Abstract
Root-knot nematode (RKN) Meloidogyne species are major polyphagous pests of most crops worldwide, and cultivars with durable resistance are urgently needed because of nematicide bans. The Ma gene from the Myrobalan plum (Prunus cerasifera) confers complete-spectrum, heat-stable, and high-level resistance to RKN, which is remarkable in comparison with the Mi-1 gene from tomato (Solanum lycopersicum), the sole RKN resistance gene cloned. We report here the positional cloning and the functional validation of the Ma locus present at the heterozygous state in the P.2175 accession. High-resolution mapping totaling over 3,000 segregants reduced the Ma locus interval to a 32-kb cluster of three Toll/Interleukin1 Receptor-Nucleotide Binding Site-Leucine-Rich Repeat (LRR) genes (TNL1–TNL3), including a pseudogene (TNL2) and a truncated gene (TNL3). The sole complete gene in this interval (TNL1) was validated as Ma, as it conferred the same complete-spectrum and high-level resistance (as in P.2175) using its genomic sequence and native promoter region in Agrobacterium rhizogenes-transformed hairy roots and composite plants. The full-length cDNA (2,048 amino acids) of Ma is the longest of all Resistance genes cloned to date. Its TNL structure is completed by a huge post-LRR (PL) sequence (1,088 amino acids) comprising five repeated carboxyl-terminal PL exons with two conserved motifs. The amino-terminal region (213 amino acids) of the LRR exon is conserved between alleles and contrasts with the high interallelic polymorphisms of its distal region (111 amino acids) and of PL domains. The Ma gene highlights the importance of these uncharacterized PL domains, which may be involved in pathogen recognition through the decoy hypothesis or in nuclear signaling.
Plant parasitic nematodes are soil-dwelling pests that cause devastating damage worldwide (Sasser and Freckman, 1987). Among them, apomictic root-knot nematode (RKN) Meloidogyne species are extremely polyphagous biotrophic pests and attack most vegetable, fruit, and ornamental crops under Mediterranean and hot climates (Trudgill and Blok, 2001). The ban of the highly toxic nematicides renders urgent environmentally friendly and pesticide-free control methods, and breeding for RKN-resistant cultivars appears as a promising alternative. Few RKN Resistance (R) genes have been identified (Williamson and Kumar, 2006), and a single one, Mi-1 from tomato (Solanum lycopersicum), has been cloned (Milligan et al., 1998).
In Prunus species, among the RKN R genes mapped (Claverie et al., 2004a), the Ma gene from the Myrobalan plum (Prunus cerasifera) confers a complete-spectrum dominant resistance with several R alleles (Esmenjaud et al., 1996b; Lecouls et al., 1997) and is used directly or through interspecific hybrids for rootstock breeding in stone fruits using marker-assisted selection (Dirlewanger et al., 2004a, 2004b). This gene is not overcome by any of the over 30 RKN species and isolates that have been tested (Esmenjaud et al., 1994). In particular, it also controls “the peach RKN,” Meloidogyne floridensis (Handoo et al., 2004), which overcomes the resistance of all commonly used peach (Prunus persica) and almond (Prunus dulcis) R sources (Lecouls et al., 1997), and the highly aggressive species Meloidogyne mayaguensis (= Meloidogyne enterolobii; Yang and Eisenback, 1983), a species uncontrolled by the Mi-1 gene that is progressively invading new territories (Brito et al., 2007; Cetintas et al., 2007; Nyczepir et al., 2008).
Most resistance genes characterized in plant-pathogen interactions belong to the intracellular Coiled Coil (CC)-Nucleotide Binding Site (NBS)-Leucine-Rich Repeat (LRR; CNL) or Toll/Interleukin1 Receptor (TIR)-NBS-LRR (TNL) class of gene or to the extracellular LRR class of plant receptors (Hammond-Kosack and Parker, 2003). CNL and TNL genes act as receptors or coreceptors of pathogen-derived elicitors. One can speculate that the Ma gene, selected from natural Myrobalan plum, targets a conserved determinant from RKN and/or depends on an original resistance mechanism. Furthermore, because of the high polyphagy of RKN, knowledge on mechanisms leading to complete-spectrum RKN resistance in Prunus is of outstanding interest. In comparison with Ma, the tomato Mi-1 gene has a more restricted spectrum, a reduced efficiency at high temperatures, and may be overcome by natural (Tzortzakakis et al., 2005) or selected (Castagnone-Sereno et al., 1996) virulent populations. Moreover, the Myrobalan plum is a perennial, near-wild, and allogamous plant. This offers the opportunity to establish the structure of a R gene locus in a perennial plant and get original data about its putative evolution, a research field still poorly documented, even if some resistance loci and genes have been sequenced in poplar (Populus spp.; Lescot et al., 2004), Citrus (Yang et al., 2003), and apple (Malus domestica; Belfanti et al., 2004).
We have previously landed at the Ma locus on a 287-kb R bacterial artificial chromosome (BAC; Claverie et al., 2004b) from the heterozygous donor accession P.2175 and constructed both the resistant (R) and the susceptible (S) contigs. Here, we report the positional cloning and characterization of the Ma gene. Additional genetic markers developed from BAC clones carrying the R and S alleles of Ma and mapping on a large segregating population of 3,000 individuals narrowed the Ma interval to a 32-kb sequence that covers three TNL candidate genes. Comparison of genomic sequences between the R and S BAC haplotypes of Ma allowed the establishment of paralogous and orthologous relationships of the TNL genes and identified one complete gene sequence (TNL1) as the best R gene candidate. We then validated this candidate gene by showing that hairy roots and composite plants transformed with its genomic sequence and native promoter region confer the same complete-spectrum, high-level resistance as the Ma donor accession P.2175 to the three major RKN, Meloidogyne incognita, Meloidogyne arenaria, Meloidogyne javanica, the peach RKN, M. floridensis, and the Mi-1-uncontrolled species M. mayaguensis (= M. enterolobii). Finally, we characterize and discuss the unique structural features of TNL1: a huge and highly polymorphic C-terminal post-LRR (PL) region and a 2.5-kb insertion within the 5′ upstream region.
RESULTS
Detection of Candidate Genes Spanning the Ma Locus
A previous positional cloning strategy allowed us to land on a single R BAC (76H19) carrying the Ma1 allele (Claverie et al., 2004b). BAC clone 76H19, 287 kb long, was then sequenced and assembled, with a mean coverage of 14 (extremes 12–17), at the Centre National de Séquençage (Evry, France) as described by Chantret et al. (2005).
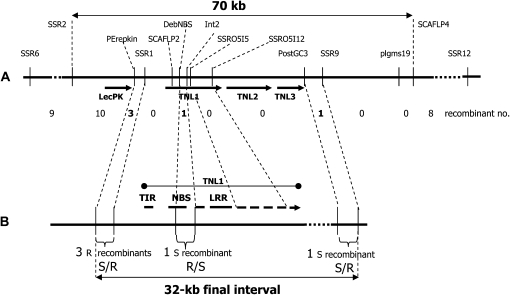
Three PCR-based genetic markers linked to Ma, SCAFLP3 (proximal; 10 recombinants with the Ma gene), SCAFLP2 (cosegregating; Lecouls et al., 2004), and SCAFLP4 (distal; a single recombinant; Supplemental Table S1), among 1,332 meioses (Claverie et al., 2004b), were placed on the sequence and determined a physical interval of 174 kb containing Ma. New simple sequence repeat (SSR) markers (Supplemental Table S1) were manually designed within this 174-kb interval and mapped on the same recombinant progeny, allowing us to reduce the interval containing the Ma gene to 70 kb flanked by the markers SSR2 and SCAFLP4 (Fig. 1A). In this interval, four candidate genes were predicted by GENESCAN and GENEMARK.hmm. These genes belonged to two distinct classes of resistance gene analogs: one lectin protein kinase (LecPK) and three TNLs (Fig. 1).
Figure 1.
Fine mapping of the Ma region within the BAC 76H19. A, Thirty-two recombinant individuals between the markers SSR6 and SSR12 were obtained from a total of 3,112 segregating individuals and further mapped with other PCR-based markers (locations and names above). Numbers of recombinants between the markers are indicated below. B, Resistant/susceptible status of a set of five recombinant individuals delineating a final 32-kb interval encompassing Ma (double arrow). Only the exon/intron structure of TNL1 is shown to indicate the positions of markers and recombination events.
Evidence of the Ma TNL Cluster as a Final Candidate Interval
In the objective to more accurately locate the candidate R genes, a final round of mapping was undertaken. A total of 1,780 additional individuals were produced from several segregating crosses (Ma1/ma × ma/ma; Supplemental Table S2). Some of these carried the RMia gene from the Nemared peach, which can be discriminated from Ma by resistance assays using M. floridensis. In order to detect individuals recombining very closely to Ma, two flanking markers, SSR6 and SSR12, separated by 209 kb and located at 0.7 centimorgan (nine recombinants) and 0.3 centimorgan (four recombinants), respectively (Fig. 1A; Supplemental Table S1), were designed and used in a duplex amplification procedure. They allowed the detection of 19 new recombinants out of the 1,780 individuals. This, added to the 13 initial recombinants of the 1,332 initial individuals, represents a total of 32 (19 + 13) recombinants out of the 3,112 (1,332 + 1,780) meioses in the SSR6-SSR12 interval, in agreement with the 1-centimorgan genetic distance previously estimated. All recombinants and control individuals were then genotyped for nine sequence-characterized amplified region or SSR markers (Supplemental Table S1) designed within the 70-kb candidate interval. Five recombinant genotypes located within the interval of candidate genes were identified and evaluated for resistance to the species M. floridensis (Supplemental Table S2). Their resistance phenotype allowed us to eliminate the LecPK candidate gene and reduced the final candidate region to the sole 32-kb sequence covering a cluster of three TNL genes (Fig. 1B). One recombination event located downstream of the TIR exon of the first TNL in a susceptible individual excluded the TIR exon as carrying alone a resistance determinant.
Comparative Analysis of the Candidate Region in the R and S BAC Clones
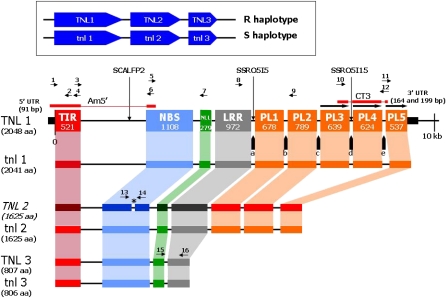
The BAC 40K9, 170 kb long and encompassing the S allele of the Ma candidate region, (Claverie et al., 2004b), was sequenced. Three TNLs were identified in the S haplotype of Ma, where sequence and structure comparison allowed the establishment of orthology relationships with those of the R haplotype. Hereafter, TNLs have been designated TNL1 to TNL3 in the R haplotype and tnl1 to tnl3 in the S haplotype (Fig. 2). Intron/exon structures were predicted, and the complete cDNA sequence of TNL1 was determined using specific and oligo(dT) primers and RACE-PCR (Fig. 2; Supplemental Table S3). 5′ and 3′ untranslated region (UTR) ends were obtained using TIRGC1-R1/TIRGC1-R2 and CT3-Fo/Ic-CT3-Ba, respectively. Final exonic sequence of the PL1 region of TNL1, which diverged highly between the predictors GENESCAN, GENEMARK.hmm, and EuGene, was determined from specific primers EpisLRR-F1 (in exon LRR) and EpisLRR-R1 (in exon PL2). The complete C-terminal fragment of TNL1, from the LRR exon up to the 3′ UTR of the gene, could be obtained in a single run from sense primer EpisLRR-F1. Sequences of the other TNLs were deduced from sequence alignments. For TNL2, we detected a frameshift mutation (deletion of a single base) in the NBS domain that makes it a truncated pseudogene. Allele-specific primers flanking the mutation in the NBS putative TNL2 exon failed to amplify the TNL2 cDNA sequence. This frameshift mutation is surprisingly absent in tnl2, a result that we have also confirmed on its transcript by sequencing the corresponding reverse transcription-PCR product. Transcription of TNL3 was shown by reverse transcription-PCR using NLR-F2 and NLR-R specific primers flanking the third intron (Fig. 2; Supplemental Table S3).
Figure 2.
Exon and intron structure and size (in bp) of the TNL cluster from the R and S haplotypes. PL1 to PL5 are the C-terminal post-LRR exons. Vertical arrows indicate polymorphic markers; a to e = SSR sequences as follows: a = (TC)33, b = (TC)18, c = (TC)21, d = (TC)21(TA)10, and e = (CT)27(CAGA)(CA)6. Direct repeats of sequences in PL3 to PL5 are indicated by horizontal arrows above the DNA sequence. Thick black lines at each end of TNL1 are the 5′ UTR (79 bp) and 3′ UTR (two transcript variants of 164 and 199 bp), respectively. Red bars above TNL1 are the TNL1-specific cDNA fragments Am5′ and CT3. Values at the left end of each gene are the total amino acids (aa) deduced from the sequenced full-length cDNA sequence or from the predicted cDNA (other genes). The TNL2 pseudogene (in italics), after correction of the single-base frameshift mutation in the NBS (indicated by the star), is shown in darker colors. Primers used for the full-length cDNA sequence and sequence verifications are shown by arrows with numbers and correspond, from 5′ to 3′, respectively, to Am5′-2F (1), TIRGC1-R1 (2), TIRNBSGC1-F (3), TIRGC1-R2 (4), NBSGC1-F (5), TIRNBSGC1-R (6), NBSNLLGC1-R (7), EpisLRR-F1 (8), EpisLRR-R1 (9), CT3-Fo (10), Ic-CT3-Ba (11), CT3-Ba (12), NBSGC2-F (13), NBSGC2-R (14), NLR-F2 (15), and NLR-R (16).
Nine, seven, and four exons were predicted for TNL1/tnl1, TNL2/tnl2, and TNL3/tnl3, respectively. TIR, NBS, NLL (defined as a TNL-specific non-LRR region, or NBS-LRR linker, by Meyers et al. [2003]), and LRR domains are each carried by separate exons (Fig. 2). The PL part of TNL1 and TNL2 showed a nonclassical TNL intron/exon structure. It comprises five C-terminal exons encoding non-LRR sequences in TNL1 and tnl1 and three in TNL2 and tnl2, of which the PL3 and PL4 exons were deleted. Another striking feature of TNL1 and TNL2 was the presence of a microsatellite motif (TC)n in the introns preceding each of the PL exons (Fig. 2). In TNL3, the C-terminal region was truncated after the seventh LRR repeat motif and was much shorter. Domain comparisons between paralogous genes showed that the TIR domain of TNL1 was the most divergent, whereas the most divergent NBS domain was found in TNL3 (data not shown). The TNL3 NBS domain was also characterized by the absence of a putative nuclear localization signal, whereas WoLF PSORT analysis (Horton et al., 2007) detected two nuclear localization signals (one bipartite and one monopartite) in the NBS domain of TNL1 and TNL2. Moreover, the C-terminal part of the LRR exon (from LRR10 to the end of the exon; Supplemental Fig. S1) and the following PL1 and PL2 exons of TNL1 were more divergent between R and S alleles (93.6% nucleotide identities; data not shown) than between the TNL1 and TNL2 or tnl1 and tnl2 paralogous genes (97.8% and 97.7% nucleotide identities, respectively; data not shown).
Between the R and S alleles of the three genes (Table I), the most polymorphic is TNL1, which showed over 5% amino acid differences with tnl1, whereas TNL2 and TNL3 exhibited less than 1% amino acid polymorphism between each other (after a frameshift correction for TNL2). TNL1 and tnl1 show 14.5% amino acid divergence in the 111 amino acids of the distal part of the LRR-encoding exon (grouping the three last LRR units [LRR10–LRR12] and the end of the exon; Supplemental Fig. S1) and 10% in their PL1 and PL2 exons (Table I). The high divergence between TNL1 and tnl1, added to the fact that TNL3 is truncated and TNL2 is a pseudogene, indicated that TNL1 is the best candidate sequence for the Ma gene.
Table I. Amino acid sequence polymorphism between the R and S haplotypes of TNL genes for each protein domain (in percentage) and for the complete gene sequence.
| Gene | TIR | NBS | NLL | LRR (First 213 Amino Acids) | LRR (Last 111 Amino Acids) | PL1 | PL2 | PL3 | PL4 | PL5 | Total TNL | Nonsynonymous Mutations | Total Length |
| TNL1/tnl1 | 0 | 1.1 | 1.1 | 0.5 | 14.5a | 9.7 | 10.6 | 6.7 | 6.3a | 3.3a | 5.1 | 104 | 2,041 |
| TNL2/tnl2 | 1.2 | 0b | 0 | 0.5 | 0.9a | 0.4 | 0 | 0 | – | – | 0.3 | 5 | 1,625 |
| TNL3/tnl3 | 0.6 | 0.3 | 1.1 | 2.3 | – | – | – | – | – | 0.9 | 7 | 806 |
Does not take into account a 3-bp insertion in TNL1 and TNL2 LRRs or a 9-bp insertion in TNL1 PL4 and PL5.
A single base has been added to correct the TNL2 frameshift mutation and calculate polymorphism.
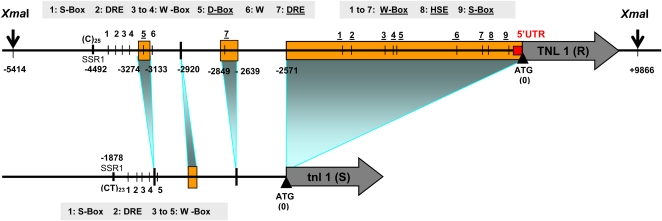
In the R and S haplotypes, sequence lengths between the LecPK gene (Fig. 1) and TNL1/tnl1, 7.1 and 4.5 kb long, respectively, differed by an approximately 2.6-kb insertion just preceding the initiation codon of TNL1 (Fig. 3). At least one part of this insertion is due to an ancient duplication of a 0.9-kb region immediately upstream of TNL1. Finally, we examined the presence of specific plant cis-acting elements (Fig. 3) such as biotic or abiotic stress-responsive elements (W box [Rushton et al., 1996], GCC-like S box [Kirsch et al., 2001], DRE [Yamaguchi-Shinozaki and Shinozaki, 1994], and HSE [Miller and Mittler, 2006]) and enhancing pathogen-responsive D-box-like elements (Rushton et al., 2002). Interestingly, the D-box-like (Supplemental Fig. S3) and HSE elements were only found in the R haplotype, and the others showed a reshuffled position (Fig. 3).
Figure 3.
Comparison of the putative promoter and regulatory regions of the R and S alleles of TNL1. The rectangles indicate major insertions/deletions between the two haplotypes. Pathogen-associated cis-acting elements (W box [Rushton et al., 1996], S box [Kirsch et al., 2001], and D box [Rushton et al., 2002]), dehydration-, cold-, and salt stress-responsive DRE element (Yamaguchi-Shinozaki and Shinozaki, 1994), and heat shock- and putatively reactive oxygen species-responsive HSE element (Miller and Mittler, 2006) are identified by numbers above TNL1 and below tnl1. Elements present in the R haplotype and absent in the S haplotype are underlined. Vertical arrows (XmaI restriction sites) delimit the 15.3-kb TNL1 genomic fragment used for validation experiments. Distances of elements from the ATG site (bp) and sizes of TNL1 and tnl1 are not drawn at scale. [See online article for color version of this figure.]
Complementation Experiments for Resistance to Several RKN Meloidogyne Species
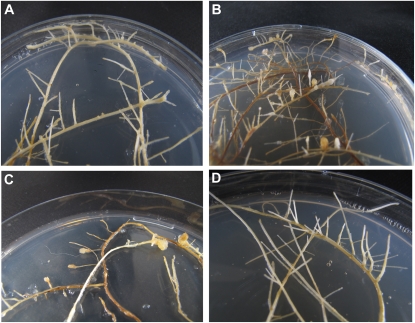
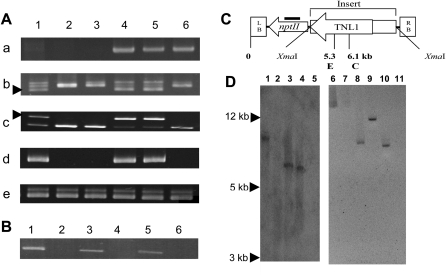
As TNL1 was the best candidate for Ma, a validation procedure for resistance complementation of this gene was undertaken. A fragment of approximately 15.3 kb containing the 9.2-kb TNL1 genomic sequence preceded by approximately 5.4 kb from the 5′ upstream region was used to transform the S accession 253 via Agrobacterium rhizogenes (Supplemental Table S2). Microplants, inoculated with agrobacteria above the base of the stem, produced roots of which the most vigorous were sectioned and grown in petri dishes. Agrobacteria were controlled by cefotaxime, but selection of transformants on kanamycin medium could not be performed because of the strong adverse effect of the antibiotics on the root growth of Prunus. Thus, roots growing autonomously and considered as hairy roots were cultured, multiplied by division at each growth time period, sampled for DNA and RNA extraction, and inoculated for evaluation of resistance to different RKN species (Bosselut et al., 2011). Fifteen independent hairy roots originating from six selected plants were infested with (1) M. incognita, (2) M. mayaguensis (= M. enterolobii), and (3) mixed populations of M. arenaria, M. javanica, and M. floridensis (5,000 J2s per individual root, two roots per transformation event and RKN species; Table II). From the 15 independent transformation events in all three separate resistance tests, 10 produced nematode galls and five were free of galls after 3 weeks, regardless of the nematode species (illustrated in Fig. 4 for M. incognita). The nine control hairy roots transformed with the empty vector pKGW, 0-253 (obtained from four vigorously rooting selected plants) were also extensively galled regardless of the RKN species (Table II; Fig. 4). Effective transformation of the hairy roots obtained from the susceptible accession 253 with the TNL1 gene (pKGW,TNL1-253) was confirmed by the amplification of the nptII gene from the vector pKGW and of the TNL1-specific genomic fragments CT3-4N and NSCALF2 (Fig. 5A) and then by the amplification of the TNL1-specific cDNA fragment CT3 (Figs. 2 and 5A). Finally, Southern-blot hybridization of EcoRV- or ClaI-digested genomic DNA with a nptII probe (Fig. 5C) showed the presence of a single copy of the T-DNA in at least two of these transformants (Fig. 5D). Finally, we showed that hairy roots that were free of galls expressed the allele-specific TNL1 transcript fragments CT3 (Fig. 5A) and Am5′ (data not shown), whereas no TNL1 transcription was detected for hairy roots that produced galls.
Table II. Validation experiments from hairy roots of accession 253 transformed with TNL1 for resistance to five RKN species.
| Vector | No. of Plantsa | No. of Independent Cultured Hairy Roots | Ranging into | Transcript Expressionb |
Gall Symptoms | |||
| Am5′ | CT3 | M. incognita | M. arenaria + M. javanica + M. floridensis | M. mayaguensis (= M. enterolobii) | ||||
| pKGW,TNL1-253 | 6 | 15 | 5 | − | − | + | + | + |
| 10 | + | + | − | − | − | |||
| pKGW,0-253 | 4 | 9 | 9 | − | − | + | + | + |
Number of selected plants with hairy roots.
Am5′ and CT3 are fragments transcribed in the 5′ UTR-TIR and the PL4 regions of TNL1, respectively.
Figure 4.
Phenotypes of the transformed hairy roots from the susceptible accession 253 inoculated with M. incognita. A, TNL1-transformed roots X2 expressing the gene (Am5′ and CT3 transcripts) are free of galls. B, Empty vector (pKGW,0)-transformed roots 3 are extensively galled. C, TNL1-transformed roots N1 lacking gene expression are galled (see Table II). D, Uninoculated control TNL1-transformed roots X2 (shown in A).
Figure 5.
Amplification gels of DNA and cDNA and Southern-blot hybridization from TNL1-transformed hairy roots. A, Genomic amplification of hairy roots in petri dishes with primers from nptII (a), CT3-4N (b), and NSCAFLP2 (c; arrowheads indicate fragments linked to the R allele of TNL1) and cDNA amplification of hairy roots in petri dishes with primers from CT3 (d) and control chalcone synthase (e). Lane 1, P.2175 (Ma donor accession); lane 2, 253 (untransformed); lane 3, P.2032 (S control accession); lanes 4 and 5, TNL1-transformed roots X2 and X5 (pKGW,TNL1-253); lane 6, empty vector-transformed roots 3 (pKGW,0-253). B, Amplification of TNL1-transformed roots from composite plants. cDNA amplification of Am5′ is shown. Lane 1, P.2175 (Ma donor accession); lane 2, 253 (untransformed); lane 3, TNL1-transformed roots of plant 8 (pKGW,TNL1-253); lane 4, empty vector-transformed roots (pKGW,0-253); lane 5, TNL1-transformed roots of plant 11 (pKGW,TNL1-253); lane 6, P.2032 (S control accession). C, Schematic representation of T-DNA from pKGW used in the transformation experiments. C, ClaI; E, EcoRV; LB, left border; RB, right border. Values represent distances between the left border and restriction sites. The thick bar represent the Southern-blot nptII probe. D, Southern-blot hybridization of EcoRV-digested (lanes 1–5) and ClaI-digested (lanes 6–11) genomic DNA with the nptII probe. Lanes 1 and 6, empty vector-transformed roots 3 (pKGW,0-253); lanes 2 and 5, 253 (untransformed); lanes 3 and 4, TNL1-transformed roots X2 and X5 (pKGW,TNL1-253), respectively; lanes 7 and 11, 253 (untransformed); lanes 8, 9, and 10, TNL1-transformed roots X2, X5, and X7 (pKGW,TNL1-253), respectively.
Independently from the hairy root transformants, sets of microplants, rooted after inoculation with agrobacteria carrying TNL1 (54 plants) or the empty vector (60 plants), were grown individually in pots as composite plants (transformed roots and untransformed aerial part) under nonsterile conditions together with control untransformed accessions (Bosselut et al., 2011). Composites microplants were infected with juveniles (J2s), and resistance was assessed after 8 weeks. Amplification of the Am5′ and CT3 TNL1-specific cDNA fragments (Fig. 2) was tested in the hairy roots and showed that TNL1 was integrated and transcribed in the roots of only two composite plants (plants 8 and 11; Fig. 5B). After infection with 20,000 M. incognita J2s per pot (Table III), only plant 8 and plants of the control Ma gene donor genotype P.2175 were free of galls. A precise examination of plant 11 showed that it had emitted new roots from the initial stem and that mixed transformed (ungalled) and untransformed (galled) roots prevented further evaluation of resistance expression. The single nongalled TNL1-transformed composite plant 11 was then reevaluated, together with appropriate control plants, to the same M. incognita isolate but under a high and steady inoculum pressure using calibrated pieces of galled tomato rootlets estimated to release approximately 3 × 105 J2s over the 8-week-long test. New roots developed on the composite plant and roots of the P.2175 resistant controls were still entirely free of galls, while the susceptible controls exhibited gall symptoms of severe attacks. In order to know whether the TNL1-transformed roots were also resistant to the two other predominant RKN species, M. arenaria and M. javanica, and to the peach RKN, M. floridensis, the M. incognita-resistant composite plant was then evaluated with the same high and steady inoculation procedure to a mixture of these three RKN (approximately 105 J2s of each species). This plant exhibited the same gall-free pattern as control P.2175 plants (Table III). Finally, this plant was rated after 8 weeks as resistant to M. mayaguensis (= M. enterolobii) using the same procedure and controls as previously.
Table III. Validation experiments from composite plants of accession 253 transformed with TNL1 through three successive tests combining the RKN species and the type of inoculation.
| RKN Species | Inoculation Type | Phenotype | Composite Plants |
Untransformed Controls |
|||||
| Total Plants | TNL1-253a | EV-253a | 253 | P.2032 | P.2175 | ||||
| CT3+b | CT3−b | ||||||||
| M. incognita | (1) Short pressure | Total plants | 54 | 2 | 52 | 30 | S | S | R |
| R | 1c | 0 | 0 | ||||||
| S | 1 | 52 | 30 | ||||||
| (2) Steady pressure | Total plants | 1 | 1c | – | 10 | Idem | Idem | Idem | |
| R | 1 | – | 0 | ||||||
| S | 0 | – | 10 | ||||||
| M. arenaria + | (3) Steady pressure | Total plants | 1 | 1c | – | 10 | Idem | Idem | Idem |
| M. javanica + | R | 1 | – | 0 | |||||
| M. floridensis | S | 0 | – | 10 | |||||
| M. mayaguensis (= M. enterolobii) | (3) Steady pressure | Total plants | 1 | 1c | – | 10 | Idem | Idem | Idem |
| R | 1 | – | 0 | ||||||
| S | 0 | – | 10 | ||||||
Composite plants of accession 253 carrying the TNL1 insert and an empty vector (EV).
Plants expressing (CT3+) or not expressing (CT3−) the TNL1 cDNA fragment CT3.
The same plant was evaluated to M. incognita successively under short (1) and steady (2) inoculum pressures and to the other RKN species under steady pressure (3).
Consequently, we demonstrated that complementation of susceptible hairy roots and composite plants with the genomic sequence of TNL1 driven by its own promoter restored the high-level (high and steady inoculum pressure) and wide-spectrum (five major RKN species) resistance conferred by the Ma gene. Thus, our results showed that TNL1 is the Ma gene.
Characterization of the TNL1/Ma Gene
The first four exons of TNL1 exhibited a classical modular TNL structure (Fig. 2), where each domain had the classical conserved motifs and was expected to be functional. They showed the highest similarity with the Gro1-4 (potato [Solanum tuberosum]; Paal et al., 2004) and N genes (tobacco [Nicotiana tabacum]; Whitham et al., 1994; data not shown). The LRR domain is composed of 12 LRR repeats, of which repeats 5 to 7 are duplicated into repeats 8 to 10 (Supplemental Fig. S1). LRRs 1 to 11 can be easily aligned, and their predicted secondary structure matched the canonical secondary structure E4C5 described by Mondragon-Palomino et al. (2002) for Arabidopsis (Arabidopsis thaliana) NBS-LRR genes.
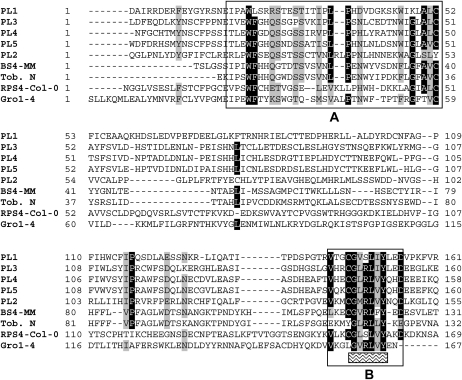
The major structural difference between TNL1 and other known TNLs resided in the presence of five PL exons instead of one. PL exons were 226, 263, 213, 208, and 178 amino acids long, for a total length of 1,088 amino acids. Among these exons, the three last ones were very similar to each other, indicating that they were recently derived by duplications (Fig. 2; Supplemental Fig. S2), whereas nucleic acid sequences from the two first exons were difficult to align with the others. Duplications of the entire C-terminal exon have already been reported for the TNL-F group of NBS-LRR genes in Arabidopsis (Meyers et al., 2003). The function of PL exons remains unknown, although they accumulated high polymorphism between the R and S haplotypes. Those five PL exons did not show a specific LRR structure (Mondragon-Palomino et al., 2002) and diverged from each other for most of their predicted secondary structures (PSIPRED prediction; Bryson et al., 2005), with alternations of putative coils, helices, and β-sheets. Alignment of the single C-terminal exons from diverse TNL genes from flax (Linum usitatissimum), Arabidopsis, and the tobacco N gene (Whitham et al., 1994) by Dodds et al. (2001) allowed the identification of seven conserved motifs, but with a lower conservation in the tobacco N gene. Four of these motifs were clearly identified in TNL1 PL sequences, which globally fitted better with the BS4 (tomato; Ballvora et al., 2001) and N (tobacco) C-terminal sequences than with most (the exception is RPS4; Gassmann et al., 1999) Arabidopsis TNL C-terminal sequences. In TNL1, the B motif (Fig. 6), a motif related to the conserved domain 7 identified by Dodds et al. (2001), is predicted to contain a β-sheet structure, with aliphatic amino acids and conserved Gly (G), Arg (R), and Tyr (Y) residues. The five PL exons from TNL1 appeared transcribed in the same RNA, given that cDNA amplifications using primers from LRR and PL domains (Fig. 2) failed to detect transcript variants in uninfected roots of P.2175 (data not shown).
Figure 6.
Alignment of all five PL (PL1–PL5) exons of TNL1/Ma with single PL exons of known R genes from the TNL family. Those genes, from diverse pathogens and selected using ψ-BLAST analysis (Altschul et al., 1997), belong to the Solanaceae family (N [Whitham et al., 1994], BS4-MM [Ballvora et al., 2001], Gro1-4 [Paal et al., 2004]) or to Arabidopsis (RPS4-Col-0 [Gassmann et al., 1999]), which is more closely related to Prunus. The conserved domains A and B can be assimilated to the conserved domains 1 to 3 and 7, respectively, defined by Dodds et al. (2001). The underlined amino acids of domain B are predicted to form a β-sheet secondary structure (PSIPRED prediction; McGuffin et al., 2000; Bryson et al., 2005) with solvent-exposed residues.
Surprisingly, among the 12 LRRs, the first nine repeats (amino acids 1–213) showed only one amino acid polymorphism between R and S alleles and four synonymous substitutions, suggesting an interallelic conservative selection (P < 0.03; Tables I and IV), unlike the remaining 111 amino acids of the LRR exon. Interestingly, the selection between alleles in the PL domains was significantly globally conservative (codon-based Z test P < 0.003 for the whole PL part of the gene), with an excess of synonymous substitutions (Table IV). Despite the small sample size (two alleles), interallelic polymorphism was also locally significantly conservative for the PL4 domain (P < 0.005). Finally, interdomain polymorphism between the three PL domains that can be aligned unambiguously (PL3–PL5) was conservative (P < 0.00005). We also noticed that the synonymous polymorphism is variable in the C-terminal part of TNL1: 12.2% in the 111 amino acids of the distal part of the LRR-encoding exon and a range between 2.8% (PL5) and 11% (PL4) in the PL domains (data not shown).
Table IV. Selection between alleles, calculated for the most variable domains of the three TNLs.
Selection pressure (Ka/Ks) is expressed by the ratio of nonsynonymous substitutions per nonsynonymous site (Ka) divided by the number of synonymous substitutions per synonymous site (Ks) and using the method of Nei and Gojobori (1986). Asterisks indicate significance for purifying selection: * P < 0.05, ** P < 0.005.
| Gene | LRR (First 213 Amino Acids) |
LRR (Last 111 Amino Acids) |
PL1 |
PL2 |
PL3 |
PL4 |
PL5 |
PL1 to PL5 |
||||||||
| Muta | Ka/Ks | Mut | Ka/Ks | Mut | Ka/Ks | Mut | Ka/Ks | Mut | Ka/Ks | Mut | Ka/Ks | Mut | Ka/Ks | Mut | Ka/Ks | |
| TNL1/tnl1 | 5 | 0.07* | 23 | 0.48 | 32 | 0.54 | 43 | 0.51 | 19 | 0.67 | 26 | 0.24** | 9 | 0.5 | 129 | 0.43** |
| TNL2/tnl2 | 1 | – | 1 | – | 1 | – | 0 | – | – | – | – | 0 | – | 1 | – | |
| TNL3/tnl3 | 7 | 0.37 | – | – | – | – | – | – | – | – | – | – | – | – | – | |
Mut indicates the total number of mutations.
DISCUSSION
A RKN R Gene from a Prunus Species
We identified the Ma resistance gene in the Myrobalan plum by an exclusive positional cloning approach. This was rendered possible by accurate fine genetic and physical mapping, comparative analysis of R and S haplotype sequences, and finally by functional complementation of susceptible Prunus accession. Fine mapping of the Ma locus, previously conducted on 1,332 adult trees by Claverie et al. (2004b) and completed here on 1,780 seed-borne plantlets, allowed the identification of the final 32-kb candidate TNL cluster. Sequence analysis of the R haplotype designated TNL1 and TNL3 as the two final candidate Ma genes and TNL2 as a young pseudogene. Comparison of the R and S haplotypes in accession P.2175 showed that TNL1 was much more polymorphic than TNL3. Moreover, the truncated LRR domain made TNL3 a less likely resistance gene; consequently, TNL1 was the most probable candidate gene for Ma. Complementation of a susceptible genetic background with the genomic sequence of TNL1 driven by its own promoter restored the high-level and wide-spectrum resistance of the Ma gene. After the Mi-1 gene from tomato (Milligan et al., 1998), the Ma gene is the second cloned gene for resistance to RKN and, to our knowledge, the first one in a perennial. This gene has the peculiarity to have been identified in a highly heterozygous and self-incompatible material from, moreover, a near-wild (unfixed) origin (Eremin, 1978). The Ma gene belongs to the TNL family of the NBS-LRR class of genes instead of the CNL family of Mi-1 (Milligan et al., 1998).
An interesting feature of the Ma gene is its wide-spectrum resistance comprising, besides the predominant species from the RKN polyphagous complex M. incognita, M. arenaria, and M. javanica, the particular species M. mayaguensis (= M. enterolobii), uncontrolled by both the Mi-1 gene in tomato and the Me genes in pepper (Capsicum annuum), and M. floridensis, uncontrolled by RKN-resistant peach rootstocks. Another major trait of interest of Ma is that resistance is not affected by high inoculum pressures from those five species. Our successful complementation with transgenesis via A. rhizogenes opens the way to the application of hairy roots transformation in other plant species in order to move across species boundaries.
Ma Structural Peculiarities
The Ma gene has the longest full-length cDNA size (2,048 deduced amino acids) of all TNL genes cloned to date. Its PL part is unique in that it represents 53% of the total cDNA length and is composed of five exons, with the latter three originating from recent duplications. We analyzed whether the complete coding sequence coexists with shorter splice variants, as shown for several TNLs (Dinesh-Kumar and Baker, 2000; Gassmann, 2008). Those alternative transcripts would be more comparable in structure and length to classical TNLs. Nevertheless, cDNA amplifications using primers from the LRR and PL domains failed to detect transcript variants. Moreover, the transcript CT3, although located close to the C-terminal end of TNL1, was always detected in the roots of P.2175 and in TNL1-transformed hairy roots of 253 at preinfectious and postinfectious dates. This suggests that the PL3 to PL5 domains, which harbor the CT3 transcript sequence, are part of a predominant splice variant of TNL1 and thus might play a key role in gene activity.
Nevertheless, the TNL family can be considered as a versatile gene family, showing different gene structures and, particularly, various C-terminal gene fusions (TNL-X, TNL-T, TNL-WRKY; Meyers et al., 2003; Ameline-Torregrosa et al., 2008; Kohler et al., 2008; Yamasaki et al., 2008). In TNL1, the five PL exons show a conserved core motif [CG(a)RL(a)Y]. The similarity of this motif with the WRKY transcription factor motif (WRKYGQK) from RRS1 (Deslandes et al., 2002; conservation of two essential residues: Arg [R] and Tyr [Y]) is strengthened by a similar C-terminal position. Furthermore, its β-sheet structure is a common feature of plant DNA-binding domains (Yamasaki et al., 2008). When considering the two putative nuclear localization signals detected in the NBS domain of TNL1 and TNL2, but not in TNL3 (which completely lacks PL exons), we could speculate that PL exons act as transcription factors for downstream signaling. This hypothesis is strengthened by the nuclear localization of N or RPS4 resistance gene products (Burch-Smith et al., 2007; Wirthmueller et al., 2007), which both possess similar C-terminal post-LRR domains (Fig. 6). These TNL genes may possess a DNA-binding function similar to the WRKY domain from RRS1 and the BEAF and DREF DNA-binding finger domains from Populus trichocarpa genes (Aravind, 2000; Tuskan et al., 2006) and would contain a new and particularly variable class of DNA-binding domains. Furthermore, the β-sheet amino acids stretches (of four to 15 amino acids long) predicted in the PL domains could lead to a structure resembling the general WRKY transcription factor secondary structure (Yamasaki et al., 2008) but with many more amino acid residues between the β-sheets. Another possibility is that those PL exons mimic conserved sites of one or several DNA-binding protein(s), such as WRKY domains, which have already been hypothesized to be targeted by pathogen avirulence (Avr) products in the TNL-WRKY gene SLH1 (Noutoshi et al., 2005). This last hypothesis would explain the high polymorphism and the poor similarities of this domain with any conserved protein domain.
Ma Polymorphism
In the Ma cluster, the most coding polymorphism accumulates in the last LRR repeats and in the PL exons of TNL1. This polymorphism is locally more elevated between alleles (TNL1 and tnl1) than between paralogous genes (TNL1 and TNL2). This observation is quite unusual (Michelmore and Meyers, 1998) and means that an ancient polymorphism that predates a TNL duplication has probably been locally maintained in TNL1. The most ancient polymorphism in TNL1 is found in the last 111 amino acids from the LRR domain (no. of synonymous substitutions per synonymous site [Ks] = 12.2%) and in the PL4 exon (Ks = 11%). The presence of another allele for RKN resistance at the same locus in the Japanese plum (Prunus salicina; Claverie et al., 2004a) suggests that resistance polymorphism is anterior to P. cerasifera and P. salicina species separation. Furthermore, the Ma locus itself is probably also ancient: in the recently sequenced species P. persica (peach genome version 1.0 [http://www.rosaceae.org/node/355]), an orthologous locus (identified between the lectin kinase and guanylate kinase genes flanking the Ma cluster) showed four TNL-like sequences. We could observe that those sequences can be predicted (GENESCAN) to contain two truncated TNL sequences, one full TNL sequence (with only one PL exon, and predicted to be fused to an N-terminal LRR domain), and one TNL gene showing the same structure (the five PL exons) as TNL1 from P. cerasifera. TNL1 shows the highest nucleotide identity with this latter gene (from 94% to 99% depending on the domains). This gene is probably the “direct” orthologous sequence of TNL1, which means that the particular structure of Ma is ancient and shared between Prunus species.
In the P.2175 accession, the high polymorphism of the upstream region of Ma suggests differences in the R and S allele gene regulations (some putative cis-acting elements lie on insertion/deletion polymorphisms in the regulatory region of Ma). Nevertheless, a susceptible recombinant obtained in the TNL1 gene (R for promoter and TIR domain and S for LRR and PL exons; Fig. 1) showed that this regulation is not sufficient to confer the resistance. The inverse recombinant would have answered whether this regulation is necessary for the resistance. Each allele may respond differently to various stresses, like drought, heat, oxidative stress, or pathogens, and the different strategies that they develop in pathogen sensing and signaling are probably crucial for an efficient and durable resistance.
Ma and Resistance to RKN
As Ma confers a complete-spectrum resistance to RKN, it is highly probable that this gene recognizes a crucial effector or guards a virulence/effector target that RKN species have in common. Myrobalan plum and the apomictic RKN (M. incognita, M. arenaria, and M. javanica) are probably not coevolving partners. The spontaneous accession P.2175 originated from continental Romania. On the contrary, Meloidogyne species and particularly those controlled by Ma1 need a Mediterranean or tropical climate to develop successfully, and it is thought that they never existed in the wild in the same areas as the plum. The Meloidogyne species from the polyphagous apomictic complex are also usually considered as unlikely coevolving partners, essentially because of their apomixis and their supposed reticulate origin (Trudgill and Blok, 2001; Blok et al., 2008). In the absence of coevolution, the wide-spectrum and high-level resistance conferred by the Ma gene may be best explained by an indirect interaction (guard hypothesis; Van der Biezen and Jones, 1998; Dangl and Jones, 2001) between the resistance gene product and the nematode Avr factors. Alternatively, in the context of the decoy hypothesis proposed by Van der Hoorn and Kamoun (2008), at least one of the PL exons might mimic a conserved host target (as hypothesized for the WRKY domain of SLH1 by Noutoshi et al. [2005]) common to Meloidogyne species and unrelated plant pathogens and lead to a direct interaction. In the TNL1/Ma gene, not only the PL domains but also the end of the LRR domain and the gene regulatory region show extensive polymorphism. In this case, we may suppose that the “guarded” host product or the host decoy is targeted by independent Avr products from one or more pathogens species that are not apomictic RKN.
If Ma is a guard protein, we can speculate that most part of its NLL+LRR region (e.g. the four LRR repeats from the NL linker and the nine first LRR repeats from the following LRR exon), which only shows two amino acid changes between TNL1 and tnl1, is involved in intramolecular and/or intermolecular (with the host target/decoy) binding. The last LRR repeats (LRR10–LRR12) and the following amino acids of this exon, which show the highest interallelic polymorphism in the gene (14.5%), would therefore be dedicated to detecting specific modifications or specific binding to the guarded target/decoy. Other examples of polymorphism accumulation in the C-terminal part of the LRR domain have already been reported for the durable resistance gene to Tomato mosaic virus, Tm-2 (CC-NBS-LRR; Lanfermeijer et al., 2003), and for the RP-1D paralogous genes (NBS-LRR) from a maize (Zea mays) rust resistance locus (Sun et al., 2001). The observations on the TNL1 gene are also congruent with those on the Rx potato gene for resistance to Potato virus X, where the N-terminal part of the LRR domain is involved in intramolecular binding with the ARC1 domain and the C-terminal part of the LRR domain is involved in recognition of Potato virus X (Rairdan and Moffett, 2006). For TNL1, if we consider that the first 213 amino acids of the LRR domain are involved in intramolecular and/or host target binding and the last 111 amino acids of the LRR domain are involved in direct or indirect pathogen recognition, the PL domains could be the target itself (decoy) or act in downstream signaling.
As hypothesized by Shen and Schulze-Lefert (2007), if nuclear action of the R protein is a widespread phenomenon, its interception points with the transcriptional machinery would constitute an Achilles’ heel for immune sabotage by pathogens. The Ma gene may prevent this sabotage: by proposing several decoys as targets to pathogens, it would constitute a sort of “trolling line” with five baits to detect diverse pathogen Avr products directed to the plant nucleus. The unusual number of baits may reduce the opportunities for the Avr product to simultaneously escape its recognition by Ma and keep its efficiency against its primary target. Finally, the Ma gene may constitute a contraction and an evolution, through intramolecular interactions and PL domain duplications, of the decoy model.
The Ma resistance has a wide spectrum (Esmenjaud et al., 1994; Lecouls et al., 1997) and is not overcome under high inoculum pressure (Esmenjaud et al., 1996a), which suggests that this resistance response cannot be easily counteracted by the nematodes. The similarity of the PL domains to the WRKY transcription factors suggests that the key targets of Meloidogyne species could be WRKY transcription factors that are known to be essential for the plant defense response and development (Rushton et al., 2010) and, more specifically, for basal response to RKN in tomato and Arabidopsis (Bhattarai et al., 2010). The finding that the TNL-WRKY protein RRS1 acts together with RPS4 (a TNL protein possessing a single PL domain; Fig. 6) for resistance against fungal and bacterial pathogens (Narusaka et al., 2009) suggests that complex defense strategies involving R proteins and WRKY transcription factors can decide the issues of plant pathogen interactions. Future research on RKN host targets will benefit from the sequencing of the M. incognita genome (Abad et al., 2008), which should provide new candidate Avr genes and maybe nematode pathogenicity factors targeted to the plant nucleus.
MATERIALS AND METHODS
Plant Material
The heterozygous resistant Myrobalan plum (Prunus cerasifera) accession P.2175 (Ma1/ma) and several susceptible clonal accessions (ma/ma) were used to produce 1,780 segregating offspring. To prevent any interference between genes or factors other than Ma, the RKN species Meloidogyne floridensis (Handoo et al., 2004), only controlled by this latter gene, was used to discriminate Ma-resistant and -susceptible individuals (Supplemental Table S2).
DNA Extraction and PCR Experiments
Genomic DNA was extracted from frozen leaves (Saghai-Maroof et al., 1984). Sequence-characterized amplified region, cleaved amplified polymorphic sequence, and SSR markers (Supplemental Table S1) amplifications were performed as described by Dirlewanger et al. (2004a). For SCAFLP2, SCAFLP4, and Perepkin markers (Supplemental Table S1), 0.3 pmol of the forward primer was [γ-33P]ATP end labeled with polynucleotide kinase, and PCR products were separated on a 5% denaturing sequencing gel in 0.5× Tris-borate/EDTA buffer and visualized following autoradiography.
BAC Sequencing and Sequence Analysis
Sequencing of BACs 76H19 (accession no. FM253563) and 40K9 (accession no. FM253564) from P.2175 was carried out at the Centre National de Séquençage as described by Chantret et al. (2005). Gene prediction was performed using GENESCAN (Burge and Karlin, 1997), GENEMARK.hmm (Lomsadze et al., 2005), and EuGene (Schiex et al., 2001). For TNL genes, sequences were aligned using ClustalW (Thompson et al., 2002) and corrected manually using BLAST results and the cDNA sequences obtained. TNL mutation rates were analyzed using MEGA version 4 (Tamura et al., 2007) with the method of Nei and Gojobori (1986), and a codon-based Z test for neutral, purifying, or positive selection was performed.
Full-Length cDNA
Total RNA was obtained from ground roots using the TQ RNA Cells & Tissues Kit (Talent). First-strand cDNA was synthesized from oligo(dT) and specific primers using the SuperScript II kit (Invitrogen). Amplification was performed using Taq DNA polymerase (Invitrogen). PCR products were cloned with pGEMT (Promega), and 5′ and 3′ UTRs were synthesized using 5′ and 3′ RACE-PCR (5′ and 3′ RACE System for RACE; Invitrogen). For both RACE-PCRs, we used the adapter primers provided in the supplier’s kit. All experiments were performed following the supplier’s instructions. Specific primers for TNL1, TNL2, and TNL3 (Fig. 2; Supplemental Table S3) were designed in polymorphic regions from the alignment of the six sequences (TNL1–TNL3 and tnl1–tnl3) predicted by GENSCAN. The full-length cDNA of TNL1 was then obtained by amplifying and assembling sequences covering the entire gene (Fig. 2).
Complementation Using Hairy Roots and Composite Plants
Plants from the vigorous accession 253, susceptible to all five RKN species, Meloidogyne incognita, Meloidogyne arenaria, Meloidogyne javanica, M. floridensis, and Meloidogyne mayaguensis (= Meloidogyne enterolobii; Supplemental Table S2), were cultured in Murashige and Skoog propagation medium (Murashige and Skoog, 1962). The binary vector pKGW,0 (Karimi et al., 2005), genomic insert of approximately 15.3 kb from BAC 76H19 and containing the TNL1 candidate gene under the control of its native promoter (Fig. 3), was transformed into Agrobacterium rhizogenes strain A4R (Tepfer, 1990) by electroporation. Microplants were inoculated by injecting the agrobacteria solution 5 to 10 mm above the base of the stem with a sterile syringe as described by Bosselut et al. (2011). Cocultivation of the plants and the agrobacteria was performed at 25°C for 5 d. Independent transformation experiments were performed in order to (1) produce hairy roots in petri dishes and (2) generate composite plants (transformed hairy roots plus nontransformed aerial part; Bosselut et al., 2011). Hairy roots were grown onto hormone-free Murashige and Skoog basal propagation medium with 500 mg L−1 cefotaxime to eliminate agrobacteria and kept dark by wrapping the dishes in aluminum foil. Roots that developed autonomously were grown for 4 weeks, divided, and transferred individually into petri dishes onto a fresh medium to increase the stock for nematode experiments. Composite plants were grown on the same propagation medium and then transferred into a container filled with perlite substrate for acclimation. During plant or root transfer, root samples were collected in liquid nitrogen and stored at −80°C until molecular analysis. In vitro screening of transformants with kanamycin was not possible because of the susceptibility of Prunus plantlets to this antibiotic (Petri and Burgos, 2005). The integration of the T-DNA binary plasmid into the roots of accession 253 was thus analyzed by amplification of genomic DNA and cDNA sequences and Southern-blot hybridization. Genomic sequences were amplified using primers in the nptII gene from the pKGW vector (forward, 5′-TCAGAAGAACTCGTCAAGAA-3′; reverse, 5′-AACAAGATGGATTGCACGCA-3′) and in the TNL1 genomic sequence within the first intron (NSCALF2) and the PL4-PL5 region (CT3-4N) of the gene (Supplemental Table S3). Transcription of TNL1 was assessed using the specific cDNA fragments CT3 and Am5′ (Fig. 2; Supplemental Table S3). Southern-blot analysis was performed with nptII probe hybridized on EcoRV- and ClaI-digested genomic DNA.
Nematode Infection of Transformed Hairy Roots and Composite Plants
For hairy roots, independent transformation events (obtained from a single initial sectioned root) of TNL1-transformed (pKGW,TNL1-253) and empty vector-transformed (pKGW,0-253) accession 253 were multiplied to generate clonal material for infection with M. incognita, M. mayaguensis (= M. enterolobii), and a mixture of individuals of M. arenaria, M. javanica, and M. floridensis. Roots were grown in petri dishes, and a minimum of two roots per transformant and RKN species were inoculated, 3 weeks after transfer, with 5,000 sterile J2s distributed onto apices, and then rated after 3 weeks. Composite plants, approximately 2 months after rooting, were transplanted into trays filled with a perlite substrate. Four weeks after acclimation, plants were transplanted individually into 200-mL peat moss:sand:silt clay pots (1:1:1, v/v/v) at 25°C in a phytotron. Together with composite plants of the TNL1-transformed and empty vector-transformed accession 253, untransformed control plants of the accessions 253, P.2032, and P.2175 (Supplemental Table S2) were evaluated successively over a series of four 8-week-long tests (i.e. two complete nematode cycles each) using the same five RKN species as for hairy root tests.
The sequences reported in this paper have been deposited in the EMBL database (accession nos. FM253563 and FM253564).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignments of repeats in the NLL and LRR exons in TNL1 (A) and duplication of the LRR repeats 5 to 7 and 8 to 10 (B).
Supplemental Figure S2. Sequence alignment of the PL3 to PL5 exons from TNL1 illustrating that these three exons derived from ancient duplications.
Supplemental Figure S3. Comparison of the putative D box from the TNL1 allele and from the parsley (Petroselinum crispum) PR2 promoter as given by Rushton et al. (2002).
Supplemental Table S1. Primer sequence, product sizes, and positions of markers of the Ma1 gene on the sequence of BAC 76H19.
Supplemental Table S2. Resistance phenotype and genotype to RKN Meloidogyne species of material used (1) as parents for Ma high-resolution mapping and (2) for complementation experiments.
Supplemental Table S3. Primers used for the cDNAs of TNL1, TNL2, and TNL3.
Supplementary Material
Acknowledgments
We thank P. Arus (Institut de Recerca i Tecnologia Agroalimentàries) and A. Abbott (Clemson University) for assistance in the initial steps of mapping, H. Duval (INRA) for his contribution to seed production, H. Etienne (CIRAD) for advice on transformation with agrobacteria, and E. Deleury (INRA) for the bioinformatics tools in sequence analyses. We are grateful to J. Polidori for his technical assistance in high-resolution mapping, transcription studies, and photographs.
References
- Abad P, Gouzy J, Aury J-M, Castagnone-Sereno P, Danchin EGJ, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, et al. (2008) Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol 26: 909–915 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB. (2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146: 5–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. (2000) The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci 25: 421–423 [DOI] [PubMed] [Google Scholar]
- Ballvora A, Pierre M, van den Ackerveken G, Schornack S, Rossier O, Ganal M, Lahaye T, Bonas U. (2001) Genetic mapping and functional analysis of the tomato Bs4 locus governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol Plant Microbe Interact 14: 629–638 [DOI] [PubMed] [Google Scholar]
- Belfanti E, Silfverberg-Dilworth E, Tartarini S, Patocchi A, Barbieri M, Zhu J, Vinatzer BA, Gianfranceschi L, Gessler C, Sansavini S. (2004) The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc Natl Acad Sci USA 101: 886–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T. (2010) WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J 63: 229–240 [DOI] [PubMed] [Google Scholar]
- Blok VC, Jones JT, Phillips MS, Trudgill DL. (2008) Parasitism genes and host range disparities in biotrophic nematodes: the conundrum of polyphagy versus specialisation. Bioessays 30: 249–259 [DOI] [PubMed] [Google Scholar]
- Bosselut N, Van Ghelder C, Claverie M, Voisin R, Onesto JP, Rosso MN, Esmenjaud D. (2011) Agrobacterium rhizogenes-mediated transformation of Prunus as an alternative for gene functional analysis in hairy-roots and composite plants. Plant Cell Rep (in press) [DOI] [PubMed] [Google Scholar]
- Brito JA, Stanley JD, Mendes ML, Kaur R, Cetintas R, Di Vito M, Thies JA, Dickson DW. (2007) Effects of the Mi-1, N, and Tabasco genes on infection and reproduction of Meloidogyne mayaguensis on tomato and pepper genotypes. J Nematol 39: 327–332 [PMC free article] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. (2005) Protein structure prediction servers at University College London. Nucleic Acids Res 33: W36– W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Castagnone-Sereno P, Bongiovanni M, Palloix A, Dalmasso A. (1996) Selection for Meloidogyne incognita virulence against resistance genes from tomato and pepper and specificity of the virulence/resistance determinants. Eur J Plant Pathol 102: 585–590 [Google Scholar]
- Cetintas R, Kaur R, Brito JA, Mendes ML, Nyczepir AP, Dickson DW. (2007) Pathogenicity and reproductive potential of Meloidogyne mayaguensis and M. floridensis compared with three common Meloidogyne spp. Nematropica 37: 21–31 [Google Scholar]
- Chantret N, Salse J, Sabot F, Rahman S, Bellec A, Laubin B, Dubois Y, Dossat C, Sourdille P, Joudrier P, et al. (2005) Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie M, Bosselut N, Lecouls AC, Voisin R, Lafargue B, Poizat C, Kleinhentz M, Laigret F, Dirlewanger E, Esmenjaud D. (2004a) Location of independent root-knot nematode resistance genes in plum and peach. Theor Appl Genet 108: 765–773 [DOI] [PubMed] [Google Scholar]
- Claverie M, Dirlewanger E, Cosson P, Bosselut N, Lecouls AC, Voisin R, Kleinhentz M, Lafargue B, Caboche M, Chalhoub B, et al. (2004b) High-resolution mapping and chromosome landing at the root-knot nematode resistance locus Ma from Myrobalan plum using a large-insert BAC DNA library. Theor Appl Genet 109: 1318–1327 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA 99: 2404–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Baker BJ. (2000) Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA 97: 1908–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirlewanger E, Cosson P, Howad W, Capdeville G, Bosselut N, Claverie M, Voisin R, Poizat C, Lafargue B, Baron O, et al. (2004a) Microsatellite genetic linkage maps of Myrobalan plum and an almond-peach hybrid: location of root-knot nematode resistance genes. Theor Appl Genet 109: 827–838 [DOI] [PubMed] [Google Scholar]
- Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howad W, Arus P. (2004b) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci USA 101: 9891–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. (2001) Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13: 163–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremin VG. (1978) Genetic potential of species Prunus cerasifera Ehr. and its use in breeding. Acta Hortic 74: 61–65 [Google Scholar]
- Esmenjaud D, Minot JC, Voisin R. (1996a) Effect of durable inoculum pressure and high temperature on root galling, nematode numbers and survival of Myrobalan plum genotypes (Prunus cerasifera Ehr.) highly resistant to Meloidogyne spp. Theor Appl Genet 92: 873–879 [Google Scholar]
- Esmenjaud D, Minot JC, Voisin R, Bonnet A, Salesses G. (1996b) Inheritance of resistance to the root-knot nematode Meloidogyne arenaria in Myrobalan plum. Fundam Appl Nematol 19: 85–90 [DOI] [PubMed] [Google Scholar]
- Esmenjaud D, Minot JC, Voisin R, Pinochet J, Salesses G. (1994) Inter- and intraspecific resistance variability in Myrobalan plum, peach and peach-almond rootstocks using 22 root-knot nematode populations. J Am Soc Hortic Sci 119: 94–100 [Google Scholar]
- Gassmann W. (2008) Alternative splicing in plant defense. Curr Top Microbiol Immunol 326: 219–233 [DOI] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ. (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20: 265–277 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE. (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14: 177–193 [DOI] [PubMed] [Google Scholar]
- Handoo ZA, Nyczepir AP, Esmenjaud D, van der Beek JG, Castagnone-Sereno P, Carta LK, Skantar AM, Higgins JA. (2004) Morphological, molecular, and differential-host characterization of Meloidogyne floridensis n. sp (Nematoda: Meloidogynidae), a root-knot nematode parasitizing peach in Florida. J Nematol 36: 20–35 [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585– W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kirsch C, Logemann E, Lippok B, Schmelzer E, Hahlbrock K. (2001) A highly specific pathogen-responsive promoter element from the immediate-early activated CMPG1 gene in Petroselinum crispum. Plant J 26: 217–227 [DOI] [PubMed] [Google Scholar]
- Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelen D, Duchaussoy F, Meyers BC, Boerjan W, Martin F. (2008) Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol 66: 619–636 [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Dijkhuis J, Sturre MJ, de Haan P, Hille J. (2003) Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-2(2) from Lycopersicon esculentum. Plant Mol Biol 52: 1037–1049 [DOI] [PubMed] [Google Scholar]
- Lecouls AC, Bergougnoux V, Rubio-Cabetas MJ, Bosselut N, Voisin R, Poessel JL, Faurobert M, Bonnet A, Salesses G, Dirlewanger E, et al. (2004) Marker-assisted selection for the wide-spectrum resistance to root-knot nematodes conferred by the Ma gene from Myrobalan plum (Prunus cerasifera) in interspecific Prunus material. Mol Breed 13: 113–124 [Google Scholar]
- Lecouls AC, Salesses G, Minot JC, Voisin R, Bonnet A, Esmenjaud D. (1997) Spectrum of the Ma genes for resistance to Meloidogyne spp. in Myrobalan plum. Theor Appl Genet 95: 1325–1334 [Google Scholar]
- Lescot M, Rombauts S, Zhang J, Aubourg S, Mathe C, Jansson S, Rouze P, Boerjan W. (2004) Annotation of a 95-kb Populus deltoides genomic sequence reveals a disease resistance gene cluster and novel class I and class II transposable elements. Theor Appl Genet 109: 10–22 [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. (2005) Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33: 6494–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang HH, Michelmore RW. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8: 1113–1130 [DOI] [PubMed] [Google Scholar]
- Miller G, Mittler R. (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot (Lond) 98: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM. (1998) The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10: 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Palomino M, Meyers BC, Michelmore RW, Gaut BS. (2002) Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res 12: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige TF, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 60: 218–226 [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y, Ito T, Seki M, Nakashita H, Yoshida S, Marco Y, Shirasu K, Shinozaki K. (2005) A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J 43: 873–888 [DOI] [PubMed] [Google Scholar]
- Nyczepir AP, Brito JA, Dickson DW, Beckman TG. (2008) Host status of selected peach rootstocks to Meloidogyne mayaguensis. HortSci 43: 804–806 [Google Scholar]
- Paal J, Henselewski H, Muth J, Meksem K, Menendez CM, Salamini F, Ballvora A, Gebhardt C. (2004) Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J 38: 285–297 [DOI] [PubMed] [Google Scholar]
- Petri C, Burgos L. (2005) Transformation of fruit trees: useful breeding tool or continued future prospect? Transgenic Res 14: 15–26 [DOI] [PubMed] [Google Scholar]
- Rairdan GJ, Moffett P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Reinstadler A, Lipka V, Lippok B, Somssich IE. (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasser JN, Freckman DW. (1987) A world perspective on nematology: the role of the society. Veech JA, Dickson DW, , Vistas on Nematology. Society of Nematologists, Hyattsville, MD, pp 7–14 [Google Scholar]
- Schiex T, Moisan A, Rouzé P. (2001) Eugène: an eukaryotic gene finder that combines several type of evidence. Paper number 2066. Gascuel O, Sagot M-F, , Computational Biology. Selected Papers from JOBIM 2000 in Lecture Notes in Computer Sciences. Springer-Verlag, Berlin, pp 118–133 [Google Scholar]
- Shen QH, Schulze-Lefert P. (2007) Rumble in the nuclear jungle: compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J 26: 4293–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Collins NC, Ayliffe M, Smith SM, Drake J, Pryor T, Hulbert SH. (2001) Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tepfer D. (1990) Genetic transformation using Agrobacterium rhizogenes. Physiol Plant 9: 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2, Unit 2.3 [DOI] [PubMed] [Google Scholar]
- Trudgill DL, Blok VC. (2001) Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu Rev Phytopathol 39: 53–77 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Tzortzakakis EA, Adam MAM, Blok VC, Paraskevopoulos C, Bourtzis K. (2005) Occurrence of resistance-breaking populations of root-knot nematodes on tomato in Greece. Eur J Plant Pathol 113: 101–105 [Google Scholar]
- Van der Biezen EA, Jones JD. (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RA, Kamoun S. (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Williamson VM, Kumar A. (2006) Nematode resistance in plants: the battle underground. Trends Genet 22: 396–403 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L, Zhang Y, Jones JD, Parker JE. (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Watanabe S, Tateno M, Seki M, Shinozaki K, Yokoyama S. (2008) Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol Biochem 46: 394–401 [DOI] [PubMed] [Google Scholar]
- Yang B, Eisenback JD. (1983) Meloidogyne enterolobii n.sp. (Meloidogynidae), a root-knot nematode parasitizing pacara earpot tree in China. J Nematol 15: 381–391 [PMC free article] [PubMed] [Google Scholar]
- Yang ZN, Ye XR, Molina J, Roose ML, Mirkov TE. (2003) Sequence analysis of a 282-kilobase region surrounding the citrus Tristeza virus resistance gene (Ctv) locus in Poncirus trifoliata L. Raf. Plant Physiol 131: 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.