1 [DET1] and PHYTOCHROME-INTERACTING FACTOR1 [PIF1]) were reported (Mustilli et al., 1999; Welsch et al., 2007; Cazzonelli et al., 2009; Toledo-Ortiz et al., 2010). In the oil palm mesocarp, contigs similar to regulators from both types of plastids were found. For example, DDB1 and a histone methyltransferase with a SET domain and Or and DET1, for chloroplasts and chromoplasts, respectively, were found to have peak transcript amounts at 140 DAP, which presumably coincides with the chloroplast-to-chromoplast transition, when carotenoids accumulate (Fig. 3). Similarly, contigs encoding type VII ethylene response factor (ERF)/RAP2.2 TFs, which may mediate PSY and PDS expression (Welsch et al., 2007), were found to be up-regulated at 140 DAP (Fig. 4B). Finally, a contig similar to the gene encoding a PIF, shown to down-regulate the accumulation of carotenoids by specifically repressing PSY (Toledo-Ortiz et al., 2010), was only expressed at 100 DAP, concomitantly with phytochromes (PHY-A, -B, and -C), prior to the onset of carotene accumulation.
Figure 4.
Ethylene production and transcript profiles of ethylene biosynthesis and core response pathway components in the oil palm mesocarp. A, Relative amounts of ethylene produced by the mesocarp from 120 to 160 DAP and transcript profiles for the Met and ethylene biosynthetic enzymes S-adenosyl Met (S-AdoMet, SAM) synthetase, ACS, ACO, and Asp aminotransferase-like (AAT-like) protein. B, Transcripts for the ethylene signaling proteins ETHYLENE RESPONSE (ETR), EIN, ERF, RAP, DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN (DREB), and CONSTITUTIVE TRIPLE RESPONSE (CTR). Oil palm type VII ERFs are highlighted in gray. Relative expression is as in Figure 2.
The Oil Palm Mesocarp Exhibits Characteristics of a Climacteric Fruit with an Autocatalytic Burst of Ethylene-Coordinated Transcriptional Activity
To determine the involvement of ethylene in the function, maturation, and ripening of the oil palm mesocarp tissue, we first analyzed ethylene produced in the mesocarp during selected stages of development (Fig. 4A; Supplemental Fig. S1). The lowest amounts of ethylene were detected in the 120-DAP mesocarp (207 nL g−1 fresh weight h−1), while the maximum amounts were detected at 160 DAP (7,263 nL g−1 fresh weight h−1), which represents a 35-fold increase in ethylene production. Within group I, we found contigs with similarity to transcripts that encode the three key enzymes involved in ethylene biosynthesis (Fig. 4A). We also identified transcripts that encoded proteins similar to 1-aminocyclopropane-1 carboxylic acid oxidase (ACO), the most prevalent of which (CL1Contig999) was similar to the Arabidopsis ACO4 and peaked at 140 DAP, which coincides with the large increase in ethylene production in the mesocarp. Notably, some ACO transcripts were more abundant at 100 DAP, while other distinct transcripts strongly increased at 140 DAP. There were also three contigs within group I with similarity to Asp aminotransferase-like (ATT), all (in particular CL1Contig505) with peak transcript abundance at 140 DAP. ACC synthase (ACS) enzymes are part of the ATT family, and based on their expression profiles, these contigs are good candidates to participate in ethylene biosynthesis in the mesocarp. We also identified two contigs with low read amounts similar to Arabidopsis ACSs. These data support the hypothesis that ethylene production in the mesocarp is controlled through a similar process as found in tomato, namely an autoinhibitory system 1 ethylene production during development, followed by a transition to a climacteric-like autocatalytic system 2 production during ripening through the transcriptional activation of specific transcripts for ACS and ACO (Nakatsuka et al., 1998; Barry et al., 2000; Yokotani et al., 2009).
To further examine the activity of ethylene-related processes in the mesocarp, we searched for contigs with similarity to transcripts that encode ethylene receptors, signal transduction factors, and ERFs. Notably, we identified contigs with similarity to key factors within group I, with the exception of ETHYLENE INSENSITIVE2 (EIN2) and CONSTITUTIVE TRIPLE RESPONSE1, which were within group II (Fig. 4B). As observed with the transcripts for ethylene biosynthesis enzymes, the transition from system 1 to system 2 ethylene production is marked by a coordinated increase in a large number of ethylene-related transcripts, in particular those encoding ERFs. Furthermore, certain components of ethylene perception and signal transduction have transcript profiles that coincide with either system 1 or 2. For example, two contigs similar to the ethylene receptors ETHYLENE RESPONSE1 and EIN4 had peak read amounts at 100 DAP, while a contig similar to the key transcriptional activator EIN3 peaked at 140 DAP. The most striking observation is the number of ERF-like transcripts with expression profile peaks at 140 DAP. Phylogenetic analysis with Arabidopsis AP2 domains distinguished the 10 major groups (Nakano et al., 2006); however, no oil palm fruit sequences were assigned to groups VI and VIII (Supplemental Fig. S6). These results indicated that the most predominant ERF found in the oil palm mesocarp was the type VII, the majority of which have peak transcript amounts at 140 or 160 DAP, during mesocarp maturation and ripening (Fig. 4B; Supplemental Fig. S6). In contrast, only a single ERF/AP2 type VII (CL1Contig256) and one type IX (CL1Contig6009) had peak transcript amounts at 100 DAP. At both 120 and 140 DAP, type I and IV ERF/AP2s were most prevalent, while a single type X was identified at 140 DAP. Overall, a large number of ethylene-related transcripts have expression profiles associated with either a system 1 basal amount of ethylene production or system 2 ethylene production during ripening. In particular, type VII ERF/AP2s represent a major transcriptional response associated with the ethylene burst measured during the maturation and ripening phases IV and V of the mesocarp.
A Diversity of MADS Box TFs Expressed in the Oil Palm Mesocarp at Keys Stages
Our pyrosequencing-based approach allowed the identification of 14 sequences that contained at minimum the MADS box domain and were retained for further phylogenetic analysis (Fig. 5; Supplemental Fig. S7). The 14 oil palm MADS box sequences belong to seven different subfamilies defined by the genes from Arabidopsis APETALA1 (AP1), PISTILLATA (PI), AP3, AGAMOUS (AG), SEPALLATA (SEP), and AGAMOUS-LIKE6 (AGL6; Becker and Theissen, 2003) in addition to the tomato TM3 subfamily (Supplemental Table S2).
Figure 5.
Phylogenetic analysis of oil palm MADS with other MADS genes. A, Phylogenetic analysis of CL1Contig3848, CL1Contig8010, CL1Contig6409, CL1Contig2367, and CL1295Contig1 with other SEP clade protein sequences using the neighbor-joining method based on the multiple alignment of the partial protein MIK sequences (139 residues). B, Phylogenetic analysis of CL1Contig5719, CL1Contig1226, and CL1Contig5512 with other AG clade protein sequences using the neighbor-joining method based on multiple alignment of the full-length MIKC protein sequences. ANR1 from Arabidopsis was used as the root. Numbers on the branches are bootstrap values for 100 replicates. The sequences included in this alignment are from Arabidopsis, maize, rice, oil palm, and snapdragon (Antirrhinum majus) and from proteins in selected fruit species such as apple, banana, grape, peach, strawberry, and tomato. The MADS box proteins from our study are in gray. Relative expression is as in Figure 2.
The class of MADS most represented in the mesocarp belongs to the AGL2/SEP subfamily, including three within group I and two within group II (Fig. 5A). Notably, there were no oil palm sequences found in the SEP subclades 1 and 5, which contain either the strawberry FaMADS9 or the tomato LeRIN, respectively, both of which perform key functions during fruit development and ripening (Vrebalov et al., 2002; Seymour et al., 2011). Overall, the five contigs within the AGL2/SEP subfamily are grouped within two distinct subclades supported by significant bootstrap values. Within the SEP3 subclade, EgAGL2-1, reported in oil palm male and female inflorescences (Adam et al., 2006), CL1Contig8010/EgAGL2-2 (100% amino acid identity), and CL1Contig6409 form a monophyletic group, while CL1Contig3848 forms a monophyletic group with the banana fruit MaMADS2 and MaMADS4 (Elitzur et al., 2010). Contig CL1Contig8010 is the most abundant MADS found in the mesocarp, with an expression profile coordinated during maturation and ripening (Fig. 5A). CL1Contig2367 and CL1Contig1295 are found within another subclade that contains uniquely monocot sequences. Notably, CL1Contig2367 peaks at 100 DAP and decreases during maturation and ripening, suggesting a function prior to phase III of mesocarp development. Furthermore, the oil palm sequences systematically form subclades with other monocot sequences and do not group with those from dicot fruit species, which suggests that a diversification of function within this subfamily has occurred within monocots since the separation from eudicots.
There were three contigs identified within the MADS box AG subfamily. CL1Contig1226 is the most abundant and peaks at 140 DAP (Fig. 5B). The three oil palm AG-like contigs analyzed constitute a monophyletic group with the banana fruit MaMADS5 (Elitzur et al., 2010; Fig. 5B). The mesocarp fruit AG-like sequences are distinct from EgAG1 and EgAG2 reported in oil palm male and female inflorescences that group together in the eudicot AG-like subclade (Adam et al., 2006). The cladogram indicates seven monophyletic groups supported by significant bootstrap values. The PLENA and eudicot AG lineages include TAGL1 and TOMATO AGAMOUS (TAG1), respectively, which have regulatory functions during tomato flower and fruit development, respectively (Seymour et al., 2008; Pan et al., 2010). Notably, these two groups contain sequences exclusively from eudicot fruit species such as peach, apple (Malus domestica), and grape.
From the GLOBOSA/PISTILLATA (GLO/PI) subfamily, a single contig from group I, CL290Contig1, shares 100% nucleic acid identity with EgGLO1 from oil palm identified previously (Supplemental Fig. S7; Adam et al., 2006). The oil palm GLO-like protein is separated from the fleshy fruit eudicot sequences and positions closest to the banana MaMADS6, the only available sequence from a monocot fleshy fruit species in this subfamily (Elitzur et al., 2010). The accumulation of CL290Contig1 is significantly increased during maturation and remains high during ripening.
Profiles of TFs and Transcription Regulators in the Mesocarp Suggest That Coordinated Transcriptional Mechanisms Occur in the Mesocarp
A total of 1,930 group I contigs (73%) were assigned INTERPRO accession annotations, including 150 contigs with functions related to transcriptional regulation, among which 127 contigs were TFs and transcription regulators (TRs) not previously described in this study (Supplemental Tables S3–S6). Overall, there are 37 TF/TR contigs with peak read amounts within cluster A, 14 contigs at 120 DAP within cluster B, 85 contigs at 140 DAP within cluster C, and 14 contigs at 160 DAP within cluster D. As observed earlier from the GO annotation enrichment analysis (Supplemental Table S1), the largest proportion of contigs related to transcriptional regulation were found at 140 DAP within cluster C.
Within cluster A, with peak transcription amounts at 100 DAP, the most abundant TF/TR is a Cys-2/His-2-type zinc finger protein found within subcluster A3 (Supplemental Table S3). Notably, there are two NAC domain proteins that are also abundant within the same subcluster A3 (Supplemental Fig. S4). Indeed, NAC domain proteins are the most represented class of proteins at 100 DAP, three within subcluster A1 and two within subcluster A3, suggesting coordinated regulation of this class of TF/TR. AUXIN RESPONSE FACTORs (ARFs) are the second most represented class, with four ARFs all within subcluster A1, suggesting strong transcriptional coordination among this class of TF/TR. Another notable feature of cluster A is the presence of contigs for genes encoding chromatin modifiers, including a condensin complex component, a DNA methyltransferase, a polycomb group protein, and two contigs for jumonji domain TFs implicated in chromatin regulation during development (Takeuchi et al., 2006). These results indicate that several factors involved in chromatin modifications that can affect changes in gene expression are present and may function at 100 DAP.
Remarkably, only 14 TF/TR contigs are found within cluster B, which suggests that restricted transcriptome regulation occurs at this stage in the mesocarp (Supplemental Table S4). In addition to EgAP2.2, the three ERF/AP2 TFs, and the MADS box described in the previous sections (Figs. 4B and 5), there are two calmodulin-related signaling proteins, one IAA/AUX protein, one zinc finger protein similar to the maize Indeterminate1, one Golden2-like protein that regulates chloroplast development and the expression of the photosynthetic apparatus, one bZIP, and a SEUSS-LIKE3 TR.
In cluster C, in addition to the type VII ERF/AP2 TFs and the MADS box TFs described above, two contigs for bZIP TF were found in subclusters C1 and C4 (Supplemental Table S5). Other classes that are well represented in cluster C include five NAC domain proteins, four zinc finger proteins, three BELL1-LIKE HOMEODOMAIN proteins, and three WRKY TF proteins. Finally, two contigs (CL1Contig6719 and CL2179Contig1) with similarity (E > 1e-30) to the homeodomain-Leu zipper protein LeHB-1, which binds to the promoter of LeACO1 to activate transcription during ripening (Lin et al., 2008), were identified.
In cluster D, 11 out of 14 contigs are found within subgroup D3 (Supplemental Fig. S4), which suggests that tight transcriptional coordination occurs at the ripening stage (Supplemental Table S6). In addition to the AGL2/SEP described above (Fig. 5A), other prevalent contigs found within subcluster D3 included a basic helix-loop-helix protein, a COP9 signalosome complex subunit 3, and a NAC domain protein. Indeed, NAC domain proteins made up the most represented class in cluster D; notably, two of the NAC domain proteins had the highest similarity (E > 1e-50) to the tomato NAC-NOR, reported as a component of the ethylene-dependent ripening regulation in tomato (Giovannoni, 2004).
Finally, no contigs for CNR or SlAP2a, regulatory factors that also directly or indirectly affect ethylene production during tomato ripening (Thompson et al., 1999; Chung et al., 2010), were found within group I.
DISCUSSION
The Oil Palm Mesocarp Is an Original Fruit Model to Examine Regulatory Mechanisms That Function during Fruit Maturation and Ripening
The oil palm mesocarp presents an original model to examine the regulatory networks in a monocot fruit tissue subject to climacteric ripening and in which high amounts of lipids accumulate. This study provides, to our knowledge for the first time, a detailed description of the original physiological and biochemical characteristics along with a thorough analysis of the underlying transcriptional activities, combined to develop a model that describes major events that occur during the defined phases of mesocarp development (Fig. 6).
Figure 6.
Scheme representing the major events that occur during the phases of mesocarp development. See text for details.
Similar to during tomato development, a decrease in auxins, GAs, and cytokinins (Gillaspy et al., 1993) is also observed prior to mesocarp ripening; however, a second auxin peak that overlaps with that of ethylene during tomato ripening is not observed in the oil palm mesocarp. ABA is also present early during tomato development, and a recent detailed study in tomato revealed that an ABA peak occurs just prior to that of ethylene during ripening (Zhang et al., 2009a). By contrast, in the oil palm mesocarp, there is a large simultaneous increase in both ethylene and ABA during fruit ripening that suggests regulatory functions and/or interactions for these two hormones in the mesocarp.
In the tomato fruit, the increase in ethylene production at the onset of ripening coincides with a transition from autoinhibitory (system 1) to autostimulatory (system 2) ethylene production. The tomato model predicts that system 1 functions prior to ripening while system 2 operates during ripening and that the two systems are regulated by coordinated developmental and ethylene-induced expression of individual ACO, ACS, and tomato ethylene receptor genes (Nakatsuka et al., 1998; Barry et al., 2000; Yokotani et al., 2009). In this study, an increase in ethylene production characteristic of a climacteric burst is observed during phases IV and V. Furthermore, the coordinated transcriptional activation of subsets of transcripts for ACO and ACS-like genes (Asp aminotransferase-like genes) suggests that a transition from system 1 to system 2 occurs in the oil palm mesocarp during the maturation phase IV (Fig. 6). Indeed, by 140 DAP, a sharp increase in transcripts for ethylene biosynthetic enzymes and ERFs in the mesocarp is observed that resembles system 2-type ethylene production. Notably, six ERFs with peak read totals at 140 or 160 DAP are type VII ERFs similar to those implicated in the ripening of tomato, kiwi (Actinidia deliciosa), and plum (Prunus domestica; Tournier et al., 2003; Wang et al., 2007; El-Sharkawy et al., 2009; Sharma et al., 2010; Yin et al., 2010). Phylogenetic analysis with the oil palm sequences and selected type VII ERF signature domains (Nakano et al., 2006) from Arabidopsis suggest conservation between monocots and dicots and that gene duplication events may have occurred for multiple type VII genes to be expressed in the mesocarp (Fig. 4; Supplemental Fig. S6).
A total of 13 NAC domain-containing proteins within clusters A, C, and D indicate that this class of TFs is one of the most highly represented in the mesocarp. Indeed, NAC domain proteins are key participants in TF networks and play central roles in plant development (Olsen et al., 2005). While NAC domain TFs were the most prominent class at 100 DAP, four ARFs were also observed that coincide with the IAA metabolite peak, suggesting auxin-related functions at this stage (Fig. 6). Furthermore, two other NAC domain contigs were identified with similarity to the tomato NAC-NOR (AAU43922.1) gene, the mutation in which has been reported to be responsible for the nor ripening mutant (Giovannoni, 2004). Notably, both of these NAC domain contigs have expression profiles associated with mesocarp ripening, which suggests functional similarity to the tomato NAC-NOR gene. While these results suggest conserved functions for some NAC domain proteins during ripening, very little is known about their roles during fruit development. In oil palm, the large number of transcripts observed for this class of TFs with different expression profiles that correspond to key phases of mesocarp development may reflect a high number of gene duplication events followed by expression domain changes that occurred in this family since the separation between monocots and eudicots.
The Expression of MADS Box Genes from the SEP, AG, and GLO Subfamilies Characterizes Key Phases of Mesocarp Development and Function
In this study, we found a diversity of MADS box transcripts with expression associated with the different phases of mesocarp development. MADS box proteins are key regulators of fruit ripening (Vrebalov et al., 2002, 2009; Ito et al., 2008; Itkin et al., 2009; Giménez et al., 2010; Jaakola et al., 2010; Seymour et al., 2011). In particular, the protein RIN within the SEP4 subclade controls the induction of the tomato ACS transcripts involved in system 2 ethylene production (Barry et al., 2000). Recently, a SEP1/2 subclade MADS box from the nonclimacteric fruit strawberry (FaMADS9) was shown to play a key role in ripening (Seymour et al., 2011). To date, very little is known about the SEP subgroup involvement in monocot fruit ripening. Three SEP3 homologs, MaMADS1, -2, and -4, expressed in ripening banana fruit tissues share low amino acid sequence similarity (55%–62%) with RIN, and none thus far complements the tomato rin mutation (Elitzur et al., 2010). However, MaMADS2 is expressed in the pulp prior to the burst of ethylene production and is not induced by ethylene treatments. In the oil palm mesocarp, there were at least five SEP homologs expressed, two of which (CL1Contig8010 and CL1Contig3848) within the SEP3 subclade increase during the ethylene burst. Interestingly, phylogenetic analysis indicated that CL1Contig3848 grouped closely to the banana MaMADS2, while neither of these homologs grouped closely with the tomato RIN or with the strawberry FaMADSS9. These results support the hypothesis that the MADS box SEP-like proteins are key factors of ripening not only in climacteric and nonclimacteric fruits but also in monocot and dicot fruits. Interestingly, FaMADS9 and RIN belong to different SEP subgroups, SEP1/2 and SEP4, respectively, and the strawberry sequence most similar to RIN (FaSEP4) is expressed at very low levels in the fruit. These results demonstrate that while SEP-like proteins indeed operate in the ripening process of both climacteric and nonclimacteric fruits, sequence divergence has occurred that may reflect differences between the two ripening models. While the downstream targets of FaMADS9 are unknown, the evidence indicates that some common functions may exist during both climacteric and nonclimacteric ripening for SEP subgroup MADS box genes in these two dicots. Our data support the hypothesis that in monocot fruits, SEP3 homologs function upstream of ripening processes analogous to the tomato RIN and the strawberry FaMADSS9, and the divergence observed within the SEP subgroup may reflect important differences between fruit ripening systems, including those of monocots.
Another tomato MADS box protein, TAGL1 within the AG subgroup, also functions upstream of ethylene production, apparently through the control of LeACS2 expression yet independently from RIN (Itkin et al., 2009; Vrebalov et al., 2009; Giménez et al., 2010). Very little is known about the role of AG clade MADS box proteins during nonclimacteric or monocot fruit ripening. In grape, there are at least three AG-like transcripts expressed in the fruit but unassociated with ripening (Boss et al., 2001, 2002; Díaz-Riquelme et al., 2009). In banana, there appears to be a least one AG-like transcript (MaMADS5/MuMADS1) expressed in the pulp that is induced by ethylene (Liu et al., 2009; Elitzur et al., 2010). In the oil palm mesocarp, we identified at least three transcripts that encode AG clade MADS box proteins. Notably, one transcript (CL1Contig5512) accumulates with no relation to the ethylene burst, while another (CL1Contig1226) accumulates during the initial burst of ethylene production and related transcriptional activity. The domains encoded by these two sequences, along with that of a third oil palm sequence and the banana MaMADS5, form a monocot monophylogenetic subclade. Furthermore, this monocot subclade is distinct from those containing the dicot tomato fruit TAGL1 and TAG1 or the oil palm EgAG1 and -2 expressed in the oil palm female flower (Adam et al., 2007a). These results suggest that gene duplications in the oil palm genome have occurred that gave rise to at least three distinct yet closely related AG-like sequences with different expression profiles during mesocarp development, maturation, and ripening, while EgAG1 and -2 expression and function may be limited to flower development. However, it is not known whether the three AG-like sequences expressed in the mesocarp are also expressed during flower development or in other oil palm tissues. Together, these results support the hypothesis that some AG clade MADS box proteins have diverged to gain expanded expression with new functions in the fruit beyond those performed during flower formation. Furthermore, as with the SEP clade MADS box proteins, a divergence of AG-like proteins between dicots and monocots is clearly observed that may reflect underlying functional differences between these two plant groups in relation to fruit development and ripening.
A transcript (EgGLO1) for a GLO/PI family MADS box was also observed with expression related to the ethylene burst. To our knowledge, EgGLO1 is the first within this class to be observed with a fruit ripening-related expression profile in any species. Indeed, while GLO members have been identified in eudicot fleshy fruit species such as grape, apple, and peach and in the monocot banana fruit, thus far none is associated with fruit ripening (Yao et al., 2001; Busi et al., 2003; Hileman et al., 2006; Poupin et al., 2007; Zhang et al., 2008; Díaz-Riquelme et al., 2009; Elitzur et al., 2010). In contrast, the oil palm mesocarp EgGLO1 was expressed in relation to ethylene production and related transcriptional activity during maturation and ripening. Notably, the EgGLO1 nucleotide sequence is identical to the transcript observed previously during flower development (Adam et al., 2006). Indeed, there were two GLO/PI family MADS box transcripts (EgGLO1 and EgGLO2) found to be expressed during flower development, but only EgGLO1 was observed in the mesocarp. Therefore, either the same gene that functions during flower development is also active and plays a role during fruit ripening or a duplication has occurred and another similar gene has diverged to acquire a new fruit-related function.
Overall, these data indicate the involvement of MADS box genes, in particular SEP-like, AG-like, and GLO-like, during maturation and ripening of the oil palm mesocarp. There appears to have been a substantial amount of functional diversification in the SEP-like and AG-like subfamilies, given the number of new transcripts found compared with those observed previously in studies of the oil palm flower structure (Adam et al., 2006, 2007a, 2007b). This diversification appears to have arisen through the expansion of the expression of genes that either have retained a function in the flower or have specialized functions within the fruit. Indeed, several sequences identified in this study, including those from the SEP-like and the GLO-like subfamilies, were identical at the nucleotide level to those observed in the oil palm flower and may be the same gene. Finally, the expression of certain MADS box transcripts either positively or negatively coincides with the burst of ethylene production that occurs by 140 and 160 DAP. This raises the question of the relationship between ethylene production and related transcriptional activity and whether these MADS box gene products play regulatory roles either upstream or downstream of the ethylene burst during mesocarp ripening.
The Last Acylating Step of TAG Assembly May Involve Several Complementary Routes
One of the most notable findings from the reconstruction of the oil synthesis pathways in the oil palm mesocarp is the possible contribution of three enzymes for the acylation of DAG to TAG (i.e. both type 1 and type 2 DGAT and PDAT). Although the literature shows that the three enzymes, encoded by three separate gene families, are all capable of catalyzing the final acylation step, their individual contributions in plant tissues that accumulate oil remain poorly understood. The similar transcript amounts detected in this study suggest that the contribution of these three enzymes could be of similar importance in the mesocarp of oil palm. Based on transcription patterns of DGAT1, DGAT2, and PDAT in developing seeds of various plants, it was suggested that DGAT2 and PDAT could be major contributors of TAG synthesis in seeds that store large amounts of epoxy and hydroxy FA (Li et al., 2010a). However, a recent study also indicates high transcription of both DGAT1 and DGAT2 in the mesocarp of olive during the stage of oil synthesis, while olive oil does not contain any unusual FA (Banilas et al., 2011). In that study, the authors hypothesize that DGAT2 could play a specific role during oil droplet enlargement, which occurs at late stages of olive mesocarp maturation. Furthermore, it was recently demonstrated that both DGAT1 and PDAT1 contribute to TAG synthesis in the developing Arabidopsis embryo (Zhang et al., 2009b). A major role for PDAT in the developing oil palm mesocarp is substantiated by the identification of a contig very similar to its necessary partner, lysophospholipid acyltransferase, whose transcript levels were comparable to those of other transcripts involved in TAG assembly.
Divergence of the FA Synthesis and TAG Assembly Transcriptional Regulation
The second major result drawn from the analysis of the lipid-related transcriptome is that the core FA synthetic machinery of the oil palm mesocarp is remarkably coordinated at the transcriptional level (Figs. 2 and 6). Large-scale transcriptomic studies focused on oil synthesis in plants remain scarce. However, since the same key result was identified in the developing embryo of Arabidopsis (Ruuska et al., 2002; Baud and Lepiniec, 2009) and the persistent living endosperm of albuminous coffee seeds (Joët et al., 2009), it is tempting to speculate that this feature is common to plant oil-storing tissues, independent of their origin. In all three cases, a sharp increase in plastidial gene transcription occurs at the onset of lipid accumulation. Similar to that observed in the Arabidopsis embryo and the coffee endosperm, ER genes involved in TAG assembly displayed an expression profile different from that of FA synthesis genes, suggesting distinct regulation programs. However, while the fold increases of most genes involved in TAG assembly were higher than those of the core FA biosynthetic machinery in the Arabidopsis embryo (Baud and Lepiniec, 2009), transcript levels observed in the oil palm mesocarp for genes responsible of TAG assembly remained low and did not show a clear increase concomitant with oil deposition, such as that observed for FA synthesis genes. Since no other tissue accumulates more oil than the oil palm mesocarp, this work clearly indicates that a marked up-regulation of TAG assembly gene transcription is not strictly required for oil accumulation and that the regulation of this pathway may differ considerably among plants and tissues. Based on our transcriptomic data here, one can hypothesize that the metabolic control of oil synthesis in the oil palm mesocarp is mostly controlled by de novo FA synthesis. This hypothesis corroborates previous studies (Ramli et al., 2002, 2009) that quantified flux control coefficients of the two blocks, FA synthesis and TAG assembly, in callus cultures of olive and oil palm using the technique of metabolic control analysis.
Phytoene Synthase and Phytoene Desaturase Are the Key Players for Carotenoid Accumulation
Our results clearly revealed a coordinated transcriptional activation of the carotenogenic genes involved in the synthesis of phytoene and subsequent desaturation steps leading to lycopene, while further steps involved in the cyclization of lycopene toward carotenes and upstream isoprenoid synthesis appear to be poorly regulated or not regulated at the transcriptional level. Despite the fact that the oil palm mesocarp stores large amounts of carotenes but not lycopene, the transcriptional regulation of carotenogenesis nevertheless resembles that of the lycopene-rich tomato fruit (i.e. through the transcriptional activation of the first two committed steps of carotenoid synthesis; Ronen et al., 1999). This important finding suggests that the control of carotenoid metabolism in oil palm may first operate by directing the metabolic flux into the carotenoid pathway. This is consistent with previous reports that showed that PSY expression was the rate-limiting step for carotenogenesis in various plant models (Fraser et al., 1994; Shewmaker et al., 1999). In addition, the constitutive overexpression of PSY was found to be sufficient to trigger the accumulation and sequestration of carotenes into crystals in plastids from nongreen tissues of Arabidopsis (Maass et al., 2009). Considering the importance of PSY and PDS up-regulation for carotenogenesis in the oil palm mesocarp and that class VII ERF/RAP2.2 TFs are known to mediate PSY and PDS expression (Welsch et al., 2007), we can speculate that the contigs encoding ERF/RAP2.2 found to be up-regulated at 140 DAP participate in the massive carotene accumulation in this fruit. Finally, the concomitant up-regulation of the Or, DDB1, and DET1 regulators suggests that carotenoid accumulation may also be controlled by enhancing sink strength (i.e. the formation of carotenoid sequestration structures during the chloroplast-to-chromoplast transition). Indeed, the Or gene was previously shown to cause high levels of β-carotene accumulation in an orange cauliflower (Brassica oleracea) mutant by enhancing sink strength (Lu et al., 2006). In addition, expression of the Or transgene in potato (Solanum tuberosum) produces deep orange-yellow flesh tubers with an over 6-fold increase of total carotenoid content. This increase in the Or transgenic tubers was found to be associated with the formation of chromoplasts and specific carotenoid sequestration structures, which serve as an effective metabolic sink to facilitate the sequestration and storage of carotenoids (Lopez et al., 2008).
The low abundance of lycopene cyclase transcripts (LCY) was at first quite puzzling, since the oil palm mesocarp does not accumulate lycopene but carotenes. However, this discrepancy can be interpreted in two ways. First, it clearly evokes the low level of transcription of the whole TAG assembly pathway, while the oil palm mesocarp is exceptionally efficient regarding TAG synthesis. One of the major lessons drawn from the combined analysis of both oil and carotenoid transcriptomes is that the coordinated up-regulation of all steps of a given pathway is not a prerequisite for the activity of the corresponding metabolism. Besides, the amount of carotenoids synthesized may also depend on the properties of the encoded enzymes and therefore may rely on the polymorphism observed at the sequence level. Indeed, a recent study carried out in cassava (Manihot esculenta) reported that a single nucleotide polymorphism in PSY2, leading to a nonconservative amino acid exchange, results in a dramatic increase of carotenoid accumulation in storage roots (Welsch et al., 2010). This polymorphism occurs in a region that is usually highly conserved among plants (the ALDRWE amino acid sequence; Supplemental Fig. S8), and the A191D exchange leads to an increased PSY enzymatic activity in cassava. Notably, this conserved region is also affected in oil palm, with an amino acid exchange in the vicinity of the Arg (the L198M exchange; Supplemental Fig. S8). This discovery opens up interesting prospects, and a more in-depth characterization of the EgPSY enzyme should allow us to test whether the exceptional carotene content of the oil palm mesocarp is also influenced by the peculiar amino acid sequence polymorphism in this region.
Toward an Understanding of FA Synthesis Regulation in the Oil Palm Mesocarp
The fact that most genes involved in the core FA biosynthetic machinery share the same temporal transcription pattern strongly suggests that they are coregulated and probably share common regulatory elements. On the basis of recent literature on the control of oil biosynthesis in seeds (Baud and Lepiniec, 2010), we hypothesize that an AP2 TF similar to WRI1 could play this key role in the oil palm mesocarp (Fig. 6). Indeed, we not only found a highly expressed WRI1-like contig, named EgAP2-2, that was perfectly coregulated with FA biosynthetic genes but also identified the same AW box sequence in several genes belonging to the core FA biosynthetic machinery, which confirms that this cis-element, considered as the direct binding site for WRI1 (Maeo et al., 2009), is highly conserved among monocots and eudicots (Pouvreau et al., 2011). To our knowledge, our work here reveals for the first time that the transcriptional regulation of FA biosynthesis via WRI1 may not be restricted to seeds but may also operate in other tissues that accumulate lipids, such as the oil palm mesocarp.
These results raise the critical question of whether EgAP2-2 expression in the mesocarp is regulated by master regulators analogous to those that function during seed maturation (Baud and Lepiniec, 2010). The absence of any LEC2-like transcript in the oil palm mesocarp transcriptome is not surprising, since LEC2 presumably exists only in dicot genomes (Li et al., 2010b). In contrast, since LEC1 was recently proposed to regulate WRI1 in maize (Shen et al., 2010), it constituted a viable candidate for EgAP2-2 regulation. However, given that LEC1 is often considered specific to seed tissues, the regulation of EgAP2-2 in the mesocarp may be completely different. Nonetheless, since LEC1 was recently shown to be expressed in fern (Adiantum capillus-veneris) leaves in response to dehydration (Xie et al., 2008), its participation in oil synthesis regulation in the mesocarp of a monocot species could not be excluded. However, we did not identify any contig showing high homology with LEC1-type HAP3, suggesting that a different regulatory cascade controls WRI1 expression in the oil palm mesocarp.
MATERIALS AND METHODS
Plant Material Production, Histology, and RNA Preparation for 454 Sequencing
Oil palm (Elaeis guineensis) fruits were harvested at Pobe Centre de Recherche Agricoles Plantes Pérennes Station (Institut National de Recherche Agricole du Bénin) from a dura parent of Deli Dabou origin, within the same self-progeny of a single palm. In this progeny, after two successive self-pollinations, the percentage of heterozygosity is estimated at only 20% to 25% (Cochard et al., 2009). For each stage of development studied, three independent bunches were collected from three distinct individuals of the same genotype. Fifteen spikelets were then collected in the center of each bunch, and five spikelets were randomly sampled from them. From all undamaged fruits withdrawn from each of the five spikelets, three fruits were then randomly sampled. Then, for each stage-bunch-spikelet combination retained, mesocarp, shell, and seed of each fruit were separated and weighed. Dry weight from all samples was measured after 1 night at 103°C. Variation in tissue weight across development was tested using three-way ANOVA and a posthoc Newman-Keuls test.
For histological analysis, samples were fixed in 4% paraformaldehyde and phosphate-buffered saline (PBS) 1× in the presence of 4% 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide for 16 h. Thereafter, samples were washed twice for 15 min in PBS 1× and Gly, then twice for 15 min in PBS 1×. Finally, samples were dehydrated in ethanol 50% for 1 h and 70°C before embedding in Technovit 7100 resin (Heraeus Kulzer). Semithin sections of mesocarp (4 μm) were stained with toluidine blue, which stains acid compounds such as pectin in blue in an acid environment. To visualize lipids in the mesocarp, fixed samples were sectioned (100 μm) using a vibratom, washed with PBS 1×, stained with Nile red, and analyzed by confocal microscopy (Zeiss LM510), with laser excitation at 458 nm and a HFT filter (548/514) using the 40×/1.2 numeric aperture. To visualize β-carotenes in mesocarp cells, fruit were sectioned (150 μm) after 16 h in fixation buffer using a vibratom, stored in PBS 1×, and then observed by confocal microscopy (Zeiss LM510 meta) with laser excitation at 488 nm and emission at 580 to 600 nm, corresponding to β-carotene emission spectra using the 63×/1.4 numeric aperture.
For RNA extractions, mesocarp tissue samples were collected and frozen immediately in liquid nitrogen. Total RNA from mesocarp was extracted as described previously (Morcillo et al., 2006). Using a titanium kit (Roche), cDNAs related to four stages of development (100, 120, 140, and 160 DAP) were tagged independently and then mixed together in one sample for 454 pyrosequencing carried out by GATC Biotech.
Hormonal Analysis
For hormonal profiling analysis, mesocarp was collected from fruit aged between 80 and 160 DAP, lyophilized, and then ground before being sent (3 × 50 mg of dry powder) to National Research Council Canada-Conseil National de Recherches Canada. ABA and ABA metabolites, cytokinins, auxins, and GAs were quantified by ultra-performance liquid chromatography-ESI-tandem mass spectrometry as described in detail by Chiwocha et al. (2003). To measure ethylene release, spikelets consisting of 10 fruits are put in hermetic jars and ethylene production is quantified after 24 h by gas chromatography (flame ionization detector, equipped with a GS-Q column). Values shown are means of duplicate determinations.
Lipid and Carotenoid Determination Analyses
Total lipids were extracted from 1-g samples of freeze-dried powder using a modified Folch method as described previously (Laffargue et al., 2007). Carotenoid extraction was adapted from the method described previously (Achir et al., 2010), which includes a preliminary TAG removal step. Briefly, oil samples (100 mg) were carefully mixed with 2 mL of acetone containing 15 mg L−1 echinenone as an internal standard and left for 30 min at –20°C, leading to TAG crystallization. TAGs were separated by rapid sampling of the upper acetone phase, which contains carotenoids, and filtered through a 0.45-mm polytetrafluoroethylene filter (Whatman). The carotenoid extract was directly injected into the HPLC column. The column was a polymeric YMC-30 (4.6 mm i.d. × 250 mm, 5 mm particle size; YMC). Elution was performed using five successive steps at a flow rate of 1 mL min−1: (1) 100% A for 15 min; (2) linear gradient from 0% B to 20% B for 10 min; (3) linear gradient from 20% B to 90% B for 10 min; (4) linear gradient from 90% B to 100% B for 5 min; and (5) 100% B for 5 min (A = methanol:water [60:40, v/v] and B = methyl tert-butyl ether:methanol:water [68:28:4, v/v/v]). A UV-visible photodiode array detector (Dionex UVD 340U) was used to analyze the chromatograms at a detection wavelength of 450 nm, except for phytofluene (348 nm), phytoene (286 nm), and ξ-carotene (401 nm). Carotenoids were identified by the combined use of their relative retention times and their wavelength absorption maxima as determined using analytical standards (Carotenature). Quantification was done by combining the standard calibration curves and correction with internal standard peak areas. All analyses were realized in triplicate (from three independent mesocarp samples).
454 Sequence Analysis, de Novo Assembly, and Contig Annotation
The 454 reads obtained from the 454 sequencing data were analyzed using a specialized version of our tool ESTtik (for EST Treatment and Investigation Kit; Argout et al., 2008) dedicated to the analysis of 454 data. To avoid problems of misassembly, we discarded sequences shorter than 120 bp and low-quality sequences. We then used the Megablast algorithm to detect and eliminate noncoding sequences by comparing the whole reads against fRNAdb (Mituyama et al., 2009) with an E value cutoff of 1e-20. To avoid chimeric consensus sequences, which contain parts of different expressed genes, we modified the parameters of the TGICL (Pertea et al., 2003) assembler to obtain good-quality contigs. We used a minimum overlap percentage identity cutoff of 94% and an overlap length cutoff of 60 bp for the clustering procedure of TGICL and a minimum overlap percentage identity cutoff of 95% and an overlap length cutoff of 60 bp for the assembly. Finally, we predicted peptide sequences from the transcriptomic data using a modified version of the prot4EST pipeline (Wasmuth and Blaxter, 2004). The peptide sequences were annotated using the InterProScan Web service (Zdobnov and Apweiler, 2001) to find protein domains and functional sites by comparing the sequences against the InterPro signature database (Hunter et al., 2009). Contigs were then annotated using the stand-alone version 2.2.16 of the BLAST software (Altschul et al., 1990). Similar sequences were searched within the TIGR6 proteome of rice (Oryza sativa japonica; Uniprot Knowledgebase Swiss-Prot and TrEMBL; Boeckmann et al., 2003), the nonredundant protein sequences database from GenBank, and the nucleotide sequences from the GenBank nucleotide collection NT. To obtain significant results for the whole BLAST analyses, we used an E value cutoff of 1e-5, and the 10 best hits of each BLAST were retained for the annotation. In addition, we used the Blast2GO software (Götz et al., 2008) to retrieve GO terms based on our BLAST analyses.
Statistical Analysis of Read Abundance Profiles
A contig was considered differentially expressed during development when at least one of the three stage transitions (100–120, 120–140, and 140–160 DAP) exhibited a highly significant difference in read abundance at P = 0.01. Differences in read abundance between two stages of development were tested using Audic-Claverie statistics (Audic and Claverie, 1997). P values obtained by the Audic-Claverie test were then transformed using the Bonferroni correction to control FDRs. Hierarchical clustering analysis was used to group contigs according to their transcription profile using the tool developed by Eisen et al. (1998; http://rana.lbl.gov/eisen/).
Phylogenetic Analysis
Phylogenetic trees were constructed based on similarity searches performed with BLASTp programs with default parameters in protein sequence databases provided by the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov). Phylogenetic analyses were performed on the Phylogeny.fr platform (http://www.phylogeny.fr; Dereeper et al., 2008). Amino acid sequences were aligned with ClustalW (version 2.0.3; Thompson et al., 1994). After alignment, ambiguous regions (i.e. containing gaps and/or poorly aligned) were removed with Gblocks (version 0.91b). The phylogenetic tree was reconstructed using the neighbor-joining method implemented in Neighbor from the PHYLIP package (version 3.66; Saitou and Nei, 1987). Distances were calculated using ProtDist. The Jones-Taylor-Thornton substitution model (Jones et al., 1992) was selected for the analysis. The robustness of the nodes was assessed by bootstrap proportion analysis (Felsenstein, 1985) computed from 100 replicates. Graphical representation and editing of the phylogenetic trees were performed with TreeDyn (version 198.3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complete data set of auxin, GA and cytokinin, ABA, and ethylene metabolite analysis.
Supplemental Figure S2. Overview of 454 pyrosequencing data.
Supplemental Figure S3. FASTA list of assembled contigs.
Supplemental Figure S4. Hierarchical cluster analysis of group I contigs differentially represented.
Supplemental Figure S5. Phylogenetic analysis of EgAP2-2, a TF similar to WRI1.
Supplemental Figure S6. Phylogenetic analysis of the AP2 domains from the ERFs identified in the mesocarp.
Supplemental Figure S7. Phylogenetic analysis of the MADS domains from mesocarp MADS box proteins.
Supplemental Figure S8. Amino acid alignments of PSY conserved domains of oil palm contig (EgPSY) and others from cassava, Narcissus pseudonarcissus, rice, tomato, and maize.
Supplemental Table S1. GO enrichment analysis between developmental stages of group I contigs.
Supplemental Table S2. Annotations of contigs discussed in the text.
Supplemental Table S3. TFs and regulators expressed in the mesocarp cluster A.
Supplemental Table S4. TFs and regulators expressed in the mesocarp cluster B.
Supplemental Table S5. TFs and regulators expressed in the mesocarp cluster C.
Supplemental Table S6. TFs and regulators expressed in the mesocarp cluster D.
Supplementary Material
Acknowledgments
We thank Myriam Collin and Jean-Luc Verdeil for their excellent assistance with the histological analysis and James W. Tregear for his support and useful discussions during the preparation of this article. We also thank Nathalie Maréchal and Sylvie Avallone for assistance with the biochemical analyses. We especially thank all the personnel at the Institut National de Recherche Agricole du Bénin, Centre de Recherche Agricoles Plantes Pérennes (Pobe, Benin), for their technical and logistical support during the collection of the genetic material used in this study.
References
- Achir N, Randrianatoandro VA, Bohuon P, Laffargue A, Avallone S. (2010) Kinetic study of β-carotene and lutein degradation in oils during heat treatment. Eur J Lipid Sci Technol 112: 349–361 [Google Scholar]
- Adam H, Jouannic S, Morcillo F, Richaud F, Duval Y, Tregear JW. (2006) MADS box genes in oil palm (Elaeis guineensis): patterns in the evolution of the SQUAMOSA, DEFICIENS, GLOBOSA, AGAMOUS, and SEPALLATA subfamilies. J Mol Evol 62: 15–31 [DOI] [PubMed] [Google Scholar]
- Adam H, Jouannic S, Morcillo F, Verdeil JL, Duval Y, Tregear JW. (2007a) Determination of flower structure in Elaeis guineensis: do palms use the same homeotic genes as other species? Ann Bot (Lond) 100: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam H, Jouannic S, Orieux Y, Morcillo F, Richaud F, Duval Y, Tregear JW. (2007b) Functional characterization of MADS box genes involved in the determination of oil palm flower structure. J Exp Bot 58: 1245–1259 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arango Y, Heise KP. (1998) Tocopherol synthesis from homogentisate in Capsicum annuum L. (yellow pepper) chromoplast membranes: evidence for tocopherol cyclase. Biochem J 336: 531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argout X, Fouet O, Wincker P, Gramacho K, Legavre T, Sabau X, Risterucci AM, Da Silva C, Cascardo J, Allegre M, et al. (2008) Towards the understanding of the cocoa transcriptome: production and analysis of an exhaustive dataset of ESTs of Theobroma cacao L. generated from various tissues and under various conditions. BMC Genomics 9: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM. (1997) The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ. (2006a) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45: 982–993 [DOI] [PubMed] [Google Scholar]
- Auldridge ME, McCarty DR, Klee HJ. (2006b) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9: 315–321 [DOI] [PubMed] [Google Scholar]
- Banilas G, Karampelias M, Makariti I, Kourti A, Hatzopoulos P. (2011) The olive DGAT2 gene is developmentally regulated and shares overlapping but distinct expression patterns with DGAT1. J Exp Bot 62: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ. (2006) Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc Natl Acad Sci USA 103: 7923–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D. (2000) The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M. (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Lepiniec L. (2009) Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem 47: 448–455 [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L. (2010) Physiological and developmental regulation of seed oil production. Prog Lipid Res 49: 235–249 [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B. (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuillème S, To A, Rochat C, Lepiniec L. (2009) Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J 60: 933–947 [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G. (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Blüthgen N, Brand K, Cajavec B, Swat M, Herzel H, Beule D. (2005) Biological profiling of gene groups utilizing Gene Ontology. Genome Inform 16: 106–115 [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Sensi E, Hua C, Davies C, Thomas MR. (2002) Cloning and characterisation of grapevine (Vitis vinifera L.) MADS-box genes expressed during inflorescence and berry development. Plant Sci 162: 887–895 [Google Scholar]
- Boss PK, Vivier M, Matsumoto S, Dry IB, Thomas MR. (2001) A cDNA from grapevine (Vitis vinifera L.), which shows homology to AGAMOUS and SHATTERPROOF, is not only expressed in flowers but also throughout berry development. Plant Mol Biol 45: 541–553 [DOI] [PubMed] [Google Scholar]
- Busi MV, Bustamante C, D’Angelo C, Hidalgo-Cuevas M, Boggio SB, Valle EM, Zabaleta E. (2003) MADS-box genes expressed during tomato seed and fruit development. Plant Mol Biol 52: 801–815 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ. (2009) Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21: 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ. (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Cernac A, Benning C. (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chen Y, Li F, Wurtzel ET. (2010) Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol 153: 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervin C, El-Kereamy A, Roustan JP, Latche A, Lamon J, Bouzayen M. (2004) Ethylene seems required for the berry development and ripening in grape, a non-climacteric fruit. Plant Sci 167: 1301–1305 [Google Scholar]
- Chervin C, Tira-Umphon A, Terrier N, Zouine M, Severac D, Roustan JP. (2008) Stimulation of the grape berry expansion by ethylene and effects on related gene transcripts, over the ripening phase. Physiol Plant 134: 534–546 [DOI] [PubMed] [Google Scholar]
- Chiwocha SD, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross AR, Kermode AR. (2003) A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J 35: 405–417 [DOI] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Cochard B, Adon B, Rekima S, Billotte N, Desmier De Chenon R, Koutou A, Nouy B, Omoré A, Razak P, Glaszmann JC, et al. (2009) Geographic and genetic structure of African oil palm diversity suggests new approaches to breeding. Tree Genet Genomes 5: 493–504 [Google Scholar]
- Coombe BG, Hale CR. (1973) The hormone content of ripening grape berries and the effects of growth substance treatments. Plant Physiol 51: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Boss PK, Robinson SP. (1997) Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol 115: 1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR. (2007) Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res (Web Server issue) 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Riquelme J, Lijavetzky D, Martínez-Zapater JM, Carmona MJ. (2009) Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol 149: 354–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elitzur T, Vrebalov J, Giovannoni JJ, Goldschmidt EE, Friedman H. (2010) The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. J Exp Bot 61: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy I, Sherif S, Mila I, Bouzayen M, Jayasankar S. (2009) Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. J Exp Bot 60: 907–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Focks N, Benning C. (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. (1994) Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol 105: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta GA, Matthews MA, Shaghasi TH, McElrone AJ, Castellarin SD. (2010) Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta 232: 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. (1993) Fruits: a developmental perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez E, Pineda B, Capel J, Antón MT, Atarés A, Pérez-Martín F, García-Sogo B, Angosto T, Moreno V, Lozano R. (2010) Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS ONE 5: e14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF. (2006) Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol Biol Evol 23: 2245–2258 [DOI] [PubMed] [Google Scholar]
- Hirschberg J. (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Howitt CA, Pogson BJ. (2006) Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ 29: 435–445 [DOI] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res (Database issue) 37: D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemefuna J, Adamson I. (1984) Chlorophyll and carotenoid changes in ripening palm fruit, Elaeis guineënsis. Phytochemistry 23: 1413–1415 [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Jaakola L, Poole M, Jones MO, Kämäräinen-Karppinen T, Koskimäki JJ, Hohtola A, Häggman H, Fraser PD, Manning K, King GJ, et al. (2010) A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol 153: 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët T, Laffargue A, Salmona J, Doulbeau S, Descroix F, Bertrand B, de Kochko A, Dussert S. (2009) Metabolic pathways in tropical dicotyledonous albuminous seeds: Coffea arabica as a case study. New Phytol 182: 146–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Josse EM, Simkin AJ, Gaffé J, Labouré AM, Kuntz M, Carol P. (2000) A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol 123: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemvong S, Suvachittanont W. (2005) Molecular cloning and expression of a cDNA encoding 1-deoxy-d-xylulose-5-phosphate synthase from oil palm Elaeis guineensis Jacq. Plant Sci 169: 571–578 [Google Scholar]
- Laffargue A, de Kochko A, Dussert S. (2007) Development of solid-phase extraction and methylation procedures to analyse free fatty acids in lipid-rich seeds. Plant Physiol Biochem 45: 250–257 [DOI] [PubMed] [Google Scholar]
- Li R, Yu K, Hildebrand DF. (2010a) DGAT1, DGAT2 and PDAT expression in seeds and other tissues of epoxy and hydroxy fatty acid accumulating plants. Lipids 45: 145–157 [DOI] [PubMed] [Google Scholar]
- Li Y, Jin K, Zhu Z, Yang J. (2010b) Stepwise origin and functional diversification of the AFL subfamily B3 genes during land plant evolution. J Bioinform Comput Biol (Suppl 1) 8: 33–45 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson M, Arondel V, Bates P, Baud S, Bird D, DeBono A, Durrett T, et al. (2010) Acyl-lipid metabolism. The Arabidopsis Book 8: e0999, doi/10.1199/tab.0999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H. (2010) Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem 48: 9–15 [DOI] [PubMed] [Google Scholar]
- Liu J, Xu B, Hu L, Li M, Su W, Wu J, Yang J, Jin Z. (2009) Involvement of a banana MADS-box transcription factor gene in ethylene-induced fruit ripening. Plant Cell Rep 28: 103–111 [DOI] [PubMed] [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J. (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AB, Van Eck J, Conlin BJ, Paolillo DJ, O’Neill J, Li L. (2008) Effect of the cauliflower Or transgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers. J Exp Bot 59: 213–223 [DOI] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M, Browse J. (2009) An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci USA 106: 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Li L. (2008) Carotenoid metabolism: biosynthesis, regulation, and beyond. J Integr Plant Biol 50: 778–785 [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O’Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al. (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass D, Arango J, Wüst F, Beyer P, Welsch R. (2009) Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS ONE 4: e6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K. (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K. (2009) The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res (Database issue) 37: D89–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo F, Gagneur C, Adam H, Richaud F, Singh R, Cheah SC, Rival A, Duval Y, Tregear JW. (2006) Somaclonal variation in micropropagated oil palm: characterization of two novel genes with enhanced expression in epigenetically abnormal cell lines and in response to auxin. Tree Physiol 26: 585–594 [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ. (2009) Oil palm: future prospects for yield and quality improvements. Lipid Technol 21: 257–260 [Google Scholar]
- Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C. (1999) Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka A, Murachi S, Okunishi H, Shiomi S, Nakano R, Kubo Y, Inaba A. (1998) Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol 118: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakkaew A, Chotigeat W, Eksomtramage T, Phongdara A. (2008) Cloning and expression of a plastid-encoded subunit, beta-carboxyltransferase gene (accD) and a nuclear-encoded subunit, biotin carboxylase of acetyl-CoA carboxylase from oil palm (Elaeis guineensis Jacq.). Plant Sci 175: 497–504 [Google Scholar]
- Ngando Ebongue GF, Dhouib R, Carrière F, Amvam Zollo PH, Arondel V. (2006) Assaying lipase activity from oil palm fruit (Elaeis guineensis Jacq.) mesocarp. Plant Physiol Biochem 44: 611–617 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Othman A, Lazarus C, Fraser T, Stobart K. (2000) Cloning of a palmitoyl-acyl carrier protein thioesterase from oil palm. Biochem Soc Trans 28: 619–622 [PubMed] [Google Scholar]
- Pan IL, McQuinn R, Giovannoni JJ, Irish VF. (2010) Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J Exp Bot 61: 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, et al. (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652 [DOI] [PubMed] [Google Scholar]
- Poupin MJ, Federici F, Medina C, Matus JT, Timmermann T, Arce-Johnson P. (2007) Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene 404: 10–24 [DOI] [PubMed] [Google Scholar]
- Pouvreau B, Baud S, Vernoud V, Morin V, Gendrot G, Py C, Pichon JP, Rouster J, Paul W, Rogowsky PM. (2011) Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramli US, Baker DS, Quant PA, Harwood JL. (2002) Use of control analysis to study the regulation of plant lipid biosynthesis. Biochem Soc Trans 30: 1043–1046 [DOI] [PubMed] [Google Scholar]
- Ramli US, Salas JJ, Quant PA, Harwood JL. (2009) Use of metabolic control analysis to give quantitative information on control of lipid biosynthesis in the important oil crop, Elaeis guineensis (oilpalm). New Phytol 184: 330–339 [DOI] [PubMed] [Google Scholar]
- Rasid OA, Wan Nur Syuhada WS, Nor Hanin A, Masura SS, Zulqarnain M, Ho CL, Sambanthamurthi R, Suhaimi N. (2008) RT-PCR amplification and cloning of partial DNA sequence coding for oil palm (Elaeis oleifera) phytoene synthase gene. Asia Pac J Mol Biol Biotechnol 16: 17–24 [Google Scholar]
- Ronen G, Cohen M, Zamir D, Hirschberg J. (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J 17: 341–351 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB. (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sambanthamurthi R, Sundram K, Tan Y-A. (2000) Chemistry and biochemistry of palm oil. Prog Lipid Res 39: 507–558 [DOI] [PubMed] [Google Scholar]
- Seymour G, Poole M, Manning K, King GJ. (2008) Genetics and epigenetics of fruit development and ripening. Curr Opin Plant Biol 11: 58–63 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K. (2011) A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria x ananassa Duch.) fruit, a non-climacteric tissue. J Exp Bot 62: 1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MK, Kumar R, Solanke AU, Sharma R, Tyagi AK, Sharma AK. (2010) Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol Genet Genomics 284: 455–475 [DOI] [PubMed] [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC. (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY. (1999) Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J 20: 401–412 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomons NW, Orozco M. (2003) Alleviation of vitamin A deficiency with palm fruit and its products. Asia Pac J Clin Nutr 12: 373–384 [PubMed] [Google Scholar]
- Ståhl U, Stålberg K, Stymne S, Ronne H. (2008) A family of eukaryotic lysophospholipid acyltransferases with broad specificity. FEBS Lett 235: 305–309 [DOI] [PubMed] [Google Scholar]
- Tadiello A, Pavanello A, Zanin D, Caporali E, Colombo L, Rotino GL, Trainotti L, Casadoro G. (2009) A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. J Exp Bot 60: 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. (2006) Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn 235: 2449–2459 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54: 733–749 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Symonds RC, Mulholland BJ, Dadswell AR, Blake PS, Burbidge A, Taylor IB. (2000) Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J 23: 363–374 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Tor M, Barry CS, Vrebalov J, Orfila C, Jarvis MC, Giovannoni JJ, Grierson D, Seymour GB. (1999) Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol 120: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Rodríguez-Concepción M. (2010) Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci USA 107: 11626–11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latché A, Pech JC, Bouzayen M. (2003) New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett 550: 149–154 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJ, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang A, Tan D, Takahashi A, Li TZ, Harada T. (2007) MdERFs, two ethylene-response factors involved in apple fruit ripening. J Exp Bot 58: 3743–3748 [DOI] [PubMed] [Google Scholar]
- Wasmuth JD, Blaxter ML. (2004) prot4EST: translating expressed sequence tags from neglected genomes. BMC Bioinformatics 5: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Arango J, Bär C, Salazar B, Al-Babili S, Beltrán J, Chavarriaga P, Ceballos H, Tohme J, Beyer P. (2010) Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 22: 3348–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P. (2007) Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol 145: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. (1995) An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270: 1807–1809 [DOI] [PubMed] [Google Scholar]
- Xie Z, Li X, Glover BJ, Bai S, Rao GY, Luo J, Yang J. (2008) Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol Biol Evol 25: 1581–1592 [DOI] [PubMed] [Google Scholar]
- Yao J, Dong Y, Morris BA. (2001) Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proc Natl Acad Sci USA 98: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB. (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y. (2009) Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J Exp Bot 60: 3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. (2001) InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- Zhang L, Xu Y, Ma R. (2008) Molecular cloning, identification, and chromosomal localization of two MADS box genes in peach (Prunus persica). J Genet Genomics 35: 365–372 [DOI] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB. (2009b) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P. (2009a) The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot 60: 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.