Abstract
Piriformospora indica is a root-colonizing basidiomycete that confers a wide range of beneficial traits to its host. The fungus shows a biotrophic growth phase in Arabidopsis (Arabidopsis thaliana) roots followed by a cell death-associated colonization phase, a colonization strategy that, to our knowledge, has not yet been reported for this plant. P. indica has evolved an extraordinary capacity for plant root colonization. Its broad host spectrum encompasses gymnosperms and monocotyledonous as well as dicotyledonous angiosperms, which suggests that it has an effective mechanism(s) for bypassing or suppressing host immunity. The results of our work argue that P. indica is confronted with a functional root immune system. Moreover, the fungus does not evade detection but rather suppresses immunity triggered by various microbe-associated molecular patterns. This ability to suppress host immunity is compromised in the jasmonate mutants jasmonate insensitive1-1 and jasmonate resistant1-1. A quintuple-DELLA mutant displaying constitutive gibberellin (GA) responses and the GA biosynthesis mutant ga1-6 (for GA requiring 1) showed higher and lower degrees of colonization, respectively, in the cell death-associated stage, suggesting that P. indica recruits GA signaling to help establish proapoptotic root cell colonization. Our study demonstrates that mutualists, like pathogens, are confronted with an effective innate immune system in roots and that colonization success essentially depends on the evolution of strategies for immunosuppression.
The success of microbes to invade plants reflects their ability to manipulate immune responses and reprogram host metabolism (O’Connell and Panstruga, 2006). These activities are antagonized in plants by a two-layered immune system. The first induced defense instance relies on the recognition of conserved microbial structures, so-called microbe-associated molecular patterns (MAMPs), and is defined as MAMP-triggered immunity (MTI). Microbes subverting MTI are confronted with the second, more costly but highly efficient effector-triggered immunity. Effector-triggered immunity is activated after recognition of virulence factors, called effectors, or the products of their activity, by extracellular or intracellular receptors (Jones and Dangl, 2006).
Bacterial flagellin, elongation factor-TU, and fungal chitin are well-defined MAMPs that are recognized by the pattern recognition receptors (PRRs) FLS2, EFR, and CERK1, respectively, leading to the activation of MTI (Gómez-Gómez et al., 1999; Kunze et al., 2004; Zipfel et al., 2004; Miya et al., 2007). MAMP perception leads to the activation of mitogen-activated protein kinase (MAPK) pathways and subsequent defense gene expression regulated by transcription factors (e.g. WRKYs). In Arabidopsis (Arabidopsis thaliana), MTI also relies on the accumulation of antimicrobial glucosinolates and camalexin (Bednarek et al., 2009; Clay et al., 2009). Ca2+ contributes to MTI as MAMP-induced Ca2+ influx activates MAPK- and Ca2+-dependent protein kinase signaling (Boudsocq et al., 2010). Ca2+-dependent protein kinases also regulate the production of reactive oxygen species, detectable as the oxidative burst (Boudsocq et al., 2010) that is thought to be generated by the plasma membrane-localized NADPH oxidase RBOHD (Zhang et al., 2007). MTI is further substantiated by the action of phytohormones like salicylic acid (SA), ethylene, and jasmonate (JA; Tsuda et al., 2009). In a simplified model, JA/ethylene protect plants against necrotrophic pathogens, whose lifestyles rely on cell death, while SA is effective against biotrophic plant colonizers (Glazebrook, 2005). Hormonal cross talk greatly affects MTI, as exemplified by GAs, which modulate the SA-JA balance (Navarro et al., 2008). In Arabidopsis, five DELLA proteins function as repressors of GA signaling, among which RGA (for Repressor of ga1-3) takes a dominant role in growth repression. Upon GA synthesis, GA binds to the GA receptor GA Insensitive Dwarf1, which results in the degradation of DELLA proteins and the activation of GA signaling (Sun, 2008; Schwechheimer and Willige, 2009). Mutants lacking DELLA protein and thus displaying constitutive GA signaling have higher SA and reduced JA defense and are more resistant against biotrophic pathogens but hypersusceptible to necrotrophic pathogens (Navarro et al., 2008). In addition, GA obviously affects cell death regulation in plants and thus supports plant colonization by necrotrophic pathogens. Achard et al. (2008) demonstrated that DELLA proteins reduce oxidative stress-induced cell death, which occurs as a result of abiotic and biotic stresses. Consistent with these findings, the negative cell death regulator BOI (for Botrytis Susceptible1 Interactor) was induced in the GA-deficient mutant ga1-3 and suppressed after GA treatment (Luo et al., 2010).
MTI warrants a basic protection, as it determines the nonhost status of plants toward most pathogens (He et al., 2006) and restricts disease progression in compatible interactions (Zipfel et al., 2004). Models of innate immunity are based almost exclusively on analyses of interactions with foliar pathogens, and it is currently unknown to what extent MTI is tissue/organ specific or nonspecific. Compared with other plant organs, roots are confronted with the highest diversity of microbes. Plant health and development are intimately associated with proper nutrient and water acquisition by roots. In crop production, root diseases are of global concern, due to a lack of resistant germplasms and their limited accessibility for chemical protection. Not until recently was the presence of a sensitive MAMP-triggered immune system described in Arabidopsis roots (Millet et al., 2010). However, the organization and protective properties of root innate immunity are almost unknown. Furthermore, we do not know if root surveillance systems discriminate between mutualists and pathogens or if colonization success rather reflects the efficiencies of microbial strategies to deactivate innate immunity.
The root-colonizing basidiomycete Piriformospora indica represents a useful model to study principles of root-symbiont interactions in terms of mutualism and root colonization. As a mutualist, it confers beneficial traits such as increased yield, abiotic stress tolerance, and disease resistance to plants (Varma et al., 1999; Waller et al., 2005). Similar to arbuscular mycorrhizal interactions, fungal phosphate supply to roots is imperative for growth promotion (Yadav et al., 2010). P. indica is an efficient root colonizer, as indicated by its exceptionally broad host range, and a nonhost plant has not yet been discovered. This suggests an enormous capacity to evade or suppress plant innate immune responses.
In this work, we aimed at elucidating the colonization strategy of P. indica and at identifying crucial components of root MTI by taking advantage of its ability to colonize Arabidopsis. By employing transmission electron microscopy and epifluorescence microscopy along with reporter and mutant plants, we describe, to our knowledge for the first time, a biotrophic colonization of Arabidopsis roots by a fungus. This biotrophic stage is followed by a cell death-associated colonization phase. Our genetic and molecular analyses demonstrate the efficiency of the root innate immune system to halt microbial colonization and indicate that mutualistic colonization success, like pathogenic colonization success, is intimately dependent on efficient immune suppression strategies. Furthermore, our data reveal similarities between leaf and root immunity.
RESULTS
The Biphasic Lifestyle of P. indica: Biotrophy followed by Cell Death
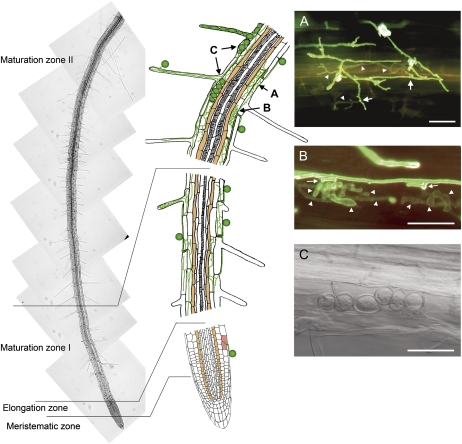
Microscopic studies of Arabidopsis roots reveal the following spatiotemporal chronology of colonization by P. indica (Fig. 1). Colonization started after chlamydospore germination with intercellular and intracellular penetration of rhizodermal and cortical tissues by 2 d after inoculation (dai). By 3 dai, intracellular colonization of rhizodermal, cortical, and root hair cells was prominent (Fig. 1, A and B). Fungal hyphae subsequently branched (Fig. 1A) and occasionally formed whorls (Fig. 1B), as reported for orchid mycorrhiza (Peterson and Massicotte, 2004; Schäfer and Kogel, 2009). Finally, external and intracellular sporulation started in the maturation zones (MZ) I and II at 7 and 14 dai, respectively (Fig. 1C). Growth of P. indica was restricted to rhizodermal and cortical cells, and colonization increased with tissue age and reached its highest level in MZ II. In contrast, meristematic and elongation zones remained free of hyphae except for the occasional infection of an epidermal cell.
Figure 1.
Colonization of an Arabidopsis root by P. indica in the meristematic zone, elongation zone, and MZ. Colonization is restricted to rhizodermal, root hair, and cortical cells, while the root endodermis (brown cells) and root vasculature are not colonized. The letters and arrows in the middle panel point to interaction sites displayed in images A to C. A, Epifluorescence image of epidermal root cells that are penetrated (arrows) without specialized penetration organs. Intracellular hyphae (arrowheads) show a branched morphology. B, Epifluorescence image of intracellular hyphae that are characterized by a distinct globular structure (arrowheads). Penetration sites are indicated by arrows. C, Bright-field image of intracellular sporulation that starts around 14 dai in MZ II. Fungal hyphae were stained with WGA-AF488. Bars = 20 μm.
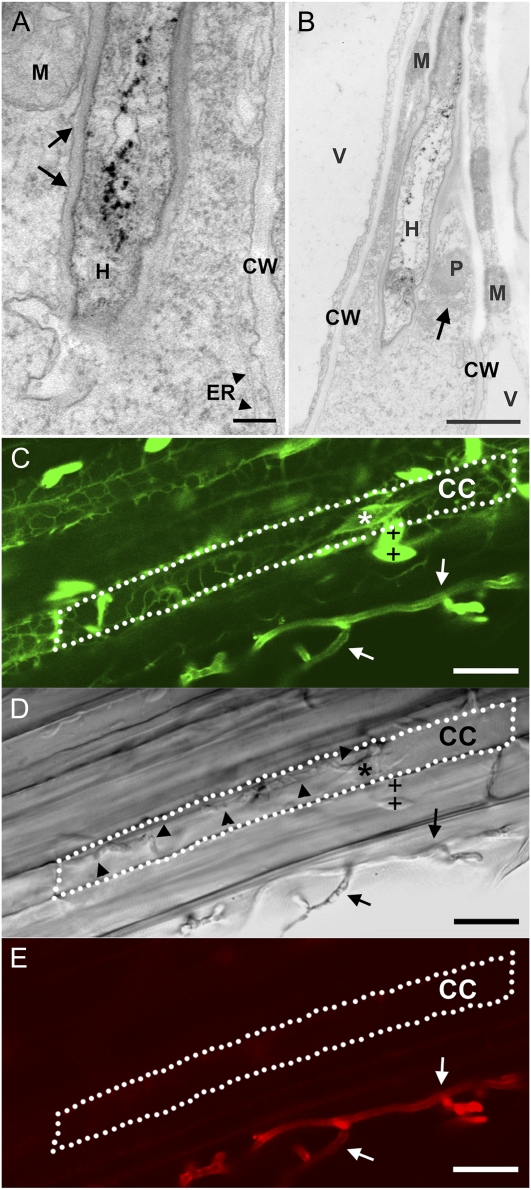
Transmission electron microscopy revealed that intracellular colonization is initiated by plasma membrane invagination (Fig. 2A). The well-preserved ultrastructure of the cytosol, plasma membrane, and all organelles at early colonization stages (3 dai) strongly suggested that colonized cells were alive (Fig. 2, A and B). Biotrophic colonization by P. indica was further substantiated by using Arabidopsis line GFP-Chi, in which the GFP is attached to a vacuolar sorting determinant, thereby visualizing endoplasmic reticulum (ER), nucleus, and ER bodies (Flückiger et al., 2003). The integrity of the ER and nucleus was confirmed in living cells of MZ II by confocal microscopy at 3 dai (Fig. 2, C–E; Supplemental Video S1). Further evidence for cell integrity was provided by the absence of staining with chitin-specific dyes WGA-AF488 and WGA-633 of intracellular hyphae (Fig. 2D), since both extracellular hyphae (Fig. 2, C and E) and intracellular hyphae in dead cells, lacking an intact plasmalemma, were stained (Supplemental Fig. S1). In contrast, at later stages of interaction (more than 3 dai), cell death was frequently observed in colonized cells of MZ II, as indicated by the absence of ER and nucleus as recorded in GFP-Chi plants (Supplemental Fig. S1). Although we could not determine whether cell death always followed biotrophic colonization, it did in most cases. Significantly, cell death was restricted to colonized cells, as adjacent noncolonized rhizodermal cells displayed intact nuclei and ER (Supplemental Fig. S1). The subcellular sequence of cell death started with the disintegration of cytoplasm and the endomembrane system (Supplemental Fig. S2A). The plasma membrane, while still present, showed inversions (Supplemental Fig. S2B) reminiscent of membrane blebbing (in sensu; Mittler et al., 1997). Adjacent noncolonized cells were intact, as demonstrated by well-preserved cytoplasm (Supplemental Fig. S2C). Tissue necrosis or browning of colonized root areas did not occur (data not shown), nor was cell death accompanied by whole cell autofluorescence.
Figure 2.
Early biotrophic stages of the Arabidopsis-P. indica interaction. A and B, Transmission electron micrographs show intact root cells of Arabidopsis ecotype Col-0 colonized with fungal hyphae (H). Root cells with dense cytosol, intact plastids (P), mitochondria (M), vacuoles (V), ER (arrowheads in A), dictyosome (arrow in B), and cell walls (CW) are shown. A, Parts of a root cell with the plasma membrane closely surrounding the hyphae (arrows). B, Biotrophic colonization by P. indica in MZ II at 3 dai. C to E, Confocal microscopy of a living cortical cell (CC) from colonized Arabidopsis line GFP-Chi with GFP-tagged ER, ER bodies (+), and nucleus (*). See Supplemental Video S1 of this interaction site. Extracellular (arrows) but not intracellular (arrowheads) hyphae are stained with WGA-AF488 in C and with WGA-AF633 in E. Bars = 0.2 μm (A), 1 μm (B), and 15 μm (C–E).
P. indica Suppresses a Conserved Set of Tissue-Nonspecific MTI Responses
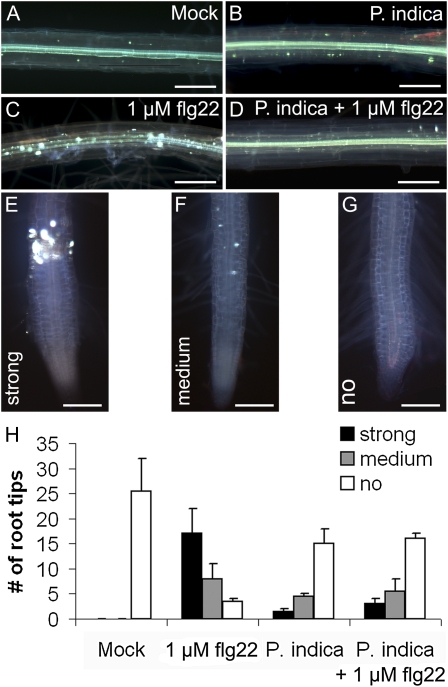
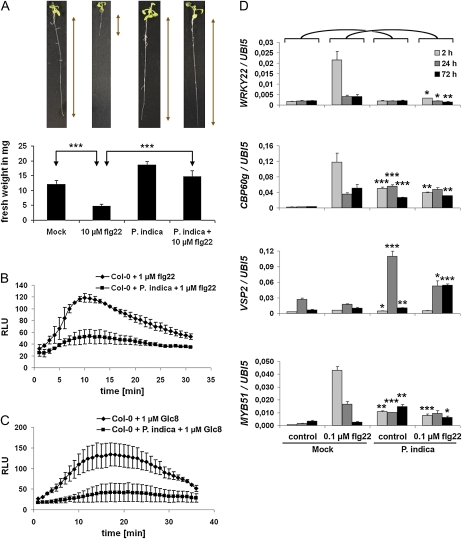
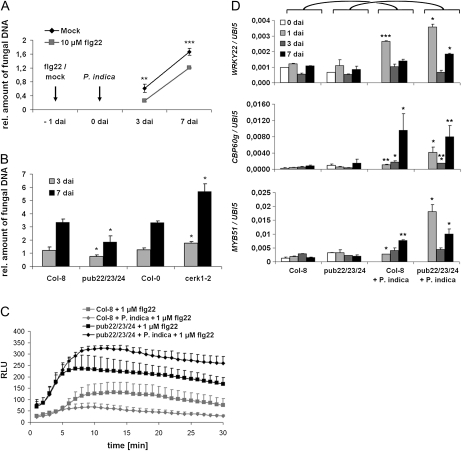
In leaves, invasion by biotrophic fungi is primarily controlled by locally confined cell wall appositions and single-cell hypersensitive responses (HRs; Lipka et al., 2005; Hückelhoven, 2007). In clear contrast, we rarely detected cell wall appositions or HR-like responses during biotrophic root colonization (Supplemental Fig. S3). This observation prompted us to test whether P. indica evades or suppresses basal immune responses. In leaves, the active epitopes of bacterial flagellin (flg22), elongation factor-TU (elf18), or the octamer of fungal chitin (N-acetylchitooctaose; Glc8) trigger the oxidative burst and induce defense gene transcription (Gómez-Gómez et al., 1999; Kunze et al., 2004; Zipfel et al., 2004; Miya et al., 2007). In addition, plantlets treated with flg22 or elf18 exhibit growth inhibition. Accordingly, a series of experiments were conducted in which P. indica-colonized roots were treated at 3 dai (biotrophic stage) with flg22. In a first assay, we treated roots that were either colonized or noncolonized by P. indica at 3 dai with 1 μm flg22. In contrast to mock (buffer)-treated controls, we observed callose deposition in all root zones, including the MZ, after flg22 treatment (Fig. 3). Callose deposition was abolished in P. indica-colonized root segments (Fig. 3D). Moreover, P. indica abolished the flg22-mediated growth inhibition, as indicated by unimpaired seedling root length and fresh weight as compared with noncolonized controls (Fig. 4A). We specifically used the MZ of roots, which are preferentially colonized by P. indica, for subsequent oxidative burst and gene expression studies. As in leaves, flg22 also induced a transient oxidative burst in noncolonized roots. Again, P. indica-colonized roots were almost completely nonresponsive to this MAMP (Fig. 4B). To assess the range of MTI suppression by the fungus, we analyzed the sensitivity of colonized roots to Glc8 and elf18. We found that P. indica is also able to abolish Glc8- and elf18-induced oxidative burst as well as elf18-mediated seedling growth inhibition (Fig. 4C; Supplemental Fig. S4, A and B). Next, we tested whether suppression of MAMP-triggered responses was also evident on the basis of defense gene expression. Flg22 was used for these experiments, since it is the best-studied MAMP in planta. To this end, roots colonized for 3 dai with P. indica were treated with flg22, harvested at 2, 24, and 72 h after treatment (hat), and analyzed for transcriptional activation of marker genes by quantitative real-time PCR (qRT-PCR). Consistent with earlier results, marker genes for MTI (WRKY22, WRKY33, WRKY53; Colcombet and Hirt, 2008), oxidative stress (OXI1; Rentel et al., 2004; Torres et al., 2005), and SA (CBP60g, SID2; Wang et al., 2009) were induced by flg22 at 2 or at 2 and 24 hat in noncolonized roots (Fig. 4D; Supplemental Fig. S5). In addition, MYB51, which participates in the biosynthesis of antimicrobial indole glucosinolates (Clay et al., 2009), was induced. In marked contrast, the flg22-induced transcription of all tested genes was suppressed by P. indica. Interestingly, however, VSP2, which is a marker for JA signaling, was strongly up-regulated in P. indica-colonized roots in the absence (at 24 hat; mock) and presence (at 24 and 72 hat) of flg22 (Fig. 4D).
Figure 3.
Suppression of flg22-triggered callose deposition by P. indica in Arabidopsis roots during biotrophic colonization (3 dai). Callose deposition in the MZ (A–D) and elongation/differentiation zone (E–G) of flg22- or mock-treated roots is shown. A to D, Noncolonized (A and C) or P. indica-colonized (3 dai; B and D) roots of 2-week-old seedlings were mock treated (A and B) or treated with 1 μm flg22 (C and D). Callose deposition was only observed in flg22-treated noncolonized roots (C) and almost absent in mock-treated (A and B) and flg22-treated (D) P. indica-colonized roots. E to G, Three types of callose deposition (strong, medium, no) were observed in root tips dependent on flg22 treatment and P. indica colonization. H, The occurrence of callose deposition in root tips in response to the various treatments was quantified. Bars = 200 μm. [See online article for color version of this figure.]
Figure 4.
P. indica suppresses MAMP-triggered growth retardation, oxidative burst, and gene transcription during biotrophic colonization (3 dai). For all analyses, MAMPs were applied to 2-week-old plants at 3 d after P. indica inoculation. A, Suppression of flg22-induced growth retardation by P. indica (se values are from 10 independent measurements of one biological experiment). Experiments were repeated three times with similar results. B and C, Suppression of flg22- and Glc8-induced root oxidative burst by P. indica. Values are given as relative light units (RLU) over time (se values are from four independent measurements per treatment in one experiment). Experiments were repeated three times with similar results. D, Suppression of flg22-induced gene transcription determined by qRT-PCR. Three days after P. indica inoculation or mock treatment, roots were treated with flg22 or mock and harvested 2, 24, and 72 h after treatments. Suppression of flg22-induced transcription was observed for WRKY22 (MTI marker), CBP60g (SA marker), and MYB51 (marker for antimicrobial glucosinolates). VSP2 (JA marker) was induced in P. indica-colonized roots after mock and flg22 treatment. See Supplemental Figure S5 for additional genes. The values represent means with se of one experiment. Experiments were repeated at least twice with similar results. Asterisks indicate significant differences at P < 0.05 (*), 0.01 (**), and 0.001 (***). For D, significant differences between individual time points of Col-0/mock and Col-0/P. indica or Col-0/flg22 and Col-0 + P. indica/flg22 were analyzed by Student’s t test. [See online article for color version of this figure.]
MAMPs of the Mutualist P. indica Can Activate a Highly Effective Root MTI
Our results revealed that P. indica efficiently suppresses MAMP-triggered responses. We subsequently wanted to elucidate the significance of root MTI in the mutualistic symbiosis. Two-week-old seedlings were flg22 or mock treated at 1 d prior to inoculation. Fungal growth was quantified during biotrophic (3 dai) and cell death-associated (7 dai) colonization stages. Flg22 treatment reduced fungal colonization at both time points (Fig. 5A), indicating that root MTI is effective against P. indica. To further substantiate this finding, we employed two mutants with altered MTI responses. First, the plant U-box-type E3 ubiquitin ligase (PUB) triple mutant pub22/23/24 was tested. In this triple mutant, although it does not exhibit a constitutive defense response, MTI is hyperactivated after MAMP application (Trujillo et al., 2008). Colonization of pub22/23/24 by P. indica was significantly reduced in comparison with its parent line ecotype Columbia-8 (Col-8; Fig. 5B). By contrast, the chitin elicitor receptor kinase1-2 (cerk1-2) mutant, which is impaired in the perception of chitin and a yet unidentified danger signal associated with bacterial attack (Gimenez-Ibanez et al., 2009), allowed higher colonization (Fig. 5B). Next, the possibility that reduced colonization of pub22/23/24 is a result of P. indica’s inability to suppress MTI in this mutant was assessed. Consistent with this possibility, the flg22-triggered oxidative burst was not abolished but rather elevated in P. indica-colonized pub22/23/24 roots as compared with the wild-type Col-8 (Fig. 5C; Supplemental Fig. S6). Furthermore, pub22/23/24 roots showed a transient oxidative burst in response to direct treatment with P. indica chlamydospores that was not observed in wild-type Col-8 (Supplemental Fig. S6C). We also analyzed the expression of marker genes in pub22/23/24 and wild-type Col-8 roots at 1, 3, and 7 d after P. indica or mock inoculation. As expected, MTI markers were not induced in noncolonized roots of pub22/23/24. While WRKY22 and OXI1 were moderately up-regulated at 1 dai, WRKY33, CBP60g, and MYB51 were most strongly up-regulated at 7 dai in wild-type Col-8. All tested genes displayed a stronger induction in pub22/23/24 that was most obvious at 1 or at 1 and 7 dai (Fig. 5D; Supplemental Fig. S7). Interestingly, in pub22/23/24, the increased expression of the SA markers CBP60g and SID2 (especially at 7 dai) coincided with the reduced expression of the JA marker VSP2 (Supplemental Fig. S7).
Figure 5.
MAMP-triggered immunity restricts colonization of Arabidopsis roots by P. indica. Three-week-old plants were inoculated with P. indica, and fungal biomass was determined during biotrophic (3 dai) and cell death-associated (7 dai) colonization stages by qRT-PCR. A, Col-0 roots were treated with 10 μm flg22 or mock treated 1 d prior to P. indica inoculation. flg22 pretreatment led to a reduced colonization at 3 and 7 dai (se values are from two independent experiments with 200 plants per treatment and time point). B, Reduced colonization of the MAMP-hyperresponsive triple mutant pub22/23/24 and enhanced root colonization of the chitin-insensitive mutant cerk1-2 [three independent experiments with 200 plants per mutant, wild type, and time point]. C, flg22-induced root oxidative burst in the pub22/23/24 mutant is not suppressed by P. indica. Values are given as relative light units (RLU) over time (se values are from four independent measurements per treatment in one experiment). The experiment was repeated three times with similar results. D, Failed suppression of root defense by P. indica in mutant pub22/23/24. Three-week-old Col-8 (wild type) or the pub22/23/24 mutant was inoculated with P. indica and analyzed at 0, 1, 3, and 7 dai with qRT-PCR for transcription of WRKY22 (MTI marker), CBP60g (SA marker), and MYB51 (marker for antimicrobial glucosinolates). Expression values were calculated by the 2−ΔCt method by relating Ct values of candidates to those of the housekeeping gene AtUbiquitin5. The values represent means with se and are based on at least two independent biological experiments. Asterisks indicate significant differences at P < 0.05 (*), 0.01 (**), and 0.001 (***) between individual time points of Col-8/mock and Col-8/P. indica or pub22/23/24/mock and pub22/23/24/P. indica and analyzed by Student’s t test.
Role of Phytohormones in Root Colonization by P. indica
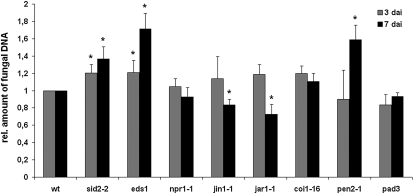
The induction of SA, JA, and glucosinolate marker genes by P. indica prompted us to determine their significance for symbiosis. To this end, fungal biomass was quantified in SA signaling mutants nonexpressor of PR1 (npr1-1) and enhanced disease susceptibility1 (eds1), the SA biosynthesis mutant salicylic acid inducible defective2-2 (sid2-2), the JA signaling mutants jasmonate insensitive1-1 (jin1-1) and coronatine insensitive1-16 (coi1-16), and the JA biosynthesis mutant jasmonate resistant1-1 (jar1-1). Except for npr1-1, all SA mutants showed higher colonization at 3 and 7 dai (Fig. 6). By contrast, root colonization was reduced in jar1-1 and jin1-1 at 7 dai (Fig. 6), while coi1-16 colonization was comparable to the wild type. coi1-16 bears a second mutation in PEN2 (Westphal et al., 2008), which might compensate the expected decrease in colonization. In support of this possibility, pen2-1 displayed higher colonization at 7 dai (Fig. 6). In contrast, phytoalexin deficient3 (pad3), which shows a marked reduction in the synthesis of antimicrobial camalexin (Glazebrook and Ausubel, 1994), was colonized as efficiently as wild-type roots.
Figure 6.
SA and glucosinolate defense restrict colonization of Arabidopsis roots by P. indica. Three-week-old plants were inoculated with P. indica, and fungal biomass was determined during biotrophic (3 dai) and cell death-associated (7 dai) colonization by qRT-PCR. For all mutant experiments, the relative amount of fungal biomass was related to the wild type (wt; set to 1). Results shown are means of at least three independent experiments. For each experiment, 200 plants were analyzed per mutant or wild type and per time point. Asterisks indicate significant differences at P < 0.05 (*) analyzed by Student’s t test.
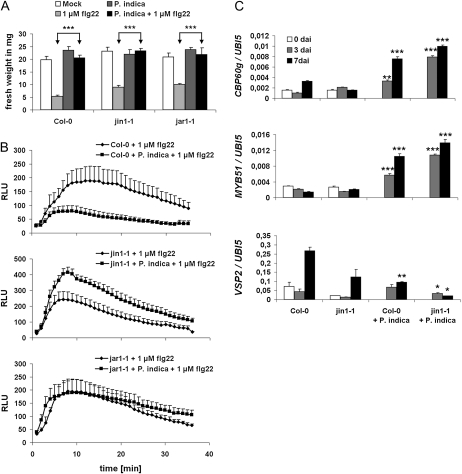
We further assessed whether reduced colonization of jar1-1 and jin1-1 was associated with an impaired ability of P. indica to suppress MAMP-triggered responses. While P. indica maintained its potential to suppress flg22-triggered growth inhibition in jin1-1 and jar1-1 (Fig. 7A), the fungus was completely unable to suppress flg22-triggered root oxidative burst in both mutants (Fig. 7B). Significantly, root oxidative burst was even higher in P. indica-colonized jin1-1 roots compared with the noncolonized flg22-treated mutant. To substantiate this finding, we tested whether reduced colonization of jin1-1 roots was associated with elevated immune-related gene expression. We found enhanced expression of MYB51 and the SA marker CBP60g in jin1-1 compared with Col-0 roots, especially during the biotrophic colonization phase (3 dai; Fig. 7C). Consistent with the induction of CBP60g, JA-mediated gene expression was suppressed, as indicated by the reduced expression of VSP2, which is known to be negatively regulated by SA (Fig. 7C).
Figure 7.
JA is required for root MTI suppression by P. indica. For all analyses, MAMPs were applied to 2-week-old plants at 3 d after P. indica inoculation. A, Suppression of flg22-induced growth retardation by P. indica in jin1-1 and jar1-1 (se values are from 10 independent measurements of one biological experiment). Experiments were repeated twice with similar results. B, P. indica is unable to suppress flg22-induced root oxidative burst in jar1-1 and elevates the oxidative burst in jin1-1 roots. Values are given as relative light units (RLU) over time (se values are from four independent measurements per treatment of one experiment). Experiments were repeated three times with similar results. C, Three-week-old Arabidopsis plants (Col-0, jin1-1) were mock treated or inoculated with P. indica and harvested at 0, 3, and 7 d after treatments. CBP60g and MYB51 transcription were enhanced in jin1-1, while VSP2 expression was reduced in P. indica-colonized roots. Expression values were calculated by the 2−ΔCt method by relating Ct values of candidates to those of the housekeeping gene AtUbiquitin5 (se values are from three technical replicates of one biological experiment). Experiments were repeated at least twice with similar results. Asterisks indicate significant differences at P < 0.05 (*), 0.01 (**), and 0.001 (***). For C, significant differences between individual time points of Col-0/mock and Col-0/P. indica or jin1-1/mock and jin1-1/P. indica were analyzed by Student’s t test.
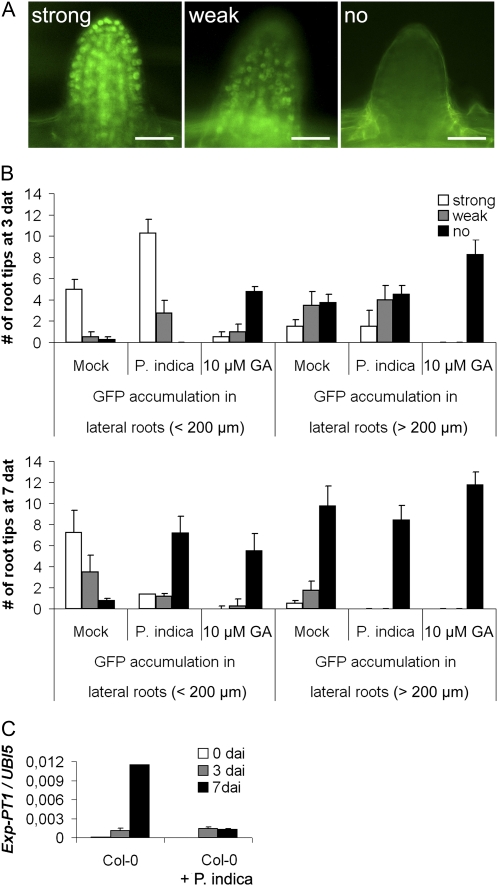
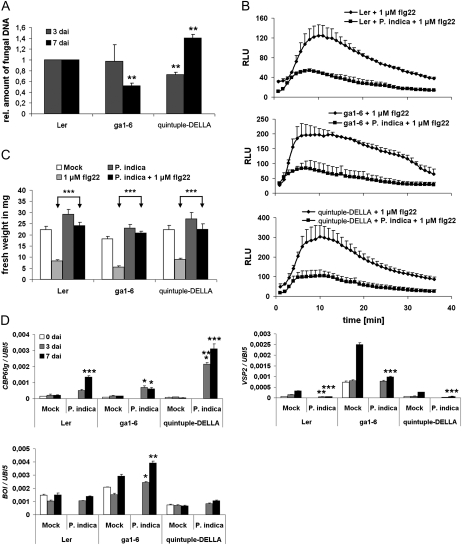
In previous studies, we found that GA-deficient barley (Hordeum vulgare) mutants showed a reduced colonization by P. indica that was accompanied by elevated defense gene expression (Schäfer et al., 2009). Consistent with the above result, GA was reported to balance SA and JA defense and affect MAMP-triggered responses (Navarro et al., 2008). These findings prompted us to analyze the impact of GA on fungal colonization and root MTI in Arabidopsis. First, we determined if P. indica affects GA signaling. Therefore, we recorded degradation of the DELLA protein RGA in response to colonization with P. indica and to GA treatment. We observed enhanced RGA degradation in root tips both after GA treatment and during cell-death associated colonization (7 dai) but not biotrophic colonization (3 dai; Fig. 8, A and B). Consistent with this, the GA marker gene Exp-PT1, which is suppressed by GA (Zentella et al., 2007), was down-regulated in P. indica-colonized roots at 7 dai (Fig. 8C). Next, the effect of GA on root colonization was examined by employing the mutants GA requiring1 (ga1-6) and quintuple-DELLA. ga1-6 is disturbed in GA synthesis but has only a mild dwarf phenotype, which favors its use in root colonization studies over mutants exhibiting strong root stunting due to high GA deficiency. Roots of ga1-6 were less colonized at 7 dai. By contrast, the quintuple-DELLA mutant, which lacks all five DELLA proteins and therefore shows constitutive GA signaling, displayed reduced colonization at 3 dai but enhanced colonization at 7 dai (Fig. 9A). Moreover, both mutants did not disturb P. indica’s ability to abolish seedling growth inhibition or to suppress the flg22-triggered root oxidative burst (Fig. 9, B and C). We observed the highest CBP60g expression in quintuple-DELLA in response to P. indica colonization, while its expression was less strongly induced in ga1-6 as compared with ecotype Landsberg erecta (Ler; Fig. 9D). Both mutants also behaved differently in JA-related defense gene expression. In quintuple-DELLA, VSP2 transcription was suppressed just as observed in wild-type Ler in response to P. indica. On the other hand, VSP2 was strongly induced in mock-treated and P. indica-colonized ga1-6 roots (Fig. 9D).
Figure 8.
GA signaling is activated during P. indica colonization. A and B, Two-week-old plants expressing the GFP fusion of the DELLA protein RGA under the control of its native promoter (RGAp::RGA-GFP) were inoculated with P. indica. The degree of GFP fluorescence was determined at 3 and 7 dai, mock treatment, or GA3 treatment. GFP fluorescence was determined in root tips (A) of short (less than 200 μm in length) and long (more than 200 μm in length) lateral roots and counted (B; se values are from 20–30 root tips per root and treatment, and four roots were analyzed). Experiments were repeated twice with similar results. Bars = 30 μm. C, The expression of the marker gene for GA signaling, Exp-PT1 (At2g45900), was quantified in P. indica-colonized and mock-treated roots at 0, 3, and 7 d after treatment (dat) by qRT-PCR. Expression values were calculated by the 2−ΔCt method by relating Ct values of candidates to those of the housekeeping gene AtUbiquitin5 (se values are from technical replicates of one biological experiment). Experiments were repeated twice with similar results.
Figure 9.
Effects of GA on MAMP-triggered responses and root colonization. A, Three-week-old Arabidopsis plants (Ler, ga1-6, quintuple-DELLA) were inoculated with P. indica and harvested at 3 and 7 dai. Colonization of ga1-6 roots was reduced at 7 dai. Colonization of quintuple-DELLA mutant roots was reduced at 3 dai and enhanced at 7 dai. P. indica colonization of roots was determined by qRT-PCR (se values are from at least two biological experiments, and 200 plants were analyzed per mutant or wild type and per time point). Asterisks indicate significant differences at P < 0.01 (**) analyzed by Student’s t test. B and C, MAMPs were applied to 2-week-old plants at 3 dai with P. indica. B, Suppression of flg22-induced growth retardation by P. indica in ga1-6 and quintuple-DELLA mutants (se values are from 10 independent measurements of one biological experiment). Experiments were repeated twice with similar results. C, Suppression of flg22-induced root oxidative burst in ga1-6 and quintuple-DELLA mutants by P. indica. Values are given as relative light units (RLU) over time (se values are from four independent measurements per treatment of one experiment). Experiments were repeated three times with similar results. D, GA metabolism affects the expression of CBP60g (SA marker), VSP2 (JA marker), as well as BOI (negative cell death regulator). Expression values were calculated by the 2−ΔCt method by relating Ct values of candidates to those of the housekeeping gene AtUbiquitin5 (se values are from technical replicates of one biological experiment). Experiments were repeated at least twice with similar results. Asterisks indicate significant differences at P < 0.05 (*), 0.01 (**), and 0.001 (***). For D, significant differences between individual time points of Ler/mock and Ler/P. indica, ga1-6/mock and ga1-6/P. indica, or quintuple-DELLA/mock and quintuple-DELLA/P. indica were analyzed by Student’s t test.
GA was formerly reported to suppress the expression of the negative cell death regulator BOI (Luo et al., 2010), which prompted us to test BOI expression in the GA mutants. We found higher BOI expression in mock-treated ga1-6 roots as compared with wild-type Ler roots, and BOI expression was further elevated during P. indica colonization in ga1-6 (Fig. 9D). In contrast, BOI expression was weakly down-regulated in quintuple-DELLA. These findings are consistent with the concept that GA might function as a proapoptotic factor that supports cell death-associated colonization of roots by P. indica.
DISCUSSION
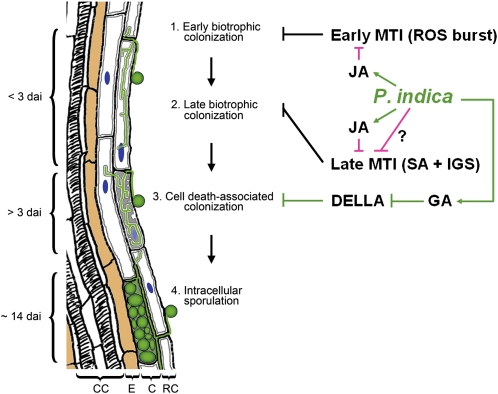
Based on the presented data, we propose a root colonization model for P. indica consisting of four consecutive stages (Fig. 10): (1) extracellular colonization of the root surface (approximately 1 dai); (2) biotrophic colonization phase (less than 3 dai), where hyphae colonize living rhizodermal and cortical cells (Fig. 2; Supplemental Video S1); (3) cell death-associated colonization phase (more than 3 dai); and (4) fungal reproduction by extracellular (approximately 7 dai) and intracellular (approximately 14 dai) sporulation. In the biotrophic colonization phase, hyphae grow into cells but remain extracytosolic through invagination of the plasma membrane. During the biotrophic phase, there are no ultrastructural changes (such as lysis of the cytosol or ruptured tonoplast); these start to occur at the cell death-associated colonization stage (Supplemental Figs. S1 and S2). This biotrophic colonization demonstrates that early fungal development is not dependent on dead cells, nor is cell death a prerequisite for penetration. Importantly, biotrophic colonization did not coincide with any defense responses. Furthermore, several findings argue against an immunity-related cell death but rather suggest a compatibility-associated role of cell death: (1) the virtual absence of whole cell autofluorescence or browning caused by the accumulation of phenolic and antimicrobial compounds, as reported for HR cells (Supplemental Fig. S3, G and H; Heath, 2000); (2) the absence of HR hallmarks such as mitochondria swelling, increased vesicle formation, vacuolization of the cytoplasm, and protoplast shrinkage (Heath, 2000; Mur et al., 2008); and (3) the nondeleterious impact of cell death on P. indica colonization, as reported for barley (Deshmukh et al., 2006) and indicated by transcellular fungal growth (Supplemental Fig. S2). Together, these findings suggest exquisite adaptation of P. indica to Arabidopsis roots.
Figure 10.
Model of the spatiotemporal colonization pattern of Arabidopsis roots. Root colonization by P. indica can be divided into four stages. After germination of the spores and extracellular growth, hyphae penetrate epidermal or cortical cells and establish an early biotrophic colonization phase. Biotrophic stages can be preceded by intercellular colonization. The early and late biotrophic stages are characterized by complete intactness of the cell organelles (e.g. nucleus; blue) and plasma membrane invagination (dark gray lines inside cells). Biotrophically colonized cells die (light gray filling of cells) during subsequent cell death-associated colonization. Host cell death is indicated by organelle disruption, while the plasma membrane (dark gray lines inside cells) still surrounds intracellular hyphae. Intracellular sporulation takes place in epidermal and cortical cells at about 14 dai. Endodermis cells (brown) are not colonized. CC, Central cylinder; E, endodermis; C, cortex; RC, rhizodermal cells. MTI is restricting root colonization by P. indica from early through late interaction stages. The fungus achieves biotrophic root colonization by the suppression of early MTI. SA-mediated defense and antimicrobial indole glucosinolates (IGS) participate in MTI. P. indica recruits JA to suppress root oxidative burst. As indicated by mutant studies (Figs. 6 and 7) and gene expression profiles (Fig. 5D), SA and indole glucosinolates might take a dominant role at later colonization stages, at which P. indica might recruit JA signaling and other yet to be defined pathways to counteract SA-supported MTI. P. indica might further induce GA signaling to achieve DELLA protein degradation, thereby elevating the proapoptotic threshold in root cells and initiating cell death-associated colonization. ROS, Reactive oxygen species.
The intracellular colonization of living cells involves an extended exposure of fungal structures to plasma membrane-localized PRRs and the subsequent activation of immune responses such as MTI. The ubiquitous expression of PRRs, such as FLS2, throughout different tissues (Robatzek et al., 2006) suggests that signaling processes are conserved in leaves and roots. The first evidence supporting this proposition was provided by Millet et al. (2010); further characterization of root MTI is presented here (Figs. 3 and 4; Supplemental Fig. S5). We demonstrate that P. indica efficiently suppressed the immune response triggered by various MAMPs (e.g. chitin, flg22, elf18). We decided to use flg22 for our studies as it represents the best-studied MAMP in leaves and thus allows a more detailed comparison of leaf with root MTI. In contrast to Millet et al. (2010), we observed flg22-induced callose deposition in all root zones, including the MZ (Fig. 3C). This might be explained by the different incubation times after flg22 treatment (18 versus 24 h) and/or the age of treated seedlings (10 versus 18 d). P. indica not only suppressed flg22-induced callose deposition in colonized root zones (e.g. MZ) but also in the elongation/differentiation zones, which generally are not colonized by the fungus. This implies that the fungus either releases highly diffusible metabolites or affects host metabolism in a systemic manner in order to suppress immune responses even in noncolonized areas. Consistent with this, we found RGA degradation in noncolonized root tips (Fig. 8, A and B). Since the jin1-1 mutant exhibited an enhanced immune response and an elevated root oxidative burst in response to P. indica colonization (Fig. 7), JA signaling contributes at least partially to the suppression of root MTI. The fungus may benefit from root-wide immune suppression, as immune activation also results in the production of antimicrobial metabolites whose diffusion to the site of penetration could disturb P. indica growth. The activation of MAMP-triggered responses in wild-type roots by flg22 (Fig. 4) or in pub22/23/24 roots by P. indica (Fig. 5) argues that leaves and roots have similar immune systems. P. indica effectively counteracts immune signaling, as seen by the abolishment of MAMP-induced seedling growth inhibition (Fig. 4A; Supplemental Fig. S4A) as well as by the suppression of the MAMP-induced oxidative burst (Fig. 4, B and C; Supplemental Fig. S4B) and gene expression (Fig. 4D; Supplemental Fig. S5). The suppression of flg22-triggered WRKY22, WRKY33, and WRKY53 transcription in colonized roots suggests that the fungus impairs a broad set of genes regulated by MAPK-mediated signaling pathways (Colcombet and Hirt, 2008). MTI suppression by P. indica affects both early (oxidative burst) and later gene induction and immune responses, which occur in distinct subcellular locations. Significantly, P. indica nullifies canonical immune responses usually triggered by both bacterial (flg22, elf18) and fungal chitin (Fig. 4; Supplemental Fig. S4). These MAMPs activate a similar set of MAPKs and induce an overlapping gene set (Wan et al., 2008). Therefore, P. indica’s target is probably an early step in the immune signaling cascade(s). One possibility would be deactivation of a major hub that integrates multiple PRR signals. Alternatively, P. indica may secrete a plethora of effectors, which are functional in distinct and evolutionary distantly related hosts, to silence multiple key components required by different MAMP-triggered responses. Since ubiquitin ligases PUB22, PUB23, and PUB24 negatively regulate leaf MTI, reduced colonization and enhanced MTI activation in pub22/23/24 argue against P. indica impairing PRR function. These U-box proteins are thought to control PRR-derived signaling upstream of the MAPK cascade. By targeting the activity of the ligases or immediate downstream signaling components, P. indica would be able to impair defense signaling. Irrespective of the existence of a major or several immune signaling integrator(s), these targets should be conserved in P. indica hosts. Supporting this view, we observed suppression of defense gene expression and chitin-induced oxidative burst by P. indica in barley roots (Schäfer et al., 2009; K.H. Kogel and P. Schäfer, unpublished data).
Only a few recent studies have described the existence of root MTI (Attard et al., 2010; Millet et al., 2010). Our study provides further evidence for the involvement of MTI in halting microbial root colonization, as shown by the reduced colonization of either flg22-pretreated roots or pub22/23/24 roots, and enhanced colonization of cerk1-2 (Fig. 5, A and B). The reduced colonization of flg22-treated Col-0 and failed inhibition of MTI in pub22/23/24 suggest that P. indica follows the strategy of suppression rather than evasion of MTI. They further indicate that the perception of MAMPs released by the mutualistic symbiont triggers immunity similar to pathogen MAMPs. Consistent with this, we observed a transient oxidative burst in pub22/23/24 roots even after application of P. indica chlamydospores (Supplemental Fig. S6C). pub22/23/24 is an appropriate tool to detect such responses, as it does not show elevated MTI under normal growth conditions (Trujillo et al., 2008; Fig. 5D). Reduced pub22/23/24 colonization correlated with an elevated oxidative burst and stronger induction of MAPK-activated WRKY22, WRKY33, and WRKY53, SA defense-related CBP60g, glucosinolate defense-associated MYB51, and OXI1, which participates in reactive oxygen species metabolism. This might reflect elevated signaling activities of existing receptors or an elevated de novo synthesis of receptors as a consequence of P. indica colonization. Alternatively, it is tempting to speculate that the PUB triplet is targeted by effector proteins, which mediate their stabilization in a similar manner to Phytophthora infestans AVR3a stabilizing the U-box protein CMPG1, thereby suppressing host cell death during biotrophic colonization (Bos et al., 2010).
In leaves, SA, JA, camalexin, and glucosinolates mediate MTI (Bednarek et al., 2009; Tsuda et al., 2009). This might also hold true for root MTI, as suggested by the improved colonization of eds1, sid2-2, and pen2 (Fig. 6). In contrast to our prior results (Stein et al., 2008), npr1-1 did not exhibit an altered colonization in five independent experiments. Since previously our system was prone to root injury during soil detachment from Arabidopsis roots prior to P. indica inoculation, we used here an agar-based screening system, thereby eliminating root injuries and also enhancing screening sensitivity. Thus, SA-mediated defense against the fungus is NPR1 independent (Fig. 6), as has been reported for dnd1, cpr1, cpr5, cpr6, and cpr30, which exert a constitutive SA defense (Clarke et al., 2000; Genger et al., 2008; Gou et al., 2009). In contrast, camalexin and JA, which are generally involved in protection against necrotrophic pathogens (Thomma et al., 1998), appear not to be root MTI components against P. indica, as indicated by unaltered pad3 and reduced (not enhanced) colonization of jar1-1 and jin1-1. Considering the inability of P. indica to suppress flg22-triggered root oxidative burst in both JA mutants and the elevated induction of MYB51 and CBP60g in P. indica-colonized jin1-1 roots (Fig. 7, B and C), the fungus might recruit JA signaling to counterbalance SA and even MTI defense. JA-SA antagonism is known from studies in coi1 and jin1, where both mutants displayed an increased SA signaling after bacterial challenge (Kloek et al., 2001; Laurie-Berry et al., 2006). In leaves, GAs were observed to balance SA-JA metabolism (Navarro et al., 2008) by reducing JA in favor of SA signaling and, accordingly, mediating hypersusceptibility against necrotrophic (and thus cell death-dependent) fungi and resistance against biotrophic pathogens (Navarro et al., 2008). We consistently found that increased GA signaling in quintuple-DELLA mutant roots resulted in the induction of the SA marker CBP60g at 3 and 7 dai, which might explain the reduced colonization of this mutant during the early biotrophic phase (3 dai). However, despite elevated SA defense, quintuple-DELLA mutant roots were hypersusceptible for P. indica at the cell death-associated colonization phase (7 dai; Fig. 9A). Recent studies indicated an essential function of DELLA proteins in plant survival under unfavorable environments (Achard et al., 2008). Interestingly, Luo et al. (2010) identified the RING E3 ligase BOI as a negative cell death regulator, whose expression was elevated in GA-deficient mutant ga1-3 and suppressed by GA application. RNA interference plants deficient in BOI showed enhanced cell death in response to the necrotrophic pathogen Botrytis cinerea and to toxin application. The absence of DELLA proteins in a quadruple-DELLA mutant correlated with increased cell death in response to salt treatment and during colonization of necrotrophic pathogens (Achard et al., 2008). Instead of disarming root MTI, P. indica might activate GA signaling in order to remove DELLA proteins and to control the activation of antiapoptotic enzymes (e.g. BOI), thereby reducing the antiapoptotic threshold in root cells and establishing host susceptibility. In fact, DELLA degradation and GA signaling were first detected at 7 dai (cell death phase). Consistent with this, we detected reduced P. indica colonization of the ga1-6 mutant (impaired in GA synthesis) at 7 dai (cell death phase), which was associated with an elevated expression of BOI (Fig. 9, A and D). Thus, our study confirms a more complex function of GA, the balance of SA-JA metabolism just representing one facet. It will be interesting to decipher in future studies to what extent DELLA protein stability and GA contribute to defense signaling and cell death regulation during the colonization of roots by P. indica. For unknown reasons and in contrast to previous reports (Navarro et al., 2008), we did not observe any effect of GA on flg22-triggered seedling growth inhibition in several independent experiments by using various flg22 concentrations (Fig. 9C). Consistently, GA did not affect flg22-triggered root oxidative burst (Fig. 9B). In contrast, we found that suppression of flg22-induced root oxidative burst by P. indica was dependent on JA signaling. In accordance with the reduced colonization of jin1-1, root innate immunity was enhanced in jin1-1 during P. indica colonization, as indicated by the enhanced expression of CBP60g and MYB51 (Fig. 7). Hence, reduced colonization of jin1-1 might reflect a hyperactivated immune response, as observed in pub22/23/24 roots (Fig. 5D; Supplemental Fig. S7).
In conclusion, we propose a model in which root MTI efficiently restricts penetration and root colonization of the mutualist P. indica (Fig. 10). This immune barrier can be overcome by P. indica through the manipulation of MAMP-triggered responses. JA signaling is recruited by P. indica to suppress early root MTI (e.g. oxidative burst). Our gene expression data (Fig. 7C) and mutant analysis (Fig. 6) further suggest that JA affects SA- and glucosinolate-associated defense, which might represent components of late MTI and efficiently control biotrophic root colonization. The elimination of root-based immunity is not only pivotal for colonization success but might also explain P. indica’s broad host range. Furthermore, the pub22/23/24 mutant studies imply that the host plant detects the mutualist P. indica, as it does pathogens, through recognition of its MAMPs. In agreement with Achard et al. (2008), our data indicate an antiapoptotic function of DELLA proteins during cell death-associated colonization. In the future, it will be interesting to elucidate to what extent the root surveillance system discriminates between pathogens and mutualists and to what extent GA/DELLA proteins affect P. indica colonization-associated cell death. Based on our studies, it is apparent that roots possess a perception system and immune repertoire similar to leaves.
MATERIALS AND METHODS
Plant Material and Inoculation
Seeds of Arabidopsis (Arabidopsis thaliana) ecotypes Col-0, Col-8, and Col-3gl1 and mutants quintuple-DELLA (N16298), ga1-6 (N3107), jin1-1 (N517005), jar1-1 (N8072), pad3 (N3805), npr1-1 (N3726), and A5 (N84735) were obtained from the Nottingham Arabidopsis Stock Centre. coi1-16 seeds were kindly provided by B. Hause, eds1 seeds by J. Parker, GFP-Chi seeds by G.P. di Sansebastiano, RGAp::GFP-RGA seeds by T.-P. Sun, and sid2-2 seeds by F.M. Ausubel. cerk1-2, pen2-1, and pub22/23/24 were described earlier (Lipka et al., 2005; Trujillo et al., 2008; Gimenez-Ibanez et al., 2009). All plants were grown on half-strength Murashige and Skoog (MS) medium without Suc at 22°C/18°C day/night (8 h of light) at 60% relative humidity. For fungal quantification and gene expression analysis, roots of 3-week-old plants were inoculated with Piriformospora indica (500,000 chlamydospores mL−1) and harvested at the indicated time points.
Quantitation of Fungal Colonization by qRT-PCR
Genomic DNA was extracted from roots with the Plant DNeasy Kit (Qiagen). Forty nanograms of DNA served as template for qRT-PCR analyses by using 20 μL of SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) with 350 nm oligonucleotides and a 7500 FAST thermal cycler with a standard protocol (Applied Biosystems). Fungal colonization was determined by the 2−ΔCt method (Schmittgen and Livak, 2008) by subtracting the raw threshold cycle (Ct) values of P. indica ITS from those of AtUBQ5 (for primer sequences, see Supplemental Table S1). Data were analyzed by Student’s t test.
Cytological Analyses
Root samples were either fixed or directly stained with chitin-specific WGA-AF488 and/or WGA-633 (Molecular Probes) as described (Deshmukh et al., 2006). Images were taken using an Axioplan 2 microscope (Zeiss). WGA-AF488 and GFP emission was detected at 505 to 530 nm (excitation, 470/20 nm). Cell autofluorescence emission was detected at 546/12 nm (excitation, 590 nm). Confocal images were recorded on a TCS SP2 microscope (Leica). WGA-AF488 and GFP were excited with a 488-nm laser line and detected at 505 to 540 nm, and WGA-AF633 was excited with a 546-nm laser line and detected at 600 to 660 nm. For ultrastructural studies, roots were embedded as described (Zechmann et al., 2007), and ultrathin sections (80 nm) were investigated after poststaining with uranyl aceate and lead citrate with a Philips CM10 transmission electron microscope. For callose assays, roots of 15-d-old seedlings were mock treated or inoculated with P. indica and thereafter treated with 1 μm flg22 or mock at 3 dai. Roots were harvested at 1 dai with flg22 or mock, and samples were fixed in alcoholic lactophenol solution (phenol:glycerol:lactic acid:water:ethanol, 1:1:1:1:2 [v/v]). Decolorization was done by vacuum infiltration in lactophenol for 15 min and subsequent incubation in fresh lactophenol at 65°C for 30 min followed by incubation at room temperature for at least 12 h. Tissue was washed in 50% (v/v) ethanol for 5 min and then rinsed several times with water prior to incubation for 30 min in darkness in 0.01% (w/v) aniline blue dissolved in 150 mm K2HPO4 (pH 9.5). Callose deposits were detected using a Zeiss Axioplan microscope (excitation, 365 nm; emission, 420 nm). For RGA degradation assays, roots of 2-week-old RGAp::GFP-RGA plants were inoculated with P. indica and mock treated or treated with 10 μm GA3. Degradation of GFP-RGA was detected in tips of lateral roots, and the number of root tips was counted as indicated in Figure 8A at 3 and 7 d after treatment by fluorescence microscopy.
Gene Expression Analysis by qRT-PCR
For gene expression analysis, 3-week-old plants and mutants were inoculated with P. indica or mock treated and harvested at 0, 1, 3, and 7 dai. For the flg22-related experiments, plants were inoculated with P. indica or mock treated. Three days later, plants were transferred to liquid MS medium containing 0.1 μm flg22 or mock solution. Roots were harvested at 2, 24, and 72 hat. For all experiments, RNA was extracted using TRIzol (Invitrogen), and aliquots were used for cDNA synthesis with the qScript cDNA synthesis kit (Quanta Biosciences). Ten nanograms of cDNA was used as template for qRT-PCR as described above for fungal quantification. The 2−ΔCt method was used to determine differential gene expression (for primer sequences, see Supplemental Table S1).
MAMP-Induced Root Oxidative Burst and Growth Retardation
Roots of 2-week-old plants were grown on solid half-strength MS medium and treated with 1 μm flg22, 1 μm N-acetylchitooctaose, or 1 μm elf18 at 3 dai with P. indica or mock treatment. For determination of the oxidative burst, roots were cut in 1-cm-long pieces (10 mg per assay) and subjected to a luminol-based assay as described (Gómez-Gómez et al., 1999). For the growth retardation assay, plants were treated with 10 μm flg22 or 1 μm elf18 at 3 dai with P. indica or mock treatment. Plant fresh weight was determined 10 d after treatment. flg22 and elf18 peptide sequences were used as described (Gómez-Gómez et al., 1999; Kunze et al., 2004).
Accession numbers of genes analyzed within this study are provided in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cell death-associated colonization by P. indica in MZ II.
Supplemental Figure S2. Cell death-associated colonization of Arabidopsis roots by P. indica.
Supplemental Figure S3. Defense responses during early stages of the Arabidopsis-P. indica interaction.
Supplemental Figure S4. Suppression of elf18-triggered responses by P. indica.
Supplemental Figure S5. Suppression of flg22-induced gene expression by P. indica in Arabidopsis roots.
Supplemental Figure S6. flg22- and P. indica chlamydospore-induced oxidative burst in pub22/23/24.
Supplemental Figure S7. Enhanced defense gene induction in pub22/23/24 roots by P. indica.
Supplemental Table S1. Primers used for qRT-PCR.
Supplemental Video S1. A, Fluorescent channel detection of the biotrophic colonization of a cortical cell by P. indica; B, transmission channel detection of the biotrophic colonization of a cortical cell by P. indica.
Supplementary Material
Acknowledgments
We thank the European Arabidopsis Stock Centre, F.M. Ausubel, B. Hause, J. Parker, G.P. di Sansebastiano, and T.-P. Sun for providing seeds. We also thank C. Neumann and R. Fensch for excellent technical assistance and D. Klessig, A. Schikora, and R. Eichmann for discussions and critical reading of the manuscript.
References
- Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P. (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Attard A, Gourgues M, Callemeyn-Torre N, Keller H. (2010) The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol 187: 449–460 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bos JI, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, et al. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Deshmukh S, Hückelhoven R, Schäfer P, Imani J, Sharma M, Weiss M, Waller F, Kogel KH. (2006) The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc Natl Acad Sci USA 103: 18450–18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flückiger R, De Caroli M, Piro G, Dalessandro G, Neuhaus JM, di Sansebastiano GP. (2003) Vacuolar system distribution in Arabidopsis tissues, visualized using GFP fusion proteins. J Exp Bot 54: 1577–1584 [DOI] [PubMed] [Google Scholar]
- Genger RK, Jurkowski GI, McDowell JM, Lu H, Jung HW, Greenberg JT, Bent AF. (2008) Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol Plant Microbe Interact 21: 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM. (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Gou M, Su N, Zheng J, Huai J, Wu G, Zhao J, He J, Tang D, Yang S, Wang G. (2009) An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J 60: 757–770 [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, Sheen J. (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575 [DOI] [PubMed] [Google Scholar]
- Heath MC. (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R. (2007) Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN. (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Berry N, Joardar V, Street IH, Kunkel BN. (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact 19: 789–800 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Luo H, Laluk K, Lai Z, Veronese P, Song F, Mengiste T. (2010) The Arabidopsis Botrytis Susceptible1 Interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol 154: 1766–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM. (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Simon L, Lam E. (1997) Pathogen-induced programmed cell death in tobacco. J Cell Sci 110: 1333–1344 [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E. (2008) The hypersensitive response: the centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- O’Connell RJ, Panstruga R. (2006) Tête à tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol 171: 699–718 [DOI] [PubMed] [Google Scholar]
- Peterson RL, Massicotte HB. (2004) Exploring structural definitions of mycorrhizas, with emphasis on nutrient-exchange interfaces. Can J Bot 82: 1074–1088 [Google Scholar]
- Rentel MC, Lecourieux D, Ouaked F, Usher SL, Petersen L, Okamoto H, Knight H, Peck SC, Grierson CS, Hirt H, et al. (2004) OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427: 858–861 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer P, Kogel KH. (2009) The sebacinoid fungus Piriformospora indica: an orchid mycorrhiza which may increase host plant reproduction and fitness. Deising HB, Esser K, , The Mycota Plant Relationships, Vol 5 Springer, Heidelberg, pp 99–112 [Google Scholar]
- Schäfer P, Pfiffi S, Voll LM, Zajic D, Chandler PM, Waller F, Scholz U, Pons-Kühnemann J, Sonnewald S, Sonnewald U, et al. (2009) Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. Plant J 59: 461–474 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Willige BC. (2009) Shedding light on gibberellic acid signalling. Curr Opin Plant Biol 12: 57–62 [DOI] [PubMed] [Google Scholar]
- Stein E, Molitor A, Kogel KH, Waller F. (2008) Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol 49: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Sun TP. (2008) Gibberellin metabolism, perception and signaling pathways in Arabidopsis. The Arabidopsis Book 6: e0103, doi/10.1199/tab.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL. (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Verma S, Sudha, Sahay N, Butehorn B, Franken P. (1999) Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl Environ Microbiol 65: 2741–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Hückelhoven R, Neumann C, von Wettstein D, et al. (2005) The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA 102: 13386–13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal L, Scheel D, Rosahl S. (2008) The coi1-16 mutant harbors a second site mutation rendering PEN2 nonfunctional. Plant Cell 20: 824–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, Tuteja N, Saxena AK, Johri AK. (2010) A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem 285: 26532–26544 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zechmann B, Müller M, Zellnig G. (2007) Membrane associated qualitative differences in cell ultrastructure of chemically and high pressure cryofixed plant cells. J Struct Biol 158: 370–377 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.