Abstract
Flavonoids represent a class of secondary metabolites with diverse functions in plants including ultraviolet protection, pathogen defense, and interspecies communication. They are also known as modulators of signaling processes in plant and animal systems and therefore are considered to have beneficial effects as nutraceuticals. The rol1-2 (for repressor of lrx1) mutation of Arabidopsis (Arabidopsis thaliana) induces aberrant accumulation of flavonols and a cell-growth phenotype in the shoot. The hyponastic cotyledons, aberrant shape of pavement cells, and deformed trichomes in rol1-2 mutants are suppressed by blocking flavonoid biosynthesis, suggesting that the altered flavonol accumulation in these plants induces the shoot phenotype. Indeed, the identification of several transparent testa, myb, and fls1 (for flavonol synthase1) alleles in a rol1-2 suppressor screen provides genetic evidence that flavonols interfere with shoot development in rol1-2 seedlings. The increased accumulation of auxin in rol1-2 seedlings appears to be caused by a flavonol-induced modification of auxin transport. Quantification of auxin export from mesophyll protoplasts revealed that naphthalene-1-acetic acid but not indole-3-acetic acid transport is affected by the rol1-2 mutation. Inhibition of flavonol biosynthesis in rol1-2 fls1-3 restores naphthalene-1-acetic acid transport to wild-type levels, indicating a very specific mode of action of flavonols on the auxin transport machinery.
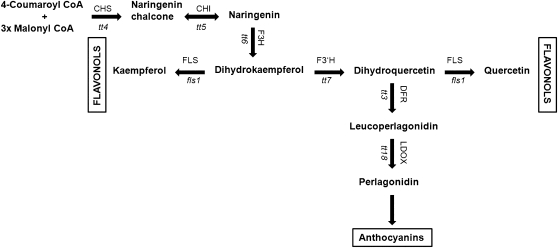
Plants produce a wide variety of secondary metabolites that cover different functions. Flavonoids are synthesized via the phenylpropanoid pathway and serve as UV protectants, in pathogen defense, for plant-microorganism communication, and the regulation of reactive oxygen species (Lepiniec et al., 2006; Buer et al., 2010). A large number of flavonoids are produced by plants and flavonols represent a subgroup of flavonoids. In Arabidopsis (Arabidopsis thaliana), the flavonols quercetin, kaempferol, and isorhamnetin are glycosylated at the C3 and C7 position mainly with Glc and Rha, resulting in a large number of different glycosidic forms (Veit and Pauli, 1999; Lepiniec et al., 2006). The biosynthesis of flavonoids and flavonols has been extensively characterized (Fig. 1) and a number of mutants affected in this pathway have been identified. The Arabidopsis transparent testa (tt) mutants were isolated based on the pale-yellow color of the seed coat due to the absence of proanthocyanidins, a final product of flavonoid biosynthesis. A number of these lines are mutated in genes coding for enzymes active in this pathway (Koornneef, 1990; Shirley et al., 1995; Routaboul et al., 2006). R2R3-MYB transcription factors control the accumulation of flavonols in an additive manner by regulating gene expression of several genes and the Arabidopsis myb11 myb12 myb111 triple mutant is devoid of flavonols (Stracke et al., 2007). Flavonols are primarily synthesized from dihydroflavonols by FLAVONOL SYNTHASE (FLS; Prescott et al., 2002; Martens et al., 2010). In Arabidopsis, of five FLS homologs, only FLS1 shows a strong and FLS3 a very moderate activity (Wisman et al., 1998; Owens et al., 2008; Preuss et al., 2009).
Figure 1.
Sketch of the flavonoid biosynthesis pathway. Bold arrows indicate the different steps leading to flavonoid formation. Names of mutants affected in the biosynthetic process are written in lowercase. CHS, Chalcone synthase; CHI, chalcone isomerase; F3H, flavonol 3-hydroxylase; F3′H, flavonol 3′-hydroxylase; DFR, dihydroflavonol-4-reductase; LDOX, leucoanthocyanidin dioxygenase. Adopted from Routaboul et al. (2006).
A function of flavonols during plant development has been demonstrated in different plant species. Blocking flavonoid biosynthesis in petunia (Petunia hybrida) results in pollen and root-hair growth defects and flavonols are important for the formation of a functional pollen tube in Zea mays (Mo et al., 1992; Taylor and Grotewold, 2005). The flavonoid-less tt4 mutant of Arabidopsis (Fig. 1) shows reduced gravitropic response and is affected in the transport of the phytohormone auxin (Buer and Muday, 2004). This observation might be related to the altered cycling of the auxin efflux facilitator PIN1 in the tt4 mutant background (Peer et al., 2004). Flavonols can compete with the auxin transport inhibitor 1-N-naphthylphthalamic acid for binding to proteins involved in auxin transport (Jacobs and Rubery, 1988; Noh et al., 2001; Murphy et al., 2002) and the kaempferol overaccumulator tt7 is affected in auxin transport (Peer et al., 2004). Hence, several experiments hint at flavonols being involved in modifying auxin transport (Peer and Murphy, 2006). At this point, however, such experiments were not performed on FLS1-less Arabidopsis mutants. One of the problems is that the stable fls1 mutants available so far are not in the Columbia (Col) genetic background. Different Arabidopsis accessions show distinct developmental properties and flavonol accumulation (Routaboul et al., 2006; Juenger et al., 2010), making the combination of several mutations in different genetic backgrounds potentially difficult to interpret (Massonnet et al., 2010).
The Arabidopsis rol1-2 (for repressor of lrx1) mutant shows a strong shoot phenotype in seedlings that seems to be under the influence of flavonols. The rol1-2 mutant is affected in the RHAMNOSE SYNTHASE RHM1. Rha is used for the biosynthesis of pectin and glycosylation of flavonols and rol1-2 mutants are affected in the pectin structure as well as the glycosylation pattern of flavonols (Diet et al., 2006; Ringli et al., 2008). While the amount of rhamnosylated flavonols is reduced in rol1-2, glucosylated flavonols are more abundant. The rol1-2 phenotype is characterized by short roots and root hairs, hyponastic cotyledons with adaxial pavement cells lacking the typical puzzle-like cell shape, and deformed trichomes on the first rosette leaves. The hyponastic cotyledons but not the pavement cell and trichome phenotype seem to be induced by the increased auxin concentration. A rol1-2 tt4 double mutant developed the rol1-2 mutant root phenotype but a wild type-like shoot where all rol1-2 shoot phenotypes were fully suppressed (Ringli et al., 2008). Hence, blocking flavonoid biosynthesis suppresses the rol1-2 shoot phenotype, indicating that flavonols accumulating in the rol1-2 mutant influence shoot development. By contrast, the effect of the rol1-2 mutation on root development seems largely independent of flavonols.
A suppressor screen was performed on the rol1-2 mutant, leading to the isolation of several new tt, myb111, and four fls1 alleles. These lines provide genetic evidence for flavonols being the inducers of the rol1-2 shoot phenotype. Localization of RHM1 and FLS1 reveal adaxial accumulation of these proteins in cotyledons that explains the asymmetric cell growth observed in rol1-2. Ectopic expression of FLS1 demonstrates that flavonols are not distributed within the plant. One function of flavonols in rol1-2 appears to be the influence on auxin transport, as exemplified by protoplast-based auxin transport.
RESULTS
Suppression of rol1-2 by Inhibition of Early Steps of Flavonoid Biosynthesis
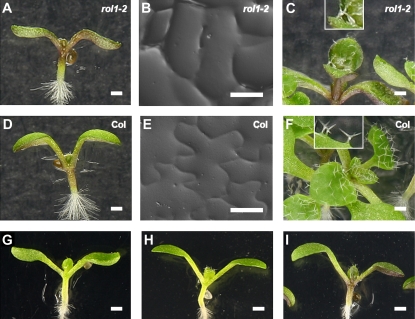
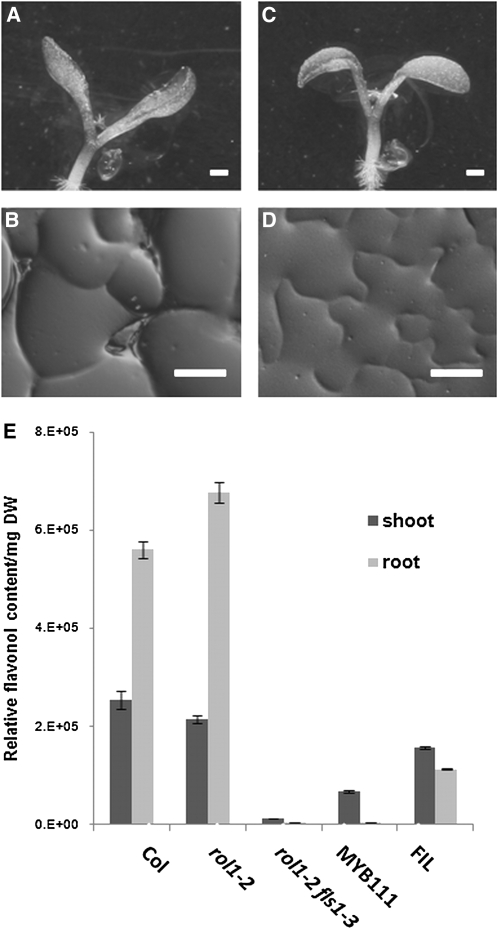
To characterize the role of flavonoids in the establishment of the rol1-2 mutant phenotype, modification of flavonoid biosynthesis in rol1-2 was taken as an approach. To this end, rol1-2 mutant seeds were subjected to mutagenesis with ethyl methanesulfonate (EMS) and propagated to the M2 generation (for details, see “Materials and Methods”). Previous work suggests that blocking the pathway prior to flavonol biosynthesis (Fig. 1) leads to suppression of the rol1-2 shoot phenotype (Fig. 2, A–C), whereas blocking the pathway further downstream does not have this effect (Ringli et al., 2008). As proof of concept, M2 seedlings with a reduced anthocyanin accumulation, based on pale-green hypocotyls, were identified. These could be divided in two classes: (1) no suppression of the rol1-2 shoot phenotype (i.e. hyponastic cotyledons; Fig. 2G) and (2) suppression of the shoot rol1-2 phenotype (i.e. epinastic cotyledons; Fig. 2H).
Figure 2.
Phenotype of rol1-2 compared to wild type and identified classes of suppressor mutants. The rol1-2 mutant shows hyponastic bending of cotyledons (A), adaxial pavement cells with straight cell borders (B), and deformed trichomes (C), whereas the wild type shows epinastic bending of cotyledons (D), a jigsaw puzzle-like structure of adaxial pavement cells (E), and normal-shaped trichomes (F). Three classes of mutants were identified in this study: no anthocyanin accumulation and no suppression (G), no anthocyanin accumulation and suppression (H), and anthocyanin formation and suppression (I). Bars = 1 mm (A, C, D, and F–I) and 40 μm (B and E). [See online article for color version of this figure.]
Mutants falling within the first class were used to sequence genes involved in flavonoid biosynthesis downstream of the branch point of flavonol biosynthesis. Among these mutants, new alleles of tt3, tt7, and tt18 were identified (Fig. 1; Table I). TT3 encodes a dihydroflavonol-4-reductase and TT18 a leucoanthocyanidin dioxygenase. The respective mutants tt3 and tt18 both lack anthocyanins but accumulate the flavonols quercetin and kaempferol. TT7 encodes the flavonol 3′-hydroxylase and the tt7 mutant only accumulates the flavonol kaempferol but not quercetin and no anthocyanins. These data confirm that TT7 and hence the accumulation of quercetin or enzymes acting downstream of TT7 are dispensable for the development of the rol1-2 shoot phenotype.
Table I. Alleles identified, type of mutation, and effect on plant development.
| Arabidopsis Genome Initiative Code | Allele Name | Mutation | Suppression of rol1-2 Shoot Phenotype | Anthocyanin Accumulation |
| At5g08640 | fls1-3 | Q71Stop | Yes | Yes |
| fls1-4 | R156K | Yes | Yes | |
| fls1-5 | P291L | Yes | Yes | |
| fls1-6 | E307K | Yes | Yes | |
| At5g49330 | myb111-2 | W89Stop | Yes | n.d.b |
| myb111-3 | E74K | Yes | n.d. | |
| At5g42800 | tt3-5 | A85T | No | No |
| tt3-6 | G130R | No | No | |
| At5g13930 | tt4-3a | G168Stop | Yes | No |
| tt4-4 | G368E | Yes | No | |
| tt4-5 | R177C | Yes | No | |
| tt4-6 | G205D | Yes | No | |
| tt4-7 | D222N | Yes | No | |
| tt4-8 | Q124Stop | Yes | No | |
| At3g51240 | tt6-6 | R128K | Yes | No |
| At5g07990 | tt7-2a | Intron border | No | No |
| At4g22880 | tt18-5 | Q46Stop | No | No |
| tt18-6 | G248S | No | No |
The effect of tt4 and tt7 alleles confirm previous findings (Ringli et al., 2008).
n.d., Not determined.
Mutants falling within the second class (suppressed rol1-2 phenotype, reduced anthocyanins; Fig. 2H) were expected to be affected in early steps of flavonoid biosynthesis. Indeed, new alleles of tt4 and tt6 were identified among these lines (Fig. 1; Table I).
Mutations in MYB111 and FLS1 Underpin the Importance of Flavonols for the rol1-2 Phenotype
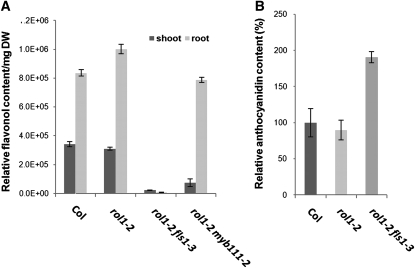
The data obtained so far point toward flavonols being involved in the establishment of the rol1-2 mutant phenotype. As inhibiting flavonol synthesis is not expected to reduce anthocyanin accumulation (Stracke et al., 2009), a third class of M2 seedlings was selected that suppress the rol1-2 shoot phenotype in the presence of anthocyanin (Fig. 2I). Among these lines, two were identified to have a mutation in the R2R3-MYB transcription factor gene MYB111. MYB111 has been shown to regulate flavonol biosynthesis specifically in cotyledons (Stracke et al., 2007). Flavonol levels were determined as the sum of the areas below the flavonol HPLC peaks per milligram dry weight of plant material. rol1-2 myb111 double mutants showed a strong reduction in flavonol levels in the shoot but not the root compared to the wild type or rol1-2 (Fig. 3A). These myb111 alleles represent one missense and one nonsense mutation (Table I). Complementation of the rol1-2 myb111-2 double mutant with a genomic clone of MYB111 resulted in plants displaying the rol1-2 shoot phenotype (data not shown). Hence, the point mutations in myb111 are indeed inducing suppression of the rol1-2 phenotype.
Figure 3.
Analysis of relative flavonol and anthocyanidin content. A, Comparison of shoot and root flavonol contents of wild type, the rol1-2 single mutant line, the rol1-2 fls1-3 suppressor line, and the rol1-2 myb111-2 suppressor line. The sum of the areas under all flavonol peaks was taken as relative measure. B, Relative anthocyanidin content (wild-type Col set to 100%) is unchanged in rol1-2 and doubled in the rol1-2 fls1-3 suppressor line.
It was speculated that mutations in the FLS1 gene, coding for the most active FLS protein of Arabidopsis, should result in a suppressed rol1-2 shoot phenotype but not inhibit anthocyanin accumulation. Due to this, M2 seedlings of the third class were sequenced at the FLS1 locus and, indeed, four fls1 alleles (fls1-3 to fls1-6) were identified. To our knowledge, these represent the first stable fls1 alleles in the Col genetic background in addition to the unstable En-1 fls1-1 allele and the fls1-2 allele that is in the No-0 genetic background (Wisman et al., 1998; Stracke et al., 2009). The FLS1 protein consists of 336 amino acids. fls1-3 introduces a stop at position Q71, fls1-4 to fls1-6 induce the changes R156K, P291L, and E307K, respectively (Table I; Supplemental Fig. S1). The missense alleles showed suppression of rol1-2 comparable to the nonsense allele fls1-3, indicating that the mutated proteins have a strongly reduced enzymatic activity. FLS1 homologs of different plant species (Arabidopsis, petunia, potato [Solanum tuberosum], citrus [Citrus unshiu]) show at least 60% identity and over 70% similarity compared to FLS1 of Arabidopsis (Supplemental Fig. S1). P291, changed to L in fls1-5, is conserved in all FLS1 homologs, while R156, mutated in fls1-4, is conserved except in AtFLS3. By contrast, the position E307, affected in fls1-6, seems more variable. In fact, the petunia FLS protein has a K at this position, as does the fls1-6 allele.
The fls1-3 allele was used for further work. In the rol1-2 fls1-3 seedlings, flavonols in shoots and roots were still detectable but strongly reduced (Fig. 3A) to a level that was insufficient to induce the rol1-2 shoot phenotype. The strong reduction in flavonol biosynthesis resulted in an increase in dihydroflavonols that were available for biosynthetic conversion. This led to a stronger accumulation of anthocyanidin in rol1-2 fls1-3 seedlings (Fig. 3B). Apart from suppressing the rol1-2 phenotype, no obvious morphological defect could be observed in the rol1-2 fls1-3 double or fls1-3 single mutant.
Since reducing the flavonol content had a strong effect on the rol1-2 phenotype, we wanted to know whether increasing the amount of flavonols also has an effect on the rol1-2 or wild-type phenotype. To this end, wild type and the rol1-2 mutant were transformed with the FLS1 genomic clone under the control of the ubiquitously active cauliflower mosaic virus 35S (35S) promoter. 35S:FLS1 transgenic plants of both lines frequently failed to show normal anthocyanin staining and rather appeared pale green, suggesting that the metabolic flux was redirected toward flavonols, leaving little intermediates for the synthesis of anthocyanins. The flavonol content analysis, however, revealed only subtle changes in 35S:FLS1-overexpressing lines compared to nontransformed plants (Supplemental Fig. S2). Also, no morphological alterations were observed in the transgenic lines, i.e. FLS1-overexpressing wild type and rol1-2 mutants showed the phenotype of their respective nontransgenic mother plants.
RHM1 and FLS1 Expression Is Predominant on the Adaxial Side of Cotyledons
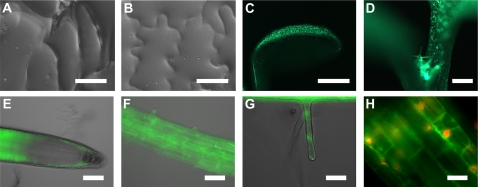
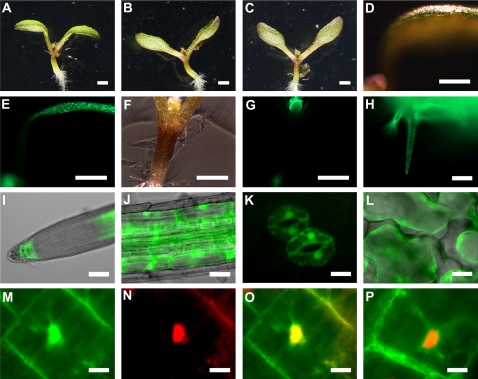
The rol1-2 hyponastic cotyledon phenotype is the result of aberrant cell growth. While adaxial epidermal cells are oversized and lack the typical jigsaw puzzle-like cell shape (Fig. 4A), abaxial epidermal cells show a wild-type phenotype (Fig. 4B). To investigate the cause of this asymmetric growth defect, RHM1 (the protein affected in the rol1-2 mutant) was localized in plants. A RHM1-GFP fusion construct under the control of the RHM1 promoter and terminator sequence was expressed in the rol1-2 mutant. This RHM1:RHM1-GFP construct led to complementation of the rol1-2 mutant, indicating that the fusion protein is biologically active. Analysis of homozygous transgenic T3 seedlings revealed predominant GFP fluorescence on the adaxial side of cotyledons, in the emerging leaves, and in trichomes, whereas no significant signal could be found on the abaxial side of cotyledons (Fig. 4, C and D). Hence, restriction of the growth defect to the adaxial side of cotyledons in the rol1-2 mutant is due to the asymmetric expression of the endogenous RHM1. In the root tip, a GFP signal was found in epidermal cells in the meristematic and elongation zone (Fig. 4E). In the later elongation and differentiation zone, the signal was detected in all cell layers (Fig. 4, E and F). GFP signal was also observed in elongating root hairs (Fig. 4G), which fits the short-root and short root-hair phenotype of the rol1-2 mutant (Diet et al., 2006). As described by others (Wang et al., 2009), RHM1 appears to be a cytoplasmic protein (Fig. 4H).
Figure 4.
The rol1-2 cell shape phenotype and localization of RHM1-GFP. Adaxial epidermal cells of 6-d-old rol1-2 seedlings lack the jigsaw puzzle-like shape (A), whereas the abaxial cells are wild type like (B). rol1-2 mutants transformed with RHM1:RHM1-GFP showed complementation and revealed fluorescence on the adaxial side of cotyledons (C) and in emerging leaves and trichomes (D). In roots, GFP fluorescence was observed in epidermal cells of the meristematic and elongation zone, whereas in the late elongation zone, RHM1-GFP was detected in all cell layers (E). F shows the differentiation zone where the GFP signal occurred in all tissues and in emerging root hairs (G). DAPI staining of DNA and merging with the GFP data indicate a cytoplasmic localization of RHM1-GFP (H). Bars = 1 mm (C and D), 40 μm (A and B), 50 μm (E and F), and 20 μm (G and H).
In a next step, a C-terminal FLS1-GFP and an N-terminal GFP-FLS1 construct was expressed in rol1-2 fls1-3 under the control of the FLS1 promoter and terminator. For each construct, three independent T3 lines were selected for further analysis. Seedlings of all lines exhibiting GFP fluorescence clearly showed a rol1-2 phenotype, i.e. complementation of the fls1-3 mutation, demonstrating that the GFP-FLS1 (Fig. 5, A and B), as well as the FLS1-GFP (not shown) fusion protein is active. In transgenic wild-type plants, GFP fluorescence was mostly found on the adaxial side of cotyledons, in emerging leaves, shoot-root transition zone, and in trichomes (Fig. 5, D–H). Also in roots, GFP-FLS1 localization overlapped well with the one of RHM1-GFP, except that GFP fluorescence was not observed in epidermal cells. In the root tip, fluorescence was observed in inner columella and adjacent root cap cells (Fig. 5, I and J). On the abaxial side of cotyledons, only guard cells showed clear GFP activity (Fig. 5K). On the subcellular level, FLS1 appears to localize to the cytoplasm as the induction of plasmolysis induced retraction of GFP fluorescence with the plasma membrane (Fig. 5L). Interestingly, GFP-FLS1 also accumulated in the nucleus. Staining of FLS1:GFP-FLS1 plants with the nuclear dye 4′,6-diamidino-2-phenylindole (DAPI) resulted in overlapping fluorescence and consequently a yellow coloring of the nucleus (Fig. 5, M–O). To elucidate a functional role of nuclear FLS1, a nuclear export signal (NES; Shen et al., 2007) was fused to the N terminus of GFP-FLS1. The analysis of three independent transgenic FLS1:NES-GFP-FLS1 lines showed the ability of this construct to rescue the fls1-3 mutation in a rol1-2 fls1-3 mutant background (Fig. 5C). Localization of NES-GFP-FLS1 in DAPI-stained Arabidopsis root cells showed effective inhibition of nuclear import of FLS1 and only a cytoplasmic distribution remaining (Fig. 5P). Taken together, these results indicate that nuclear-localized FLS1 is dispensable with respect to the induction of the rol1-2 phenotype.
Figure 5.
Complementation of fls1-3 and localization of GFP-FLS1. The shoot phenotype of rol1-2 is suppressed in rol1-2 fls1-3 (A). The introduction of a GFP-FLS1 construct (B) as well as the introduction of FLS1:NES:GFP:FLS1 (C) in a rol1-2 fls1-3 background led to the induction of the rol1-2 phenotype. In wild-type plants, GFP-FLS1 was detected on the adaxial side of cotyledons (D and E), in emerging leaves, the root-shoot transition zone (F and G), and in trichomes (H). In the root tip (I), GFP signal was restricted to inner columella cells, adjacent inner lateral root cap cells, and late elongation zone. J shows GFP-FLS1 fluorescence in the differentiation zone. On the abaxial side of cotyledons, fluorescence was specifically detected in guard cells (K). L shows plasmolysis of epidermal cells induced by 0.6 m Man treatment. Magnification of a root cortex cell (M), staining of the nucleus with DAPI (N), and merging of both signals (O) indicate also a nuclear localization of FLS1-GFP. In NES-GFP-FLS1 transgenic lines, FLS1 is located in the cytoplasm, whereas nuclear accumulation is prevented as merged GFP and DAPI data (P) show no overlay. Bars = 1 mm (A–G), 50 μm (H and I), 20 μm (J–L), and 5 μm (M–P).
Flavonols Do Not Easily Diffuse between Tissues
Previous reports on flavonol feeding experiments suggested that dihydroflavonols, i.e. dihydrokaempferol and dihydroquercetin, are transported within Arabidopsis seedlings. By contrast, no significant transport of flavonols was observed (Buer et al., 2007). To investigate this issue by a pure in vivo approach, flavonol biosynthesis was induced in rol1-2 fls1-3 double mutants by targeted expression of FLS1-GFP followed by monitoring of flavonol distribution. FLS1-GFP was expressed under the control of the promoters MYB111 (active in cotyledons; Stracke et al., 2007) and FIL (active on the abaxial side of cotyledons; Sawa et al., 1999). For each construct, several transgenic lines were produced and two independent homozygous T3 lines were analyzed in detail. MYB111:FLS1-GFP complemented the fls1-3 mutation, resulting in seedlings exhibiting a rol1-2 phenotype (Fig. 6, A and B). Analysis of the flavonol content in these lines showed an increase in the shoot, which was expected since MYB111 regulates flavonol biosynthesis in cotyledons and thus is likely to be expressed in the same cells as FLS1. By contrast, the flavonol content in roots was not increased, indicating that the flavonols produced in the shoot are not transported to a measurable extent to the root. The FIL promoter, inducing expression on the abaxial side of cotyledons, resulted in the accumulation of flavonols (Fig. 6E), but complementation of the fls1-3 mutation was not observed (Fig. 6, C and D). Surprisingly, the amount of flavonols in the shoot was doubled compared to the MYB111:FLS1-GFP transgenic lines. Hence, flavonols can accumulate in tissues where they are normally not found, but transport from these cells seems not to take place.
Figure 6.
Ectopic expression of FLS1-GFP in a rol1-2 fls1-3 background. A and B show shoot phenotypes of a rol1-2 fls1-3 line transformed with a MYB111:FLS1-GFP construct. C and D, Phenotype of a FIL:FLS1-GFP transgenic rol1-2 fls1-3 line. E, The flavonol content is specifically increased in shoots of MYB111:FLS1-GFP transgenic lines in a rol1-2 fls1-3 background. In the case of FIL:FLS1-GFP transgenic lines, flavonols accumulate in shoots and roots alike. Bars = 1 mm (A and C) and 40 μm (B and D).
Auxin Transport in the rol1-2 Mutant Is Affected by Flavonols
Previous work had revealed that the rol1-2 mutant phenotypes are partly caused by increased auxin concentration in cotyledons and that this effect can be reverted by inhibiting flavonoid biosynthesis (Ringli et al., 2008). Therefore, the effect of flavonols on auxin transport was determined. Because the rol1-2 mutant develops significantly different from the wild type, which might cause aberrant results in whole-seedling auxin transport measurements, a protoplast-based experimental system was chosen. In a first step, expression of the relevant genes had to be assessed in protoplasts. Reverse transcription (RT)-PCR with primers specific for RHM1/ROL1 and FLS1 on RNA isolated from protoplasts indicated that both genes are expressed (Supplemental Fig. S3), which is in agreement with microarray data on protoplasts (Zimmermann et al., 2004).
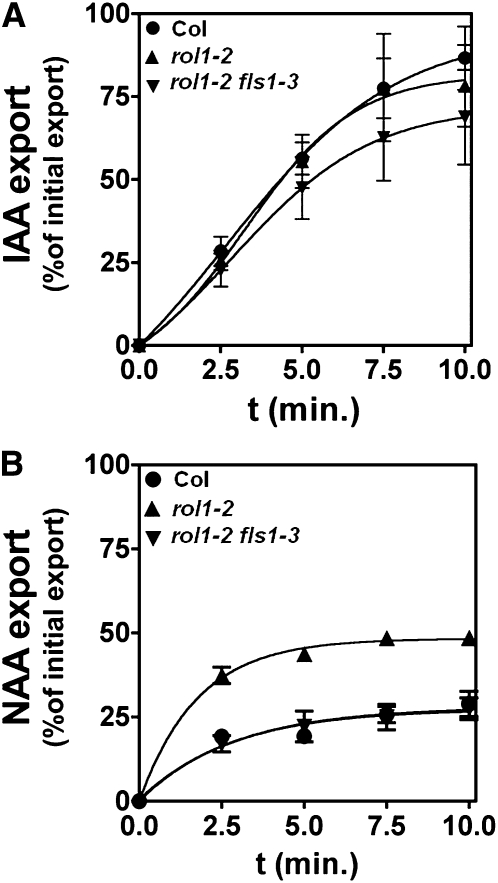
Protoplasts of 6-week-old rosette leaves were isolated, loaded with the radioactively labeled auxins, indole-3-acetic acid (IAA) or naphthalene-1-acetic acid (NAA), and subsequent export of IAA and NAA into the medium was measured. IAA transport was comparable between protoplasts derived from wild-type, rol1-2, and rol1-2 fls1-3 mutant plants (Fig. 7A). By contrast, NAA export was increased in rol1-2 compared to wild-type protoplasts. However, protoplasts of a rol1-2 fls1-3 double mutant showed wild-type NAA export activity (Fig. 7B). Hence, the rol1-2 mutation induces an increase in NAA transport, and inhibiting flavonol accumulation by the fls1-3 mutation relieves this inhibition and leads to wild-type NAA transport levels.
Figure 7.
Auxin export from Arabidopsis mesophyll protoplasts. A, IAA export is not significantly altered in rol1-2 and rol1-2 fls1-3 compared to the Col-0 wild type. B, NAA export is significantly increased in rol1-2 and reduced to wild-type levels in rol1-2 fls1-3.
DISCUSSION
An EMS-mutagenesis screen was performed to genetically characterize the cause of the rol1-2 shoot mutant phenotypes. Mutagenized rol1-2 plants with a suppressed, i.e. wild type-like shoot phenotype and a lack of anthocyanin accumulation were affected in an enzymatic step prior to flavonol synthesis such as TT4 and TT6. Plants lacking anthocyanin but still displaying the rol1-2 shoot phenotype were affected in steps downstream of flavonol accumulation such as TT3, TT7, and TT18. This confirms previous findings on tt4 and tt7 (Ringli et al., 2008). It also shows the opportunities of this screen to distinguish between mutants affected in either the upper or the lower part of the flavonoid biosynthesis pathway, taking flavonol biosynthesis as the line separating the two halves of the pathway.
The identification of fls1 and myb111 mutants confirms that flavonols are the causing agents of the developmental defects observed in the rol1-2 mutant. Interestingly, myb111 and fls1 mutants allow for the accumulation of flavonols to 20% and 5%, respectively, compared to rol1-2. Hence, a complete block of flavonol synthesis as caused by the tt4 mutation (Ringli et al., 2008) is not required for suppression of rol1-2, as no apparent difference in suppression efficiency was detectable between tt4, myb111, and fls1. Similar to tt4, mutations in MYB111 and FLS1 did not have an effect on the rol1-2 root phenotype. Hence, the rol1-2 root phenotype seems induced by the modification in cell wall structures caused by the rol1-2 mutation (Diet et al., 2006).
The residues affected in the fls1 alleles appear to alter protein activity to a similar extent since all suppress the rol1-2 shoot phenotype in a comparable way. FLS proteins belong to the group of 2-oxoglutarate-dependent dioxygenases (Prescott and John, 1996; Martens et al., 2010), whose core domain spans the FLS1 amino acid residues 196 to 296. None of the missense mutations are within this domain. The alignment of FLS proteins from different plant species revealed that R156, mutated to K in fls1-4 is completely conserved between FLS1 and FLS of other plant species, but not in FLS3 of Arabidopsis where this position corresponds to I127. FLS3 has been shown to be the only FLS protein beside FLS1 with measurable, yet low activity (Preuss et al., 2009). Even though FLS3 lacks homology in different regions that are otherwise conserved among 2-oxoglutarate-dependent dioxygenase-type enzymes (Owens et al., 2008) it is possible that the I127 residue also affects FLS3 activity. It is interesting to note that the petunia FLS contains a K residue encoded by the fls1-6 allele. Hence, an amino acid that strongly affects the activity of the Arabidopsis FLS1 is present in a genuine and active FLS protein of petunia (Holton et al., 1993). It is possible that this amino acid functions in relation with other parts of the protein that are different between the petunia FLS and Arabidopsis FLS1.
The amount of anthocyanin in rol1-2 fls1-3 is doubled compared to the wild type, which was shown previously for fls1-2 in the No-0 genetic background (Stracke et al., 2009). This suggests a redirection of the metabolic flux toward the formation of anthocyanins. On the other hand, this opens the possibility for directing the metabolic flux toward flavonols by reducing anthocyanin formation, as these processes compete for the same substrates. Arabidopsis seedlings have the capacity of accumulating more flavonols as shown by the overexpression of MYB12, which encodes a R2R3-MYB transcription factor regulating flavonol biosynthesis genes, leading to a 3- to 4-fold increase in flavonol content (Mehrtens et al., 2005). Here, overexpression of FLS1 was chosen as an alternative approach to manipulate the flavonol content in Arabidopsis. Surprisingly, the transgenic lines neither showed an obvious phenotypic change except loss of anthocyanin accumulation nor an increase in flavonol content. This was quite unexpected in the light of the fact that accumulation of anthocyanins was strongly inhibited, suggesting a redirection of the metabolic flux toward flavonols. A model for metabolome formation and metabolic channelling in flavonoid biosynthesis proposed by Winkel (2004) helps to shed light on this phenomenon. According to this model, FLS1 competes with enzymes, leading to anthocyanin formation that therefore could be blocked by excessive amounts of FLS1. At the same time, FLS1 appears not to be the limiting factor for flavonol biosynthesis.
The analysis of the cell-specific protein localization by protein-GFP fusion constructs provides an explanation for the asymmetric cell-growth phenotype observed in rol1-2 mutants (Diet et al., 2006; Ringli et al., 2008) as expression of RHM1 is limited to the adaxial side of cotyledons. This finding appears to be contradictory to previous studies with RHM1 promoter-GUS fusion constructs (Diet et al., 2006; Wang et al., 2009), which could be explained by the known diffusion of the GUS product. Further, the whole genomic RHM1 sequence was used for GFP localization studies that most likely reflect expression of the endogenous RHM1 more accurately. The expression patterns of RHM1-GFP and GFP-FLS1 are largely overlapping. On the abaxial side, GFP-FLS1 could only be detected in guard cells, where flavonols might have a particular function. Within roots, RHM1-GFP and GFP-FLS1 fluorescence occurs in late elongation and early differentiation zone alike. Hence, RHM1 and FLS1 expression correlate not only on the tissue but also on the cellular level, supporting the finding that RHM1 might be the main supplier of Rha for flavonol glycosylation (Yonekura-Sakakibara et al., 2008).
Grafting experiments with reciprocal combination of tt4 and wild-type shoots and roots revealed that endogenous flavonoids are transported from shoot to root and vice versa (Buer et al., 2008). Flavonoid transport experiments performed by external flavonoid application showed that dihydroflavonols are transported in planta, whereas flavonol aglycones are not (Buer et al., 2007). In this study, targeted in situ biosynthesis of flavonols in the rol1-2 fls1-3 genetic background was chosen as a complete in vivo approach to elucidate a potential transport of flavonols. Expressing FLS1-GFP under the control of the MYB111 promoter was expected to be efficient and indeed resulted in complementation of the rol1-2 fls1-3 double mutant, i.e. development of the rol1-2 shoot phenotypes. Interestingly, driving FLS1-GFP expression with the FIL promoter resulted in even higher levels of flavonols compared to MYB111:FLS1-GFP lines, yet no complementation of the rol1-2 fls1-3 double mutant. Hence, little—if any—flavonols are being transported between the abaxial and adaxial side of cotyledons. The reason for this might be that flavonols remain in the tissue where they are produced. Alternatively, abaxial epidermal cells are not supposed to synthesize flavonols and thus might not be competent to release them to neighboring tissues.
Flavonoids are known to be nonessential regulators of polar auxin transport. They accumulate transiently in epidermal cells of the root elongation zone and act in multifunctional ways on polar auxin transport (Murphy et al., 2000; Brown et al., 2001; Peer et al., 2001, 2004; Buer and Muday, 2004; Peer and Murphy, 2006, 2007). Besides being versatile modulators of the distribution of auxin efflux facilitating PIN proteins (Santelia et al., 2008), recent work points toward flavonols as modulators of transporters of the B-group ATP-binding cassette transporter superfamily (ABCB), another class of auxin efflux transporters (Geisler et al., 2005; Blakeslee et al., 2007). This regulation is thought to be mediated by various processes like protein phosphorylation, protein-protein interaction, vesicular trafficking, and binding to the ATP-binding pocket of ABCBs (Jacobs and Rubery, 1988; Di Pietro et al., 2002; Titapiwatanakun and Murphy, 2009). Previous analyses have shown that auxin distribution is altered in rol1-2, an effect that correlates with flavonoid accumulation and is absent in the flavonoid-less rol1-2 tt4 double mutant (Ringli et al., 2008). Here, we demonstrate that it is most likely the changed flavonol accumulation pattern in rol1-2, leading to a modified rate of auxin transport and thus an altered auxin distribution. The increased auxin transport from rol1-2 protoplasts is reduced to wild-type levels in rol1-2 fls1-3 protoplasts. The polar localization of PIN proteins has been shown to be altered following modifications of the cell wall (Feraru et al., 2011), which reveals a limitation of the cell wall-free protoplast assay system. Yet, the general transport activity is likely to be correctly reflected and the data fits the general picture of flavonol-modified auxin transport. Interestingly, only export of NAA but not of the native auxin IAA is affected. This suggests that flavonols target a specific subset of the auxin transport machinery as NAA export is not caused by diffusion but is an active process (Delbarre et al., 1998). In recent years, ABCBs were identified that seem to be involved in NAA transport (Geisler et al., 2005; Cho et al., 2007; Lewis et al., 2009) and these proteins are thus potential targets of flavonols. Although flavonol aglycones are thought to effectively interfere with auxin transport, those were not detectable in our extracts. It is possible, however, that flavonol aglycones accumulate only transiently and to levels that are below the detection limit.
Flavonols accumulate in the nucleus and cytoplasm as are the flavonoid biosynthetic enzymes CHS and CHI (Saslowsky et al., 2005; Fig. 1). GFP studies presented in this work show FLS1 being a cytoplasmic and nuclear protein alike. This suggests that the flavonol biosynthetic machinery is also active in the nucleus where flavonols might modulate gene expression (Naoumkina and Dixon, 2008; Gilbert and Liu, 2010). However, excluding FLS1 from the nucleus did not prevent complementation of fls1-3, suggesting that nuclear FLS1 activity is not relevant with respect to the rol1-2 phenotype. In future experiments, it will be necessary to investigate the exact role of FLS1 in the nucleus.
MATERIALS AND METHODS
Plant Material and EMS Mutagenesis
The rol1-2 allele used in this study is described by Diet et al. (2006). All lines described in this study are in the Col-0 genetic background. Seeds were surface sterilized with 1% sodium hypochlorite, 0.03% Triton X-100, stratified for 4 d at 4°C, and plated on half-strength Murashige and Skoog medium containing 0.6% phytagel, 2% Suc. Plates were put in a vertical orientation in a 16-h-light, 8-h-dark cycle at 22°C. For propagation and crossings, 10-d-old plants were transferred to soil and grown in a 16-h-light, 8-h-dark cycle at 22°C and irradiated with 100 μmol m−2 s−1 white light (Biolux, Osram).
EMS-mutagenized rol1-2 seeds were germinated in 10 families of 250 M1 plants each and M2 seeds of each family were pooled. In total, 75,000 M2 seedlings were grown on agar plates in a vertical orientation and screened for their phenotype. Selected mutants (all recessive) were backcrossed with the nonmutagenized rol1-2 at least three times prior to detailed characterization.
DNA Constructs, Plant Transformation, and Molecular Markers
For the FLS1 complementation construct, the FLS1 genomic clone, including the 1.7-kb promoter and the 350-bp terminator sequence, was PCR amplified with the primers 5′-AATTTCTACTGAATTCGACAGAG-3′ and 5′-TAATAGCGAATGTGTCGGTTTG-3′. The resulting fragment (FLS1:FLS1) was cloned into pGEM-T easy (Promega) for sequencing.
For the GFP fusion constructs, a BamHI was introduced into FLS1:FLS1 clone by PCR either 3′ of the ATG (N-terminal fusion) or 5′ of the TGA (C-terminal fusion). A previously produced GFP construct flanked by BamHI sites (Leiber et al., 2010) was cloned into these BamHI sites, resulting in FLS1:GFP-FLS1 and FLS1:FLS1-GFP, respectively. These constructs were cloned into pART27 by NotI (Gleave, 1992) and plants were transformed as described (Diet et al., 2006).
For the construction of FLS1:NES-GFP-FLS1, the NES sequence (AACGAGCTTGCTCTTAAGTTGGCTGGACTTGATATTAACAAG) was introduced at the 5′ end of GFP via PCR. For plant transformation, the construct was cloned into pART27 by NotI.
For the RHM1-GFP fusion construct, the genomic clone of RHM1 encompassing 1.9-kb promoter and 500-bp terminator sequence was amplified by PCR using the primers 5′-CACTAAAGATAGAGCATTGAGAAG-3′ and 5′-GTTGGTATCGAATCCTTTGAGTTC-3′ and cloned into pGEM-T easy for sequencing. The GFP clone (Leiber et al., 2010) was introduced 5′ adjacent to the stop codon. For plant transformation, the RHM1:RHM1-GFP construct was cloned into pART27 by NotI.
The molecular marker for rol1-2 was described previously (Diet et al., 2006). All alleles were identified by sequencing of candidate genes and comparison with the wild-type DNA. Cleaved-amplified polymorphic sequence markers were established for some of the alleles: fls1-3, 5′-GATCTAAGCGATCCCGACGAAG-3′ and 5′-CAGAATCTGTAATTGACGCATGAC-3′, digested with DdeI; myb111-2, 5′-GTCTCATGTGTGTTTTGTGTAC-3′ and 5′-CTCCAATGTTATCTCTCCAATATC-3′, digested with BclI; myb111-3, point mutations had to be introduced in one primer (underlined positions) to create a restriction site polymorphism; 5′-AAAGAGGAAATATTACTTCGTAC-3′ and 5′-CAGCATTAACAGTCACTATTCAC-3′, digested with BsiWI.
RT-PCR
Total RNA was extracted from protoplasts or from seedlings grown for 10 d in a vertical orientation using the TRIzol method (Gibco BRL). Two micrograms of each RNA sample were reverse transcribed using oligo(dT) primer and a Superscript II RNase H reverse transcriptase kit (Invitrogen) following the manufacturer’s recommendations. The resulting cDNA was used to perform a RT-PCR with the primers 5′-GATTCGAAAGACATTGAAGGATACG-3′ and 5′-CTCCGATAGCTTCTTCACATGCAC-3′, or 5′-CTCGGAA-TTTTCCAATTGTCTTC-3′ and 5′-CTTGAAGTTCGGAGAGTGCTTAG-3′ specific for FLS1 and ROL1/RHM1, respectively. Both primer pairs are flanking intronic sequence to distinguish in the RT-PCR experiment between cDNA and contamination with genomic DNA.
Microscopic Analysis
Light microscopic observations were made using a Leica DM6000 stereomicroscope. Gel prints of epidermal cells were produced following an established protocol (Horiguchi et al., 2006) and observed by differential interference contrast microscopy using a Leica DMR microscope.
Flavonol and Anthocyanidin Content Analysis
For the analysis of the flavonol accumulation profile, seedlings were grown in a vertical orientation for 6 d as described. One hundred intact seedlings were cut in the hypocotyl region, and roots and shoots were pooled separately, frozen in liquid nitrogen, and lyophilized to determine the dry weight. The dried material was incubated in 500 μL of 80% methanol overnight at 4°C and subsequently macerated with a pestle, followed by vigorous vortexing. After pelleting the cell debris by centrifugation, the supernatant was transferred to a fresh tube and evaporated in a Speed-Vac centrifuge, with the temperature being limited to a maximum of 42°C. After evaporation, the pellet was resuspended in 100 μL of fresh 80% methanol and used for analysis. HPLC-electrospray ionization-mass spectrometry (MS) and tandem MS experiments were performed on a Acquity UPLC (Waters) connected to a Bruker maXis high-resolution quadrupole time-of-flight mass spectrometer (Bruker Daltonics). An Acquity BEH C18 HPLC column (1.7 μm, 2.1 × 100 mm fitted with a 2- × 2-mm guard column) was used with a gradient of solvent A (water, 0.1% [v/v] HCOOH) and solvent B (CH3CN, 0.1% [v/v] HCOOH; 0.45 mL flow rate, linear gradient from 5% to 95% B within 30 min).
The mass spectrometer was operated in the negative electrospray ionization mode at 3,500 V capillary voltage, −500 V endplate offset, with a N2 nebulizer pressure of 1.3 bar and dry gas flow of 8.0 L/min at 200°C. MS acquisitions were performed in the full scan mode in the mass range from mass-to-charge ratio (m/z) 50 to 2,000 at 25,000 resolution (full width at half maximum) and 2 scans per second. The MS instrument was optimized for maximum signal intensities of quercitrine at m/z 447. Masses were calibrated with a 2 mm solution of sodium formate over m/z 180 up to 1,472 mass range prior to analysis. The lock mass signal of hexakis (1H,1H,2H-perfluoroethoxy)phosphazine at m/z 556.00195 was further used as lock mass during the HPLC run. The area under each flavonol peak was used for relative quantification. The sum of all peak areas represented the relative total amount of flavonols, which was divided by the amount of plant material used for extraction. These values were compared between different plant lines.
Spectrophotometric determination of anthocyanidin levels was performed according to Solfanelli et al. (2006) except some minor changes. Material of 10-d-old seedlings was lyophilized overnight. Three independent biological replicates were used for every genotype. Per milligram dry weight, 200 μL extraction solution containing 18% 2-propanol and 1% HCl was added and the samples incubated 10 min at 90°C. After centrifugation for 5 min, 13,000 rpm, the extracts were recovered and absorption determined at 535 nm.
Auxin Transport Experiments
Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts were prepared from rosette leaves of plants grown on soil under 100 μmol m−2 s−1 white light, 8-h-light, 16-h-dark cycle at 22°C. Intact protoplasts were isolated as described (Geisler et al., 2003) and loaded by incubation with 1 μL mL−1 3H-IAA (specific activity 20 Ci mm−1; American Radiolabeled Chemicals), or 4-31H-naphthalene acetic acid (25 Ci mm−1; American Radiolabeled Chemicals) on ice. External radioactivity was removed by separating protoplasts using a 50%-30%-5% percoll gradient. Samples were incubated at 25°C and efflux halted by silicon oil centrifugation. Retained and effluxed radioactivity was determined by scintillation counting of protoplast pellets and aqueous phases. Efflux experiments were performed with three to five independent protoplast preparations with four replicas for each time point.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RHM1, At1g78570; for all other genes, see Table I.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of FLS protein sequences of different plant species.
Supplemental Figure S2. RT-PCR data, anthocyanidin content, and flavonol content of 35S:FLS1 transgenic lines.
Supplemental Figure S3. Expression of RHM1/ROL1 and FLS1 in protoplasts.
Supplementary Material
Acknowledgments
We thank Anja Meury and Vincent Vincenzetti for valuable technical assistance and Bruno Müller for providing plant material for protoplast preparations.
References
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK. (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA. (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK. (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK, Djordjevic MA. (2007) Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol 145: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK, Djordjevic MA. (2008) Implications of long-distance flavonoid movement in Arabidopsis thaliana. Plant Signal Behav 3: 415–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M, Lee SH, Cho HT. (2007) P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Guern J. (1998) Short-lived and phosphorylated proteins contribute to carrier-mediated efflux, but not to influx, of auxin in suspension-cultured tobacco cells. Plant Physiol 116: 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A, Conseil G, Pérez-Victoria JM, Dayan G, Baubichon-Cortay H, Trompier D, Steinfels E, Jault JM, de Wet H, Maitrejean M, et al. (2002) Modulation by flavonoids of cell multidrug resistance mediated by P-glycoprotein and related ABC transporters. Cell Mol Life Sci 59: 307–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet A, Link B, Seifert GJ, Schellenberg B, Wagner U, Pauly M, Reiter WD, Ringli C. (2006) The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18: 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E, Feraru MI, Kleine-Vehn J, Martinière A, Mouille G, Vanneste S, Vernhettes S, Runions J, Friml J. (2011) PIN polarity maintenance by the cell wall in Arabidopsis. Curr Biol 21: 338–343 [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, Frangne N, Koncz-Kalman Z, Koncz C, Dudler R, et al. (2003) TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol Biol Cell 14: 4238–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert ER, Liu D. (2010) Flavonoids influence epigenetic-modifying enzyme activity: structure-function relationships and the therapeutic potential for cancer. Curr Med Chem 17: 1756–1768 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Holton TA, Brugliera F, Tanaka Y. (1993) Cloning and expression of flavonol synthase from Petunia hybrida. Plant J 4: 1003–1010 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H. (2006) Large-scale histological analysis of leaf mutants using two simple leaf observation methods: identification of novel genetic pathways governing the size and shape of leaves. Plant J 48: 638–644 [DOI] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH. (1988) Naturally occurring auxin transport regulators. Science 241: 346–349 [DOI] [PubMed] [Google Scholar]
- Juenger TE, Sen S, Bray E, Stahl E, Wayne T, McKay J, Richards JH. (2010) Exploring genetic and expression differences between physiologically extreme ecotypes: comparative genomic hybridization and gene expression studies of Kas-1 and Tsu-1 accessions of Arabidopsis thaliana. Plant Cell Environ 33: 1268–1284 [DOI] [PubMed] [Google Scholar]
- Koornneef M. (1990) Mutations affecting the testa color in Arabidopsis. Arabidopsis Inf Serv 19: 113–115 [Google Scholar]
- Leiber RM, John F, Verhertbruggen Y, Diet A, Knox JP, Ringli C. (2010) The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 22: 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Wu GS, Ljung K, Spalding EP. (2009) Auxin transport into cotyledons and cotyledon growth depend similarly on the ABCB19 Multidrug Resistance-like transporter. Plant J 60: 91–101 [DOI] [PubMed] [Google Scholar]
- Martens S, Preuss A, Matern U. (2010) Multifunctional flavonoid dioxygenases: flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L. Phytochemistry 71: 1040–1049 [DOI] [PubMed] [Google Scholar]
- Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GTS, Meyer RC, Messerli G, Gronlund JT, et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152: 2142–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo YY, Nagel C, Taylor LP. (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A, Peer WA, Taiz L. (2000) Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211: 315–324 [DOI] [PubMed] [Google Scholar]
- Murphy AS, Hoogner KR, Peer WA, Taiz L. (2002) Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol 128: 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M, Dixon RA. (2008) Subcellular localization of flavonoid natural products: a signalling function? Plant Signal Behav 3: 573–575 [Google Scholar]
- Noh B, Murphy AS, Spalding EP. (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BSJ. (2008) Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol 147: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS. (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS. (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126: 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS. (2006) Flavonoids as signal molecules. Grotewold E, , The Science of Flavonoids. Springer-Verlag, Berlin, pp 239–268 [Google Scholar]
- Peer WA, Murphy AS. (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Prescott AG, John P. (1996) Dioxygenases: molecular structure and role in plant metabolism. Annu Rev Plant Physiol Plant Mol Biol 47: 245–271 [DOI] [PubMed] [Google Scholar]
- Prescott AG, Stamford NPJ, Wheeler G, Firmin JL. (2002) In vitro properties of a recombinant flavonol synthase from Arabidopsis thaliana. Phytochemistry 60: 589–593 [DOI] [PubMed] [Google Scholar]
- Preuss A, Stracke R, Weisshaar B, Hillebrecht A, Matern U, Martens S. (2009) Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Lett 583: 1981–1986 [DOI] [PubMed] [Google Scholar]
- Ringli C, Bigler L, Kuhn BM, Leiber RM, Diet A, Santelia D, Frey B, Pollmann S, Klein M. (2008) The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. Plant Cell 20: 1470–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L. (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224: 96–107 [DOI] [PubMed] [Google Scholar]
- Santelia D, Henrichs S, Vincenzetti V, Sauer M, Bigler L, Klein M, Bailly A, Lee Y, Friml J, Geisler M, et al. (2008) Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J Biol Chem 283: 31218–31226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky DE, Warek U, Winkel BSJ. (2005) Nuclear localization of flavonoid enzymes in Arabidopsis. J Biol Chem 280: 23735–23740 [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K. (1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, De Vos RCH, Bartelniewoehner L, Ishihara H, Sagasser M, Martens S, Weisshaar B. (2009) Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229: 427–445 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8: 317–323 [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B, Murphy AS. (2009) Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot 60: 1093–1107 [DOI] [PubMed] [Google Scholar]
- Veit M, Pauli GF. (1999) Major flavonoids from Arabidopsis thaliana leaves. J Nat Prod 62: 1301–1303 [DOI] [PubMed] [Google Scholar]
- Wang JF, Ji QM, Jiang L, Shen SD, Fan YL, Zhang CY. (2009) Overexpression of a cytosol-localized rhamnose biosynthesis protein encoded by Arabidopsis RHM1 gene increases rhamnose content in cell wall. Plant Physiol Biochem 47: 86–93 [DOI] [PubMed] [Google Scholar]
- Winkel BSJ. (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. (1998) Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA 95: 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.