Abstract
We examined ways in which the Brown planthopper induced008a (Bphi008a; AY256682) gene of rice (Oryza sativa) enhances the plant’s resistance to a specialist herbivore, the brown planthopper (BPH; Nilaparvata lugens). Measurement of the expression levels of ethylene synthases and of ethylene emissions showed that BPH feeding rapidly initiated the ethylene signaling pathway and up-regulated Bphi008a transcript levels after 6 to 96 h of feeding. In contrast, blocking ethylene transduction (using 1-methylcyclopropene) reduced Bphi008a transcript levels in wild-type plants fed upon by BPH. In vitro kinase assays showed that Bphi008a can be phosphorylated by rice Mitogen-activated Protein Kinase5 (OsMPK5), and yeast two-hybrid assays demonstrated that the carboxyl-terminal proline-rich region of Bphi008a interacts directly with this kinase. Furthermore, bimolecular fluorescence complementation assays showed that this interaction occurs in the nucleus. Subsequently, we found that Bphi008a up-regulation and down-regulation were accompanied by different changes in transcription levels of OsMPK5, OsMPK12, OsMPK13, and OsMPK17 in transgenic plants. Immunoblot analysis also showed that the OsMPK5 protein level increased in overexpressing plants and decreased in RNA interference plants after BPH feeding. In transgenic lines, changes in the expression levels of several enzymes that are important components of the defenses against the BPH were also observed. Finally, yeast two-hybrid screening results showed that Bphi008a is able to interact with a b-ZIP transcription factor (OsbZIP60) and a RNA polymerase polypeptide (SDRP).
Plants are sessile and constantly exposed to diverse external biotic stresses during their life cycle (e.g. viruses, pathogens, and pest infestations). Plant defenses to external biotic stresses differ from those of mammals, which have evolved specialized mobile defense lymphocytes and a somatic acquired immune system that provide protection against external stimuli. Instead, plants depend on a preexisting innate immune system in each cell (Ausubel, 2005). The innate immunity of plants can provide them with nonhost resistance and host resistance against external biotic stresses. In addition, mutant and transcription analyses have shown that several low-molecular-mass signal molecules, including salicylic acid (SA), jasmonic acid (JA), and ethylene (Et), play important roles in the regulation of signal transduction pathways involved in plant innate immune responses (Zhu-Salzman et al., 2004; Glazebrook, 2005). Among the signaling pathways involved in innate immune responses of plants, canonical mitogen-activated protein kinase (MAPK) signaling cascades act as bridges between diverse effectors or signal sensors and downstream defense genes (Ausubel, 2005; Pitzschke et al., 2009).
In the ongoing battle between plants and pathogens, the innate immunity of plants is induced by the conserved microbial feature microbe- or pathogen-associated molecular patterns (PAMPs) and special effectors, which are recognized by the plant’s Leu-rich repeat receptor kinase, and then initiate PAMP-triggered immunity (PTI) and effector-triggered immunity, respectively (Chisholm et al., 2006; Jones and Dangl, 2006). This host-pathogen defense model, which describes the direct or indirect recognition of pathogen Avr effectors and receptor proteins, has been genetically characterized as gene-for-gene resistance (Flor, 1971). A number of resistance (R) genes and their direct or indirect target effectors have been cloned from many plant species and pathogens; their interaction mechanisms have also been elucidated in some cases (Chisholm et al., 2006). However, only two insect R genes have been cloned to date, Mi-1 and Bph14, which provide resistance to the root-knot nematode (Meloidogyne incognita) and the brown planthopper (BPH; Nilaparvata lugens), respectively. Furthermore, the mechanisms whereby plants recognize the nematode or BPH Avr effectors remain unknown (Rossi et al., 1998; Du et al., 2009), although RNA profiles of plant-herbivore interactions indicate that the signal perception and transduction pathways involved may be similar to those of plant-pathogen interactions (Voelckel and Baldwin, 2004; Kaloshian and Walling, 2005; Wang et al., 2008).
Plant-herbivore interactions generally induce complex responses in plants, including the following. Various housekeeping genes that are involved in photosynthesis and general metabolism are suppressed in plant interactions with pathogens as well as herbivores (Scheideler et al., 2002; Kaloshian and Walling, 2005). This implies that plants possess mechanisms that improve their defenses against external stress by “sacrificing” some of the carbon and nitrogen resources required for growth and development. In addition, several important signaling pathways are activated, including JA, Et, and/or SA signaling pathways, following the production of reactive oxygen species. For example, JA contents increase in response to some necrotrophic fungal pathogens and arthropod herbivores, such as the tissue-damaging herbivores Manduca sexta and Trichoplusia ni (Farmer et al., 2003; Howe and Jander, 2008). SA is also required for the activation of systemic acquired resistance, a hypersensitive response induced by reactive oxygen species that triggers a type of programmed cell death thought to limit the access of biotrophs to water and nutrients (Glazebrook, 2005). However, it should be noted that (unlike pathogens) herbivores can avoid barren cells that have been stripped of resources by programmed cell death, since they are mobile and can thus simply move to other parts of the plant. This suggests that coinduced signaling pathways may synergistically protect plants against herbivore attack. For instance, Et sometimes acts synergistically with JA in the regulation of both stress and developmental responses, in which the transcription factor Ethylene Response Factor1 (ERF1) serves as a genetic bridge between the JA and Et signaling pathways (Lorenzo et al., 2003). Further typical responses to herbivory include the activation of calcium flux and MAPK cascades, in which the calcium ions (Ca2+) act as important second messengers (Maffei et al., 2007; Howe and Jander, 2008). Finally, the signaling pathways, calcium fluxes, and MAPK cascades collectively activate a series of downstream defense-related genes and the production of various important protein and secondary metabolites, including pathogen-related proteins, proteinase inhibitors, polyphenol oxidases, and compounds that target physiological processes in the attacking insect (Ussuf et al., 2001; Baldwin et al., 2002; Chen et al., 2005; Howe and Jander, 2008). Resistance mechanisms to phloem-feeding insects also include the induction of forisome dispersion, callose deposition, and (thus) phloem plugging, which prevent insects from continuously ingesting phloem sap from plants (Will et al., 2007; Hao et al., 2008).
MAPK cascades constitute a group of evolutionarily conserved Ser/Thr-specific protein kinases in eukaryotic organisms. These protein kinases can transduce extracellular stimuli into intracellular responses by either phosphorylating a target protein in the cytoplasm or directly translocating into the nucleus and regulating gene expression through the phosphorylation of specific transcription factors. These kinase cascade-mediated defense mechanisms are important signaling modules that are involved in the regulation of numerous abiotic and biotic stress responses (Jonak et al., 2002; Hamel et al., 2006; Howe and Jander, 2008). Recognition of PAMPs by plants may result in the activation of PTI via a MAPK cascade, but some bacterial effectors can block MAPK cascade-mediated innate immunity by suppressing the cascade upstream of MAPKKK (Asai et al., 2002; He et al., 2006). CTR1, a pivotal regulator involved in the Et signaling transduction pathway, is actually a Raf-like MAPKKK, and its regulation of downstream targets is mediated by a MAPK cascade (Kendrick and Chang, 2008). Furthermore, the AtMKK3-AtMPK6 cascade plays an important role in JA signaling in Arabidopsis (Arabidopsis thaliana; Takahashi et al., 2007). Although no complete MAPK signaling pathway that provides insect resistance has been described, there is evidence that MAPK cascades play important roles in plant-insect interactions (Kandoth et al., 2007; Wu et al., 2007). The cited studies indicate that the signaling events initiated by a diverse range of stresses have converged into a conserved MAPK cascade.
Phloem-feeding insects have a highly specialized feeding mode and cause complex damage to plants. Not only do these pests feed directly on the amino acids and carbohydrates of the host with a “stealthy” feeding mechanism but they may also act as vectors for plant viruses or pathogens, and they may deposit honeydew, thereby encouraging the growth of mold (Brown and Czosnek, 2002; Kempema et al., 2007). The BPH is a typical phloem-feeding insect that is a significant pest of rice (Oryza sativa), causing considerable crop losses globally, especially in Asia. However, little is known about how rice perceives and resists the BPH through its innate immune system, and few genes have been reported with roles in plant defenses against BPH infestation. An exception is Brown planthopper induced008a (Bphi008a; AY256682), a gene induced by both BPH feeding and ethephon (Yuan et al., 2004). The aim of this study was to explore the defense mechanism of rice against the BPH, in particular to clarify the roles of the Bphi008a gene in the rice innate immunity system and resistance to the BPH.
RESULTS
Overexpression and RNA Interference of Bphi008a Affect Rice Resistance to the BPH
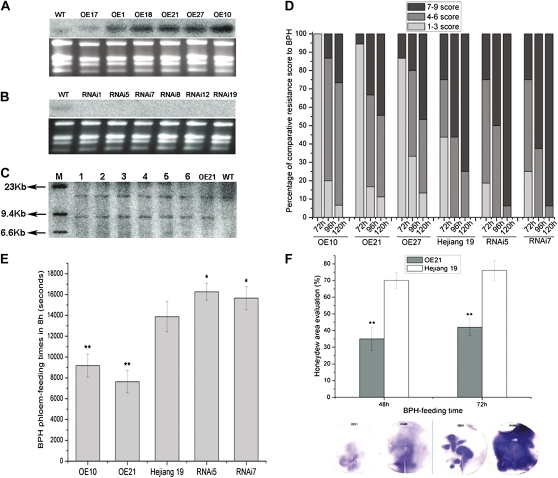
To clarify the role of Bphi008a in rice responses to the BPH, it was overexpressed (OE) or suppressed by double-stranded (ds) RNA interference (RNAi) in rice cv Hejiang 19. Northern-blot analysis showed that its expression was enhanced in six OE lines compared with wild-type plants (Hejiang 19) when they were not exposed to the BPH (Fig. 1A). From the T1 progeny of OE lines, we selected a homozygous transgenic plant, designated OE21-15, to generate offspring for further analyses; all of these offspring had a single-copy insertion, according to Southern-blot analysis (Fig. 1C). Since the endogenous expression level of Bphi008a in wild-type plants was relatively low under normal growth conditions, the effectiveness of RNAi in six T0 transgenic lines was examined in plants that had been infested with the BPH for 48 h. The results showed that Bphi008a expression was blocked in RNAi plants (Fig. 1B). None of the OE lines or RNAi lines exhibited obvious morphological changes (data not shown).
Figure 1.
Northern- and Southern-blot analysis of Bphi008a in wild-type and transgenic plants, and evaluation of its contribution to resistance to the BPH. A, Bphi008a expression levels in normal wild-type (WT) and six T0 OE plants. OE17 was a three-copy insertion plant, OE10 was a two-copy insertion plant, and the rest were one-copy insertion plants. B, Bphi008a expression levels in normal wild-type and six T0 RNAi plants after the BPH had been feeding for 48 h. C, Southern-blot analysis of OE21 T2 progeny. Lanes 1 to 6, Samples from six randomly chosen T2 plants; lane M, λDNA/HindIII marker. D, Evaluation of resistance to the BPH. Fifty plants of each line were chosen for analysis of percentage of severity score to BPH after the BPH had been feeding for 72, 96, and 120 h. Five independent experiments were performed with similar results, and the results of one representative experiment are shown. E, Electronic penetration graph evaluation of BPH phloem-feeding times on OE10, OE21, wild-type, RNAi5, and RNAi7 plants. Each bar represents the mean time ± se of five replicates. F, Honeydew area evaluation after the BPH had been feeding for 48 and 72 h on OE21 and wild-type plants. The size of the honeydew area and the intensity of the honeydew color indicate the BPH feeding activity. Standard bars indicate mean values from eight replicates. Significant differences in E and F are indicated with asterisks (* P < 0.05, ** P < 0.01, by Student’s t test). [See online article for color version of this figure.]
Fifteen-day-old seedlings of the OE, RNAi, and wild-type lines were infested with second- to third-instar BPH nymphs at a level of seven insects per seedling. After the BPH had been feeding for 72, 96, and 120 h, all plants of each wild-type and transgenic line were assigned a severity score of 0, 1, 3, 5, 7, or 9 (Huang et al., 2001), and the average scores (4.2 and 7, respectively, at 96 h) clearly indicated that the OE plants had greater resistance to the BPH than wild-type plants (Fig. 1D). In contrast, the change in susceptibility of rice to the BPH caused by RNAi of Bphi008a was not obvious, but we hypothesized that this may be because cv Hejiang 19 has a high susceptibility to the BPH (with a severity score exceeding 8.0). Hence, it may be difficult to differentiate subtle differences in susceptibility between the wild-type and RNAi lines. Therefore, to evaluate further the differences in susceptibility among the wild-type, OE, and RNAi plants, we used a real-time electronic penetration graph technique to monitor the feeding behavior of the BPH on them (Tjallingii, 2006; Hao et al., 2008). The results showed that the duration of phloem ingestion was approximately 40% lower on the two OE lines and approximately 15% higher on the two RNAi lines compared with the wild-type plants (Fig. 1E). These findings strongly indicate that the level of Bphi008a expression affects BPH feeding on the phloem of rice. Finally, the honeydew area was measured after the BPH had been feeding for 48 and 72 h. Clear differences were observed between the OE and wild-type plants (Fig. 1F), supporting the suggestion that Bphi008a expression enhances rice resistance to the BPH by impairing BPH feeding.
Selection of Housekeeping Genes for Real-Time PCR during BPH Feeding on Rice
As mentioned in the introduction, some housekeeping (HK) genes are suppressed in host-herbivore interactions. The current consensus is that multiple stably expressed HK genes are required for accurate and robust normalization (Schmittgen and Zakrajsek, 2000; Vandesompele et al., 2002). Therefore, to characterize changes in gene expression following BPH feeding on the wild-type and transgenic lines as accurately as possible by real-time PCR, we chose the most stably expressed candidates for normalization from the following 10 commonly used rice HK genes: β-actin, LSD1, GAPDH, eEF1α, SDHA, TBP, RPS27α, HSP, β-tubulin, and ubiquitin. These genes all belong to different functional classes, reducing the chances of coregulation (detailed information on the HK genes is provided in Supplemental Tables S1 and S2). The comparative expression levels of each HK gene at seven time points (0, 6, 12, 24, 48, 72, and 96 h after the BPH had been feeding on leaf sheaths) were determined by real-time PCR.
Using the geNorm algorithm, we ranked these 10 HK genes according to their comparative expression stability at the seven time points (Vandesompele et al., 2002). As shown in Figure 2, the results indicated that the most stably expressed combinations of HK genes were as follows: β-tubulin, HSP, and ubiquitin (in wild-type plants alone); TBP, ubiquitin, and HSP (in OE and wild-type plants); TBP, ubiquitin, and β-tubulin (in RNAi and wild-type plants); and LSD1 and ubiquitin (in wild-type controls and wild-type plants pretreated with the Et competitive inhibitor 1-methylcyclopropene [1-MCP]). To confirm the results of the geNorm algorithm, we also used another algorithm, NormFinder (Andersen et al., 2004), to examine quantitative PCR data of these 10 HK genes. This gave almost identical ranking trends for the genes to those of the geNorm algorithm, except for some minor differences that we attributed to the different emphases of the algorithms (Supplemental Fig. S1). After we had identified the most stably expressed HK genes in the studied lines, we used the Biogazelle qBasePlus relative quantification framework for further analysis of the real-time PCR data (Hellemans et al., 2007).
Figure 2.
Determination of the optimal numbers of control genes for normalization. Average expression stability values (M) of the remaining HK genes during stepwise exclusion of the least stably expressed HK gene in leaf sheath cDNA samples after BPH feeding for 0, 6, 12, 24, 48, 72, and 96 h. A, HK gene stability evaluation and choice in wild-type lines. B, HK gene stability evaluation and choice in OE and control wild-type lines. C, HK gene stability evaluation and choice in RNAi and control wild-type lines. D, HK gene stability evaluation and choice in 1-MCP-treated wild-type and control wild-type lines. E, Pairwise variation (Vn/n + 1) analysis between the normalization factors NFn and NFn + 1 to determine the number of control genes required for accurate normalization in our study. This experiment was repeated with two biological replicates, and we obtained the same results.
Bphi008a Is a Downstream Gene of the Et Signaling Pathway in Rice
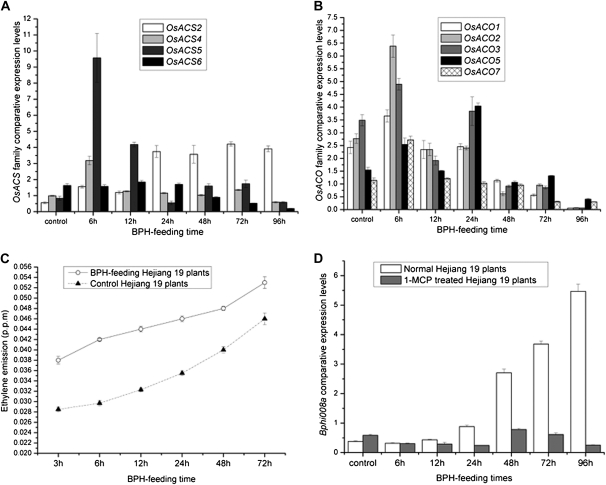
In a previous study, Bphi008a expression was found to be induced by both BPH feeding and spraying plants with ethephon, which slowly releases Et (Yuan et al., 2004). Therefore, we investigated changes in the expression of genes involved in Et biosynthesis in rice plants that had been fed upon by the BPH. In rice, a large gene family controls Et biosynthesis, composed of six 1-aminocyclopropane-1-carboxylic acid (ACC) synthase genes (designated OsACS1–OsACS6) and seven ACC oxidase genes (designated OsACO1–OsACO7; Yang and Hoffman, 1984; Iwai et al., 2006). We found that expression levels of all of the Et biosynthesis genes were increased after BPH feeding, except OsACS1, OsACS3, and OsACO4, whose transcription was not detected in rice cv Hejiang 19 (data not shown). Expression levels of three members of the ACC synthase subfamily (OsACS2, OsACS4, and OsACS5) were increased after BPH feeding for 6 h and then OsACS2 and OsACS5 expression levels remained high until 96 h, while OsACS4 expression levels were increased 2-fold after 6 h but then decreased to control levels. OsACS6 expression levels were almost unaffected by BPH feeding before 48 h but then began to decrease (Fig. 3A). In addition, expression levels of four members of the ACC oxidase subfamily (OsACO1, OsACO2, OsACO3, and OsACO7) had all increased by 6 h, but then they began to decrease and reached their lowest levels after 96 h of BPH feeding. OsACO5 expression levels were increased until 24 h of BPH feeding but then subsequently declined to just one-quarter of control levels after 96 h (Fig. 3B). Furthermore, as shown in Figure 3C, Et measurements at 3, 6, 12, 24, 48 and 72 h time points showed that levels of Et emitted from wild-type plants rapidly increased upon BPH feeding and were maximal at the end of the monitoring period (72 h). These results suggest that BPH feeding rapidly initiated Et biosynthesis.
Figure 3.
Bphi008a is a downstream gene of the Et signaling pathway. A and B, Comparative expression levels of members of the OsACS and OsACO family after BPH feeding for between 0 and 96 h on wild-type plants. C, Profiles of Et emissions from wild-type plants after BPH feeding for between 3 and 72 h. Values shown are means ± sd based on three independent experiments. The experiment was repeated two times with similar results. D, Bphi008a expression patterns with and without 1-MCP treatment in response to BPH feeding from 0 to 96 h on Hejiang 19 plants.
In addition, we used 1-MCP (C4H6), a volatile gas and Et inhibitor that competitively binds to the Et receptor in plants, to block the transduction of Et signals. We pretreated wild-type plants for 48 h with 1-MCP (10 μL L−1) and then investigated changes in the expression levels of Bphi008a after 0, 6, 12, 24, 48, 72, and 96 h of BPH feeding in these and control plants (which received no 1-MCP treatment). Bphi008a expression levels increased from 0 to 96 h in the controls, but in Hejiang 19 plants pretreated with 1-MCP, Bphi008a expression levels were very low (Fig. 3D). We also found no significant differences in the expression levels of OsACOs and OsACSs between the wild-type and transgenic plants, except that OsACO1 expression was increased after BPH feeding for 96 h and OsACO2 after BPH feeding for 72 and 96 h in the OE plants (Supplemental Fig. S2). Together, these results indicate that Bphi008a transcription is stimulated by active Et and that Bphi008a is a downstream gene of the Et signaling pathway in rice.
Detection of Bphi008a Expression Patterns and Localization in Rice
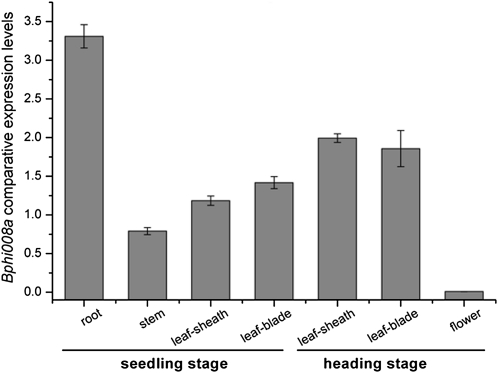
We examined relative expression levels of Bphi008a in leaf sheaths, stems, leaf blades, and roots of seedlings and in leaf sheaths, leaf blades, and flowers of plants at the heading stage. Using the NormFinder algorithm, we chose LSD1 as the most stably expressed HK gene in these seven types of samples (Supplemental Fig. S3). We found that Bphi008a expression levels were highest in the roots of seedlings, followed (in order) by leaf blades and leaf sheaths at the heading stage, then leaf sheaths, leaf blades, and stems of seedlings. The lowest detected level of Bphi008a expression was in the flowers (Fig. 4).
Figure 4.
Bphi008a expression patterns in rice from the seedling and heading stages. The HK gene used for normalization in this study was LSD1.
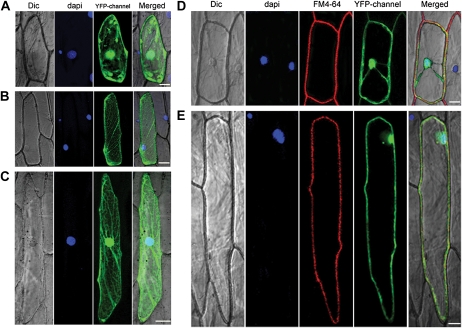
SMART sequence analysis by the Bork Group (http://www.bork.embl.de/j/) has shown that Bphi008a has an N-terminal transmembrane region somewhere between amino acid positions 7 and 29. To obtain preliminary indications of the function of Bphi008a, we determined its subcellular localization by translationally fusing full-length Bphi008a to the C terminus of the yellow fluorescent protein (YFP) and expressing the chimeric protein under the control of the cauliflower mosaic virus 35S promoter. In addition, we truncated the gene by removing the sequence encoding the 20 N-terminal amino acids, which might be the main transmembrane region of Bphi008a, and cloned the rest of the coding sequence (designated tnBph8a) into the C terminus of the YFP. Onion (Allium cepa) epidermal cells transiently expressing the YFP without Bphi008a showed signals throughout cells (Fig. 5A). We found that the YFP-tnBph8a fusion protein lost its plasma membrane localization and was mainly localized in the cytosol or in both the cytosol and the nucleus (Fig. 5, B and C), while cells transiently expressing the YFP-Bphi008a fusion protein predominantly showed YFP signals at the plasma membrane and the nucleus (Fig. 5D). These results suggest that Bphi008a has dual localization and that its N-terminal amino acids are likely to be involved in its localization.
Figure 5.
Subcellular localization of Bphi008a, tnBph8a, and OsMPK5. A, YFP protein localization in onion epidermal cells. B and C, YFP-tnBph8a fusion protein localization in onion epidermal cells. D, Dual YFP-Bphi008a protein localization in onion epidermal cells. E, Dual YFP-OsMPK5 protein localization in onion epidermal cells. Dic, Differential interference contrast. Bar = 50 μm.
Bphi008a Is Phosphorylated by OsMPK5 in Vitro
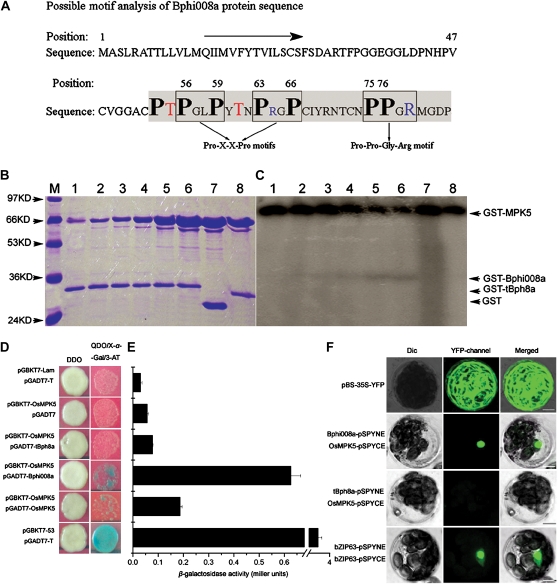
In attempts to elucidate the function of Bphi008a in signal transduction processes, we first analyzed its possible motifs. In the C-terminal Pro-rich region of Bphi008a, we found several Pro-related motifs, including two Pro-X-X-Pro motifs and one Pro-Pro-Gly-Arg motif (Fig. 6A; Kay et al., 2000; Kofler and Freund, 2006). In addition, two robust, comprehensive motif search algorithms, Scansite 2.0 and Eukaryotic Linear Motif, showed that it has a putative MAPK phosphorylation site and a conserved docking site for a Pro-directed MAPK at positions between amino acids 52 and 58 (Obenauer et al., 2003; Puntervoll et al., 2003). These findings suggested that it might be posttranslationally regulated through phosphorylation. OsMPK5, which is also involved in disease resistance and abiotic stress tolerance, was identified as a candidate target kinase for Bphi008a (Xiong and Yang, 2003). In addition, we cleaved the C-terminal 29 amino acids of Bphi008a to use the remaining coding sequence (designated tBph8a) as a negative control (Fig. 6A). We produced glutathione S-transferase (GST) fusion proteins containing OsMPK5, Bphi008a, and tBph8a in Escherichia coli and purified them. We then assayed their kinase activity in vitro, after checking the amounts of GST, GST-MPK5, GST-Bphi008a, and GST-tBph8a proteins by 12% (w/v) SDS-PAGE analysis (Fig. 6B). The results showed that GST-MPK5 has concentration-dependent autophosphorylation ability and the capacity to phosphorylate GST-Bphi008a. When less than 4 μg of GST-MPK5 was used in the assays, it showed strong autophosphorylation ability, and we only detected weak GST-Bphi008a phosphorylation (Fig. 6C, lanes 1-4). However, when we increased the amounts of GST-MPK5 to 16 μg, we detected a strong GST-Bphi008a phosphorylation band, accompanied by abatement of GST-MPK5 autophosphorylation (Fig. 6C, lane 6). Furthermore, no phosphorylation interaction between GST-MPK5 and GST-tBph8a was detected (Fig. 6C, lane 8). These results indicate that phosphorylation of Bphi008a’s C-terminal motif might have a function in vivo.
Figure 6.
Bphi008a interacts with OsMPK5 in vitro and in vivo. A, Possible motifs of the Bphi008a protein. Enclosed squares are two possible Pro-X-X-Pro motifs and one Pro-Pro-Gly-Arg motif. Bold black letters represent Pro residues in possible motifs; red letters represent possible Thr phosphorylated sites before the Pro-X-X-Pro motifs; blue letters represent positively charged Arg in motifs. The shaded region of the C terminus was also cleaved, and the remaining region was used as a negative control. B and C, Phosphorylation of the Bphi008a by OsMPK5 in vitro. Lanes 1 to 6, Kinase activities obtained from incubating 3 μg of GST-Bphi008a protein with 0.5, 1, 2, 4, 8, and 16 μg of GST-OsMPK5 protein, respectively, in an in vitro assay; lanes 7 and 8, kinase activities obtained from incubating 3 μg of GST and GST-tBph8a protein, respectively, with 16 μg of GST-OsMPK5 protein in an in vitro assay; lane M, protein masses in kD. D, Yeast two-hybrid assay of the interaction between Bphi008a or tBph8a and OsMPK5. The diploids were grown on DDO plates and transferred to QDO/X-α-Gal/3-amino-1,2,4-triazole (3-AT) plates with 20 μg mL−1 X-α-Gal and 10 μm 3-AT for 3 d. E, Relative β-galactosidase activities obtained were consistent with the results observed on QDO/X-α-Gal/3-AT plates. F, BiFC visualization of the Bphi008a-OsMPK5 and tBph8a-OsMPK5 interaction in transiently coexpressed Arabidopsis mesophyll protoplasts. The pBS-35S-YFP vector served as a transfected control, and the Arabidopsis nuclear protein bZIP63 served as a BiFC positive control. Dic, Differential interference contrast. Bars = 10 μm.
Bphi008a Interacts with OsMPK5 in the Nucleus
Since the interaction between OsMPK5 and Bphi008a involved phosphorylation in vitro, we wanted to determine whether these two proteins could interact in vivo. For this purpose, complete Bphi008a, tBph8a,and OsMPK5 coding sequences were separately cloned into pGADT7 and pGBKT7 vectors to create a yeast two-hybrid system. Among all 17 mating combinations, we found one (designated no. 15) in addition to the positive control that showed a clear positive interaction on the QDO/X-α-Gal plate, which corresponded to pGBKT7-OsMPK5 mating with pGADT7-Bphi008a (Fig. 6D; Supplemental Table S3). In addition, combinations 12 and 16 (corresponding to pGBKT7-Bphi008a mating with pGADT7-OsMPK5 and pGBKT7-OsMPK5 mating with pGADT7-OsMPK5, respectively) showed comparatively weak interactions. These findings imply that OsMPK5 has weak ability to interact with itself in vivo, in contrast to the strong autophosphorylation capacity observed in vitro (Fig. 6, C and D). Relative β-galactosidase activities were also measured and provided results consistent with those from the QDO/X-α-Gal/3-amino-1,2,4-triazole plate (Fig. 6E).
Following the confirmation that Bphi008a can interact with OsMPK5 in yeast cells, we tested the possibility that these two proteins interact in plant cells using a particle gun-mediated system. We found that OsMPK5 also had dual localization, coinciding with that of Bphi008a (Fig. 5E), and postulated that if these two proteins interact in plant cells, they presumably do so in the nucleus and/or the plasma membrane. To test this hypothesis, we transiently coexpressed Bphi008a (or tBph8a) tagged with pSPYNE (split YFP N-terminal fragment expression) and OsMPK5 tagged with pSPYCE (split YFP C-terminal fragment expression) using Arabidopsis protoplast transfection (Walter et al., 2004; Yoo et al., 2007). As anticipated, the results indicated that Bphi008a and OsMPK5 can interact with each other in the nucleus but not in the plasma membrane. Meanwhile, we detected no interaction between tBph8a and OsMPK5 in protoplasts (Fig. 6F). Similar bimolecular fluorescence complementation (BiFC) results were observed in onion epidermal cells using the particle gun-mediated system (Supplemental Fig. S5).
Bphi008a Overexpression and RNAi Result in Different OsMPK Expression Patterns
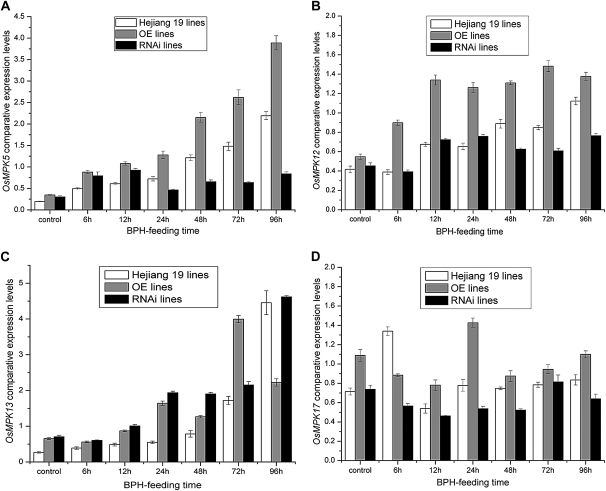
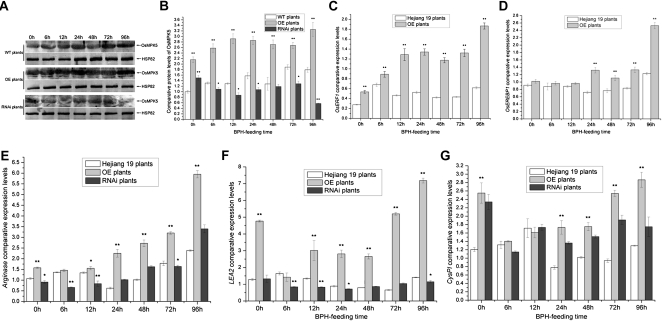
The BiFC and kinase assay results showed that OsMPK5 can interact with Bphi008a by mediating its C-terminal Pro-directed phosphorylation and that this interaction occurs in the nucleus. We speculated that phosphorylated Bphi008a plays a role in MAPK signaling pathways. In rice, we had identified 17 members of the MAPK family (OsMPK1–OsMPK17), four of which (OsMPK5, OsMPK12, OsMPK13, and OsMPK17) can be induced by both treatment with ACC (an Et precursor) and pathogen (Magnaporthe grisea) infection (Reyna and Yang, 2006). We found that expression levels of these four genes also increased in response to BPH feeding (most strongly after 96 h, except for OsMPK17, which was induced after the BPH had been feeding for only 6 h; Fig. 7). These findings suggested that the conserved MAPK signaling pathways serve as bridges in response to hormones and biotic stresses. In the OE plants (OE21-15), we found that expression levels of these four genes increased compared with the wild type in both controls and plants fed on by the BPH (Fig. 7). However, we found that expression levels of both OsMPK12 and OsMPK17 remained unchanged in Bphi008a-suppressed (RNAi7) plants that had not been exposed to the BPH, but they declined after BPH feeding, compared with levels in wild-type and OE plants (Fig. 7, B and D). Furthermore, OsMPK5 expression levels increased before the BPH had been feeding for 24 h, compared with levels in wild-type plants, but decreased notably in response to BPH feeding from 24 to 96 h in RNAi plants (Fig. 7A). Unexpectedly, OsMPK13 expression levels were increased in both OE and RNAi plants compared with wild-type plants (Fig. 7C). In addition, immunoblot analysis showed that the OsMPK5 protein level was increased after BPH had been feeding for 6 h and remained at a high level until 96 h in the wild-type plants. Compared with the wild-type plants, OE plants had consistently higher OsMPK5 protein levels, and RNAi plants had significantly lower levels, after BPH feeding (Fig. 8, A and B). These results suggest that Bphi008a-induced resistance to BPH is directly related to the MAPK signaling pathway and that one or more other genes may mediate the regulation of MAPK expression after BPH feeding. This could partly compensate for the reduction in Bphi008a expression caused by RNAi, such as OsMPK13. The expression levels of the OsMPKs increased in OE lines following BPH feeding; therefore, some downstream transcription factors and defense-related genes might be activated, finally resulting in increased resistance to the BPH.
Figure 7.
OsMPK5, OsMPK12, OsMPK13, and OsMPK17 expression patterns in transgenic and wild-type plants after the BPH had been feeding for between 0 and 96 h. Corresponding Bphi008a expression levels in transgenic and wild-type plants are shown in Supplemental Figure S4.
Figure 8.
A, Immunoblot analysis of OsMPK5 in transgenic and wild-type (WT) plants after the BPH had been feeding for between 0 and 96 h. Protein (50 μg) was separated by SDS-PAGE, electroblotted, and probed with rabbit OsMPK5 polyclonal antibody (1:1,000). After incubation with goat anti-rabbit IgG (1:10,000) conjugated to alkaline phosphatase, the complex was visualized using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium solution. Rice HSP82 (Q69QQ6) mouse monoclonal antibody (1:10,000) was used as a loading control. B, Comparative protein levels of OsMPK5 in wild-type and transgenic plants from three replicates. The western-blot signals of OsMPK5 were analyzed with Quantity One (Bio-Rad) and are shown with a histogram. C to G, Comparative expression levels of OsERF1, OsEREBP1, Arginase, LEA2, and CysPI after the BPH had been feeding for between 0 and 96 h in wild-type and transgenic plants. Each bar represents the mean value ± se of three replicates. Significant differences in B to G are indicated with asterisks (* P < 0.05, ** P < 0.01, by Student’s t test).
Bphi008a Overexpression Enhances Levels of AP2/ERF Transcription Factors and Defense-Related Genes
Recently, there have been claims that Et inactivates the negative regulator CTR1 in Arabidopsis, thereby activating a positive AtMKK9-AtMPK3/6 cascade that targets EIN3 through two MAPK phosphorylation sites (Yoo et al., 2008). Transcription factor EIN3/EIL is a key component of the primary Et signaling pathway, and the AP2/ERF transcription factor ERF1 serves as an immediate target for EIN3 in Arabidopsis (Guo and Ecker, 2004). Overexpression of ERF1 in Arabidopsis is sufficient to confer resistance to several necrotrophic fungi, such as Botrytis cinerea and Plectosphaerella cucumerina (Berrocal-Lobo et al., 2002). These findings have established that a linear AtMPK3/6-AtEIN3-AtERF1 signaling pathway in the nucleus participates in responses to external biotic stresses. The rice gene OsMPK5 has high similarity to AtMPK3, and both are associated with diverse biotic and abiotic stress responses (Mizoguchi et al., 1996; Xiong and Yang, 2003). In addition, OsMPK12, originally named OsBWMK1, mediates the expression of pathogen-related genes by phosphorylation-mediated activation of an AP2/ERF transcription factor, OsEREBP1, and enhances resistance of tobacco (Nicotiana tabacum) to the virulent oomycete pathogen Phytophthora parasitica var nicotianae (Cheong et al., 2003). This evidence suggests that a linear OsMPK5/12-OsERF1/OsEREBP1 signaling pathway may mediate responses to external biotic stresses in rice. We found that expression levels of OsERF1 and OsEREBP1 were also enhanced in OE plants compared with wild-type plants (Fig. 8, C and D), suggesting that Bphi008a-mediated enhancement of plant resistance to the BPH might be correlated with the activation of defense-related genes by some specific transcription factors.
We hypothesized that the suppression of BPH feeding in the Bphi008a OE lines described above (Fig. 1, E and F) may be ultimately related to the up-regulation of some defense-related genes, which could thwart BPH feeding. Some well-studied JA-inducible proteinase inhibitors (PIs) in tomato (Solanum lycopersicum) plants are reportedly involved in plant resistance to herbivorous insects by disrupting digestive processes in their gut. These PIs include arginase, Thr deaminase, Cys protease inhibitor (CysPI), and Leu aminopeptidase A (LEA). Arginase catabolizes the essential amino acid Arg in the M. sexta midgut and thus interferes with digestive processes, thereby increasing the resistance of tomato to M. sexta larvae (Chen et al., 2005). Our analyses showed that Arginase expression levels were increased in OE plants and somewhat decreased generally in RNAi plants when fed on by the BPH (Fig. 8E). Furthermore, we found that the expression levels of (especially) LEA2 and CysPI were also enhanced in the OE plants (Fig. 8, F and G). These results may explain why BPH feeding behavior was suppressed in OE plants.
Bphi008a Interacts with OsbZIP60 and SDRP in Yeast Cells
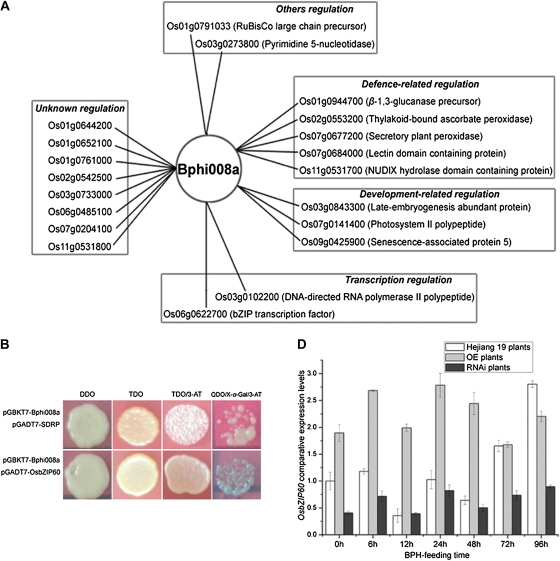
Bphi008a is clearly not an upstream gene of MAPK signaling pathways, so we speculated that the function of phosphorylated Bphi008a could be related to transcriptional regulation. Since Bphi008a has no DNA-binding domain, unlike many traditional transcriptional factors, we surmised that it may be a key member of a transcriptional complex, and thus could regulate the transcription of OsMPKs, via interactions between the Pro-X-X-Pro or Pro-Pro-Gly-Arg motifs and other components of the transcription machinery. To test our conjecture, Bphi008a was used as a bait to screen a prey cDNA rice library. The yeast two-hybrid screening resulted in 20 unique genes encoding putative Bphi008a-interacting proteins. The predicted functions of Bphi008a-interacting proteins are quite diverse but can be classified into five categories (Fig. 9A; Supplemental Table S4). Among them, we found two clones (Os03g0102200 and Os06g0622700) related to transcription regulation. Os03g0102200 is a 14.5-kD polypeptide that is similar to DNA-directed RNA polymerase II, and we renamed it SDRP, while Os06g0622700 is a bZIP (for basic Leu zipper) transcription factor that shows the highest homology to AtbZIP60 in Arabidopsis (Supplemental Fig. S6), so we renamed it OsbZIP60.
Figure 9.
Bphi008a-interacting proteins identified by yeast two-hybrid screening and its interaction with OsbZIP60 or SDRP in vivo. A, Bphi008a-interacting proteins identified by yeast two-hybrid screening. The Bphi008a-interacting proteins were classified into five categories (as indicated in boxes) based on their putative functions. B, Yeast two-hybrid assay of the interaction between Bphi008a and OsbZIP60 or SDRP. TDO/3-AT, Plates containing 5 mm 3-AT; QDO/X-α-Gal/3-AT, plates containing 10 mm 3-AT and 20 μg mL−1 X-α-Gal. C, Comparative expression levels of OsbZIP60 after the BPH had been feeding for between 0 and 96 h in wild-type and transgenic plants.
Complete Bphi008a and OsbZIP60 (or SDRP) coding sequences were cloned into pGBKT7 and pGADT7 vectors, respectively, to create a yeast two-hybrid system. As shown in Figure 9B, we detected a clear interaction between Bphi008a and OsbZIP60. In addition, we detected a weak interaction between Bphi008a and SDRP. Furthermore, analysis of OsbZIP60 expression levels in wild-type and transgenic plants after BPH feeding from 0 to 96 h (Fig. 9C) indicated that Bphi008a (possibly phosphorylated) might form a transcriptional complex with OsbZIP60 and SDRP in vivo that activates the transcription of target genes.
DISCUSSION
Rice-BPH Interactions Are Complicated and Might Involve Multiple Signaling Pathways
What signaling pathway(s) activated by extracellular stress are essential for plants to turn on the appropriate defense pathways in their innate immune systems? Our results indicate that when the BPH feeds on rice, upstream genes involved in Et production are rapidly up-regulated, within 6 h, and Et emission is continuously enhanced thereafter until 72 h (Fig. 3, A–C). In addition, transcript levels of two JA synthesis-related genes, AOS2 (encoding allene oxide synthase 2) and DoX2, and two SA synthesis-related genes, PAD4 (for phytoalexin deficient 4) and PAL (encoding Phe ammonia lyase), are continuously enhanced for at least 96 h during BPH feeding (Zhou et al., 1998; Glazebrook, 2005; Wei et al., 2009; Supplemental Fig. S7A). ERF1 is a member of the AP2/ERF transcription factor and plays a key role in the integration of JA and Et signals, thereby activating Et/JA-dependent responses to pathogens (Lorenzo et al., 2003). Our findings show that the expression of OsERF1 increases after BPH feeding, implying that a combined Et/JA signaling pathway might be involved in the defense of rice against the BPH (Fig. 8B). Thus, these results collectively indicate that BPH feeding not only induces the Et signaling pathway but might also activate JA and SA signaling pathways. However, as the BPH feeding time increased, especially after the BPH had been feeding for 24 h, we found that expression levels of all OsACO genes and OsACS4 began gradually to decrease, and all were lowest at the end of the monitoring period (96 h), compared with wild-type levels (Fig. 3, A and B). These changes in relative expression levels are very different from the pattern displayed by genes encoding components of the SA/JA pathways, whose expression levels consistently increased during BPH feeding (Supplemental Fig. S7A). These findings suggest that the BPH can protect itself from defenses induced by the Et signaling pathway by preventing the expression of some genes that participate in this pathway in the later stages of its feeding. Several researchers have reported analogous findings. For example, caterpillar (Helicoverpa zea) and silverleaf whitefly (Bemisia tabaci type B) can suppress JA defenses by inducing SA defenses (Musser et al., 2002; Zarate et al., 2007). These results suggest that the BPH might introduce effector proteins to plant cells that suppress certain signaling pathways and thus protect itself, in a similar fashion to the suppression of PTI by microbial pathogens (Chisholm et al., 2006; Jones and Dangl, 2006). Moreover, we also observed increased expression of many downstream pathogen-related genes, which was maximal at the end of the monitoring period, after 96 h of BPH feeding (Supplemental Fig. S7, B and C). In conclusion, when the BPH feeds on rice, it can immediately activate the Et signaling pathway; rice-BPH interactions are complicated and might involve JA and SA signaling pathways.
OsMPKs Are Involved in Immune Responses of Rice to the BPH
The way in which signals are transduced is key in the activation of the correct downstream targets of defense pathways in plants. Phosphorylation is an important posttranslational modification and a major regulatory mechanism that is estimated to affect one-third of the proteome and that controls many cellular signaling pathways (Ptacek et al., 2005). Conserved MAPK cascades play a key role (through series of phosphorylations) in the establishment of resistance to external stimuli in all eukaryotes (Ausubel, 2005). On the basis of experiments using transient expression in Arabidopsis protoplasts, the MAPK cascade MEKK1-MKK4/MKK5-MPK3/MPK6-WRKY29, in which WRKY29 could bind to W-box DNA elements (TGAC core sequence) that are found in the promoters of many defense-related genes, has been proposed to be responsible for the flg22-induced defense response (Asai et al., 2002). Seventeen MAPK genes have been identified in the rice genome, yet little is known about how these OsMPKs are involved in defense responses to herbivore attack through the innate immune system in rice. In our research, the expression levels of four MAPK genes (OsMPK5/12/13/17), which were also induced by ACC and M. grisea, were enhanced in response to BPH feeding (Fig. 7). In addition, the protein levels of OsMPK5 were also increased in wild-type and OE plants exposed to BPH feeding (Fig. 8, A and B). Although OsMPK1/5 has a high similarity to AtMPK6/3, we found that OsMPK1 does not respond to introduction of the pathogen M. grisea or to BPH feeding (Reyna and Yang, 2006; J. Hu and G. He, unpublished data). These findings suggest that regulation patterns in response to biotic stresses differ between Arabidopsis and rice. Furthermore, OsMPK5/12 can activate some downstream transcription factors, such as OsERF1 and OsEREBP1, through phosphorylation, which enhances their ability to bind to the target sequence AGCCGCC (i.e. the GCC box) contained in some defense-related gene promoters and further activate their transcription (Fig. 8, C and D; Cheong et al., 2003; Gutterson and Reuber, 2004). In this study, we found that OsMPK5 could also phosphorylate Bphi008a in vivo. Finally, we found enhanced expression levels of some downstream defense-related genes, such as Arginase, LEA2, and CysPI, which can hinder digestive processes in the insect midgut (Fig. 8, E–G). Since it is unclear whether there is a direct relationship between OsERF1/OsEREBP1 or Bphi008a and the activation of Arginase, CysPI, or LEA2, this warrants further investigation. However, our results demonstrate that several OsMPK genes play an important role in the response of rice to the BPH.
Bphi008a Might Be Involved in the Stress Response through Interaction with OsbZIP60
The yeast two-hybrid screening results showed that Bphi008a can interact with the transcription factor OsbZIP60, a homolog of AtbZIP60 in Arabidopsis and NtbZIP60 in tobacco (Supplemental Fig. S6). The membrane-bound transcription factor AtbZIP60, the activity of which is controlled by proteolytic cleavage, is considered to function as a sensor for endoplasmic reticulum stress and may regulate the expression of many endoplasmic reticulum chaperone genes by activating plant-UPR and ERSE cis-elements (Iwata and Koizumi, 2005; Iwata et al., 2008). Furthermore, size-exclusion chromatography analysis has shown that the nuclear form of AtbZIP60 exists as a protein complex of approximately 260 kD (Iwata et al., 2009). There has been limited research on the link between biotic stress defense and stress response in plants, but NtbZIP60 is reportedly involved in defense responses to the nonhost pathogen Pseudomonas cichorii and plays an important role in plant innate immunity (Tateda et al., 2008). Our result demonstrates that BPH feeding could suppress OsbZIP60 expression levels before 72 h; this is a different outcome from that of treatment with tunicamycin in Arabidopsis or inoculation with the nonhost pathogen P. cichorii in tobacco (Fig. 9C; Iwata et al., 2008; Tateda et al., 2008). These differences hint that the BPH may protect itself by introducing effector proteins that suppress stress responses. In the OE and RNAi plants, we also detected the up-regulation and down-regulation of OsbZIP60, respectively (Fig. 9C). We suggest that Bphi008a might involve the transcription of some OsMPKs through combination with OsbZIP60 and SDRP (Fig. 9B). Interestingly, we found that the expression levels of OsMPK13 and CysPI were also enhanced in RNAi plants (Figs. 7C and 8F). We hypothesize that other genes, besides Bphi008a, may also mediate defense responses of rice to the BPH through MAPK cascades, which could compensate for the RNAi-induced reduction of Bphi008a expression. Isolation and characterization of proteins that interact with the nuclear form of OsbZIP60 and examination of the mechanism whereby the phosphorylated (or nonphosphorylated) form of Bphi008a binds to OsbZIP60 in plant cells should yield valuable insights in this context. In addition, we should unravel how the OsbZIP60 transcription complex activates cis-elements of OsMPKs through chromatin immunoprecipitation analysis. Research in this area is currently under way.
MATERIALS AND METHODS
Gene Constructs and Rice Transformation
To make an overexpression construct, the Bphi008a coding sequence was cloned into the plasmid vector pAHC17. The resulting recombinant pAHC17-Bphi008a vector was sequenced to ensure the correct insert. After digestion of the pAHC17-Bphi008a vector with EcoRI and HindIII, the insert was ligated in frame into the plasmid vector pCAMBIA1301 to make the overexpression construct (Bphi008a-OE).
To make a dsRNAi construct, the complete Bphi008a sequence was cloned into the XhoI-EcoRI restriction enzyme sites in the pHANNIBAL vector (Wesley et al., 2001), thus creating pHAN-Bph8a1. The PCR fragment generated using the XbaI-HindIII pair of primers (to form the antisense fragment) was also XbaI-HindIII digested and cloned into the same sites in pHAN-Bph8a1 to create pHAN-Bph8a2. The pHAN-Bph8a2 vector was digested with NotI, and the insert was ligated in frame into the plasmid vector pART27 to form the dsRNAi construct (Bphi008a-RI). Overexpression and dsRNAi constructs were introduced into Agrobacterium tumefaciens (strain LBA4404) by electroporation using vigorously growing calli derived from mature embryos of rice (Oryza sativa ‘Hejiang 19’) following standard procedures (Hiei et al., 1994). T0 transgenic plants were grown in a greenhouse under 14-h-light/10-h-dark cycles at 28°C.
Plants, Insects, and BPH Resistance Evaluation
Japonica rice cv Hejiang 19, a line that is susceptible to the BPH (Nilaparvata lugens) with a severity score exceeding 8.0 in seedling bulk tests, was used in this study. Unless otherwise stated, the BPH insects were maintained on TN1 plants in the Genetics Institute at Wuhan University, and second to third instar nymphs were used in experiments (all of which were carried out on rice plants at the three-leaf stage).
Electronic penetration graph recordings were carried out for 8 h per insect per plant, with at least five replicates for each variety (using fresh seedlings and insects in each case), and the data acquired were analyzed as described previously (Hao et al., 2008). Honeydew area evaluation was carried out for 48 and 72 h per four insects per plant, with eight replicates for each variety. After the experiment was finished, the filter papers were rinsed with 0.25% ninhydrin and color developed in a 55°C drying oven. Photoshop 7.0 was used to calculate the percentage of colored area to total area.
DNA Isolation and Southern-Blot Analysis
Genomic DNA was extracted from rice leaves of wild-type (cv Hejiang 19) and transgenic plants using the cetyltrimethyl ammonium bromide method. The genomic DNA was digested with the restriction endonuclease DraI and then electrophoretically separated on a 0.8% agarose gel, transferred onto a Hybond N+ membrane (Amersham-Pharmacia), and probed with Bphi008a cDNA labeled with [α-32P]dCTP using the Prime-a-Gene labeling system (Promega). The blots were hybridized for more than 10 h at 65°C with the labeled probe, washed at 65°C for 15 min in 1× SSC and 0.2% SDS, and then kept at 65°C for 15 min in 0.5× SSC and 0.1% SDS. The membranes were finally exposed to x-ray film for autoradiography.
RNA Isolation, Northern-Blot Analysis, and First-Strand cDNA Synthesis from RNA
Total RNA was extracted from 15-d-old seedling tissue frozen in liquid nitrogen using TRIzol reagent (Invitrogen) followed by isopropyl alcohol precipitation. The RNA was then dissolved in diethyl pyrocarbonate-treated water. The concentration of RNA was determined by spectrophotometric measurements (Perkin-Elmer). For northern-blot analysis, total RNA (10 μg) was separated on a 1.5% formaldehyde agarose denaturing gel and then blotted onto Hybond N+ nylon membranes. Full-length Bphi008a cDNA probes were labeled with [α-32P]dCTP. Hybridization was performed for more than 10 h at 55°C and then membranes were washed with 1× SSC and 0.2% SDS at 55°C for 15 min followed by 0.5× SSC and 0.1% SDS at 55°C for 15 min. The membranes were finally exposed to x-ray film for autoradiography.
cDNA was synthesized using the oligo(dT)18 primer and a RevertAid First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer’s instructions, using 3 μg of total RNA from each sample for reverse transcription. The cDNA was diluted 10-fold and amplified for real-time PCR analysis.
Protein Extraction and Immunoblot Analysis
Samples (0.125 g) of 15-d-old seedlings were ground to powder with liquid nitrogen and homogenized in 750 μL of extraction buffer (0.1 m Tris-HCl, pH 7.5, 5 mm MgCl2, 1 mm EDTA, 0.05% Triton X-100, and 2 mm dithiothreitol [DTT]). After 2 h of incubation on ice, the homogenate was centrifuged for 15 min at 20,000g at 4°C, and the resulting supernatant was used for immunoblot analysis. Fifty micrograms (about 12 μL) of supernatant was mixed with 4 μL of 4× SDS sample buffer (250 mm Tris-HCl, pH 6.8, 40% glycerol, 6% SDS, 20% β-mercaptoethanol, and 0.04% bromphenol blue) and boiled for 8 min. The samples were then analyzed by 12% (w/v) SDS-PAGE. After electrophoresis, proteins were electroblotted to nitrocellulose membranes. Membranes were blocked in 5% nonfat dried milk in TBS-T (20 mm Tris-HCl, pH 7.6, 137 mm NaCl, and 0.1% Tween 20) overnight at 4°C and then washed three times for 8 min each time in TBS-T. The membranes were incubated with the rabbit OsMPK5 polyclonal antibody (1:1,000) or with the mouse HSP82 monoclonal antibody (1:10,000) diluted with 1% bovine serum albumin in TBS-T for 1.5 h at room temperature. Next, the membranes were washed in TBS-T three times for 8 min each time and incubated for 1.5 h with goat anti-rabbit IgG or goat anti-mouse IgG (1:10,000 dilution in TBS-T and 1% bovine serum albumin) conjugated to alkaline phosphatase, followed by a further three times for 8 min each time in TBS-T. Finally, OsMPK5 was detected using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium as the substrate.
Primer Design and Real-Time PCR Analysis
The primers used for real-time PCR were designed using Beacon Designer 7.5 software according to mRNA sequences obtained from the National Center for Biotechnology Information GenBank. Reactions were carried out using a Rotorgene 6000 real-time PCR system (QiaGene) with the following temperature profile: 95°C for 2 min, followed by 34 cycles of 95°C for 5 s, 55°C for 5 s, and 72°C for 10 s. After the amplification, a melting curve was determined for each primer pair across a temperature range from 72°C to 98°C to verify that only one specific product was present. A standard curve for each gene was also determined. The reactions were performed in triplicate, and the results were averaged. All of the primer data from this study can be found in Supplemental Tables S1, S2, and S5. Last, we also determined comparative expression levels of all genes normalized to the same transcripts (TBP and HSP) among the wild-type, OE, and RNAi lines. The results showed almost the same comparative expression levels as those from OE, RNAi, and wild-type lines separately (Supplemental Fig. S8; Supplemental Table S6).
Measurement of Et Emission
Twenty 15-d-old seedlings were placed in a gas-proof transparent plastic housing (diameter, 15 cm; height, 50 cm) and left in a growth cabinet at 28°C ± 2°C for 12 h under light. At 3, 6, 12, 24, 48, and 72 h after the start of BPH feeding, 1 mL of gas was withdrawn from the air space of each plastic housing using a gas-tight syringe (Agilent) and injected into a gas chromatograph (Agilent GC-6890N) equipped with an aluminum column (Shumpak-A; Shimazu) and a flame ionization detector for Et determination.
1-MCP Treatment
For the 1-MCP treatment, 15-d-old seedlings were transferred into a hermetic glass house (approximately 5 m3) and sprayed with 1-MCP at a final concentration of 10 μL L−1. After 2 d, the pretreated plants were transferred into a greenhouse under a 14-h-light/10-h-dark cycle at 28°C for BPH feeding.
Subcellular Localization and 4′,6-Diamino-Phenylindole and FM4-64 Staining
For analysis of the subcellular localization of Bphi008a and tnBph8a, the Bphi008a, tnBph8a, and OsMPK5 coding sequences were cloned in frame to a C-terminal Bsp1407I site and a XbaI site of YFP, respectively, using the pBS-35S-YFP vector. Onion (Allium cepa) epidermal cells were then bombarded with recombined constructs using a particle gun-mediated system (PDS-1000/He; Bio-Rad) and analyzed by confocal microscopy (FV10-ASW; Olympus).
For 4′,6-diamino-phenylindole (DAPI) staining, onion epidermis was dyed for 10 min in DAPI (1 μg mL−1) and then rinsed in phosphate-buffered saline (PBS) solution before being examined with the confocal microscope. For DAPI and FM4-64 counterstaining, after DAPI staining and rinsing, onion epidermis was redyed with FM4-64 (10 nm) for 10 min and examined with the confocal microscope.
Expression and Purification of Recombinant Proteins
For expression and purification of GST-MPK5, GST-Bphi008a, and GST-tBph8a proteins, the coding sequences were cloned into the BamHI and XhoI sites of the pGEX-6P-1 expression vector (GE Healthcare). Escherichia coli BL21 cells were transformed with the three recombinant plasmids and grown to an optical density at 600 nm of 0.6. Expression of the recombinant proteins was then induced with 0.5 mm isopropyl β-d-thiogalactopyranoside for 5 h at 37°C. Bacterial culture (100 mL) was centrifuged at 12,000g for 10 min, and the sedimented bacteria were resuspended in PBS buffer (1× PBS, 1 mm phenylmethylsulfonyl fluoride, 1 mm DTT, 0.1 mm MgCl2, and 1% Triton X-100) for ultrasonication. Most of the GST-Bphi008a and GST-tBph8a fusion proteins were insoluble, being localized in inclusion bodies (data not shown). Therefore, the inclusion bodies were resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 0.1 mm EDTA, 5% glycerol, 0.1 mm DTT, and 0.1 m NaCl) containing 0.3% sodium lauroylsarcosine. The proteins (GST-Bphi008a and GST-tBph8a) were then renatured in dialysis bags at 4°C for 24 h in refolding buffer (50 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 5% glycerol, 0.1 mm DTT, and 0.05 m NaCl). Soluble GST-MPK5 and renatured GST-Bphi008a and GST-tBph8a proteins were purified by passage through High-Affinity GST Resin (GenScript), and their concentrations were determined using a Bradford Protein Assay Kit (Beyotime).
In Vitro Kinase Assay
Phosphorylation assays were performed by incubating (at 30°C for 2 h) 30-μL reaction mixtures containing 0.5 to 16 μg of GST-MPK5 kinase and 4 μg of GST, GST-Bphi008a, or GST-tBph8a with 20 mm Tris-HCl, pH 7.5, 1 mm DTT, 10 mm MgCl2, 50 μm unlabeled ATP, 0.1 mm NaVO3, and 5 μCi of [γ-32P]ATP (6,000 Ci mmol−1; Amersham Pharmacia Biotech). The reaction was stopped by adding 10 μL of 4× SDS sample buffer and boiling for 8 min. The samples were then analyzed by 12% (w/v) SDS-PAGE and subjected to autoradiography.
Yeast Two-Hybrid Assay and Screening
The Matchmaker gold yeast two-hybrid system (Clontech) was used to confirm the interactions between Bphi008a (or tBph8a) and OsMPK5 and the interaction between Bphi008a and OsbZIP60 (or SDRP). Complete Bphi008a and tBph8a coding sequences were separately cloned in frame to the EcoRI and BamHI sites of the pGADT7 and pGBKT7 vectors, respectively. The complete OsMPK5 (AF479883) coding sequence was amplified from mRNA by nested PCR using the first-round primers 5′-ATTAGGTTGGTCAATTCG-3′ with 5′-CAACAAACAAACAAATGC-3′ and the second-round primers 5′-GAATTCATGGACGGGGCGCCGGTGGCG-3′ (EcoRI) with 5′-GGATCCCTAGTACCGGATGTTTGGGTT-3′ (BamHI). The OsMPK5 coding sequence was then cloned in frame to the EcoRI and BamHI sites of the pGADT7 and pGBKT7 vectors, respectively. The complete OsbZIP60 (Os06g0622700) coding sequence was amplified from mRNA by nested PCR using the first-round primers 5′-CATGGATGTAGAGTTCTTCG-3′ with 5′-AGACTGGAAACAACTTTGC-3′ and the second-round primers 5′-CGGAATTCATGGATGTAGAGTTCT-3′ (EcoRI) with 5′-CGGGATCCCTAGCAAGCAGCTGCT-3′ (BamHI). The complete SDRP (Os03g0102200) coding sequence was amplified from mRNA using the primers 5′-CGGAATTCATGAGCACCATGAAGT-3′ (EcoRI) with 5′-CGGGATCCTCATTCCCTCCATCGG-3′ (BamHI). Both were then cloned in frame to the EcoRI and BamHI sites of the pGADT7 vector. At least three independent experiments were performed, and the results of one representative experiment are shown.
The Make Your Own “Mate & Plate” Library System (Clontech) was used for yeast two-hybrid screening. A prey cDNA rice library was constructed by fusing cDNAs with the GAL4 activation domain in the pGADT7-Rec vector. The yeast strain Y2HGold was transformed with the bait plasmid, and the strain Y187 was transformed with the plasmid DNA of the prey cDNA library, according to the manufacturer’s instructions. A total of 5.45 × 106 diploids were screened on DDO plates, and the mating efficiency was 5%. A total of 385 clones appearing within 7 d were picked out on TDO plates containing 5 mm 3-amino-1,2,4-triazole and 20 μg mL−1 X-α-Gal.
BiFC Analysis
The complete Bphi008a (or tBph8a) coding sequence was cloned in frame to the XbaI and BamHI sites of the pSPYNE vector, and the complete OsMPK5 coding sequence was cloned in frame to the XbaI and BamHI sites of the pSPYCE vector. Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts isolated from 4-week-old wild-type plants were cotransfected with constructs expressing Bphi008a-pSPYNE (or tBph8a-pSPYNE) and OsMPK5-pSPYCE by previously described procedures (Yoo et al., 2007). Onion epidermal cells were also bombarded with the same constructs as those used to cotransfect the protoplasts. Finally, the protoplasts (or onion epidermal cells) were imaged using a confocal microscope (FV10-ASW; Olympus).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY256682 (Bphi008a), X85747 (OsACO1), AF049888 (OsACO2), AF049889 (OsACO3), Os05g0149400 (OsACO5), AK102472 (OsACO7), Os04g0578000 (OsACS2), NM_001048814 (OsACS4), X97066 (OsACS5), Os06g0130400 (OsACS6), AF479883 (OsMPK5), AF177392 (OsMPK12), AY524973 (OsMPK13), AAT39148 (OsMPK17), Os04g0106300 (Arginase), Os01g0270100 (CysPI), and Os06g0622700 (OsbZIP60).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. NormFinder algorithm results for HK gene ranking and selection in different lines.
Supplemental Figure S2. Comparative expression levels of OsACS2, OsACO1, and OsACO2 after BPH feeding for between 0 and 96 h on the wild-type, OE, and RNAi plants.
Supplemental Figure S3. geNorm algorithm versus NormFinder algorithm results for the ranking and selection of HK genes in seven different parts of rice.
Supplemental Figure S4. Bphi008a expression levels in OE, RNAi, and wild-type plants after BPH feeding for different times.
Supplemental Figure S5. BiFC visualization of the Bphi008a-OsMPK5 interaction in transiently coexpressing onion epidermal cells.
Supplemental Figure S6. Comparison of rice OsbZIP60 with homologous proteins from other plant species.
Supplemental Figure S7. Expression patterns of SA, JA synthesis-related, and pathogen-related genes after BPH feeding for 0 to 96 h.
Supplemental Figure S8. Average expression stability values (M) of the remaining HK genes in 21 cDNA samples of wild-type, OE, and RNAi plants.
Supplemental Table S1. Detailed information about the HK genes in our study.
Supplemental Table S2. HK gene primer sequences used for real-time PCR.
Supplemental Table S3. Yeast two-hybrid mating combinations and results.
Supplemental Table S4. Bphi008a-interacting proteins identified by yeast two-hybrid screening.
Supplemental Table S5. Real-time PCR gene information in our study.
Supplemental Table S6. Comparative expression levels of all genes normalized to the same transcripts.
Supplementary Material
Acknowledgments
We thank Zhidan Luo and Zhibiao Ye (Huazhong Agricultural University) for measuring Et emissions. We thank Lizhong Xiong (Huazhong Agricultural University) and Guozhen Liu (Agricultural University of Hebei) for providing the OsMPK5 and HSP82 protein antibodies.
References
- Andersen CL, Jensen JL, Ørntoft TF. (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Ausubel FM. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Kessler A, Halitschke R. (2002) Volatile signaling in plant-plant-herbivore interactions: what is real? Curr Opin Plant Biol 5: 351–354 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Brown JK, Czosnek H. (2002) Whitefly transmission of plant viruses. Adv Bot Res 36: 65–100 [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA. (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102: 19237–19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, Park CY, Kim JC, Park BO, Koo SC, et al. (2003) BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol 132: 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, et al. (2009) Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci USA 106: 22163–22168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Alméras E, Krishnamurthy V. (2003) Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol 6: 372–378 [DOI] [PubMed] [Google Scholar]
- Flor HH. (1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9: 275–296 [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7: 465–471 [DOI] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, et al. (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Hao P, Liu C, Wang Y, Chen R, Tang M, Du B, Zhu L, He G. (2008) Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol 146: 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, Sheen J. (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575 [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: 19.1–19.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Huang Z, He G, Shu L, Li X, Zhang Q. (2001) Identification and mapping of two brown planthopper resistance genes in rice. Theor Appl Genet 102: 929–934 [Google Scholar]
- Iwai T, Miyasaka A, Seo S, Ohashi Y. (2006) Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol 142: 1202–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Yoneda M, Yanagawa Y, Koizumi N. (2009) Characteristics of the nuclear form of the Arabidopsis transcription factor AtbZIP60 during the endoplasmic reticulum stress response. Biosci Biotechnol Biochem 73: 865–869 [DOI] [PubMed] [Google Scholar]
- Jonak C, Okrész L, Bögre L, Hirt H. (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol 5: 415–424 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kaloshian I, Walling LL. (2005) Hemipterans as plant pathogens. Annu Rev Phytopathol 43: 491–521 [DOI] [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, Jayanty S, Walla MD, Miller W, Howe GA, Lincoln DE, Stratmann JW. (2007) Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc Natl Acad Sci USA 104: 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 14: 231–241 [PubMed] [Google Scholar]
- Kempema LA, Cui X, Holzer FM, Walling LL. (2007) Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs: similarities and distinctions in responses to aphids. Plant Physiol 143: 849–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick MD, Chang C. (2008) Ethylene signaling: new levels of complexity and regulation. Curr Opin Plant Biol 11: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler MM, Freund C. (2006) The GYF domain. FEBS J 273: 245–256 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei ME, Mithöfer A, Boland W. (2007) Before gene expression: early events in plant-insect interaction. Trends Plant Sci 12: 310–316 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. (1996) A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser RO, Hum-Musser SM, Eichenseer H, Peiffer M, Ervin G, Murphy JB, Felton GW. (2002) Herbivory: caterpillar saliva beats plant defences. Nature 416: 599–600 [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. (2003) Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31: 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H. (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. (2005) Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- Puntervoll P, Linding R, Gemünd C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al. (2003) ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res 31: 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna NS, Yang Y. (2006) Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol Plant Microbe Interact 19: 530–540 [DOI] [PubMed] [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM. (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95: 9750–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hoheisel JD. (2002) Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem 277: 10555–10561 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA. (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46: 69–81 [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. (2007) The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda C, Ozaki R, Onodera Y, Takahashi Y, Yamaguchi K, Berberich T, Koizumi N, Kusano T. (2008) NtbZIP60, an endoplasmic reticulum-localized transcription factor, plays a role in the defense response against bacterial pathogens in Nicotiana tabacum. J Plant Res 121: 603–611 [DOI] [PubMed] [Google Scholar]
- Tjallingii WF. (2006) Salivary secretions by aphids interacting with proteins of phloem wound responses. J Exp Bot 57: 739–745 [DOI] [PubMed] [Google Scholar]
- Ussuf KK, Laxmi NH, Mitra R. (2001) Proteinase inhibitors: plant-derived genes of insecticidal protein for developing insect-resistant transgenic plants. Curr Sci 80: 847–853 [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 0034.1–0034.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel C, Baldwin IT. (2004) Herbivore-induced plant vaccination. Part II. Array-studies reveal the transience of herbivore-specific transcriptional imprints and a distinct imprint from stress combinations. Plant J 38: 650–663 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang YY, Wang XL, Yuan HY, Chen RZ, Zhu LL, He RF, He GC. (2008) Responses of two contrasting genotypes of rice to brown planthopper. Mol Plant Microbe Interact 21: 122–132 [DOI] [PubMed] [Google Scholar]
- Wei Z, Hu W, Lin QS, Cheng XY, Tong MJ, Zhu LL, Chen RZ, He GC. (2009) Understanding rice plant resistance to the brown planthopper (Nilaparvata lugens): a proteomic approach. Proteomics 9: 2798–2808 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, van Bel AJ. (2007) Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci USA 104: 10536–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT. (2007) Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y. (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–75912615946 [Google Scholar]
- Yang SF, Hoffman NE. (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Chen X, Zhu L, He G. (2004) Isolation and characterization of a novel rice gene encoding a putative insect-inducible protein homologous to wheat Wir1. J Plant Physiol 161: 79–85 [DOI] [PubMed] [Google Scholar]
- Zarate SI, Kempema LA, Walling LL. (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Salzman K, Salzman RA, Ahn JE, Koiwa H. (2004) Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.