Abstract
Growth of the aerial part of the plant is dependent upon the maintenance of the shoot apical meristem (SAM). A balance between the self-renewing stem cells in the central zone (CZ) and organogenesis in the peripheral zone (PZ) is essential for the integrity, function, and maintenance of the SAM. Understanding how the SAM maintains a balance between stem cell perpetuation and organogenesis is a central question in plant biology. Two related BELL1-like homeodomain proteins, PENNYWISE (PNY) and POUND-FOOLISH (PNF), act to specify floral meristems during reproductive development. However, genetic studies also show that PNY and PNF regulate the maintenance of the SAM. To understand the role of PNY and PNF in meristem maintenance, the expression patterns for genes that specifically localize to the peripheral and central regions of the SAM were examined in Arabidopsis (Arabidopsis thaliana). Results from these experiments indicate that the integrity of the CZ is impaired in pny pnf plants, which alters the balance of stem cell renewal and organogenesis. As a result, pools of CZ cells may be allocated into initiating leaf primordia. Consistent with these results, the integrity of the central region of pny pnf SAMs can be partially restored by increasing the size of the CZ. Interestingly, flower specification is also reestablished by augmenting the size of the SAM in pny pnf plants. Taken together, we propose that PNY and PNF act to restrict organogenesis to the PZ by maintaining a boundary between the CZ and PZ.
Postembryonic shoot development is dependent upon the shoot apical meristem (SAM), a highly organized group of self-renewing cells, which initiates leaves, axillary meristems, and structures such as internodes (Steeves and Sussex, 1989; Lyndon, 1998). The SAM is subdivided into cytohistological domains including the central zone (CZ), which is located at the apical tip of the SAM and is the site at which stem cells are maintained. Lateral organs are initiated at the peripheral zone (PZ), which surrounds the CZ on its flanks, while the rib meristem (RM) located beneath the CZ produces cells that differentiate into the internal stem tissue (Bernier et al., 1981; Steeves and Sussex, 1989; Lyndon, 1998). The maintenance of the SAM is achieved by a balance of stem cell renewal in the CZ and the allocation of cells into primordia in the PZ (Vollbrecht et al., 2000). To date, little is known about how the SAM regulates the balance of these two interdependent processes in the CZ and PZ.
In Arabidopsis (Arabidopsis thaliana), recent studies show that the integrity of the CZ and RM is maintained by the CLV-WUS negative feedback system (Sablowski, 2007; Tucker and Laux, 2007; Bleckmann and Simon, 2009). The WUSCHEL (WUS) homeobox gene, expressed in a small number of cells in the core of the SAM, acts to maintain stem cell identity in the CZ (Laux et al., 1996; Mayer et al., 1998). The CLAVATA (CLV) genes encode receptors, CLV1 and CLV2, and a secreted ligand, CLV3 (Clark, 2001; Fletcher, 2002). The CLV3 expression domain marks the CZ, while CLV1 is expressed in the core of the meristem (Clark et al., 1997; Fletcher et al., 1999). The CLV pathway functions to down-regulate and restrict the WUS expression domain to the cells in the core of the SAM. At the same time, WUS somehow signals to the apical cells to promote CLV3 expression in the CZ. The negative feedback interaction displayed by CLV3 and WUS acts to maintain a stable population of stem cells (Brand et al., 2000, 2002; Schoof et al., 2000). Mathematical modeling predicts that an additional signaling mechanism(s) is required to maintain stem cells in the CZ and the WUS expression domain in the RM (Jönsson et al., 2005; Geier et al., 2008). Recent studies indicate that stem cells produce active cytokinins (CKs; Kurakawa et al., 2007), which regulate the WUS expression domain through CLV-dependent and independent pathways (Gordon et al., 2009). At the same time, WUS functions to down-regulate the CK negative RESPONSE REGULATOR5 (ARR5), ARR6, ARR7, and ARR15, creating a region of high CK response in the RM (Leibfried et al., 2005). Thus, CK and WUS form a positive feedback loop, which functions to specify the RM and the stem cells during shoot growth (Gordon et al., 2009).
The Arabidopsis KNOTTED1-like HOMEOBOX (KNOX) protein SHOOT MERISTEMLESS (STM) regulates the maintenance of the SAM during shoot development (Long et al., 1996). Phenotypic analysis of weak stm alleles indicates that this homeodomain protein maintains the central region of the SAM (Endrizzi et al., 1996) as well as organ boundaries (Barton and Poethig, 1993; Endrizzi et al., 1996; Kanrar et al., 2006). Experimental studies indicate that STM regulates lateral organ boundaries via the interplay between cytokinin and gibberellin (Jasinski et al., 2005; Yanai et al., 2005). Genetic analyses demonstrate that loss of CLV1 and CLV3 function partially restores shoot development in stm mutants (Clark et al., 1996). At the same time, stm suppresses the enlarged meristems produced in clv1 and clv3 plants. Therefore, results from this study indicate that STM and CLV proteins act in an opposite manner to regulate meristem maintenance and cell proliferation (Clark et al., 1996). In maize (Zea mays), the interplay between knotted1 and thick tassel dwarf1, the orthologs of STM and CLV1, respectively, appear to also act oppositely in regulating SAM homeostasis, but only during reproductive development (Lunde and Hake, 2009). Interestingly, the hypomorphic mutant called gorgon represents a novel stm allele that leads to an increase in the size of the SAM due to the expansion of the central region, which includes the CLV3 and WUS expression domains (Takano et al., 2010). It was postulated that the increase in the size of the central region of the SAM is attributed to a reduction in the allocation of cells into lateral organs and axillary meristems. Taken together, STM acts to control multiple processes that maintain the integrity and function of the SAM.
Genetic and molecular studies indicate that KNOX proteins are regulated in part by the interaction with specific BELL1-like homeodomain (BLH) proteins (Hake et al., 2004). In Arabidopsis, BLH proteins play crucial roles in regulating meristem function and fate during plant development (Hamant and Pautot, 2010). Genetic studies show that the BLH gene called PENNYWISE (PNY; also known as BELLRINGER, REPLUMLESS, VAAMANA, and BLH9) is required for internode patterning as well as flower and fruit development (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bao et al., 2004; Bhatt et al., 2004). Mutations in a gene closely related to PNY called POUND-FOOLISH (PNF) have no visible phenotypes (Smith et al., 2004; Rutjens et al., 2009). However, the pny pnf plants initiate compact shoots that fail to form flowers even though a subset of inflorescence meristem identity genes are expressed in the SAM (Smith et al., 2004; Rutjens et al., 2009). In the SAM, PNY is mainly expressed in the PZ (Smith and Hake, 2003; Cole et al., 2006), while PNF transcripts localize to the CZ and PZ (Smith et al., 2004). Genetic analyses show that PNY and PNF also act to regulate meristem maintenance. For example, the SAM of pny pnf plants frequently terminates during early stages of vegetative growth, which corresponds to a time at which the meristem is small (Smith et al., 2004; Rutjens et al., 2009). After termination of the main shoot, the outgrowth of axillary shoots from the axils of the rosette leaves restores shoot development. Once the development of the axillary shoots is established, the frequency of SAM termination decreases. Histological analyses demonstrate that pny pnf SAMs are smaller and narrower than wild-type SAMs, indicating that the integrity of the CZ and/or PZ is altered. The fact that internode patterning is impaired in pny and pny pnf plants suggests that these homeodomain proteins also regulate RM integrity (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bao et al., 2004; Bhatt et al., 2004). Genetic evidence indicates that the association of PNY and PNF with STM regulates meristem maintenance during shoot growth (Byrne et al., 2003; Bhatt et al., 2004; Kanrar et al., 2006; Rutjens et al., 2009). Currently, the role of PNY and PNF in regulating meristem maintenance is not understood.
In this paper, the role of PNY and PNF in SAM integrity was investigated. Localization of a PZ-expressed gene indicates that the integrity of the meristem’s central region is impaired in pny pnf. Consistent with this result, the localization pattern for genes expressed in the central region of the SAM, including CLV3 and WUS, is altered in pny pnf SAMs. Interestingly, the expression patterns for WUS and CLV3 are reestablished in pny pnf plants lacking CLV function. The reproductive potential is also restored in pny pnf plants, with a reduction in CLV function or an increase in WUS function. We propose a model in which PNY and PNF regulate the integrity of the central region of the SAM by maintaining a boundary between the CZ and PZ, which restricts the allocation of cells into leaf primordial to the PZ.
RESULTS
A Role for PNY and PNF in Regulating the Integrity of the Central Region of the SAM
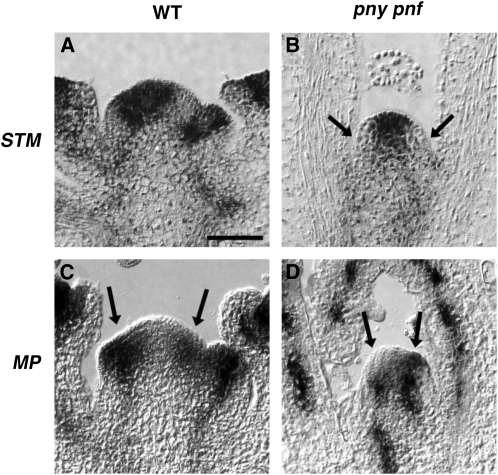
Histological studies indicate that the SAM of pny pnf plants is narrower than that of wild-type plants (Smith et al., 2004). To determine if PNY and PNF regulate the integrity of the SAM, the localization pattern of genes expressed in specific regions of the SAM was analyzed in 30-d-old wild-type and pny pnf SAMs. In wild-type inflorescence meristems, STM was expressed in the PZ and CZ (Fig. 1A; Long et al., 1996). On the flanks of wild-type shoot meristems, STM is down-regulated in a small group of cells that correspond to initial cells, which give rise to lateral organs (Fig. 1A; Long et al., 1996). In pny pnf plants, STM was also expressed in the SAM (Fig. 1B; Smith et al., 2004). In contrast to the mRNA localization pattern in wild-type SAMs, the expression domain for STM was narrower in pny pnf (Fig. 1B; Smith et al., 2004). In addition, the cells in the periphery of pny pnf shoot meristems that fail to express STM appeared to impinge on the central region of the SAM, indicating that a higher proportion of meristem cells are allocated into leaf primordia (Fig. 1B, arrows). The fact that the size of the SAM and the expression domain for STM was narrower in pny pnf compared with the wild type indicates that the integrity of the central and/or peripheral regions of the SAM may be impaired. To address this, we first localized the transcripts for MONOPTEROS (MP), which was expressed in the PZ in wild-type inflorescence meristems (Fig. 1C; Hardtke and Berleth, 1998). MP encodes an auxin response factor, which regulates the positioning and organization of vascular cells required for patterning of the reproductive shoot (Przemeck et al., 1996). In pny pnf plants, cells expressing MP were detected on the flanks of the SAM (Fig. 1D). However, in contrast to the wild type, the central region separating the MP expression domains was reduced (Fig. 1D). These results indicate that the integrity of the central region of the SAM in pny pnf is impaired.
Figure 1.
The integrity of the meristem’s central region is impaired in pny pnf. Localization is shown for STM (A and B) and MP (C and D) transcripts in 30-d-old wild-type (WT) inflorescence (A and C) and pny pnf shoot (B and D) apices. In B, the arrows point to the leaf initial cells. In C and D, the arrows point to the MP-expressing cells. Bar = 50 μm.
PNY and PNF Regulate Gene Expression Patterns in the Core of the SAM
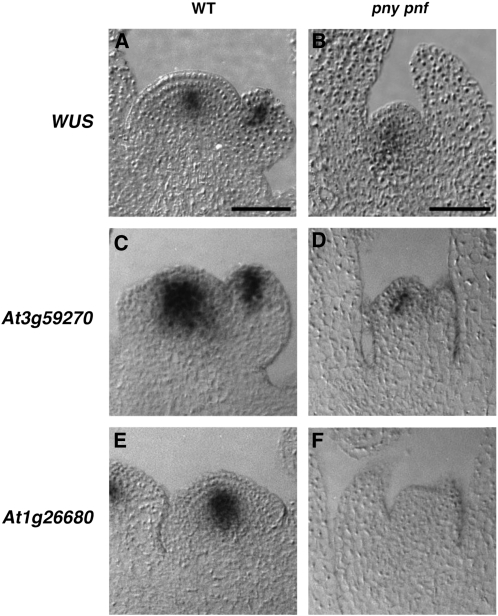
To gain insight into the role of PNY and PNF in regulating the central region of the SAM, the expression pattern for three genes expressed in the core of the SAM was compared between wild-type and pny pnf shoot apices. In wild-type inflorescence meristems, WUS localizes to a small number of cells located in the core of the SAM, overlapping with the apical cells of the RM (Mayer et al., 1998; Fig. 2A). In contrast to the wild type, the WUS expression domain was diffuse and slightly expanded in pny pnf SAMs (Fig. 2B). Slight alterations in the WUS expression domain are observed during the floral transition (Geier et al., 2008). However, the perturbation of the WUS domain in pny pnf SAMs was not likely due to periodic fluctuations, since the diffuse pattern of expression for WUS was displayed in all the pny pnf shoot meristems analyzed (data not shown). To further characterize SAM integrity in pny pnf, two additional genes, At3g59270 and At1g26680, expressed in the core of wild-type inflorescence meristems were examined (Yadav et al., 2009; Fig. 2, C and E). At3g59270 and At1g26680 encode a syntaxin-like protein and a B3 transcription factor, respectively. Unlike the diffuse localization pattern displayed by WUS, the At3g59270 expression pattern was confined to fewer cells in the core of pny pnf SAMs (Fig. 2D). Unlike WUS or At3g59270, transcripts for At1g26680 were not detected in pny pnf SAMs (Fig. 2F) even after a long period of exposure (data not shown). Although the functions of At3g59270 and At1g26680 have not been determined, this study shows that genes expressed in the core of the SAM respond differentially to the loss of PNY and PNF.
Figure 2.
Alteration in the expression patterns for genes transcribed in the core pny pnf shoot meristems. In 30-d-old plants, in situ hybridization was utilized to determine the spatial expression patterns for WUS (A), At3g59270 (C), and At1g26680 (E) in wild-type (WT) inflorescence apices. The mRNA localization patterns for WUS (B), At3g59270 (D), and At1g26680 (F) were examined in 30-d-old pny pnf apices. Bars = 50 μm.
PNY and PNF Regulate the Integrity of the CZ
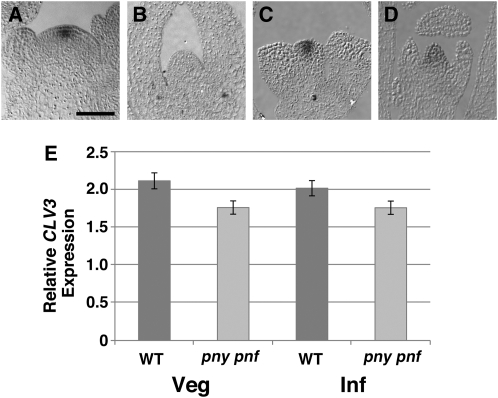
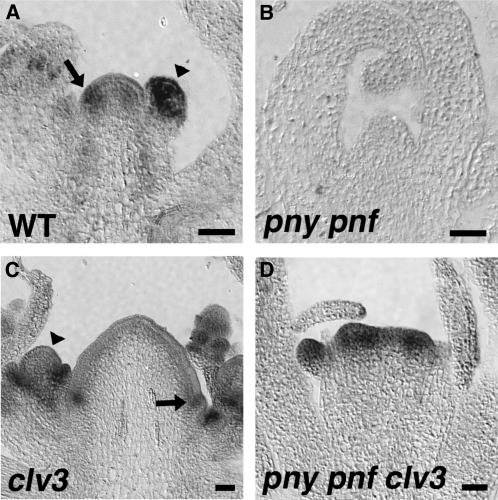
To determine if the integrity of the CZ is altered in pny pnf plants, the localization pattern of CLV3 was determined. In wild-type inflorescence SAMs, CLV3 transcripts localized to the CZ (Fig. 3A; Fletcher et al., 1999). In contrast to the wild type, CLV3 transcript accumulation was not detected by in situ hybridization in pny pnf SAMs (Fig. 3B). The inability to detect CLV3 transcript in the apical cells of pny pnf SAMs was surprising, since the shoot meristems of pny pnf plants are smaller than those of wild-type plants (Smith et al., 2004; Rutjens et al., 2009). A recent study indicates that the stability of GUS activity allows for the spatial detection of low-abundance transcripts that cannot be visualized by mRNA in situ hybridization (Shuai et al., 2002). Therefore, to examine the CLV3 expression pattern further, pCLV3:GUS was crossed into pny pnf. In wild-type SAMs, the pattern of pCLV3:GUS activity was detected in the CZ of the SAM (Brand et al., 2002; Fig. 3C). In contrast to the wild type, pCLV3:GUS activity was detected throughout the SAM in pny pnf plants (Fig. 3D). The broad expression pattern detected for pCLV3:GUS in pny pnf plants was not due to a diffusion of the signal, as the conditions used for this experiment were identical to those used to detect the expression of organ boundary genes (Shuai et al., 2002). To determine if the relative levels of CLV3 were similar in wild-type and pny pnf SAMs, quantitative (q)-PCR was performed. The results showed that transcript levels for CLV3 were comparable between wild-type and pny pnf apices (Fig. 3E). Taken together, we propose that a decrease in the integrity of CZ in pny pnf SAMs alters the expression domain of CLV3. Furthermore, the impaired CZ may explain the aberrant expression patterns for genes expressed in the core of pny pnf SAMs, including WUS.
Figure 3.
The CLV3 expression domain is altered in pny pnf SAMs. A and B, CLV3 transcripts were localized in 30-d-old wild-type inflorescence (A) and pny pnf shoot (B) apices. C and D, GUS activity was visualized in the apices of 30-d-old wild-type (C) and pny pnf (D) plants containing the CLV3:GUS transgene. Bar = 50 μm. E, The relative levels of CLV3 transcripts were determine by q-PCR in the shoot apices of wild-type (WT) and pny pnf plants during vegetative (Veg) and inflorescence (Inf) development. In the q-PCR analysis, UBIQUITIN10 (AT4G05320) was used as a reference.
Restoration of Floral Specification in pny pnf clv3 Plants
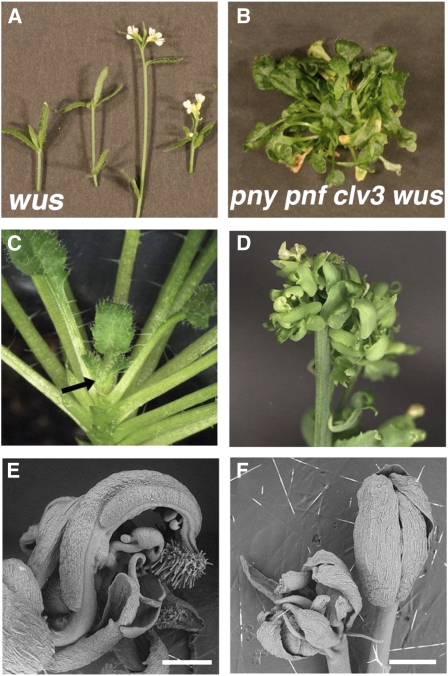
Mutations in clv genes result in a lateral expansion of the CZ and RM during shoot development (Brand et al., 2000; Schoof et al., 2000). Therefore, to determine if the alteration in the integrity of the CZ is attributed to the pny pnf phenotype, clv3-2 was crossed to pny pnf and the pny pnf clv3 plants were examined. In the wild type, pny, and clv3, the inflorescences initiated flowers (Fig. 4, A–C). The inflorescence phenotypes of pny clv3 plants are additive (Byrne et al., 2003), and the loss of these genes had no effect on floral specification (Fig. 4D). In addition, the inflorescence shoots produced by pnf clv3 were morphologically similar to clv3 (data not shown). In pny pnf plants, the compact shoots initiated leaves and displayed a non-flower-producing phenotype when plants were grown in long or short days (Smith et al., 2004; Rutjens et al., 2009; Fig. 4E). Interestingly, under long-day photoinductive conditions, the SAMs of pny pnf clv3 underwent the floral transition, producing compact inflorescence shoots (Fig. 4, F–I) that initiated flower-like structures composed mostly of unfused carpels and some sepals (Fig. 4, J–L). The inflorescence shoots of pny pnf clv3 initiated under short-day growth conditions were similar in morphology to those in long-day conditions (data not shown). Floral specification also occurred in pny pnf clv1; however, the shoots were less compact and petals as well as stamen-like organs were initiated in addition to carpels and sepals (Supplemental Fig. S1; Supplemental Materials and Methods S1). These results demonstrate that loss of CLV function partially restored the floral specification potential of pny pnf shoots.
Figure 4.
Restoration of floral specification during pny pnf clv3 shoot development. A to D, Inflorescence shoots of wild-type (A), pny (B), clv3 (C), and pny clv3 (D) plants. E and F, Images of pny pnf (E) and pny pnf clv3 (F) plants. In F, the arrow points to the compact inflorescence shoot produced in pny pnf clv3 plants. G, Closeup of a pny pnf clv3 inflorescence. H and I, Scanning electron microscopy images of a reproductive pny pnf clv3 apex. In H, bar = 1 mm. In I, the arrow points to the shoot meristem. Bar = 100 μm. J, Scanning electron microscopy image of unfused carpels produced during later stages of reproductive development. The arrow points to one of the unfused carpels. Bar = 200 μm. K, A mature reproductive shoot of pny pnf clv3 was dissected with a razor blade to display the growth patterns. The arrows point to the reproductive shoots that produce the carpel-like organs. L, Closeup view of a carpel-bearing shoot. The arrow points to a carpel-like organ.
LEAFY Is Up-Regulated in the Reproductive Shoots of pny pnf clv3
In Arabidopsis, environmental and endogenous cues converge and activate the key flower meristem identity gene LEAFY (LFY; Blázquez and Weigel, 2000; Yamaguchi et al., 2009). LFY functions downstream of PNY and PNF, as flower specification is restored in pny pnf plants overexpressing LFY (Kanrar et al., 2008). Therefore, to determine if the floral specification phenotype of pny pnf clv3 is attributed to the up-regulation of LFY, the expression pattern of this flower meristem identity gene was examined. In wild-type inflorescence apices, LFY was expressed in a small group of cells in the PZ of the SAM as well as in the floral meristem (Weigel et al., 1992; Fig. 5A). During clv3 inflorescence development, LFY was expressed in the floral meristem and small groups of cells in the peripheral region near the base of the SAM (Fig. 5C). In addition, LFY expression was not detected in the central region of the clv3 inflorescence meristem, which corresponds to the enlarged CZ (Fig. 5C). In contrast to the wild type and clv3, LFY transcripts failed to accumulate in pny pnf shoot meristems under conditions that promote reproductive development (Kanrar et al., 2008; Fig. 5B). In pny pnf clv3, LFY was expressed in the reproductive shoot meristems, demonstrating that the floral specification potential was restored in pny pnf clv3 SAMs (Fig. 5D).
Figure 5.
Expression of LFY in pny pnf clv3 reproductive shoots. The spatial expression pattern for LFY was determined in wild-type (WT; A), pny pnf (B), clv3 (C), and pny pnf (D) apices after 30 d of growth. The inflorescence meristem of clv3 (C) is much larger than the wild-type meristems (A), due to the expanded CZ. In A and C, the arrows point to LFY-expressing cells in the PZ of the shoot meristem. The arrowheads point at the floral meristems. Bars = 50 μm.
The Integrity of the WUS Expression Domain Is Restored in pny pnf clv3 Plants
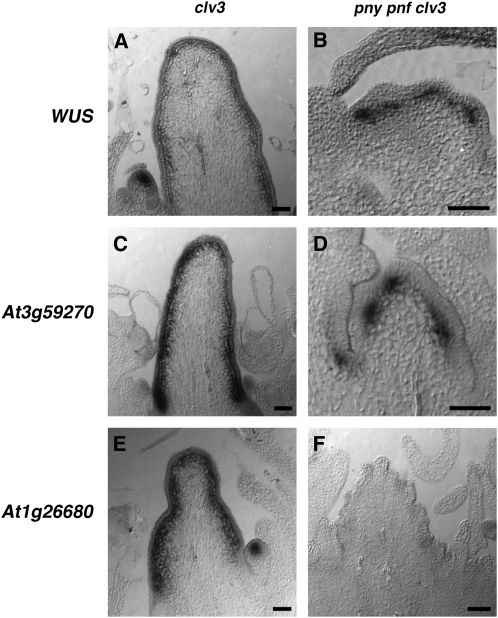
To determine if PNY and PNF control the expression patterns for WUS, At3g59270, and At1g26680 by maintaining the integrity of the CZ, the mRNA localization patterns for these genes were examined in the pny pnf clv3 triple mutant. Mutations in clv3 led to the formation of enlarged shoot meristems in which the WUS domain expanded laterally in the SAM (Fig. 6A; Brand et al., 2000, 2002; Schoof et al., 2000). In clv3 SAMs, the expression domains also expanded for At3g59270 (Fig. 6C; Aggarwal et al., 2010) and At1g26680 (Fig. 6E). Interestingly, in pny pnf clv3, the WUS expression domain appeared to be restored such that WUS transcripts were easily detected in a small group of cells in the core of the SAM (Fig. 6B). In addition, the mRNA localization pattern for At3g59270 expanded in pny pnf clv3 SAMs compared with the expression pattern observed in pny pnf (Fig. 6D). However, unlike WUS and At3g59270, transcripts for At1g26680 could not be detected in pny pnf clv3 SAMs (Fig. 6F) even after a long exposure period (data not shown). Taken together, these results indicate that functional integrity of the central region of the SAM is partially restored in pny pnf clv3 plants. The fact that the WUS expression domain was reestablished in pny pnf clv3 indicates that PNY and PNF regulate the WUS expression domain by maintaining the integrity of the CZ. That the expression pattern for At1g26680 was not restored in pny pnf clv3 SAMs suggests that PNY and PNF control a subset of “core expressed genes” independent of CZ regulation.
Figure 6.
Reestablishment of the WUS and At3g59270 expression domains in pny pnf clv3 shoot meristems. In 30-d-old plants, in situ hybridization was utilized to determine the spatial expression patterns for WUS (A), At3g59270 (C), and At1g26680 (E) in clv3 inflorescence apices. The mRNA localization patterns for WUS (B), At3g59270 (D), and At1g26680 (F) were examined in 30-d-old pny pnf clv3 apices. Bars = 50 μm.
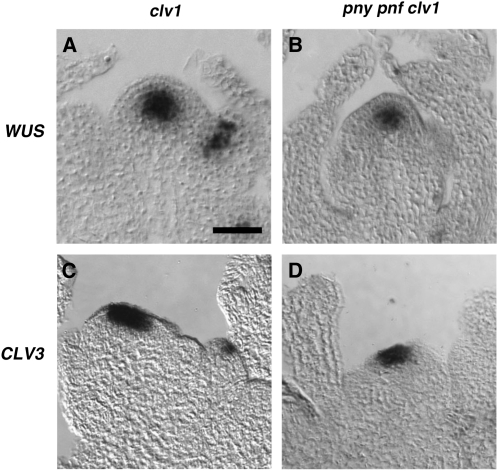
Restoration of the CLV3 Expression Domain in pny pnf clv1
Although the expression domains do not completely coincide, experimental observations indicate that WUS positively regulates CLV3 (Brand et al., 2000; Schoof et al., 2000; Yadav et al., 2010). Since clv1 produces a similar phenotype as clv3 (Clark et al., 1995), we asked if restoration of the WUS expression domain correlated with the accumulation of CLV3 transcripts in the apical cells of pny pnf clv1-8 shoot meristems. In the SAMs of clv1, both the WUS and CLV3 expression domains were expanded compared with the wild type (Fig. 7, A and C). Similar to pny pnf clv3, transcripts for WUS were detected at relatively high levels in the cells of the RM in pny pnf clv1-8 (Fig. 7B). Moreover, the CLV3 expression domain was also reestablished in the apical cells of the SAM in pny pnf clv1-8 (Fig. 7D). Therefore, results from this experiment show that the loss of CLV signaling in pny pnf restores the ability of WUS to positively activate CLV3 in the apical cells of the SAM.
Figure 7.
Restoration of the CLV3 expression domain in pny pnf clv1 plants. The spatial expression pattern for WUS was determined in 30-d-old clv1 (A) and pny pnf clv1 (C) inflorescence shoot apices. An expanded CLV3 expression domain is detected in clv1 (B) and pny pnf clv1 (D) inflorescence apices. Bar = 50 μm.
An Increase in WUS Function Promotes Reproductive Development in pny pnf Plants
The fact that loss of CLV signaling in pny pnf restored floral specification and the WUS expression domain led us to examine whether the levels of this homeobox gene could trigger reproductive development in pny pnf plants. Loss-of-function wus plants produced terminal shoots during vegetative and reproductive development due to the failure to maintain stem cells in the shoot and floral meristems (Fig. 8A; Laux et al., 1996; Mayer et al., 1998). Genetic studies showed that clv3 and clv1 phenotypes are dependent upon WUS function, since wus clv3 and wus clv1 mutants display a phenotype similar to wus single mutants during vegetative and reproductive development (Schoof et al., 2000). To determine if WUS function is required for reproductive development in pny pnf clv3, pny pnf clv3 wus-1 plants were created and analyzed. During the vegetative mode of development, the plants of pny pnf clv3 wus-1 produced terminal shoots that were morphologically similar to wus-1 single mutants (data not shown). However, in contrast to wus-1, inflorescence shoots and flowers were never produced in pny pnf clv3 wus-1 (Fig. 8B). Thus, these results showed that WUS function was required for reproductive development in pny pnf clv3 plants.
Figure 8.
The levels of WUS control floral specification in plants with reduced PNY and PNF function. A, Image of wus inflorescence shoots. B, The shoots of pny pnf clv3 wus fail to initiate flowers. C, The shoot apex of a 35S:WUS-GR pny pnf plant treated with a mock solution without DEX. The arrow points to the leaf-bearing shoot apex. D, Repeated applications of DEX to 35S:WUS-GR pny pnf plants promotes inflorescence development. E, Closeup of a carpel-like floral organ in a 35S:WUS-GR pny pnf inflorescence. F, A single application of DEX to WUS-GR pny pnf induced the formation of terminal flowers. Bars = 0.5 mm.
The fusion of WUS to the glucocorticoid receptor-binding domain (GR) has been successfully used to induce WUS function in order to identify genes regulated by this homeodomain protein in a dexamethasone (DEX)-dependent manner (Brand et al., 2002; Lenhard et al., 2002; Leibfried et al., 2005; Busch et al., 2010). Applications of 1 μm DEX to wild-type inflorescences have mild effects on flower development (Kieffer et al., 2006; data not shown). To determine if an increase in WUS function can promote the floral specification in pny pnf plants, 35S:WUS-GR was crossed to pny pnf to create 35S:WUS-GR pny pnf plants. In the absence of DEX, the shoots of 35S:WUS-GR pny pnf plants treated with a control solution continued to produce leaves without any visible signs of flower formation (Fig. 8C). However, repeated applications of 1 μm DEX to 35S:WUS-GR pny pnf produced inflorescences that terminated with sepal- and carpel-like organs approximately 43% of the time (Fig. 8, D and E). In addition, a terminal flower often formed after 35S:WUS-GR pny pnf plants were treated with a single application of 1 μm DEX (Fig. 8F). These results showed that an increase in WUS function promotes the floral specification in pny pnf plants.
DISCUSSION
The function of the SAM is dependent upon the balance between the self-renewing activities of the stem cells and the allocation of PZ cells into lateral organs and axillary meristems (Vollbrecht et al., 2000). An alteration in the balance between stem-cell perpetuation and organogenesis is predicted to disrupt meristem function. In this paper, we provide evidence that PNY and PNF act to control meristem maintenance by regulating the integrity of the central region of the SAM. First, the expression pattern for MP indicates that the central region of the SAM is reduced in pny pnf plants. Second, the expression patterns for WUS, At3g59270, At1g26680, and CLV3 are altered. Third, the loss of CLV signaling, which increases the size of the central region of the SAM, restores the expression patterns for WUS, At3g59270, and CLV3 in pny pnf plants. Experimental evidence indicates that PNY/PNF-STM complexes act to regulate meristem maintenance (Byrne et al., 2003; Bhatt et al., 2004; Kanrar et al., 2006; Rutjens et al., 2009). Furthermore, genetic analyses indicate that STM also regulates the central region of the SAM (Clark et al., 1996; Endrizzi et al., 1996; Long et al., 1996). Therefore, the association of PNY and PNF with STM may act to regulate the integrity of the CZ during shoot development.
During early stages of vegetative growth, the SAM of pny pnf plants frequently terminates after several leaves are initiated (Smith et al., 2004; Rutjens et al., 2009). Shoot development is reestablished by the outgrowth of axillary shoots, which results in a bushy plant phenotype. The frequency of meristem termination decreases as the shoots grow and mature, presumably due to an increase in meristem size, which may render the SAM to be less susceptible to termination. This hypothesis is supported by studies showing that a null allele of the STM ortholog in maize called knotted1 (kn1) displays a terminal shoot phenotype in inbred lines that typically produce smaller embryonic meristems (Vollbrecht et al., 2000). However, in inbred lines that initiate larger meristems, the SAM is readily maintained in kn1 mutants (Kerstetter et al., 1997; Vollbrecht et al., 2000). How might PNY and PNF regulate the integrity of the central region of the SAM? mRNA in situ hybridization experiments indicate that PNY and PNF expression patterns overlap in the PZ (Smith and Hake, 2003; Smith et al., 2004; Cole et al., 2006). Therefore, these homeodomain proteins may act in the PZ to mediate the allocation of cells into initiating leaf primordia. Based on the expression patterns of STM, leaf initial cells appear to impinge on the central region of pny pnf shoot meristems. Thus, in the absence of PNY and PNF, the balance of organogenesis and stem cell renewal is altered, leading to the allocation of CZ cells into developing leaf primordia. Based on genetic analyses between clv and stm mutants, it was hypothesized that STM functions to maintain a boundary between the CZ and PZ in the SAM (Clark et al., 1996). In support of this hypothesis, we propose that PNY/PNF-STM complexes act to maintain a boundary between the CZ and PZ, which restricts organogenesis to the peripheral region of the SAM. During early stages of vegetative development, the small size of the pny pnf vegetative SAM combined with the failure to maintain the CZ/PZ boundary may result in the allocation of all of the CZ or stem cells into leaf primordia, resulting in the termination of the SAM. However, once shoot growth is established, the SAM is able to maintain itself, because the size of the meristem increases to a point at which a small pool of CZ or stem cells can be readily maintained. In pny pnf clv mutants, the central region of the meristem expands, leading to an increase in the size of the SAM. Expression analyses of WUS, At3g59270, and CLV3 in pny pnf clv3 suggest that the integrity of the SAM’s central region is partially restored, indicating that the CZ/PZ boundary is reestablished.
The CLV3-WUS negative feedback loop plays a pivotal role in maintaining a stable population of stem cells in the CZ (Sablowski, 2007; Tucker and Laux, 2007; Bleckmann and Simon, 2009). To date, the mechanism by which WUS controls the spatial expression pattern of CLV3 is poorly understood. However, the CZ/PZ boundary may provide positional cues that direct the transcription of CLV3 to the CZ. In this study, experimental evidence suggests that the transcript levels for CLV3 are similar between wild-type and pny pnf apices. However, we were unable to detect the CLV3-expressing cells by in situ hybridization in pny pnf SAMs. Furthermore, analysis of the CLV3:GUS expression pattern in pny pnf shoot meristems suggests that the CLV3 expression domain is expanded throughout the SAM. Based on our results, we propose that in the absence of PNY and PNF, the CZ/PZ boundary is not properly maintained. Therefore, the positional cues required for confining the spatial expression pattern of CLV3 to the CZ are absent. As a result, WUS activates CLV3 in all the cells of the meristem. However, since the CLV pathway acts to negatively regulate WUS, transcript levels for CLV3 are expected to be significantly lower in the meristem cells of pny pnf than in the CZ cells of the wild type. Therefore, the cells of pny pnf shoot meristems express CLV3 at a level that cannot be detected by in situ hybridization.
Recent studies show that a mutation in the BLH gene ARABIDOPSIS THALIANA HOMEOBOX1 (ATH1) enhances the terminal shoot phenotype of pny pnf that occurs shortly after germination (Rutjens et al., 2009). ATH1 appears to be structurally and phylogenetically distinct from PNY and PNF (Quaedvlieg et al., 1995; Mukherjee et al., 2009). Genetic studies indicate that ATH1 acts to maintain the basal boundary between the stem and lateral organ (Gómez-Mena and Sablowski, 2008; Rutjens et al., 2009). Similar to PNY and PNF, ATH1 interacts with STM to regulate meristem maintenance events (Hackbusch et al., 2005; Rutjens et al., 2009). However, it is unclear if ATH1 acts redundantly with PNY and PNF. As postulated by Cole et al. (2006), distinct KNOX-BLH complexes may regulate specific meristem maintenance pathways in discrete regions of the SAM. Therefore, the function of STM may be modulated by interaction with specific BLH proteins in the SAM. Consequently, ATH1-STM complexes may control the basal organ boundaries, while PNY/PNF-STM complexes act to restrict organogenesis to the PZ. Thus, the terminal SAM phenotype of pny pnf ath1 plants may be due to combinatorial loss of multiple meristem maintenance pathways controlled by STM.
The fact that pny pnf clv mutants specify floral cell fate is quite intriguing. In pny pnf plants, leaf organogenesis is not perturbed, while the formation of flowers and the development of internodes are severely impaired. Clonal analysis studies indicate that a small number of meristem initial cells give rise to lateral organs (Stewart and Dermen, 1975; Poethig, 1989). During the allocation processes, PNY and PNF may function in the PZ to establish positional cues in the initial cells that give rise to floral meristems and internodes. In the absence of PNY and PNF, the initial cells may not be competent for floral specification and internode patterning cues. In addition, fewer initial cells may be produced due to the smaller size of the pny pnf SAM. As a result, a reduction in the number of initial cells combined with a loss of positional information may have a dramatic effect on floral specification and internode patterning. However, in pny pnf clv shoots, the increase in the size of the SAM may augment the number of cells that are responsive to or competent for the cues that promote floral specification. Future studies are aimed at understanding how PNY and PNF act to establish positional cues in the PZ of the SAM.
MATERIALS AND METHODS
Expression Analyses
Wild-type inflorescence and pny pnf apices from Arabidopsis (Arabidopsis thaliana) were harvested and fixed 30 d after germination under long-day growth conditions. Tissue fixation, processing, and in situ hybridization were performed as described previously (Jackson, 1991). Preparation of WUS, CLV3, At3g59270, and At1g26680 in situ probes was described previously (Yadav et al., 2009). Preparation of STM and MP in situ probe was published previously (Long et al., 1996; Zhao et al., 2010). For each genotype examined by in situ hybridization, at least three biological replicas were fixed and sectioned. Two to three sectioned apices from each genotype were adhered to a single microscope slide. In this analysis, we performed at least one hybridization experiment for each replica. Therefore, each genotype was hybridized with a specific in situ probe six or more times.
For q-PCR, apices from wild-type and pny pnf plants were dissected before and after floral induction. Note that transcripts for the flower meristem identity gene, APETALA1, were only detected in the inflorescence shoot apices of the wild type (data not shown). RNA was isolated using the RNeasy kit from Qiagen. cDNA synthesis and q-PCR procedures were described by Kanrar et al. (2008). The CLV3-F (5′-ATGGATTCGAAGAGTTTTCTG-3′) and CLV3-R (5′-CAAGGGAGCTGAAAGTTGTTTC-3′) primers were used to amplify CLV3. UBIQUITIN10 (AT4G05320) was used as a reference gene (Kanrar et al., 2008). Genotype determination for pny-40126 and pnf-33879 was described previously (Smith and Hake, 2003; Smith et al., 2004). The procedure used to visualize GUS activity in plant tissues was described previously (Sundaresan et al., 1995; Springer, 2000).
Genetic Analyses
The pny-40126 and pnf-33879 alleles used in this study were identified previously in the Columbia ecotype (Smith and Hake, 2003; Smith et al., 2004). clv1-8, which is in the Columbia background, was crossed to pny PNF/pnf in order to characterize the pny pnf clv1-8 phenotype. The clv3-2 and wus-1 alleles, both in the Landsberg ecotype, were backcrossed to wild-type Columbia plants two times before crossing to pnf, pny, pny PNF/pnf, and pnf clv PNY/pnf plants. PNF and CLV3 are located on chromosome 2 approximately 255 kb apart. To generate pny pnf clv3 plants, we first crossed pnf with clv3 and screened F3 for clv3 plants that were heterozygous for pnf. After allowing these plants to self-pollinate, pnf clv3 plants were crossed to pny. The phenotypes for pny pnf clv3, pny pnf clv1, and pny pnf clv3 wus plants were characterized by maintaining pnf clv3 PNY/pnf, pny clv1 PNF/pnf, and pnf clv3 PNY/pny WUS/wus plants. F3 pny clv3 WUS/wus and pny WUS/wus plants were used to characterize pny clv3 wus and pny wus plants. The 35S:WUS-GR pny pnf plants were characterized by maintaining 35S:WUS-GR pny PNF/pnf plants. To examine the spatial patterns of CLV3, wild-type plants containing pCLV3:GUS were crossed to pny PNF/pnf plants. Seeds derived from pny PNF/pnf plants containing pCLV3:GUS were used to analyze the expression pattern for CLV3 in pCLV3:GUS pny pnf plants. CLV3:GUS was created by fusing the CLV3 promoter and 3′ enhancer region to GUS (Brand et al., 2002).
All DEX induction experiments were performed 4 weeks after germination, to allow sufficient time for shoot maturation. In these experiments, 1 μm DEX was applied once or every other day for a 10-d period. The TM-1000 Tabletop Microscope from Hitachi (www.hitachi.com) was used for scanning electron microscopy (Kanrar et al., 2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Flower specification occurs in pny pnf clv1 plants.
Supplemental Materials and Methods S1. Formation of inflorescence-like structures in pny pnf clv1.
Supplementary Material
Acknowledgments
We are grateful to Drs. Patricia Springer, Sarah Hake, Venugopala Reddy, and Alon Samach for critical reading of the manuscript. We thank Jennifer Fletcher for the clv1-8 allele, Rudiger Simon for CLV3:GUS, and Detlef Weigel for the pDW124 plasmid used to synthesize the LFY in situ probe. We also thank Venugopala Reddy for providing us with the wus-1 allele, 35S:WUS-GR seed, and plasmids containing CLV3, WUS, At3g59270, and At1g26680, which were used to synthesize the antisense UTP-digoxigenin-labeled probes. We thank Dr. David Carter for help with scanning electron microscopy.
References
- Aggarwal P, Yadav RK, Reddy GV. (2010) Identification of novel markers for stem-cell niche of Arabidopsis shoot apex. Gene Expr Patterns 10: 259–264 [DOI] [PubMed] [Google Scholar]
- Bao X, Franks RG, Levin JZ, Liu Z. (2004) Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Poethig RS. (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Bernier G, Kinet JM, Sachs RM. (1981) The Physiology of Flowering, Vol 2 CRC Press, Boca Raton, FL [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H. (2004) VAAMANA, a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328: 103–111 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Weigel D. (2000) Integration of floral inductive signals in Arabidopsis. Nature 404: 889–892 [DOI] [PubMed] [Google Scholar]
- Bleckmann A, Simon R. (2009) Interdomain signaling in stem cell maintenance of plant shoot meristems. Mol Cells 27: 615–620 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Brand U, Grünewald M, Hobe M, Simon R. (2002) Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol 129: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. (2010) Transcriptional control of a plant stem cell niche. Dev Cell 18: 849–861 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Groover AT, Fontana JR, Martienssen RA. (2003) Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER Development 130: 3941–3950 [DOI] [PubMed] [Google Scholar]
- Clark SE. (2001) Meristems: start your signaling. Curr Opin Plant Biol 4: 28–32 [DOI] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. (1996) The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067 [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cole M, Nolte C, Werr W. (2006) Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res 34: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. (1996) The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J 10: 967–979 [DOI] [PubMed] [Google Scholar]
- Fletcher JC. (2002) Shoot and floral meristem maintenance in Arabidopsis. Annu Rev Plant Biol 53: 45–66 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Geier F, Lohmann JU, Gerstung M, Maier AT, Timmer J, Fleck C. (2008) A quantitative and dynamic model for plant stem cell regulation. PLoS ONE 3: e3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Mena C, Sablowski R. (2008) ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 20: 2059–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Müller J, Salamini F, Uhrig JF. (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA 102: 4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hamant O, Pautot V. (2010) Plant development: a TALE story. C R Biol 333: 371–381 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, editor (1991) In Situ Hybridisation in Plants. Oxford University Press, Oxford [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M. (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jönsson H, Heisler M, Reddy GV, Agrawal V, Gor V, Shapiro BE, Mjolsness E, Meyerowitz EM. (2005) Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics (Suppl 1) 21: i232–i240 [DOI] [PubMed] [Google Scholar]
- Kanrar S, Bhattacharya M, Arthur B, Courtier J, Smith HM. (2008) Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J 54: 924–937 [DOI] [PubMed] [Google Scholar]
- Kanrar S, Onguka O, Smith HMS. (2006) Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224: 1163–1173 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. (1997) Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B. (2006) Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18: 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G. (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Jürgens G, Laux T. (2002) The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129: 3195–3206 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Lunde C, Hake S. (2009) The interaction of knotted1 and thick tassel dwarf1 in vegetative and reproductive meristems of maize. Genetics 181: 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndon RF. (1998) The Shoot Apical Meristem: Its Growth and Development. Cambridge University Press, Cambridge, UK [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Brocchieri L, Bürglin TR. (2009) A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol 26: 2775–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig S. (1989) Genetic mosaics and cell lineage analysis in plants. Trends Genet 5: 273–277 [DOI] [PubMed] [Google Scholar]
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. (1996) Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200: 229–237 [DOI] [PubMed] [Google Scholar]
- Quaedvlieg N, Dockx J, Rook F, Weisbeek P, Smeekens S. (1995) The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell 7: 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AH, Ferrándiz C, Yanofsky MF. (2003) The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol 13: 1630–1635 [DOI] [PubMed] [Google Scholar]
- Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. (2009) Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J 58: 641–654 [DOI] [PubMed] [Google Scholar]
- Sablowski R. (2007) The dynamic plant stem cell niches. Curr Opin Plant Biol 10: 639–644 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Peña CG, Springer PS. (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM, Campbell BC, Hake S. (2004) Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr Biol 14: 812–817 [DOI] [PubMed] [Google Scholar]
- Smith HM, Hake S. (2003) The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer PS. (2000) Gene traps: tools for plant development and genomics. Plant Cell 12: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. (1989) Patterns in Plant Development, Ed 2 Cambridge University Press, Cambridge, UK [Google Scholar]
- Stewart RN, Dermen H. (1975) Flexibility in ontogeny as shown by the contribution of the shoot apical layers to leaves of periclinal chimeras. Am J Bot 62: 935–947 [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Takano S, Niihama M, Smith HM, Tasaka M, Aida M. (2010) gorgon, a novel missense mutation in the SHOOT MERISTEMLESS gene, impairs shoot meristem homeostasis in Arabidopsis. Plant Cell Physiol 51: 621–634 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Laux T. (2007) Connecting the paths in plant stem cell regulation. Trends Cell Biol 17: 403–410 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S. (2000) Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. (2009) Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Tavakkoli M, Reddy GV. (2010) WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137: 3581–3589 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. (2009) The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N. (2005) Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. (2010) Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.