Abstract
FAR-RED INSENSITIVE219 (FIN219) in Arabidopsis (Arabidopsis thaliana) is involved in phytochrome A-mediated far-red (FR) light signaling. Previous genetic studies revealed that FIN219 acts as an extragenic suppressor of CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1). However, the molecular mechanism underlying the suppression of COP1 remains unknown. Here, we used a transgenic approach to study the regulation of COP1 by FIN219. Transgenic seedlings containing ectopic expression of the FIN219 amino (N)-terminal domain in wild-type Columbia (named NCox for the expression of the N-terminal coiled-coil domain and NTox for the N-terminal 300-amino acid region) exhibited a dominant-negative long-hypocotyl phenotype under FR light, reflected as reduced photomorphogenic responses and altered levels of COP1 and ELONGATED HYPOCOTYL5 (HY5). Yeast two-hybrid, pull-down, and bimolecular fluorescence complementation assays revealed that FIN219 could interact with the WD-40 domain of COP1 and with its N-terminal coiled-coil domain through its carboxyl-terminal domain. Further in vivo coimmunoprecipitation study confirms that FIN219 interacts with COP1 under continuous FR light. Studies of the double mutant fin219-2/cop1-6 indicated that HY5 stability requires FIN219 under darkness and FR light. Moreover, FIN219 levels positively regulated by phytochrome A can modulate the subcellular location of COP1 and are differentially regulated by various fluence rates of FR light. We conclude that the dominant-negative long-hypocotyl phenotype conferred by NCox and NTox in a wild-type background was caused by the misregulation of COP1 binding with the carboxyl terminus of FIN219. Our data provide a critical mechanism controlling the key repressor COP1 in response to FR light.
Light affects almost every aspect of plant development from seed germination to flowering, especially seedling development. In the absence of light, seedlings undergo so-called skotomorphogenesis and show long hypocotyls, unexpanded cotyledons, undeveloped chloroplasts, and blocked expression of light-regulated genes. In the presence of light, they undergo photomorphogenesis and show short hypocotyls, expanded cotyledons, well-developed chloroplasts, and expression of light-responsive genes, which give plants the ability to perceive light energy and signals to survive in environments (Kendrick and Kronenberg, 1994).
Plants contain multiple photoreceptors to sense light environments, including light quality, quantity, duration, and direction. Genetic and molecular studies of Arabidopsis (Arabidopsis thaliana) have identified numerous light-signaling intermediates, including positive and negative regulators (Ni et al., 1998; Soh et al., 1998, 2000; Hoecker et al., 1999; Hudson et al., 1999; Bolle et al., 2000; Fairchild et al., 2000; Fankhauser and Chory, 2000; Hsieh et al., 2000; Ballesteros et al., 2001; Desnos et al., 2001; Dieterle et al., 2001; Zeidler et al., 2001; Wang and Deng, 2002). Some regulators are located in the nucleus (Ni et al., 1998; Hoecker et al., 1999; Hudson et al., 1999; Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000; Ballesteros et al., 2001; Wang and Deng, 2002), others are in the cytoplasm (Bolle et al., 2000; Hsieh et al., 2000) or in both subcellular locations (Desnos et al., 2001; Zeidler et al., 2001). A group of components downstream of light signaling, named COP/DET/FUS as repressors of photomorphogenesis in darkness, play central roles in the integration of far-red (FR), red, and blue light signaling (Kwok et al., 1996). Especially CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), composed of three recognizable structural domains (an N-terminal RING-finger domain, a coiled-coil domain, and a C-terminal WD-40 repeat domain), can partition between the nucleus and cytosol in response to darkness or light (Deng et al., 1992; von Arnim and Deng, 1994). COP1 has E3 ligase activities to target positive regulators, such as ELONGATED HYPOCOTYL5 (HY5), LONG AFTER FAR-RED LIGHT1, phytochrome A (phyA), and LONG HYPOCOTYL IN FAR-RED LIGHT1, for ubiquitin-mediated protein degradation by the 26S proteasome to repress photomorphogenic development (Osterlund et al., 2000b; Saijo et al., 2003; Seo et al., 2003; Duek et al., 2004; Jang et al., 2005; Yang et al., 2005). SUPPRESSOR OF PHYA1 (SPA1) can interact with COP1 to negatively regulate the accumulation of HY5 for repressing light signaling (Saijo et al., 2003). Moreover, SPA1 down-regulates HY5 protein levels via direct interaction without changes in COP1 levels under continuous far-red (cFR) light (Saijo et al., 2003). However, the molecular mechanisms that regulate negative key players such as SPA1 and COP1 in photomorphogenesis are largely unknown.
A molecular genetic approach to isolate genetic modifiers of COP1 revealed two genes, HY5 and FAR-RED INSENSITIVE219 (FIN219), as extragenic suppressors of COP1; these have been characterized at the molecular level (Ang and Deng, 1994; Ang et al., 1998; Hsieh et al., 2000). HY5, a basic Leu zipper transcription factor, functions as a positive regulator downstream of COP1 and is regulated by multiple photoreceptors to control various light responses (Osterlund et al., 2000a), including gravitropism, wavy root growth, and hypocotyl elongation (Oyama et al., 1997), as well as the integration of light signals and various phytohormones to regulate germination and seedling development (Vandenbussche et al., 2007; Alabadí et al., 2008; Chen et al., 2008). HY5 interacting with COP1 and SPA1 leads to its degradation to trigger skotomorphogenic development (Saijo et al., 2003). In contrast, its accumulation at high levels leads to photomorphogenic development. Thus, HY5 also plays a key role in light signaling.
FIN219 was cloned by a map-based method and found to encode a 575-amino acid GH3-like protein; its transcripts are rapidly induced by auxin. The fin219 mutant (renamed fin219-1 in this study) exhibits a long-hypocotyl phenotype only under cFR light and was thought to be an epiallele containing changes of methylation status in the promoter region (Hsieh et al., 2000; H.-L. Hsieh, unpublished data). Transgenic seedlings overexpressing FIN219 display a hypersensitive phenotype under cFR light (Hsieh et al., 2000). Thus, FIN219 might play a role in the phyA-mediated FR signaling pathway. Staswick et al. (2002) reported that JASMONATE RESISTANT1 (JAR1), isolated by positional cloning from the jar1-1 mutant, with the substitution of Phe for a conserved Ser-101 in its open reading frame, is in the same locus as FIN219 and belongs to the firefly luciferase family of adenylate-forming enzymes. Further studies indicated that JAR1 is actually a jasmonic acid (JA)-amino synthetase and mediates the conjugation of JA with various amino acids, especially Ile (Staswick and Tiryaki, 2004). Interestingly, the JA-Ile conjugate can complement the JA insensitivity of jar1-1, which has been shown to have no active JAR1 enzyme (Staswick and Tiryaki, 2004; Suza and Staswick, 2008). Recently, we showed that jar1-1 has a long-hypocotyl phenotype under weak FR light and a phenotype comparable to that of fin219-1 (Chen et al., 2007).
To further understand the FIN219 function in the integration of light and JA signaling pathways and the molecular mechanism underlying the suppression of COP1 by FIN219 in modulating hypocotyl elongation of Arabidopsis seedlings, we characterized transgenic seedlings with ectopic expression of the N-terminal region of FIN219/JAR1 and a putative coiled-coil domain. Our data reveal that FIN219/JAR1 negatively regulates COP1 levels via physical interaction and is required for HY5 stability to modulate photomorphogenic responses of Arabidopsis seedlings.
RESULTS
Transgenic Seedlings in the Wild Type Expressing the FIN219 N Terminus Exhibit a Dominant-Negative Long-Hypocotyl Phenotype Specifically under cFR Light
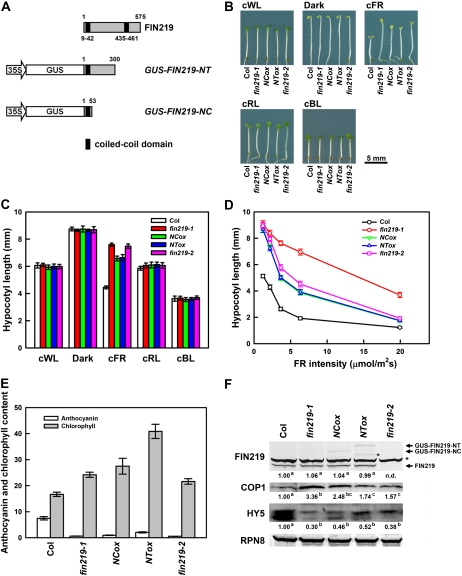
The fin219-1 mutant displays a long-hypocotyl phenotype specifically under cFR light (Hsieh et al., 2000). FIN219 isolated by a map-based method encodes a GH3-like protein containing two putative coiled-coil domains, one in the N terminus and the other in the C terminus (Fig. 1A), which are believed to be involved in protein-protein interaction. To further understand the functional roles of these domains in phyA-mediated FR light signaling, we generated GUS-fused constructs containing the N-terminal 53 amino acid residues (35S::GUS-FIN219-NC) or 300 amino acid residues (35S::GUS-FIN219-NT) of FIN219 that harbor a putative coiled-coil domain at the very end of the N terminus of FIN219 (named NCox for the expression of the N-terminal coiled-coil domain and NTox for the N-terminal 300-amino acid region; Fig. 1A) and ectopically expressed them in wild-type Columbia (Col) and fin219 mutants. NCox and NTox seedlings in Col displayed a longer hypocotyl phenotype than did the wild type specifically under cFR light but not other light conditions, including darkness (Fig. 1, B and C). In addition, we included a null mutant of FIN219 (fin219-2; SALK_059774) obtained from the Arabidopsis Biological Resource Center that has no FIN219 transcripts or proteins (Chen et al., 2007; also see Figs. 1–3). The fin219-2 mutant exhibited a hypocotyl length similar to that of fin219-1 under cFR light, especially weak FR light (Fig. 1, B and C). Further examination under different fluence rates of FR light revealed NCox and NTox transgenic seedlings in Col with hypocotyl lengths similar to that of fin219-1 at 1 to 2 μmol m−2 s−1 low-fluence FR and intermediate lengths between that of fin219-1 and the wild type under 20 μmol m−2 s−1 FR light (Fig. 1D). The fin219-2 mutant displayed hypocotyl lengths similar to those of NCox and NTox transgenic seedlings in Col under the same conditions (Fig. 1D). However, NCox and NTox in a fin219-2 or fin219-1 mutant background did not show hypocotyls longer than those of fin219-2 or fin219-1 (data not shown), which suggests that the dominant-negative phenotype conferred by ectopic expression of the FIN219 N terminus in Col requires the presence of functional FIN219.
Figure 1.
Phenotypic analysis of transgenic seedlings overexpressing the N terminus of FIN219 in Arabidopsis wild-type Col. A, Schematic diagram of the N-terminal domain of FIN219. FIN219 contains two coiled-coil domains shown in black blocks. Numbers above and below the diagram represent numbers of amino acids. B, Transgenic seedlings containing 35S::GUS-FIN219-NT (NTox) or 35S::GUS-FIN219-NC (NCox) exhibit a long-hypocotyl phenotype specifically under cFR light. Seedlings of wild-type Col, fin219-1, NCox, NTox, and fin219-2 were grown in various light conditions for 3 d. cBL, Continuous blue light, 5.08 μmol m−2 s−1; cFR, 1.47 μmol m−2 s−1; cRL, continuous red light, 6.34 μmol m−2 s−1; cWL, continuous white light, 5.88 μmol m−2 s−1. C, Measurement of hypocotyl lengths of the seedlings shown in B (n = 30). Each value represents an average of two independent experiments. Error bars indicate se. D, Quantitative measurement of hypocotyl lengths of the seedlings under various FR fluence rates. Wild-type Col, fin219-1, NCox, NTox, and fin219-2 were grown under various FR fluence rates for 3 d, and the hypocotyl lengths of seedlings were measured (n = 30). E, Anthocyanin accumulation and chlorophyll content in NCox and NTox transgenic seedlings. Seedlings of wild-type Col, fin219-1, NCox, and NTox transgenic lines and fin219-2 were grown on GM plates under cFR for 3 d, and anthocyanin accumulation was measured. The same samples were grown in cFR for 3 d, followed by white light treatment for 1 d, and then chlorophyll content was measured. F, Protein gel-blot analyses of FIN219, COP1, and HY5 levels in various fin219 mutants and FIN219 N-terminal transgenic seedlings shown in B. The arrows indicate the transgene fusion proteins GUS-FIN219-NT and GUS-FIN219-NC and endogenous FIN219. The asterisk indicates artificial bands. RPN8, a subunit of the regulatory complex of the 26S proteasome, was a loading control. The FR fluence rate is 1.47 μmol m−2 s−1. The number below each blot represents the level of the indicated protein. The level of wild-type Col was arbitrarily set to 1. Different lowercase letters represent significant differences by ANOVA at P < 0.05.
Figure 2.
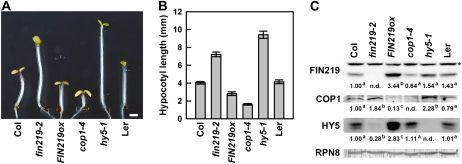
Levels of COP1 are negatively regulated by FIN219. A, Phenotypes of FIN219-overexpressed transgenic seedlings under cFR light. Phenotype examination of 3-d-old wild-type Col, fin219-2, FIN219ox, cop1-4, hy5-1, and Landsberg erecta (Ler) seedlings is shown (n = 30). B, Quantitative analysis of hypocotyl lengths of the seedlings shown in A. Data represent results of at least two replicates (n = 30). C, Protein gel-blot analysis of COP1 and HY5 in FIN219-overexpressed seedlings grown under cFR light. Protein extracts were isolated from the seedlings shown in A and then underwent protein gel-blot analysis with FIN219, COP1, and HY5 polyclonal antibodies. Each lane contained 100 μg of total protein. RPN8 was a loading control. The numbers below each blot represent relative expression ratios normalized to RPN8 expression; the level of wild-type Col was arbitrarily set to 1. Different lowercase letters represent significant differences by ANOVA at P < 0.05. The asterisk indicates artificial bands. The FR fluence rate is 1.47 μmol m−2 s−1. [See online article for color version of this figure.]
Figure 3.
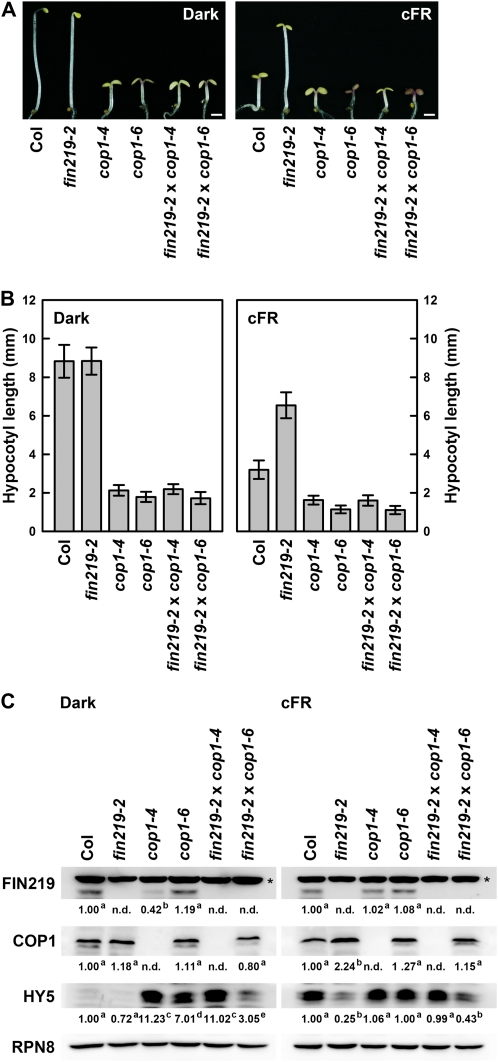
Phenotype and protein gel-blot analyses of the double mutants between fin219 and cop1 alleles under darkness and cFR light. A, Phenotypes of the double mutant fin219-2/cop1 under darkness and cFR conditions. Three-day-old seedlings of wild-type Col, fin219-2, cop1-4, cop1-6, fin219-2/cop1-4, and fin219-2/cop1-6 underwent phenotype examination (n = 30). B, Quantitative analysis of hypocotyl lengths of the seedlings shown in A. Data represent results of at least two replicates (n = 30). C, Protein gel-blot analysis of FIN219, COP1, and HY5 in the double mutant fin219-2/cop1 seedlings grown under darkness and cFR light. Protein extracts were isolated from the seedlings shown in A and used for protein gel-blot analysis. Each lane contained 100 μg of total protein. RPN8 was a loading control. The numbers below each blot represent relative expression ratios normalized to RPN8 expression; the level of wild-type Col was arbitrarily set to 1. Different lowercase letters represent significant differences by ANOVA at P < 0.05. The asterisks indicate artificial bands. The FR fluence rate is 1.47 μmol m−2 s−1. [See online article for color version of this figure.]
Ectopic Expression of the FIN219 N Terminus in the Wild Type Was Reflected by Reduced Photomorphogenic Responses and Changes in Expression of Key Regulators in Light Signaling
To further understand whether the reduced photomorphogenic development resulting from ectopic expression of the FIN219 N terminus in the wild type under cFR light affects phyA-mediated responses, we examined anthocyanin levels of NCox and NTox transgenic seedlings in Col grown under cFR light for 3 d. NCox and NTox transgenic seedlings in Col showed substantially lower anthocyanin levels than did the wild type under the same conditions (Fig. 1E), which is consistent with significantly lower expression of the gene chalcone synthase (CHS) than in the wild type under the same conditions (Supplemental Fig. S1A); moreover, anthocyanin levels in fin219-2 were comparable to those of fin219-1 (Fig. 1E). In addition, NCox and NTox transgenic seedlings in Col displayed a defect in phytochrome A-mediated FR blockage of greening response, but NTox transgenic seedlings in Col were more prominent in this respect (Fig. 1E). Interestingly, fin219-2 showed a degree of defect similar to that of fin219-1 in the FR-mediated blockage of greening (Fig. 1E), which suggests that FIN219 may play a role in the process of FR blockage of greening response. However, how various deletions or mutations of FIN219 result in different effects of greening after FR irradiation remains to be elucidated. In addition, the fin219-1 mutant displayed a slightly delayed flowering phenotype under long-day conditions (Hsieh et al., 2000); we further found that the delayed-flowering phenotype of both NCox and NTox transgenic plants in Col was even enhanced in terms of the number of rosette leaves and days to flowering under long-day conditions (Supplemental Fig. S1B). The fin219-2 mutant showed a trend similar to that of NCox and NTox in Col in the control of flowering (Supplemental Fig. S1B).
Since FIN219/JAR1 has been shown to be a JA-conjugating enzyme (Staswick and Tiryaki, 2004), NCox and NTox transgenic seedlings in Col were examined to see whether they show altered responses to exogenous JA. The response of NCox and NTox in Col to methyl jasmonate (MeJA) or coronatine was similar to that of the wild type (Supplemental Fig. S2). Furthermore, we examined the level of JA-Ile in NCox and NTox transgenic seedlings in Col under FR light by the gas chromatograph/mass spectrometer method. There is no significant difference between wild-type and NCox and NTox transgenic seedlings, but the fin219-2 mutant contains only a small amount of JA-Ile (Table I), which implies that the dominant-negative phenotype of NCox and NTox in Col under FR light is not due to changes in the level of JA-Ile and their responses to MeJA or coronatine. Therefore, the ectopic expression of the FIN219 N terminus in Col results in pleiotropic effects on Arabidopsis development.
Table I. Levels of JA-Ile in Arabidopsis wild-type and fin219 mutant seedlings under FR light.
Values (pmol g−1 fresh weight) shown here are means ± sd of three biological replicates of 3-d-old seedlings under cFR condition (1.47 μmol m−2 s−1). Different lowercase letters represent significant differences by ANOVA at P < 0.05.
| Col | fin219-1 | NCox | NTox | fin219-2 |
| 10.4 ± 2.1 a | 10.7 ± 3.2 a | 11.1 ± 2.3 a | 11.0 ± 3.0 a | 1.3 ± 0.4 b |
Furthermore, to understand the molecular mechanisms underlying the long-hypocotyl phenotype of NCox and NTox in Col under cFR light, we found GUS-FIN219-NC and GUS-FIN219-NT transcript overexpression in NCox and NTox transgenic seedlings in Col, respectively (Supplemental Fig. S1A). The transcripts of two key modulators, the repressor of photomorphogenesis COP1 and the positive regulator of light signaling HY5, in NCox and NTox transgenic seedlings in Col remained largely the same as in the wild type (Supplemental Fig. S1A). We further examined the protein expression in these seedlings grown under the same conditions. GUS-FIN219 fusion proteins were detected by FIN219 polyclonal antibodies raised against the N terminus of FIN219, and their levels were substantially lower than that of their respective transcripts (Fig. 1F; Supplemental Fig. S1A), which implies that the expression of the transgenes may involve posttranscriptional regulation. Endogenous FIN219 level was largely unaffected in the NCox and NTox transgenic seedlings in Col grown under cFR light (Fig. 1F), whereas the FIN219 level was abolished in fin219-2. In addition, COP1 protein levels were increased in NTox and fin219-2 and especially fin219-1 and NCox as compared with the wild type (Fig. 1F). HY5 protein level was substantially reduced in fin219-1, NCox, NTox, and fin219-2 transgenic seedlings. Intriguingly, fin219-1 and fin219-2 showed a significantly reduced level of the lower-band, unphosphorylated form of HY5 (Fig. 1F), shown to be the active form in light signaling (Hardtke et al., 2000).
Because NCox and NTox transgenic seedlings in Col exhibited a hypophotomorphogenic phenotype under cFR light but not under other light conditions (Fig. 1), we wondered whether reduced HY5 protein in NCox and NTox transgenic seedlings in a wild-type background occurred specifically under cFR light. Thus, we determined the level of HY5 protein in seedlings grown in white light (Supplemental Fig. S3A). The result supported the notion that the HY5 level remains constant in fin219-1, NCox, NTox, and fin219-2 as well as the wild type. The levels of COP1 were largely equal among these seedlings (Supplemental Fig. S3A). In contrast, COP1 levels were slightly reduced in all fin219 mutants and NTox transgenic lines but not in NCox under continuous red light (Supplemental Fig. S3B). HY5 levels were slightly increased in all the samples examined, but the lower-band, unphosphorylated form of HY5 was slightly lower in level as compared with the wild type (Supplemental Fig. S3B). Under blue light, the expression of COP1 was slightly reduced as well in all samples, and that of HY5 was largely the same as compared with the wild type (Supplemental Fig. S3C). In the dark, the expression of FIN219 remained the same in all samples examined as compared with the wild type (Supplemental Fig. S3D); however, the COP1 level in fin219-1, NCox, NTox, and fin219-2 remained largely the same as in the wild type, thus leading to barely detectable differences in level of HY5 (Supplemental Fig. S3D). Thus, FIN219 may differentially regulate both COP1 and HY5 under various light conditions, with substantial effect in cFR light.
FIN219 Modulates COP1 Levels in Response to FR Light to Regulate Hypocotyl Elongation
To further confirm the authenticity of the regulation between FIN219 and COP1, we examined COP1 levels in FIN219-overexpressing transgenic seedlings (FIN219ox) under cFR light. Transgenic FIN219ox seedlings exhibited a hypersensitive hypocotyl phenotype as compared with the wild type under cFR light (Fig. 2, A and B). In contrast, fin219-2 and hy5-1 seedlings displayed a hyposensitive long hypocotyl; cop1-4 seedlings with expression of the N-terminal 282 amino acids of COP1 proteins showed a markedly short hypocotyl under the same condition (Fig. 2, A and B; McNellis et al., 1996). Further examination revealed that FIN219 protein was highly expressed in FIN219ox; COP1 level was increased in fin219-2 and substantially reduced in FIN219ox (Fig. 2C). Intriguingly, COP1 level was increased in hy5-1, which is consistent with a previous report of the hy5 mutant isolated as an extragenic suppressor of cop1-6 (Ang and Deng, 1994; Ang et al., 1998). By contrast, HY5 levels were greatly reduced in fin219-2 and increased in FIN219ox (Fig. 2C).
Analysis of the Double Mutant fin219-2/cop1-6 Reveals That HY5 Stability Requires Functional FIN219 in Darkness and cFR Light
To further understand whether COP1 serves as a direct target of FIN219, we generated the double mutant for fin219-2 and cop1-4 or cop1-6, a weak allele of COP1 with a mutation at the splicing junction of its intron 4, which leads to detectable COP1 proteins of wild-type size and abundance (McNellis et al., 1994). Surprisingly, the double mutants fin219-2/cop1-4 and fin219-2/cop1-6 exhibited a short-hypocotyl phenotype similar to respective single mutants cop1-4 and cop1-6 under both darkness and cFR light conditions (Fig. 3, A and B), which is distinct from previous results showing that the original double mutant fin219-1/cop1-6 exhibited a long-hypocotyl phenotype in the dark (Hsieh et al., 2000). Thus, suppression of cop1-6 by the fin219 mutation might require physical interaction of FIN219 and COP1. Further examination revealed the FIN219 protein level reduced in the cop1-4 mutant as compared with Col in the dark but not in cFR light (Fig. 3C), which implies that COP1 in the dark might directly affect FIN219 stability or influence the stability of another factor that regulates FIN219 expression. However, studies of the double mutants fin219-2/cop1-4 and fin219-2/cop1-6 also revealed that the effect of fin219 mutation on COP1 suppression was allele specific, such as with the fin219-2 and cop1-6 combination but not with cop1-4, which is consistent with previous results showing allele-specific genetic interaction between fin219-1 and cop1-6 instead of cop1-5, a null mutant of cop1 (Hsieh et al., 2000). This allele-specific regulation was substantiated with significant reduction of HY5 in fin219-2/cop1-6 but not fin219-2/cop1-4 under darkness and cFR light conditions (Fig. 3C), which suggests that FIN219 can protect HY5 against protein degradation by COP1. Thus, FIN219 may function as a positive regulator of hypocotyl elongation under FR by negatively regulating COP1 level, thus leading to an accordingly increased HY5 level.
FIN219 May Exist as a Dimer via Intermolecular Interactions between Its N-Terminal and C-Terminal Domains to Regulate Photomorphogenic Development
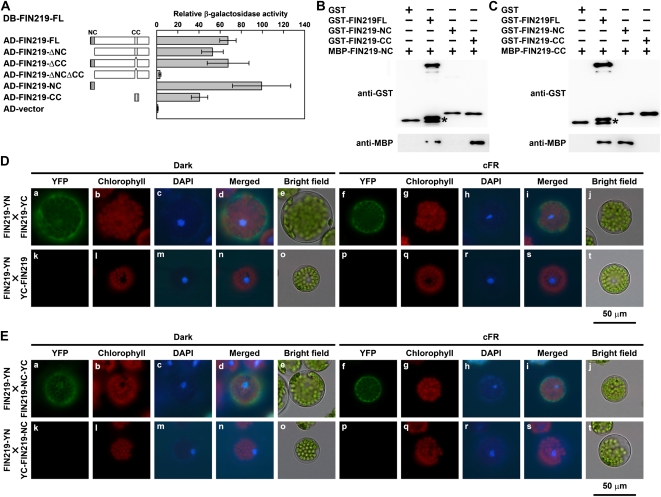
Ectopic expression of the N terminus of FIN219 results in a hypophotomorphogenic phenotype under cFR light, with increased level of COP1 and reduced level of HY5 (Fig. 1). To further understand the mechanisms causing the changes in levels of the vital regulators COP1 and HY5 leading to a long-hypocotyl phenotype of NCox and NTox seedlings in a wild-type background specifically under cFR light, we performed interaction studies by yeast two-hybrid and pull-down assays to determine whether the N-terminal coiled-coil domain of FIN219 can bind to the endogenous full-length FIN219. The β-galactosidase activities in yeast two-hybrid assays revealed that the full-length FIN219 could interact with itself and both the N-terminal and C-terminal coiled-coil domains of FIN219, but with higher affinity to the N-terminal coiled-coil domain (Fig. 4A). Moreover, the recombinant MBP-FIN219-NC, GST-FIN219-NC, GST (or MBP)-FIN219-CC, containing the C-terminal coiled-coil domain with 47 amino acids of FIN219, and GST-full-length FIN219 (GST-FIN219FL) fusion proteins were purified to be homogeneous and used for binding assays. The binding affinity of MBP-FIN219-NC was strong with GST-FIN219-CC, weak with GST-FIN219FL, and absent with GST-FIN219-NC or GST alone (Fig. 4B). Further tests demonstrated that MBP-FIN219-CC could bind with GST-FIN219FL and GST-FIN219-NC (Fig. 4C). To further confirm the interaction between the full-length FIN219 and itself or its N-terminal coiled-coil domain, we performed bimolecular fluorescence complementation (BiFC) assay using the mesophyll protoplasts isolated from fin219-2. The full-length FIN219 interacted with itself in the cytoplasm under both darkness and cFR light (Fig. 4D, top row). The control with the combination of FIN219-YN and YC-FIN219 showed no fluorescence signal (Fig. 4D, bottom row). Other combinations, such as YN-FIN219 and YC-FIN219, also showed interaction in the cytoplasm in both darkness and cFR light (Supplemental Fig. S4). Further BiFC assays indicated that the full-length FIN219 could interact with the N-terminal coiled-coil domain (FIN219-NC) in the dark and cFR light (Fig. 4E, top row). Thus, the full-length FIN219 can interact with the N-terminal coiled-coil domain of FIN219 through its C-terminal coiled-coil domain.
Figure 4.
FIN219 is able to interact with itself in vitro and in vivo. A, Interaction assay of FIN219 and FIN219 by the yeast LexA two-hybrid system. The value of interaction by β-galactosidase activity was the average of five individual yeast colonies. Error bars represent sd. The level of activation domain (AD) vector was arbitrarily set to 1. FIN219-ΔNC, FIN219FL minus the N-terminal coiled-coil domain; FIN219-ΔCC, FIN219FL minus the C-terminal coiled-coil domain; FIN219-ΔNCΔCC, FIN219FL minus the N-terminal coiled-coil domain and the C-terminal coiled-coil domain; FIN219-NC, the N-terminal coiled-coil domain of FIN219; FIN219-CC, the C-terminal coiled-coil domain of FIN219. DB, DNA-binding domain. B, The N-terminal coiled-coil domain of FIN219 can bind with the C-terminal coiled-coil domain on pull-down assay. Different recombinant proteins, including FIN219 fused with a GST tag (GST-FIN219FL, GST-FIN219-NC, and GST-FIN219-CC) or with maltose-binding peptide (MBP-FIN219-NC) purified from E. coli, were used. MBP-FIN219-NC was mixed with various recombinant proteins indicated above the blots and immunoprecipitated with GST resins. The immunoprecipitated products were probed with GST (top panel) and MBP (bottom panel) antibodies. The asterisk indicates a breakdown product of GST-FIN219FL. C, The interaction of the N-terminal coiled-coil domain of FIN219 and the C-terminal coiled-coil domain was confirmed by switch-tag fused proteins. The C-terminal coiled-coil domain of FIN219 fused with MBP (MBP-FIN219-CC) was mixed with various recombinant proteins indicated above the blot and used for pull-down assay as shown in B. The asterisk indicates a breakdown product of GST-FIN219FL. D and E, Assays of the interaction of the full-length FIN219 and itself or its N-terminal coiled-coil domain (FIN219-NC) by BiFC. The BiFC plasmids containing the full-length FIN219 and itself or FIN219-NC were cotransfected into Arabidopsis mesophyll protoplasts isolated from fin219-2. The transfected protoplasts were cultured overnight under darkness (a–e and k–o) or cFR light (f–j and p–t). The yellow fluorescent protein (YFP) channel is shown as the interaction signal of BiFC, the chlorophyll channel as autofluorescence of chlorophyll, the 4′,6-diamidino-2-phenylindole (DAPI) channel as the nucleus with DAPI staining, and the merged channel as the combination of YFP, chlorophyll, and DAPI channels.
To further understand whether this binding involves intermolecular or intramolecular interaction, we introduced constructs harboring the C-terminal 171 or 275 amino acid residues of FIN219 fused with GUS (FIN219-C171ox or FIN219-C275ox) into NCox and NTox transgenic plants in Col. The resulting transgenic seedlings did not show even longer hypocotyls than their parental background under cFR light (Supplemental Fig. S5), which suggests that the binding of the N-terminal coiled-coil domain of FIN219 with its C terminus may occur in an intermolecular, not intramolecular, manner.
FIN219 Can Interact with COP1 in the Cytoplasm under Darkness and cFR Light
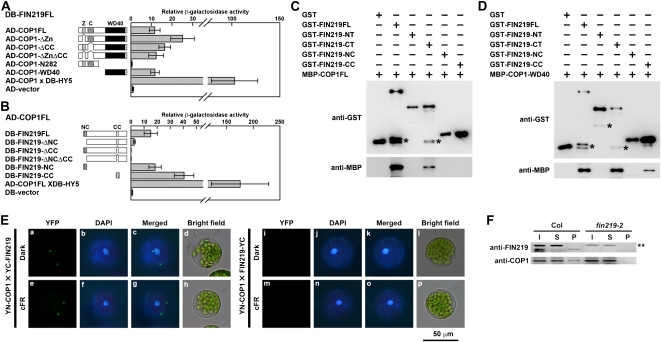
Here, we demonstrate that FIN219 negatively regulates COP1 levels, which raises the possibility that the suppression of COP1 by FIN219 may directly involve physical interaction, which might contribute to the dominant-negative phenotype conferred by ectopic expression of the FIN219 N terminus. Yeast two-hybrid assay revealed that FIN219 could interact with the C-terminal WD-40 domain of COP1 via its coiled-coil domain in the N or C terminus (Fig. 5, A and B). The zinc-finger domain and the coiled-coil domain of COP1 seem to fine-tune the binding affinity between FIN219 and COP1 (Fig. 5A). The coiled-coil domain in the C terminus of FIN219 appears to have higher affinity with COP1 than the one in the N terminus (Fig. 5B), which implies that the N terminus may negatively regulate interacting activities of the C-terminal domain binding with COP1. Since HY5 stability requires functional FIN219, determining whether both proteins interact with each other was of interest. However, the two proteins did not interact by yeast two-hybrid assay (data not shown). Furthermore, in pull-down assays, various GST-FIN219 fusion proteins and MBP-COP1 fusion proteins (MBP-COP1FL and MBP-COP1-WD-40) were used to confirm the respective interacting domain. GST-FIN219 full-length fusion (GST-FIN219FL) and the C-terminal 275-amino acid fusion (GST-FIN219-CT) were able to bind with the MBP-COP1 full-length fusion (MBP-COP1FL; Fig. 5C); other fusions, including the N terminus (GST-FIN219-NT), N-terminal coiled-coil domain (GST-FIN219-NC), and C-terminal coiled-coil domain (GST-FIN219-CC), were not able to bind with MBP-COP1FL (Fig. 5C). GST-FIN219-CC might not be able to bind with MBP-COP1FL because of the structural mask of MBP-COP1FL fusions. This speculation was supported by results showing that GST-FIN219FL, GST-FIN219-CT, and GST-FIN219-CC were able to bind with the MBP-WD-40 domain of COP1 fusion (MBP-COP1-WD-40; Fig. 5D).
Figure 5.
FIN219 can physically interact with COP1. A and B, Interaction of FIN219 and COP1 by the yeast LexA two-hybrid system. Assays of protein-protein interactions are shown by β-galactosidase activities. Values are averages of five individual yeast colonies, and error bars represent sd. The level of activation domain (AD) vector was arbitrarily set to 1. COP1FL, Full-length COP1; COP1-ΔZn, COP1FL minus the zinc-finger domain; COP1-ΔCC, COP1FL minus the coiled-coil domain; COP1-ΔZnΔCC, COP1FL minus the zinc-finger domain and the coiled-coil domain; COP1-N282, the N-terminal 282 amino acid residues of COP1; COP1-WD-40, the WD-40 repeat domain of COP1; COP1FL × HY5FL, a positive control; HY5FL, full-length HY5. Various deletion constructs of FIN219 were described in Figure 4A. DB, DNA-binding domain. C, Interaction of various FIN219 recombinant proteins and full-length COP1 by pull-down assay. Different recombinant proteins, 5 μg each, indicated above the blots were used for pull-down assays. The reaction mixture was precipitated with glutathione-Sepharose 4B beads, and then pellets underwent protein gel-blot analysis with anti-GST or anti-MBP antibodies. Asterisks indicate breakdown products of GST-FIN219FL and GST-FIN219-CT. D, Interaction of various FIN219 recombinant proteins and the WD-40 domain of COP1 by pull-down assay. Asterisks indicate breakdown products of GST-FIN219FL, GST-FIN219-NT, and GST-FIN219-CT. E, Assay of FIN219 and COP1 interaction by BiFC assay. The YN-COP1 and YC-FIN219 plasmids were cotransfected into Arabidopsis mesophyll protoplasts isolated from fin219-null. The transfected protoplasts were cultured overnight in the dark (a–d and i–l) or cFR light (e–h and m–p). F, Coimmunoprecipitation analysis of FIN219 interaction with COP1 in the wild type (Col). Two milligrams of protein extracts isolated from Col and fin219-2 seedlings grown in cFR light for 3 d was mixed with the FIN219 polyclonal antibodies and then coimmunoprecipitated. The pellets were used for SDS-PAGE and protein gel-blot analysis. The probes are FIN219 and COP1 antibodies. I, Input proteins; S, supernatants after precipitation; P, pellets after precipitation. The double asterisk indicates artificial bands.
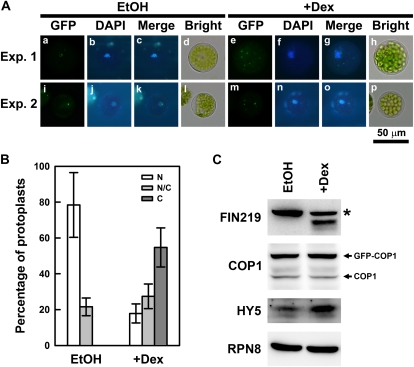
Moreover, BiFC assay with protoplasts isolated from fin219-2 further confirmed the interaction of FIN219 and COP1 (Fig. 5E). Coimmunoprecipitation studies using wild-type Col and fin219-2 seedlings grown in cFR light for 3 d showed that FIN219 did interact with COP1 (Fig. 5F). This interaction occurred in the cytoplasm under both darkness and cFR light (Fig. 5E). Intriguingly, the interaction of FIN219 and COP1 was shown with two or more specific spots that appear to have similar features to the formation of nuclear speckles due to COP1 and HY5 interaction in the nucleus (Jang et al., 2005). In fact, we observed a high ratio of similar cytoplasmic speckle structure when both FIN219 and COP1 were cotransfected into protoplasts in darkness and far-red light (data not shown). We hypothesized that FIN219 levels might modulate the subcellular location of COP1. Thus, transgenic plants harboring a glucocorticoid-inducible FIN219 construct were generated. Homozygous FIN219-inducible transgenic seedlings treated with 1 μm dexamethasone (Dex) in darkness and cFR light for 3 d showed a markedly short-hypocotyl phenotype as compared with the wild type under the same conditions (Supplemental Fig. S6, A and C), in which FIN219 levels were highly induced by Dex up to a 10-fold increase (Supplemental Fig. S6, B and D). Moreover, when we introduced GFP-COP1 into FIN219 protoplasts, followed by Dex induction in darkness, most of the GFP-COP1 was excluded out of the nuclei and become cytoplasm localized (Fig. 6, A and B). Further examination indicated that FIN219 was highly induced in the protoplasts treated with Dex, and COP1 protein level remained largely the same in both control and Dex-treated samples, whereas HY5 level was significantly increased in Dex-treated protoplasts (Fig. 6C), which suggests that induction of FIN219 protein levels can mediate COP1 transition from the nucleus to the cytoplasm, thus resulting in an increase of nuclear HY5 levels even in the dark and a photomorphogenic response.
Figure 6.
FIN219 can exclude COP1 out of the nuclei into the cytoplasm. A, Subcellular localization of GFP-COP1 in the protoplasts isolated from glucocorticoid-inducible FIN219 (pGR:FIN219) transgenic plants. The transfected protoplasts were cultured on W5 solution containing ethanol (EtOH) or 1 μm Dex and incubated in the dark overnight. The experiments were repeated once and shown in Exp.1 and Exp. 2. B, Percentage of transfected protoplasts with different subcellular locations of GFP-COP1. Thirty transfected protoplasts were counted for the subcellular localizations of GFP-COP1. C, Cytoplasm; N, nuclei; N/C, both locations. C, Protein gel-blot analysis of FIN219, COP1, and HY5 levels in the protoplasts with (+Dex) or without (EtOH) Dex treatment. The asterisk indicates a nonspecific band. RPN8 was a loading control.
FIN219 Protein Levels Show a FR Light Fluence-Dependent Regulation
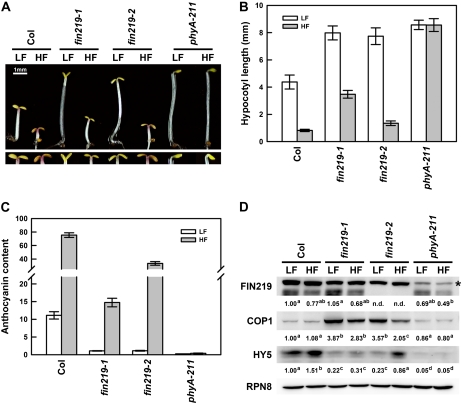
Because the original fin219-1 mutant showed a substantial long-hypocotyl phenotype with both low and high fluence rates of FR and fin219-2 exhibited a significant phenotype only in the low-fluence FR (Fig. 1D), we wondered whether this discrepancy was due to a change in COP1 or HY5 level. Further examination of fin219-1 and fin219-2 mutants under low and high fluence rates of FR (LF and HF, respectively) indicated consistent phenotypic responses similar to those shown in Figure 1D (Fig. 7, A and B), which was also reflected in high accumulation of anthocyanin in wild-type Col under HF, an intermediate level in fin219-2, and a lesser level in fin219-1 (Fig. 7, A, bottom panel, and C), whereas the phyA-211 mutant showed an insensitivity of hypocotyl elongation and no anthocyanin accumulation in both conditions (Fig. 7, A–C). Interestingly, FIN219 levels in Col and fin219-1 were slightly reduced under HF as compared with LF. Moreover, FIN219 level in the phyA-211 mutant was reduced under LF and HF conditions as compared with Col in respective fluences. In contrast, COP1 level in Col did not show a significant difference between LF and HF; it increased greatly in both fin219-1 and fin219-2 but had less increase in fin219-2 under HF and showed a comparative level in phyA-211. Surprisingly, HY5 level in fin219-2 under HF was significantly increased up to the wild-type level under LF (Fig. 7D), which is consistent with the short-hypocotyl phenotype for fin219-2 under the HF condition. However, the level of HY5 in fin219-2 under LF was greatly reduced to low levels similar to those in fin219-1 under both LF and HF conditions (Fig. 7D), which might be why the fin219-1 mutant still showed a substantial long-hypocotyl phenotype under HF. In addition, the level of HY5 was much reduced in phyA-211 under both LF and HF conditions as compared with Col under the same conditions (Fig. 7D). Thus, these data suggest that FIN219 may have a differential role in the regulation of HY5 in LF and HF to modulate hypocotyl elongation.
Figure 7.
FIN219 shows differential expression levels and regulation on HY5 in fin219 mutants under low and high fluence rates of FR light. A, Phenotypic examination of wild-type Col, fin219-1, fin219-2, and phyA-211 mutant seedlings under LF and HF light conditions. Three-day-old seedlings were used for phenotypic examination. The bottom panel shows a closeup image of cotyledons and anthocyanin accumulation. LF, 1.42 μmol m −2 s−1; HF, 20.54 μmol m−2 s−1. B, Quantitative analysis of hypocotyl lengths of the seedlings shown in A (n = 30). C, Quantitative measurement of anthocyanin accumulation of the seedlings shown in A (n = 6). D, Protein gel-blot analysis of Col, fin219-1, fin219-2, and phyA-211 seedlings shown in A. The asterisk indicates artificial bands. RPN8, a subunit of the regulatory complex of the 26S proteasome, was a loading control. The number below each blot represents the relative level of the indicated protein. The level in wild-type Col was arbitrarily set to 1. Different lowercase letters represents significant differences by ANOVA at P < 0.05.
DISCUSSION
COP1 functions as a repressor of photomorphogenesis in darkness and has an E3 ligase activity involved in the protein degradation of light signaling players, including the photoreceptors phytochromes and cryptochromes (Wang et al., 2001; Yang et al., 2001; Seo et al., 2004; Jang et al., 2010); however, the information on the regulation of COP1 activities is limited. Here, we demonstrate that FIN219/JAR1 negatively regulates COP1 levels through physical interaction under cFR light and can modulate the subcellular location of COP1 even in the dark. Moreover, we also provide evidence showing that HY5 protein stability requires FIN219. Thus, our data reveal a new finding that FIN219/JAR1 functions as a positive regulator of phyA-mediated FR light signaling by controlling the levels of COP1 and HY5 to modulate photomorphogenic development in Arabidopsis.
Our recent data demonstrated that jar1-1 showed a long-hypocotyl phenotype under a FR light fluence rate below 20 μmol m−2 s−1 (Chen et al., 2007). Especially, it exhibited a hypocotyl length similar to that of fin219-1 at 1.47 μmol m−2 s−1 FR light. Moreover, wild-type Col seedlings overexpressing FIN219 displayed a hypersensitive phenotype under LF light (Hsieh et al., 2000), which implies that FIN219 may play a vital role under LF light. Here, we further show that a fin219-2 allele and NCox, as well as NTox, with ectopic expression of the FIN219 N-terminal domain in the wild type, produce a hyposensitive long-hypocotyl phenotype under cFR light (Fig. 1), which is consistent with features of reduced photomorphogenic development, including decreased expression of CHS and reduced anthocyanin accumulation in various fin219 mutant seedlings grown under the same conditions (Fig. 1; Supplemental Fig. S1A). In addition, the phyA null mutant, phyA-211, resulted in reduced levels of FIN219 under LF and HF conditions (Fig. 7D). All these data strongly indicate that FIN219/JAR1 participates in phyA-mediated FR signaling.
To further illustrate the molecular mechanism underlying the long-hypocotyl phenotype of NCox and NTox in a wild-type background specifically under cFR light, we found that the expression of COP1, a repressor of photomorphogenesis in the dark (Deng et al., 1992), was differentially increased in level in fin219, NCox, and NTox seedlings in a wild-type background under cFR light (Fig. 1F); however, it was much reduced in the FIN219-overexpressed transgenic line FIN219ox (Fig. 2C), which implies that FIN219 negatively regulates COP1 level under cFR light. Moreover, HY5, a positive regulator of light signaling existing in both phosphorylated and unphosphorylated forms in Arabidopsis (Hardtke et al., 2000), was substantially decreased in level in fin219-1, NCox, NTox, and fin219-2 mainly under cFR light but not other light conditions (Fig. 1F; Supplemental Fig. S3). This specificity of HY5 reduction is consistent with the specific phenotype that fin219-1, NCox, NTox, and fin219-2 conferred. Intriguingly, fin219-1 and fin219-2 showed greatly reduced levels of the unphosphorylated form of HY5 (Fig. 1F), previously shown to be active and unstable in light signaling (Hardtke et al., 2000), which implies that FIN219 may affect HY5 stability. In addition, COP1 works synergistically with SPA1, the Arabidopsis suppressor of phyA-105, to negatively control HY5 abundance under cFR light (Saijo et al., 2003; Yang and Wang, 2006). Here, we found that the double mutant fin219-2/cop1-6 showed greatly reduced HY5 level in the dark and cFR light (Fig. 3C) as compared with that in the parental cop1-6 under the same conditions, which indicates that the presence of FIN219 somehow stabilizes HY5 level. The cop1-6 mutant contains the wild-type size and abundance of COP1 (McNellis et al., 1996); the double mutant fin219-2/cop1-6 might increase functional COP1 level, thus leading to HY5 degradation in cFR light and darkness. Moreover, FIN219 is induced at high levels by Dex treatment and can move COP1 to the cytoplasm even in darkness (Figs. 5E and 6), whereas in the absence of FIN219, the wild-type size of COP1 in cop1-6 might stay in the nucleus, thus resulting in increased degradation of HY5 in both darkness and cFR light (Fig. 3C). In addition, SPA1 was shown to enhance COP1 E3 ligase activity to negatively influence HY5 levels under cFR light (Saijo et al., 2003). Our preliminary data also showed up-regulated expression of SPA1 transcripts in the fin219-1 mutant under cFR light (Supplemental Fig. S7). The double mutant fin219-2/cop1-6 likely contains increased levels of SPA1, thus giving rise to enhanced COP1 E3 ligase activity, which leads to increased HY5 degradation. However, we still do not have a good explanation for the comparable level of COP1 in the double mutant fin219-2/cop1-6 and the wild type (Fig. 3C), which might involve feedback regulation. Therefore, FIN219 plays an important role in phyA-mediated FR light signaling to negatively regulate COP1 and positively regulate HY5 levels for photomorphogenic development of Arabidopsis seedlings.
Furthermore, FIN219 was shown to interact with the WD-40 domain of COP1 through its coiled-coil domain in the C terminus (Fig. 5), where the N-terminal coiled-coil domain of FIN219 bound in a manner of intermolecular interaction (Fig. 4). The intermolecular interaction of FIN219-NC/-NT and FIN219 was further indicated, with the results showing that the introduction of the C-terminal domain of FIN219 (FIN219-C171ox or FIN219-C275ox) into NCox or NTox transgenic plants in a wild-type background did not result in an even longer hypocotyl phenotype than NCox or NTox (Supplemental Fig. S5). These results suggest that the dominant-negative phenotype conferred by NCox or NTox involves intermolecular binding of the N terminus of FIN219/JAR1 and the endogenous full-length FIN219/JAR1, instead of intramolecular interaction of its N and C termini. However, we still could not rule out the possibility of intramolecular interaction, with misregulation of normal FIN219/JAR1 complex function or failure of forming an accurate structure of the FIN219/JAR1 complex between the NCox or NTox construct and the FIN19 C-terminal construct FIN219-C171 or FIN219-C275, resulting in a hypophotomorphogenic phenotype in cFR light. Therefore, ectopic expression of the N terminus of FIN219 in the wild type likely competes with endogenous COP1 to bind with the C-terminal coiled-coil region of endogenous FIN219, thus leading to a reduction in HY5 level and a long-hypocotyl phenotype in cFR light. However, in the wild type under cFR light, FIN219 exists as a homodimer or heterodimer with COP1 in the cytoplasm, which leads to an increase of HY5 level in the nucleus and further photomorphogenic development. This notion was substantiated by the ectopic expression of the C terminus of FIN219, which entailed COP1 in the cytoplasm, thus stabilizing HY5 in the nucleus, which leads to a hypersensitive short-hypocotyl phenotype in cFR light (Supplemental Fig. S8).
Our previous studies indicated that the fin219-1 mutation can suppress the short-hypocotyl phenotype of cop1-6 in darkness; that is, the double mutant fin219-1/cop1-6 exhibited a long-hypocotyl phenotype in darkness, but the double mutant fin219-1/cop1-5 still showed the cop1-5 mutant phenotype with short hypocotyls, which indicates an allele-specific suppression by FIN219 (Hsieh et al., 2000). In contrast, our double mutant studies revealed that the fin219-2 mutation did not suppress cop1-6 or cop1-4 in darkness and cFR light; moreover, fin219-2/cop1-6 but not fin219-2/cop1-4 resulted in significant reduction of HY5 protein level in darkness and cFR light. This allele-specific effect may involve interaction of FIN219 in fin219-1 and the WD-40 domain of COP1. The fin219-2/cop1-6 double mutant does not contain FIN219 protein, which leads to COP1 or partial COP1 activity action in the nuclei that results in HY5 level reduction. However, fin219-2/cop1-4 does not have FIN219, and cop1-4 only contains the N-terminal 282 amino acids without the WD-40 domain; thus, HY5 is not degraded. Taken together, these data indicate that the interaction of FIN219 and COP1 is critical to modulate the level and stability of HY5 for photomorphogenic development.
JAR1 has been well documented to play a role in plant defense responses (Staswick et al., 1998; Ferrari et al., 2003; Kazan and Manners, 2008) and acts as a JA-amino acid synthetase with a preferred generation of JA-Ile (Guranowski et al., 2007; Suza and Staswick, 2008). A recent report showed that the JA-Ile conjugate but not other JA derivatives such as JA or MeJA could enhance the physical interaction between COI1 and JASMONATE ZIM-DOMAIN PROTEIN1 (JAZ1), thus leading to JAZ1 degradation and JA signaling (Chini et al., 2007; Thines et al., 2007); therefore, the JA-Ile conjugate acts as an active form of JA to fulfill physiological functions in plant defense responses and development. Here, we demonstrated that ectopic expression of the N terminus of FIN219/JAR1 resulted in a dominant-negative long-hypocotyl phenotype under cFR light, which did not alter endogenous levels of JA-Ile as well as the root and hypocotyl responses to exogenous MeJA or coronatine (Table I; Supplemental Fig. S2); therefore, coronatine, a phytotoxin with a structure mimicking that of JA-Ile, can initiate jasmonate responses once it is perceived by the jasmonate receptor COI1, even in fin219-2 and jar1-1 mutants. Interestingly, the coi1-16 mutant as a control in this study exhibited a long-hypocotyl phenotype in cFR light in the absence of coronatine and remained with an unchanged phenotype even in the presence of coronatine (Supplemental Fig. S2, E and F). Therefore, jasmonate perception contributes to FR-mediated inhibition of hypocotyl elongation, which is consistent with the results of Robson et al. (2010) showing that COI1 can modulate responses to light, including photomorphogenic responses to FR light, flowering, and shade responses. Here, we further found that FIN219 exhibits a FR fluence rate-dependent expression pattern with a more abundant level in LF and a reduction in level in HF, leading to a high level of HY5 in HF (Fig. 7D). It appears that FIN219 in wild-type Col under LF and HF can negatively regulate COP1 and entrap it in the cytoplasm through physical interaction; however, the FIN219 in fin219-1 was unable to suppress COP1 level by interaction, leading to COP1 in the nuclei and, accordingly, HY5 degradation under LF and HF. In contrast, in fin219-2 under LF, COP1 stayed in the nuclei, giving rise to HY5 degradation, but under HF, some of the COP1 proteins moved to the cytoplasm, thus leading to an increase of HY5 level. HY5 increase in level in fin219-2 under HF may involve another factor responsible for COP1 reduction and increase in HY5 level (Fig. 7D), whereas, in the dark, FIN219 mainly affects COP1 partition, but not COP1 levels, between the nucleus and the cytoplasm (Fig. 6C; Supplemental Fig. S3D). Thus, FIN219 levels responding to different fluence rates of FR may provide a molecular mechanism to modulate shade responses for the production of JA-Ile in response to the ratio of red and FR light.
Recent evidence revealed that the Arabidopsis phytochrome chromophore mutants hy1 and hy2 show elevated levels of JA and constant activation of COI1-dependent JA responses; moreover, JA inhibits the expression of a group of light-inducible photosynthetic genes (Zhai et al., 2007). Thus, phytochrome chromophore-mediated light signaling and the JA signaling pathway may have a mutually antagonistic relation. As well, phytochrome inactivation by FR can strongly reduce plant sensitivity to jasmonates (Moreno et al., 2009), and a recent report indicated that phyA is required for JA- and wound-mediated JAZ1 degradation (Robson et al., 2010). Moreover, phyA in rice (Oryza sativa) requires jasmonate for photodestruction (Riemann et al., 2009). Hence, phyA- and jasmonate-mediated signals are mutually regulated. Here, we showed that phyA positively regulates FIN219/JAR1 level under LF and HF (Fig. 7D), which may have a good link to negatively control JAZ1 level. In addition, FIN219/JAR1 also shows a gradual down-regulated expression pattern in the dark-FR light transition (data not shown), possibly giving rise to reduced levels of JA-Ile, which allows for rapid induction of photosynthesis-related genes such as RbcS and Cab to undergo photomorphogenic development. This raises an interesting question: how does FIN219/JAR1, a jasmonate-conjugating enzyme, work in the integration of FR light and JA signaling? We propose that FIN219/JAR1 may have a dual function, one with JA-conjugating enzyme activity and the other as a component of phyA-mediated FR signaling. This dual function of FIN219/JAR1 might have a synergistic or independent effect. For instance, FIN219/JAR1 as a JA-conjugating enzyme may trigger COP1 out of the nuclei, as shown in Figure 6, and then serve as a component of FR signaling to bind with the WD-40 domain of COP1 in the cytoplasm, thus resulting in an increase of HY5 levels in the nuclei; however, the molecular mechanism underlying FIN219/JAR1 dual functions remains to be elucidated. Intriguingly, several other members of the GH3 gene family in Arabidopsis, such as GH3.2, GH3.5, GH3.6, and GH3.10, have been reported to participate in light signaling (Nakazawa et al., 2001; Tanaka et al., 2002; Takase et al., 2003, 2004) and categorized as group II or group I (GH3.10) based on sequence homology and enzymatic activity (Staswick et al., 2002; Staswick and Tiryaki, 2004). These GH3 members with functional roles in light signaling are either IAA-amido synthetase (group II) or JA-amido synthetase (group I) and contain a coiled-coil domain either in the N terminus or C terminus. FIN219/JAR1 has two coiled-coil domains, one in the N terminus and the other in the C terminus that has been shown to interact with FIP1 (Chen et al., 2007). Here, we showed that the C-terminal coiled-coil domain of FIN219 is able to interact with COP1. It will be worthy to see how the binding of FIP1 with FIN219 affects the FIN219-COP1 interaction. Therefore, these GH3 members may act in a similar manner as FIN219 with a dual function; that is, they may have enzymatic activities and nonenzymatic functions via protein-protein interacting manner. The characteristic of FIN219/JAR1 with a dual function was indicated by a recent paper showing that a 2C-type protein phosphatase, PP2CA, can directly interact with the protein kinase OST1, leading to inhibition of the kinase independently of its phosphatase activity in response to low levels of abscisic acid, which results in the activation of the SLAC1 anion channel with basal activity (Lee et al., 2009). Thus, FIN219/JAR1 can serve as a good candidate to understand the integration between FR light and JA signaling pathways.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The mutants fin219-1 (Hsieh et al., 2000), jar1-1 (Staswick et al., 2002), fin219-null (SALK_059774 from the Arabidopsis Biological Resource Center; named fin219-2 in this study), cop1 alleles (cop1-4 and cop1-6), and two transgenic plants, NCox and NTox, are in the Arabidopsis (Arabidopsis thaliana) Col ecotype; hy5-1 is in the Landsberg ecotype background. Arabidopsis seedlings were grown in an E30LEDL3 growth chamber (Percival Scientific) for phenotype analysis. The wavelengths of FR, red, and blue light-emitting diode light sources (Percival Scientific) were 739 ± 31 nm, 669 ± 25 nm, and 470 ± 25 nm, respectively. Light fluence rates were measured by the LI-200SA pyranometer sensor (LI-COR). The light fluence rates used for FR, blue, red, and white light were 1.47, 5.08, 6.34, and 5.88 μmol m−2 s−1, respectively. Arabidopsis seedlings were grown on GM (Murashige and Skoog salts plus vitamins and 0.5 g/L MES buffer, pH 5.7) agar plates containing 0.3% Suc for phenotype and molecular analysis as described previously (Hsieh et al., 2000).
Gene Constructs and Transgenic Plants
To construct GUS-FIN219-N53 and GUS-FIN219-N300 plasmids, the cDNA sequences corresponding to the N-terminal 53 and 300 amino acids (NC and NT, respectively) of FIN219 were amplified with primers (Supplemental Materials and Methods S1) and the full-length FIN219 cDNA clone 11-2 as a template. Both PCR products were subcloned into the BglII site of the pRTL2-GUS plasmid. The PstI DNA fragments were released from pRTL2-GUS-FIN219-NC and pRTL2-GUS-FIN219-NT constructs, subcloned into the binary vector pPZP221, and then introduced into plants by the floral dip method (Clough and Bent, 1998). To clone the coiled-coil domain and the 275 amino acids in the C-terminal region of FIN219 (FIN219-CC and FIN219-CT, respectively), the cDNA clone 11-2 was used for PCR amplification. For recombinant protein constructs, various BglII FIN219 fragments were cloned into pGEX-4T-1 (Amersham) and pMAL-C2X (New England Biolabs).
Generation of Double Mutants of fin219-2 and cop1-4 or cop1-6
For double mutant production, cop1-4 or cop1-6 as male was crossed with fin219-2 as female. The resulting F2 generation progeny were used for PCR-based genotyping by using the combinations of T-DNA primer (LBa1, 5′-TGGTTCACGTAGTGGGCCATCG-3′) and FIN219-specific primer (FIN219N299_R, 5′-CTTTGCGTTGGGGAAGAGC-3′) for the FIN219 locus, mutant-specific primer (cop1-4-Rv-1669m, 5′-CAGTTGACTGATTCAAACTTTA-3′) and wild-type COP1-specific primer (cop1-4-Fw-326, 5′-CCCTGTTGTAGCCAACACCTCACC-3′) for the cop1-4 mutant locus, and mutant-specific primer (cop1-6-Rv-2295m, 5′-ACATTCTTGTAAATCATTGAACC-3′) and wild-type COP1-specific primer (cop1-6-Fw-1274, 5′-ACGTATAACCCCTGAATGTTAG-3′) for the cop1-6 mutant locus. For identification of homozygous double mutants, the following wild-type-specific primers were used for genomic genotyping: for FIN219, FIN219N299_R and FIN219F_XhoI (5′-CAGCTCGAGATGTTGGAGAAGGTTGAA-3′); for cop1-4, cop1-4-Fw-326 and cop1-4-Rv-1669 (5′-CAGTTGACTGATTCAAACTTTG-3′); for cop1-6, cop1-6-Fw-1274 and cop1-6-Rv-2295 (5′-ACATTCTTGTAAATCATTGAACT-3′). One candidate was identified as homozygous for the fin219-2 mutant locus but heterozygous for the cop1-4 or cop1-6 mutant locus. This line was further subjected to phenotyping in the dark, thus leading to a ratio of 3:1 long-hypocotyl:short-hypocotyl phenotype. Thus, the short-hypocotyl seedlings were transferred to white light for the next generation and further genotyping until homozygous loci for the fin219-2 and cop1-4 or cop1-6 mutations were identified.
Analysis of Pigments
Seedlings of wild-type Col, fin219/jar1, NCox, and NTox transgenic plants were grown in cFR light for 3 d for investigation of anthocyanin accumulation or followed by another 1-d exposure of continuous white light for analysis of chlorophyll content. The measurements of chlorophyll and anthocyanin were described previously (Hsieh et al., 2000).
Protein Extraction and Protein Gel-Blot Analysis
Seedlings of wild-type Col, fin219/jar1, NCox, and NTox transgenic plants grown in cFR light at the indicated fluence rate for 3 d were harvested and frozen immediately for protein extraction. Total protein extraction was described previously (Hsieh et al., 2000). In total, 100 μg of total protein was separated on a 12% SDS-polyacrylamide gel and blotted onto an Immobilon-P membrane (Millipore). Protein gel-blot analysis involved standard methods (Sambrook and Russell, 2001), and detection was with the use of anti-FIN219, anti-COP1 (Hsieh et al., 2000), anti-HY5 (Osterlund et al., 2000a), and anti-RPN8 (Yang et al., 2004) specific antibodies. Protein expression data are means of at least two independent replicates. Data were analyzed by ANOVA with Scheffe’s test. The differences were considered significant at P < 0.05.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays involved use of the LexA-based two-hybrid system. Various COP1 fragments were cloned into pJG4-5 as described previously (Ang et al., 1998). Full-length and different deletions of FIN219 were generated by PCR and cloned into pEG202. All constructs were transformed into the yeast strain EGY48 with pSH18-34 encoding a LacZ reporter gene, and yeast cells were grown on minimal medium/−His/−Trp according to the manufacturer’s instructions (Clontech). Transformed colonies were plated onto minimal medium/−His/−Trp/−Leu/+Gal/+raffinose/5-bromo-4-chloro-3-indolyl-ββ-d-galactopyranoside. Yeast cell selection and β-galactosidase activity assay were essentially the same as standard methods (Clontech).
Protein Interactions by Pull-Down Assays
Various MBP and GST fusion proteins were purified from Escherichia coli as described previously (Saijo et al., 2003; Chen et al., 2007). Five micrograms of recombinant protein was mixed with 1 mL of binding buffer (50 mm Tris, pH 7.5, 100 mm NaCl, 0.2% glycerol, 0.6% Triton X-100, and 0.5 mm β-mercaptoethanol). After incubation at 4°C for 1 h, the reaction mixture was further incubated with glutathione-Sepharose 4B beads (Amersham) at 4°C for another 1 h. The beads were washed five times with washing buffer (50 mm Tris, pH 7.5, 100 mm NaCl, and 0.6% Triton X-100), then the pellet was boiled with 2× sample buffer and underwent SDS-PAGE. Western-blot analysis was performed by standard methods (Chen et al., 2007).
Protoplast Transfection and BiFC Analysis
Arabidopsis mesophyll protoplast isolation and transfection were as described (Yoo et al., 2007). Four-week-old well-expanded leaves were cut into 1-mm pieces and incubated in enzyme solution (20 mm MES, 1.5% cellulase R10, 0.4% macerozyme R10, 0.4 m mannitol, 20 mm KCl, 10 mm CaCl2, 5 mm β-mercaptoethanol, and 0.1% bovine serum albumin, pH 5.7) for 3 h. Protoplasts were collected by centrifugation at 100g and washed twice with W5 solution (2 mm MES, 154 mm NaCl, 125 mm CaCl2, and 5 mm KCl, pH 5.7). Protoplasts were resuspended in W5 solution and incubated on ice for at least 30 min, then washed with MMG solution (4 mm MES, 0.4 m mannitol, and 15 mm MgCl2, pH 5.7) and resuspended at 2 × 105 cells mL−1 in MMG solution. For protoplast transfection, 200 μL of protoplasts was mixed with 20 μL of DNA and 220 μL of PEG solution (40% polyethylene glycol 4000, 0.2 m mannitol, and 100 mm CaCl2), then transfected protoplasts were washed twice, resuspended in W5 solution, and incubated under experimental conditions. We constructed BiFC plasmids as described (Walter et al., 2004). Various DNA fragments of FIN219 and COP1 were cloned into 35p-YFP-N155/pRTL2 and 35p-YFP-C84/pRTL2, respectively. The nuclei of protoplasts were stained with 4′,6-diamidino-2-phenylindole. All fluorescence images were obtained by use of a BX51 fluorescence microscope with a DP-72 digital camera (Olympus) and processed by use of Adobe Photoshop.
Protein-Protein Interactions by Coimmunoprecipitation Analysis
Coimmunoprecipitation analysis was performed as described (Chen et al., 2007). The seedlings grown in cFR light for 3 d were ground with grinding buffer (50 mm Tris-HCl, pH 5.7, 150 mm NaCl, 10 mm MgCl2, 0.1% bovine serum albumin, 0.1% Nonidet P-40, 1 mm phenylmethanesulfonyl fluoride, 1× Protease Inhibitor Cocktail [Sigma], and 40 μm MG132). A total of 2 mg of proteins was mixed with beads and incubated at 4°C for 4 h, then washed three times with grinding buffer without bovine serum albumin. Pellets were analyzed by standard SDS-PAGE and subjected to western-blot analysis.
Generation of the Glucocorticoid-Inducible FIN219 Construct and Dex Treatment
The full-length fragment of FIN219 was subcloned into the XhoI site of the pTA7002 plasmid that contains a glucocorticoid-inducible promoter as described previously (Aoyama and Chua, 1997). For phenotype and protein gel-blot analyses, homozygous T4 transgenic seeds were grown on 0.3% GM plates containing ethanol or 1 μm Dex under darkness and cFR light for 3 d. For protoplast analysis, the transfected protoplasts were cultured in W5 solution containing ethanol or 1 μm Dex in the dark.
Production of FIN219 Polyclonal Antibodies
A cDNA fragment corresponding to the N-terminal 300 amino acids of FIN219 was amplified by PCR with primers containing the BamHI site (Supplemental Materials and Methods S1) and the cDNA clone 11-2 as a template and then cloned into the expression vector pCAL-n for protein expression in the E. coli host BL21(DE3). The resulting recombinant proteins were purified by gel electroelution and then sent to the animal center for raising the polyclonal antibodies from rabbits.
Measurement of JA-Ile Levels in Arabidopsis Seedlings
The isolation and quantification of JA-Ile was performed as described (Staswick and Tiryaki, 2004; Suza and Staswick, 2008). About 1.5 to 2 g of 3-d-old FR light-grown seedlings was ground in liquid nitrogen and extracted with 10 mL of 80% methanol containing 10 nm dihydrojasmonic acid (Tokyo Chemical Industry) as internal control and then centrifuged for 10 min at 3,000g. The samples were run through DEAE Sephadex A-25 resins (GE Healthcare) and the SiOH column (Macherey-Nagel). The sample was eluted with 1 column volume of 0.2% acetic acid in ethylacetate and dried in a stream of nitrogen, then dissolved in the isooctane, which was analyzed by an Agilent Technologies 6890N Gas Chromatograph and 5975 Mass Selective Detector with a DB-1 capillary column (30 m × 0.25 mm i.d., 0.25 μm film thickness; J&W Scientific), and the column gradient was 60°C to 300°C in 25.4 min. The standard curve of JA-Ile was obtained from the integrated peak area of the serial concentrations of synthetic JA-Ile and compared with the internal standard. The level of JA-Ile in the samples was then determined by comparing with the standard curve.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Investigation of light-responsive genes and flowering time for FIN219 N-terminal transgenic plants.
Supplemental Figure S2. Physiological responses of the FIN219 N-terminal transgenic seedlings in the presence of MeJA or coronatine.
Supplemental Figure S3. Protein gel-blot analyses of FIN219, COP1, and HY5 levels in various fin219 mutants and FIN219 N-terminal transgenic seedlings under different light conditions.
Supplemental Figure S4. The full-length FIN219 interacts with itself by BiFC.
Supplemental Figure S5. Hypocotyl lengths of transgenic seedlings harboring ectopic expression of the C terminus of FIN219/JAR1 in the NCox or NTox genetic background under cFR light condition.
Supplemental Figure S6. Phenotype and protein gel-blot analysis of FIN219-inducible transgenic seedlings under cFR light.
Supplemental Figure S7. RNA gel-blot analysis of SPA1 transcript levels in wild-type and fin219-1 mutant seedlings.
Supplemental Figure S8. Phenotype and FIN219 expression in the FIN219 C-terminal transgenic seedlings under cFR condition.
Supplemental Table S1. A list of oligonucleotide sequences used in this study.
Supplemental Materials and Methods S1. Gene constructs used in this study, and quantification of northern and western blots.
Supplementary Material
Acknowledgments
We are grateful to the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for the fin219-2 and jar1-1 seeds. We thank Xing-Wang Deng (Yale University) and Hongyong Fu (Academia Sinica, Taiwan) for kind gifts of COP1 and RPN8 antibodies, respectively. Many thanks are given to Tsan-Piao Lin (Institute of Plant Biology, National Taiwan University) for technical assistance with fluorescent images and also to Shih-Han Chou (Taiwan Forestry Research Institute) for technical assistance in the measurement of JA-Ile levels in Arabidopsis. We also thank Huai-Ju Chen (Institute of Plant Biology, National Taiwan University) for critically reading the manuscript.
References
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Ang LH, Deng XW. (1994) Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6: 613–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH. (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14: 1269–1278 [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, Xiong L. (2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA 105: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IC, Huang IC, Liu MJ, Wang ZG, Chung SS, Hsieh HL. (2007) Glutathione S-transferase interacting with far-red insensitive 219 is involved in phytochrome A-mediated signaling in Arabidopsis. Plant Physiol 143: 1189–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP. (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15: 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T. (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C. (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. (2000) RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol 124: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM. (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J 35: 193–205 [DOI] [PubMed] [Google Scholar]
- Guranowski A, Miersch O, Staswick PE, Suza W, Wasternack C. (2007) Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett 581: 815–820 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. (1999) SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499 [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW. (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14: 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13: 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2008) Jasmonate signaling: toward an integrated view. Plant Physiol 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. (1994) Photomorphogenesis in Plants. Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- Kwok SF, Piekos B, Misera S, Deng XW. (1996) A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol 110: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, Torii KU, Deng XW. (1996) Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell 8: 1491–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng XW. (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballaré CL. (2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25: 213–221 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. (2000a) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW. (2000b) The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol 124: 1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M, Bouyer D, Hisada A, Müller A, Yatou O, Weiler EW, Takano M, Furuya M, Nick P. (2009) Phytochrome A requires jasmonate for photodestruction. Planta 229: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Robson F, Okamoto H, Patrick E, Harris SR, Wasternack C, Brearley C, Turner JG. (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW. (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001) SDS-poylacrylamide gel electrophoresis of proteins. Sambrook J, Russell DW, , Molecular Cloning: A Laboratory Manual, Ed 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp A8.40–A8.45 [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. (1998) Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J 16: 411–419 [DOI] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. (2000) REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12: 2061–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML. (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC. (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15: 747–754 [DOI] [PubMed] [Google Scholar]
- Suza WP, Staswick PE. (2008) The role of JAR1 in jasmonoyl-L: -isoleucine production during Arabidopsis wound response. Planta 227: 1221–1232 [DOI] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M. (2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J 37: 471–483 [DOI] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Manabe K, Matsui M. (2003) DFL2, a new member of the Arabidopsis GH3 gene family, is involved in red light-specific hypocotyl elongation. Plant Cell Physiol 44: 1071–1080 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Mochizuki N, Nagatani A. (2002) Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol 43: 281–289 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, Ahmad M. (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J 49: 428–441 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW. (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW. (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR. (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H. (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang H. (2006) The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J 47: 564–576 [DOI] [PubMed] [Google Scholar]
- Yang P, Fu H, Walker J, Papa CM, Smalle J, Ju YM, Vierstra RD. (2004) Purification of the Arabidopsis 26 S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem 279: 6401–6413 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeidler M, Bolle C, Chua NH. (2001) The phytochrome A specific signaling component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol 42: 1193–1200 [DOI] [PubMed] [Google Scholar]
- Zhai Q, Li CB, Zheng W, Wu X, Zhao J, Zhou G, Jiang H, Sun J, Lou Y, Li C. (2007) Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsis. Plant Cell Physiol 48: 1061–1071 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.