Abstract
Chlamydomonas reinhardtii possesses a CO2-concentrating mechanism (CCM) that allows the alga to grow at low CO2 concentrations. One common feature seen in photosynthetic organisms possessing a CCM is the tight packaging of Rubisco within the cell. In many eukaryotic algae, Rubisco is localized to the pyrenoid, an electron-dense structure within the chloroplast. In order to identify genes required for a functional CCM, insertional Bleomycin resistance (BleR) mutants were generated and screened for growth on minimal medium under high CO2 conditions (5% CO2 in air) but only slow or no growth under very low CO2 conditions (0.01% CO2 in air). One mutant identified from this screen was named cia6. Physiological studies established that cia6 grows poorly on low levels of CO2 and has an impaired ability to accumulate inorganic carbon. The inserted BleR disrupted a gene encoding a protein with sequence similarity to proteins containing SET domain methyltransferase, although experiments using overexpressed CIA6 failed to demonstrate the methyltransferase activity. Electron microscopy revealed that the pyrenoid of cia6 mutant cells is highly disorganized. Complementation of the mutant restored the pyrenoid, the ability to grow under low-CO2 conditions, and the ability to concentrate inorganic carbon. Quantitative reverse transcription-polymerase chain reaction data from a low-CO2 induction time-course experiment demonstrated that the up-regulation of several CCM components is slower in cia6 compared with the wild type. This slow induction was further confirmed at the protein level using western blots. These results indicated that CIA6 is required for the formation of the pyrenoid and further supported the notion that the pyrenoid is required for a functional CCM in C. reinhardtii.
Most aquatic photosynthetic organisms have a CO2-concentrating mechanism (CCM) that increases the CO2 concentration around the carboxylating enzyme Rubisco when CO2 is limiting (Badger et al., 1998; Giordano et al., 2005; Moroney and Ynalvez, 2007). Two problems faced by organisms with CCMs are the potential leakage of CO2, which can readily cross most biological membranes (Gutknecht et al., 1977), and the rate of diffusion of CO2 in water, which is 104 times slower than in air. Since HCO3− crosses biological membranes at about 10−6 times the rate of CO2, a strategy that concentrates HCO3− would greatly minimize the loss of inorganic carbon (Ci). The cyanobacterial CCM adopts a system that concentrates HCO3− in the cytoplasm using a series of HCO3− transporters and packages Rubisco in a proteinaceous structure called the carboxysome (Price et al., 2008). The HCO3− accumulated in the cytoplasm could then diffuse into the carboxysome through carboxysomal pores, be converted to CO2 by carboxysomal carbonic anhydrases, and finally be fixed by Rubisco (Price et al., 2008). The importance of the carboxysome in the cyanobacterial CCM is illustrated by the fact that the ectopic expression of carbonic anhydrase in the cyanobacterial cytosol short-circuits the CCM, as the HCO3− in the cytosol is converted into CO2 that diffuses out of the cell without fixation (Price and Badger, 1989).
In the eukaryotic green algae Chlamydomonas reinhardtii, the CCM appears to have strong similarities to the cyanobacterial system (Moroney and Ynalvez, 2007). In C. reinhardtii, bicarbonate is concentrated in the chloroplast stroma through the cooperation of multiple Ci transporters and carbonic anhydrases. Rubisco is packaged in a specialized chloroplast microcompartment, called the pyrenoid, where active Ci fixation takes place (Borkhsenious et al., 1998). In the pyrenoid of C. reinhardtii, Rubisco is the predominant protein (Kuchitsu et al., 1988; Morita et al., 1997; Borkhsenious et al., 1998), and nonsense mutations in the Rubisco large subunit totally abolish the formation of the pyrenoid (Rawat et al., 1996). C. reinhardtii’s pyrenoid is also penetrated by a network of thylakoid tubules (Henk et al., 1995; Borkhsenious et al., 1998). The carbonic anhydrase CAH3, which has been localized to the thylakoid lumen, is further enriched in the thylakoid tubules inside the pyrenoid (Karlsson et al., 1998; Mitra et al., 2005). Mutants that lack CAH3 have a nonfunctional CCM in which HCO3− accumulates intracellularly, but the mutant cells cannot grow on low and very low levels of CO2 (Spalding et al., 1983; Moroney et al., 1986; Pronina and Semenenko, 1992; Karlsson et al., 1998). In addition, a putative Ci transporter, LCIB, is localized around the pyrenoid (Yamano et al., 2010). Mutants with defects in LCIB expression result in the unusual air-dier phenotype, which dies at air level of CO2 but survives under very low CO2 (Wang and Spalding, 2006; Duanmu et al., 2009).
A number of current models have proposed that the pyrenoid is critical to the optimal function of the CCM in C. reinhardtii, with the structure serving a role similar to the cyanobacterial carboxysomes (Morita et al., 1999; Moroney and Ynalvez, 2007; Spalding, 2008; Yamano et al., 2010). However, very few mutations affecting pyrenoid structure have been described. Among the few reports, Rawat et al. (1996) demonstrated that the loss of Rubisco in C. reinhardtii resulted in the loss of the pyrenoid. Recently, Genkov et al. (2010) described the creation of C. reinhardtii strains with hybrid Rubisco, in which a higher plant small subunit gene (RbcS) was transformed into a C. reinhardtii RbcS mutant background. The resultant C. reinhardtii strain contained hybrid Rubisco with native RbcS and higher plant Rubisco large subunit (RbcL). However, strains containing the hybrid Rubisco could not grow as well as wild-type cells at low CO2 levels. Strikingly, in the resultant C. reinhardtii strains, although the catalytic properties of the hybrid Rubisco were similar to wild-type Rubisco, and the Rubisco was expressed, it was found that these strains lacked pyrenoids as well as an active CCM.
In this report, we describe a novel mutant of C. reinhardtii in which a disruption in a nuclear gene other than in Rubisco resulted in a disrupted pyrenoid structure and a high-CO2-requiring phenotype. This work provides further evidence that the organization of the pyrenoid itself is important in the functioning of the CCM.
RESULTS
cia6 Needs a High-CO2 Environment to Grow Optimally
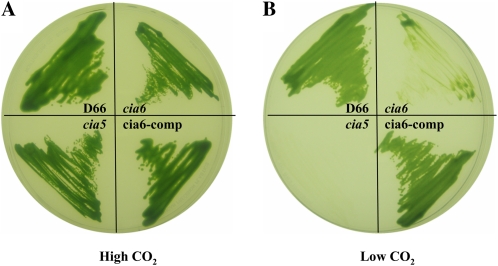
C. reinhardtii strain D66 (Schnell and Lefebvre, 1993) was transformed with the pSP124s plasmid, which contains the modified Bleomycin resistance (BleR) cassette conferring bleomycin resistance (Lumbreras et al., 1998). A total of 42,000 BleR insertional mutants were selected and screened for a high-CO2-requiring phenotype (Colombo et al., 2002; Pollock et al., 2003). Strains that could grow well on elevated CO2 but grew slowly on low CO2 were subsequently tested for their ability to accumulate Ci. One of those transformants also had a reduced ability to accumulate Ci and was named cia6. Figure 1 shows the photoautotrophic growth characteristics in high (Fig. 1A) and low (Fig. 1B) CO2 environments of the parent strain, D66, the insertional mutant, cia6, and a known high-CO2-requiring mutant, cia5, a strain defective in a transcription factor and that fails to induce the CCM (Moroney et al., 1989). In high-CO2 conditions, all three strains displayed similar growth characteristics. However, in the low-CO2 environment, compared with the D66 strain, which displayed a growth pattern similar to the one under high-CO2 conditions, the cia6 mutant grew poorly, as did the CCM-defective strain cia5. The results presented here show that cia6 requires an elevated CO2 environment to grow photoautotrophically.
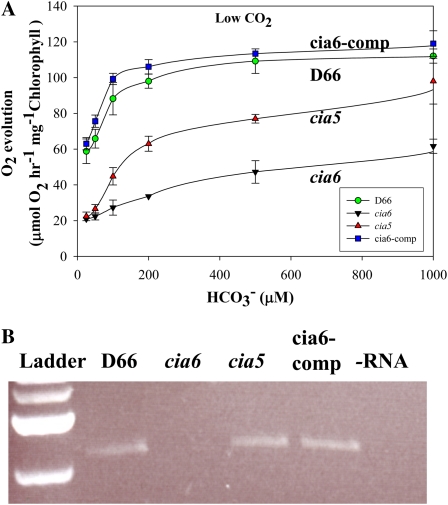
Figure 1.
The mutant cia6 demonstrated a typical CCM-deficient phenotype. The ability of cia6 to grow photoautotrophically was tested by comparing its growth on minimal plates under high-CO2 (5% [v/v] CO2; A) and very-low-CO2 (0.01% [v/v] CO2; B) conditions. A, cia6 grew well in high CO2 compared with its parental strain D66. B, cia6 grew poorly under low-CO2 conditions, similar to the CCM-deficient mutant cia5. The complemented cia6, cia6-comp, in which the wild-type CIA6 gene was put back into cia6, exhibited a phenotype similar to that of the wild-type D66. [See online article for color version of this figure.]
cia6 Has a Reduced Affinity for Ci
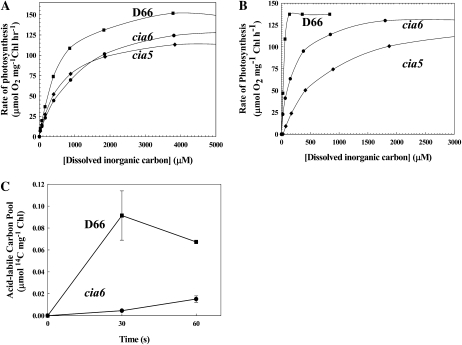
To determine the apparent affinity of cia6 cells for Ci, the rate of photosynthesis as a function of the dissolved inorganic carbon (DIC) concentration was determined. When grown on elevated CO2, D66, cia5, and cia6 all had maximum photosynthesis rates of greater than 100 μmol CO2 fixed mg−1 chlorophyll h−1. However, cia5 and cia6 had affinities for DIC that were 1.5 to two times lower than D66 (Fig. 2A; Table I). Air-acclimated cells also exhibited similar maximum rates of photosynthesis, but in contrast, both cia5 and cia6 had an affinity for DIC approximately 10 times lower when compared with D66 cells (Fig. 2B; Table I). The rate of photosynthesis was also lower in air-grown cia6 compared with the wild type at low DIC concentrations. In order to measure the ability of cia6 to accumulate Ci, light-dependent Ci uptake was estimated using the silicone oil centrifugation method (Fig. 2C). The amount of Ci accumulation by cia6 was one-fifth that of D66 during a 60-s time course. In summary, these photosynthetic characteristics led to the naming of this mutant, cia6, for Ci accumulation-deficient mutant, following the nomenclature described by Moroney et al. (1986).
Figure 2.
The mutant cia6 showed a reduced affinity for Ci. A and B, The rates of photosynthesis of D66 (squares), cia6 (circles), and cia5 (diamonds) as a function of the DIC concentration for high-CO2-grown (A) and low-CO2-grown (B) cells were measured. C, The Ci accumulation in low-CO2-acclimated D66 (squares) and cia6 (circles) was measured during the time course of [14C]DIC accumulation. Each point represents the mean and se of three separate experiments.
Table I. The K0.5(DIC) value of cia6-comp was recovered to the wild-type (D66) level.
K0.5(DIC) is calculated by regression analysis from three individual experiments using the Michaelis-Menten kinetics equation V = Vmax[S]/(Km + [S]), in which [S] equals the bicarbonate concentration, V equals the oxygen evolution rate at each bicarbonate concentration, Vmax equals the maximum oxygen evolution rate when bicarbonate concentration is saturated, and Km equals K0.5(DIC).
| Condition | D66 | cia6 | cia5 | cia6-comp |
| High CO2 | 179 ± 23 | 296 ± 24 | 444 ± 46 | 106 ± 8 |
| Low CO2 | 29 ± 7 | 106 ± 17 | 138 ± 26 | 25 ± 1 |
cia6 Has an Insertion in a Novel Gene
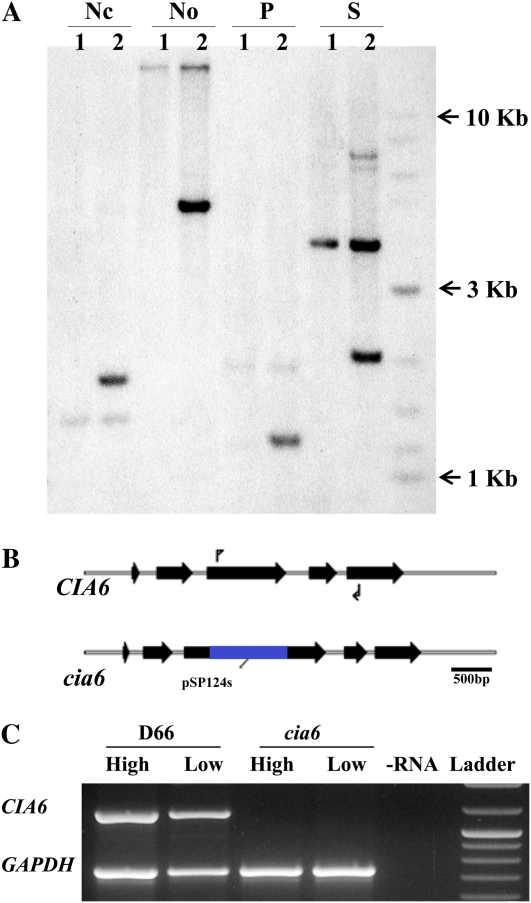
Southern-blot analysis of this mutant showed that a single insertion event had taken place during transformation, as evidenced by a single intense hybridization band in cia6 genomic DNA digested with different restriction enzymes that do not have cleavage sites within the predicted transgene (Fig. 3A). Using an adaptor-mediated PCR strategy (Siebert et al., 1995), the DNA flanking one side of the pSP124s insert was cloned and sequenced. Comparison of this sequence with the Joint Genomics Initiative’s C. reinhardtii genome sequence database (Merchant et al., 2007) yielded a match to a region on chromosome 10. No ESTs were aligned with this region. However, the gene structure prediction programs Genewise (http://www.ebi.ac.uk/Tools/Wise2/index.html) and GeneMark (http://exon.biology.gatech.edu/) identified putative open reading frames (ORFs) in this region of the chromosome. Using primers designed to bracket this region, a cDNA was amplified using RNA from wild-type cells as the template. RACE was employed to obtain a full-length cDNA, and DNA sequencing verified its identity. The CIA6 gene consists of five exons spanning 2.9 kb (Fig. 3B; GenBank accession no. JF288753). Subsequent sequence analysis demonstrated that the BleR cassette insertion disrupted the third exon. Primers spanning the BleR insertion (Fig. 3B) were utilized to determine whether the CIA6 mRNA was present in high- and low-CO2-grown D66 and cia6, and primers amplifying glyceraldehyde 3-phosphate dehydrogenase were used as an internal control (Fig. 3C). In D66, the CIA6 message was present in both high- and low-CO2-grown cells, while the message was absent in cia6. Tetrad analysis confirmed the linkage between the CCM-deficient phenotype and the BleR insertion (data not shown). These results are consistent with the hypothesis that the single pSP124s insertion disrupted the CIA6 locus and abolished the transcription of CIA6.
Figure 3.
The mutant cia6 has an insertion in the CIA6 locus. A, Southern-blot analysis using the bleomycin resistance gene as a probe. D66 (lanes 1) and cia6 (lanes 2) genomic DNA was digested with NcoI (Nc), NotI (No), PstI (P), and SacI (S) and blotted onto a nylon membrane. The membrane was then probed with a 32P-labeled pSP124s-specific fragment containing BleR DNA and the Rbcs2 intron. The weak bands present in both D66 and cia6 correspond to the RbcS2 intron, which is present in both the BleR mutant as well as the endogenous RbcS2 gene. B, Genomic structure of the CIA6 locus in the wild type and cia6 showing the introns and exons, the site of the pSP124s insertion in cia6, and the cDNA primer sites (arrows) used for the RT-PCR analysis in C. C, RT-PCR analysis of the D66 and cia6 strains using poly(A) RNA from high- and low-CO2-grown cells as templates for RT. The primer-binding sites for CIA6 are shown in B and amplify an 1,100-bp product from cDNA. The internal control fragment (730 bp) is the product of primers designed to amplify glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The last lane contains the 2-Log DNA Ladder (NEB). [See online article for color version of this figure.]
cia6 Has a Disorganized Pyrenoid
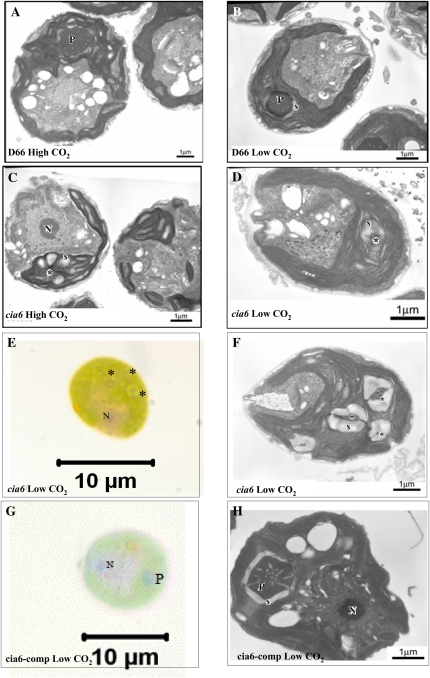
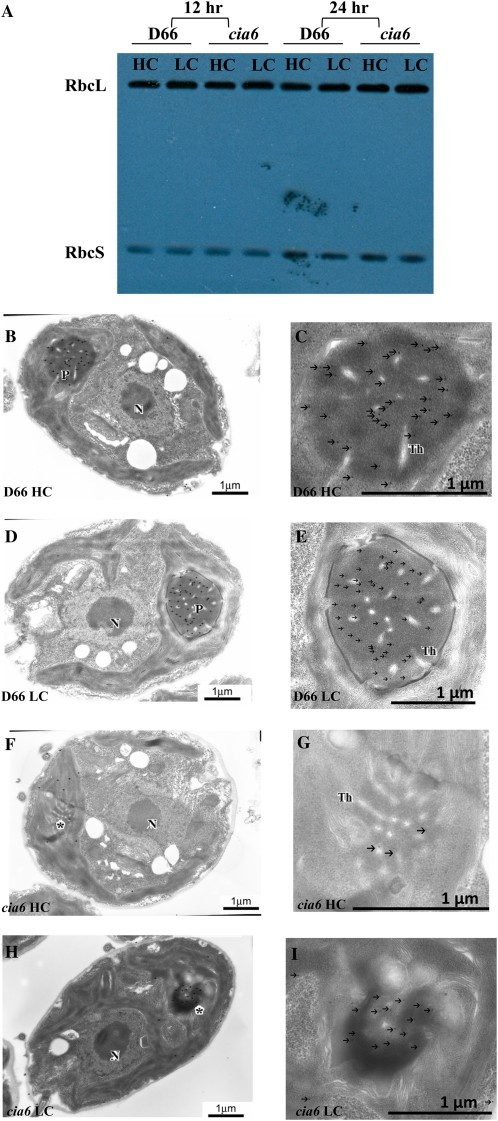
In C. reinhardtii, the pyrenoid is a spheroid, electron-dense, Rubisco-containing body inside the chloroplast (Fig. 4, A and B, where the pyrenoid is labeled as “P”). The pyrenoid is penetrated by numerous thylakoid membranes and is estimated to occupy a volume of 2.4 μm3, approximately 1/16th of the entire chloroplast (Griffiths, 1970; Lacoste-Royal and Gibbs, 1987). In transmission electron microscopy (TEM) thin sections, a normal spherical pyrenoid is often encountered at a chance of 35% out of all wild-type cell thin sections (Henk et al., 1995). During the cell’s acclimation to a low-CO2 environment, the pyrenoid undergoes dramatic structural changes as a ring of starch accumulates around the pyrenoid (Fig. 4B, where the starch sheath is labeled as “s”). Using TEM, cia6 mutant cells were examined and were shown to have disorganized pyrenoids in both high-CO2-grown cells (Fig. 4C, where a possible pyrenoid region is labeled with an asterisk) and-low-CO2 grown cells (Fig. 4D, where a possible pyrenoid region is labeled with an asterisk). In contrast to the normal sphere-shaped, electron-dense pyrenoid with an average of 2.3 μm2 in area seen in wild-type cells (Fig. 4, A and B), it was revealed that the pyrenoid was either absent or highly disorganized in all the cia6 ultrathin cell sections examined. Although the electron-dense region could still be observed in the mutant cells, it was significantly smaller in size, with an average of 1.2 μm2, highly irregular in shape, and often associated with the starch sheath. Over 100 cia6 cell thin sections were observed, but no normal pyrenoid could be detected in cia6. Under light microscopy (100×), however, the possible presence of small pyrenoid-like structures was observed in the cia6 mutant cells, and oftentimes multiple small pyrenoids were observed inside the chloroplast of a single cell (Fig. 4, F and G).
Figure 4.
Electron micrographs (A–D, F, and H) and light micrographs (E and G) of C. reinhardtii cells grown in minimal medium in high-CO2 (A and C) and low-CO2 (B and D–H) conditions. A, Wild-type D66 grown under high-CO2 conditions. B, Wild-type D66 grown under low-CO2 conditions. C, Mutant cia6 grown under high-CO2 conditions. D, Mutant cia6 grown under low-CO2 conditions. E, Light microscopy of cia6 cells showing multiple pyrenoid or pyrenoid-like structure (asterisks). F, Electron micrograph showing multiple pyrenoid or pyrenoid-like structures (asterisks) in cia6. G, Pyrenoid (P) and nucleus (N) are observed as dark staining in the complemented cia6 grown under low-CO2 conditions. H, A positive pyrenoid in the complemented cia6 grown under low-CO2 conditions. S, Starch; asterisks indicate the locations of pyrenoid-like bodies in mutant cells. [See online article for color version of this figure.]
cia6 Can Be Complemented
To confirm whether the CIA6 mutation is responsible for the CCM-deficient phenotype and the disorganized pyrenoid, complementation was attempted by transforming a 3.7-kb genomic DNA fragment containing the wild-type CIA6 locus into the cia6 mutant. cia6 cells were transformed and selected under very-low-CO2 conditions (0.007% [v/v] CO2 in air) on minimal plates. A total of 50 transformants that survived under very-low-CO2 conditions were further examined for the restoration of the pyrenoid, as observed by light microscopy. Twenty-two out of 49 transformants selected in this manner showed positive pyrenoid staining, as indicated by strong staining using the HgCl2-bromphenol blue reagent (Kuchitsu et al., 1988) in the pyrenoid and nucleus. One transformant, named cia6-comp, was selected for growth and photosynthetic kinetics study. Under both high- and low-CO2 conditions, cia6-comp grew as well as the wild-type cells (Fig. 1), showing that the growth defects had been complemented. Figure 5A shows that the complemented cia6 had a higher affinity for Ci as compared with the mutant. The calculated affinity of cia6-comp for DIC was also close to the wild-type level (Table I). In addition, the RNA sample isolated from cia6-comp also showed that the CIA6 mRNA is recovered in the complemented strain, based on reverse transcription (RT)-PCR analysis (Fig. 5B). The recovery of the wild-type pyrenoid structure (Fig. 4, G and H) together with the restoration of its Ci-concentrating ability demonstrated that the phenotype of the cia6 mutant can be complemented with a 3.7-kb genomic fragment containing the wild-type CIA6 locus.
Figure 5.
Complementation rescued the CCM deficiency in cia6. By transforming genomic DNA containing the wild-type CIA6 gene into the cia6 mutant, the resulting complemented strain cia6-comp exhibited a growth phenotype similar to the wild type (Fig. 1). A, The rate of photosynthesis as a function of the DIC was measured using low-CO2-grown D66 (green circles), cia6 (black triangles), cia5 (red triangles), and the complemented strain cia6-comp (blue squares). B, RT-PCR analysis of D66, cia6, cia5, and the complemented cia6 (cia6-comp) using RNA isolated from low-CO2-grown cells as templates for RT. Primers were designed to amplify the entire ORF. The reappearance of the CIA6 full-length ORF in the complemented strain (cia6-comp) was observed on a DNA agarose gel. The last lane contained the no-RNA control. [See online article for color version of this figure.]
CIA6 Is Predicted to Encode a 72-kD Protein Containing a SET Domain
The predicted CIA6 ORF encodes for a 72-kD protein that has sequence similarity with a group of SET domain proteins. Using the BLASTP program searching the National Center for Biotechnology Information (NCBI) protein database, 13 homologs of CIA6 were identified (E value ≤ 8e−20, maximum score ≥ 100; Fig. 6). Among organisms possessing CIA6 homologs, the multicellular green alga Volvox carteri f. nagariensis has the highest similarity, followed by the unicellular photosynthetic green alga Chlorella variabilis NC64A and Micromonas pusilla CCMP1545, which also possess pyrenoid and an active CCM (Shiraiwa and Miyachi, 1983; Ikeda and Takeda, 1995; Worden et al., 2009). In addition, the nonvascular plant Selaginella moellendorffii has two genes similar to CIA6. The genomes of the early-branching land plant Physcomitrella patens as well as the land plants Vitis vinifera, Populus trichocarpa, Arabidopsis (Arabidopsis thaliana), and Oryza sativa all contained a gene predicted to encode a protein with significant homology to CIA6. To date, among all the CIA6 homologs identified in the NCBI database, none has been characterized.
Figure 6.
Evolutionary relationships of CIA6 among 13 taxa. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007). Each protein is designated by the abbreviation of its scientific name and its NCBI accession number. The species are as follows: Chlamydomonas reinhardtii, Volvox carteri f. nagariensis, Micromonas pusilla CCMP1545, Physcomitrella patens subsp. patens, Vitis vinifera, Ricinus communis, Selaginella moellendorffii, Micromonas sp. RCC299, Populus trichocarpa, Arabidopsis thaliana, Arabidopsis lyrata subsp. lyrata, Sorghum bicolor, and Oryza sativa.
Assessment of CIA6’s Methyltransferase Activity in Vitro
In order to test the putative methyltransferase activity of CIA6, the protein was overexpressed in Escherichia coli using the pMALc2x overexpression system. The entire CIA6 ORF was fused to the maltose-binding protein in pMALc2x and overexpressed as a fusion protein in E. coli cells. C. reinhardtii Rubisco, calf thymus histone, and C. reinhardtii whole cell extracts were used as possible substrates. Positive controls in which pea (Pisum sativum) Rubisco large subunit methyltransferase (RLsMT; Klein and Houtz, 1995) was incubated with either the spinach (Spinacia oleracea) or the C. reinhardtii Rubisco isolated from both the wild type and the cia6 mutant showed significant methyltransferase activity. In contrast, no in vitro methyltransferase activity was detected when the purified CIA6 fusion protein was incubated with the same array of substrates. The detected activity of pea RLsMT with the C. reinhardtii Rubisco is probably achieved by methylating on the Lys-14 site, as the pea RLsMT does in vivo with pea Rubisco. The observation is consistent with previous reports that the isolated Rubisco could be in vitro substrates, even though their Lys-14 is not methylated in vivo (Houtz et al., 1992; Raunser et al., 2009).
cia6 Has Normal Levels of Rubisco, But Rubisco Fails to Associate and Form a Pyrenoid
Rawat et al. (1996) demonstrated that the pyrenoid could not be found in the RbcL nonsense strain 18-7G (Spreitzer et al., 1985), which indicates that the pyrenoid formation requires the presence of Rubisco. Thus, to address why the cia6 mutant cells lacked a pyrenoid, Rubisco protein levels from D66 and cia6 were compared using western blot (Fig. 7A). However, when using anti-Rubisco polyclonal antibody (Borkhsenious et al., 1998) that recognizes both the RbcL and the RbcS, it was found that the protein levels of both Rubisco subunits from both strains were not different. Another possible explanation regarding the disruption of pyrenoid in the mutant cells could be that, even though the amount of Rubisco protein was not changed, the directing of the Rubisco holoenzyme into the correct position (pyrenoid) in the mutant chloroplast was impaired. Immunolocalization of Rubisco was performed in cells from the wild-type and mutant strains under both high- and low-CO2 conditions. As seen for the high-CO2-grown (Fig. 7B) and low-CO2-grown (Fig. 7C) wild-type cells, the pyrenoid contained the majority of immunogold particles, with almost all the immunological particles found inside of the wild-type pyrenoid upon low-CO2 induction (Fig. 7C). This result agrees with earlier published reports from Morita et al. (1997) and our laboratory (Borkhsenious et al., 1998) that more than 90% of the Rubisco resides inside the pyrenoid in wild-type cells grown under low-CO2 conditions. In cia6, however, Rubisco particles were no longer accumulating inside the pyrenoid in either high-CO2-grown (Fig. 7D) or low-CO2-grown (Fig. 7E) cells. Using the estimated volume of the stroma and pyrenoid (Lacoste-Royal and Gibbs, 1987), it was estimated that the fraction of Rubisco in the pyrenoid-like structures was about 35% in cia6, which is similar to the percentage seen in wild-type cells grown in high-CO2 conditions (Table II). Hence, the total Rubisco concentration in the mutant cia6 was not changed, while the localization of this CO2 fixation enzyme was greatly altered as a result of the pyrenoid disruption.
Figure 7.
Rubisco content in the wild-type D66 and mutant cia6 cells was investigated using western blotting (A) and immunogold labeling (B–I) against anti-Rubisco antibody. A, Total protein isolated from D66 and cia6 grown under high- and low-CO2 conditions (HC and LC, respectively) for 12 and 24 h. Using western blotting, the amount of dissociated Rubisco large and small subunits (RbcL and RbcS) was estimated to be normal in the mutant compared with the wild type. B and C, High-CO2-grown wild-type D66 cells grown on minimal medium probed with an antibody raised against Rubisco. D and E, Low-CO2-grown wild-type D66 cells grown on minimal medium probed with an antibody raised against Rubisco. F and G, High-CO2-grown cia6 cells grown on minimal medium probed with an antibody raised against Rubisco. H and I, Low-CO2-grown cia6 cells grown on minimal medium probed with an antibody raised against Rubisco. N, Nucleus; P, pyrenoid; Th, thylakoid. Asterisks indicate pyrenoid-like structures in cia6. Bars = 1 μm. [See online article for color version of this figure.]
Table II. Comparison of the Rubisco fraction found in the pyrenoid and in the chloroplast stroma between the wild type and cia6.
The data shown are averages of 25 C. reinhardtii electron microscopy thin sections and a total of 300 particles counted.
| C. reinhardtii Strain | Immunogold Density |
Rubisco in the Pyrenoid | |
| Pyrenoid | Stroma | ||
| particles μm−2 | % | ||
| D66 | 23.3 | 0.00 | 100 |
| cia6 | 7.5 | 1.3 | 35.8 |
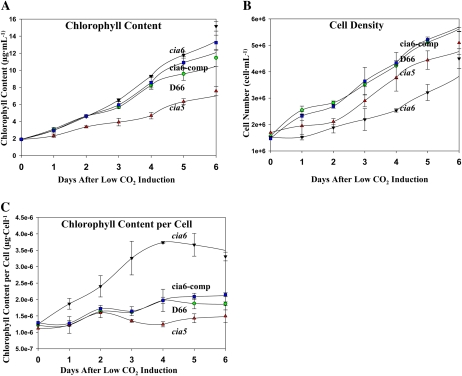
cia6 Has a Higher Chlorophyll Content per Cell
In C. reinhardtii wild-type cells, the chlorophyll per cell ratio remains constant during log phase growth, at around 2.8 × 10−6 μg chlorophyll cell−1 and 1.7 × 10−6 μg chlorophyll cell−1 under photoautotrophic and heterotrophic conditions, respectively (Fig. 8; Supplemental Fig. S1). However, in the mutant cia6, it was observed that the chlorophyll per cell ratio is significantly higher in photoautotrophically grown cells (minimal medium bubbled with air; Fig. 8), especially after switching from Tris-acetate-phosphate (TAP) medium to minimal medium. In addition, the chlorophyll per cell ratio is also higher in minimal medium even under high-CO2 concentrations (Supplemental Fig. S1). When cia6 was grown under photoheterotrophic conditions, where acetate was used as a carbon source, the chlorophyll per cell ratio was similar to that in wild-type cells. The complemented strain cia6-comp exhibited the same growth characteristics and chlorophyll per cell ratio as the wild-type strain in all growth conditions.
Figure 8.
A higher chlorophyll content per cell was observed in the mutant cia6. Cultures of D66 (green circles), cia6 (black triangles), cia5 (red triangles), and cia6-comp (blue squares) were started at the same chlorophyll concentration and subjected to low-CO2 stress. For the subsequent 6 d, samples were collected every 24 h and chlorophyll concentration (A) and cell density (B) were measured and the chlorophyll concentration per cell was calculated (C). A, Cell density was determined by direct counting using a hemacytometer. B, Chlorophyll concentration was measured, and the mutant cia6 had the highest chlorophyll concentration at the end of the time course. C, Chlorophyll content per cell value was plotted, so that it was clear that the mutant cia6 had an increased chlorophyll content per cell compared with the other tested strains. Each point represents the mean and sd of three separate experiments. [See online article for color version of this figure.]
cia6 Acclimates Slowly to Low-CO2 Conditions
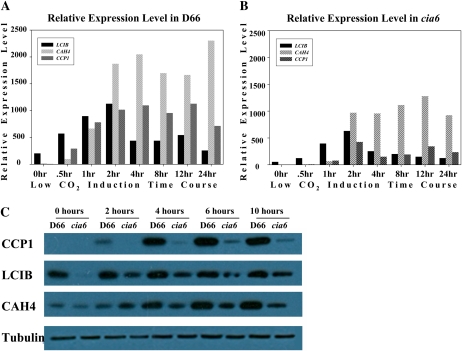
To test whether the disruption of pyrenoid structure in cia6 might have other pleiotropic effects on the expression of the CCM, changes in the expression of other key CCM components, including LCIB, CAH4, and CCP1, were investigated by performing quantitative RT analysis using RNA samples collected during a low-CO2 induction time course (Fig. 9A). As shown in Figure 9A, the relative transcript abundance of those key CCM genes was lower in the mutant cells than in the wild-type cells. In addition, protein samples collected at the same time points were probed with CCP1, LCIB, and CAH4 antibodies to estimate the protein levels by western blotting (Fig. 9B). For most of the CCM-related proteins, the amount of the protein was close to wild-type levels after 4 h (Fig. 9B). However, the abundance of CCP1, a chloroplast envelope protein, was reduced the most among the CCM proteins that were examined and remained at low levels even after 4 h.
Figure 9.
Time course of LCIB, CAH4, and CCP1 levels during the low-CO2 induction process as measured by quantitative RT-PCR (A and B) and western blotting (C). A, Wild-type D66 cells subjected to low-CO2 induction were sampled at the time points indicated. The CBLP gene was used as the internal control. B, Mutant cia6 cells subjected to low-CO2 induction were sampled at the time points indicated. The CBLP gene was used as the internal control. C, Wild-type D66 and mutant cia6 protein samples were collected at the time points indicated and then subjected to western-blot analysis using antibody raised against CCP1, LCIB, and CAH4. Anti-α-tubulin antibody was used as a loading control. [See online article for color version of this figure.]
DISCUSSION
In this report, we described the isolation and characterization of a novel C. reinhardtii mutant in which a nuclear gene named CIA6 was disrupted. The mutation in CIA6 resulted in the dysfunction of the CCM (Figs. 1 and 2), the disruption of the chloroplast pyrenoid (Figs. 4 and 7), and a higher than normal chlorophyll concentration (Fig. 8). Transformation of the cia6 mutant with a functional wild-type CIA6 gene restored a functional CCM (Figs. 1 and 5B; Table I), a normal pyrenoid phenotype (Fig. 4, G and H), and its normal levels of chlorophyll (Fig. 8). This work revealed that the nuclear gene product CIA6 is required for the normal formation of pyrenoid and also provided evidence that the presence of pyrenoid is essential for the functioning of CCM in C. reinhardtii.
Most eukaryotic photosynthetic algae have pyrenoids (Griffiths, 1970; Bold and Wynne, 1985), while pyrenoids are almost absent from higher plants, with the exception of some species of hornworts (Anthocerotophyta; Griffiths, 1970). In electron micrographs of a C. reinhardtii cell, the pyrenoid appears as an electron-dense microcompartment embedded inside the chloroplast stroma. A network of modified thylakoid tubules is present inside the pyrenoid body (Griffiths, 1970), and the accumulation of the starch plates surrounding the pyrenoid is often observed, especially when the environmental CO2 level is low (Ramazanov et al., 1994; Henk et al., 1995). The dense matrix of the pyrenoid mainly consists of Rubisco (Kuchitsu et al., 1988; Rawat et al., 1996; Morita et al., 1997; Borkhsenious et al., 1998). Rubisco activase has also been shown to be present in the pyrenoid (McKay et al., 1991).
The specific localization of Rubisco to the pyrenoid in C. reinhardtii is considered to be a key element in the optimal functioning of its CCM (Moroney and Ynalvez, 2007; Spalding, 2008; Yamano et al., 2010). Analogous to the packaging of Rubisco in cyanobacterial carboxysomes (Badger et al., 1998), the pyrenoid in C. reinhardtii could likewise be the location of the high CO2 concentration generated by CCM. Since this high CO2 concentration is in the vicinity of Rubisco, the carboxylation reaction will be enhanced at the expense of the oxygenation reaction, hence reducing the photorespiration. In the pyrenoid-containing hornworts, there is also good correlation between the operation of a CCM and the presence of a pyrenoid (Vaughn et al., 1990; Hemsley and Poole, 2004). Besides the role in physical compartmentalization of Rubisco away from the rest of chloroplast stroma, the dynamic nature of pyrenoid in response to environmental changes adds extra complexity to the CCM. One notable observation made using immunolocalization was that in response to a switch from high-CO2 to low-CO2 environment, the Rubisco in the wild-type chloroplast started to redistribute and accumulate in the pyrenoid after 2 h in low CO2 (Borkhsenious et al., 1998). The percentage of the pyrenoid-Rubisco labeling over the total labeling rose from 40% on high CO2 to a maximum of 90% within 4 h on low CO2. This process is tightly correlated with the time required for the appearance of a starch plate surrounding the pyrenoid (Henk et al., 1995), for the induction of most of the CCM genes (Miura et al., 2004), and for the maximal increase in Ci affinity (Borkhsenious et al., 1998). The rapid biochemical and morphological rearrangement indicates that the pyrenoid is a dynamic structure and is actively involved in the cell’s acclimation to low-CO2 conditions.
One prediction based on the present CCM models is that, if the pyrenoid structure is disrupted, the CCM should also be adversely affected. However, among the few mutants with disrupted pyrenoid, either Rubisco was not present (Rawat et al., 1996), or chloroplast ribosomes were mutated, leading to the absence of Rubisco (Goodenough and Levine, 1970), or the entire chloroplast structure was highly disrupted (Inwood et al., 2008). One piece of direct evidence supporting the role of the pyrenoid in an active CCM comes from a recent study by Genkov et al. (2010). In that work, the C. reinhardtii Rubisco holoenzyme was engineered so that the C. reinhardtii native RbcL assembled with higher plant RbcS. Although the hybrid Rubisco had similar kinetic properties and was present in amounts equivalent to wild-type Rubisco, the resultant C. reinhardtii cells were found to lack the chloroplast pyrenoid and failed to grow on low CO2. This is strong evidence that the pyrenoid is playing an important role in the CCM.
In this study, the characterization of the cia6 mutant with disorganized pyrenoids provides additional evidence that the pyrenoid is a necessary part of the CCM. Additionally, to our knowledge, the CIA6 mutation is the first mutation in a gene other than the Rubisco large or small subunit genes that disrupts the pyrenoid structure while not affecting the entire chloroplast. Here, we show that the pyrenoid is disorganized in the cia6 mutant cells. We also show that Rubisco is mislocalized in cia6, as approximately 65% of the Rubisco was found in the stroma rather than in the “pyrenoid-like” structures. This result is reminiscent of the Synechococcus sp. PCC 7942 strain EK6, described by Schwarz et al. (1995), where a 30-amino acid extension in the RbcS caused the Rubisco to mislocalize to the cytoplasm. The EK6 strain had empty carboxysomes and was unable to grow under low-CO2 conditions. However, the mutation in cia6 is not within RbcS, as in the EK6 strain, but rather in the CIA6 gene on chromosome 10. By using both denaturing gels and nondenaturing gels, the Rubisco holoenzymes in wild-type and mutant cells have been examined. Cell extracts from both the wild-type and mutant cells were analyzed by western blot on a denaturing gel using an anti-Rubisco antibody, but no significant concentration differences in the two subunits could be revealed between the strains (Fig. 7A). Rubisco isolated from both the wild-type and mutant cells was also subjected to electrophoresis on a nondenaturing gel and stained with Coomassie blue. However, no significant differences were noticed between the two Rubisco preparations, as evidenced by the appearance of the same three high-Mr bands at the same position, which were all recognized by anti-Rubisco antibody after using western blotting (data not shown). The combination of both the denaturing and the nondenaturing gel electrophoresis analysis indicated the normal composition of the Rubisco holoenzyme in the mutant cells. On the other hand, the appearance of the four high-Mr bands in both C. reinhardtii Rubiscos on a nondenaturing gel is similar to the reported four bands observed when the pea RLsMT-bound spinach Rubisco was subjected to nondenaturing gel electrophoresis (Raunser et al., 2009), with each band representing the addition of one RLsMT onto the Rubisco while the lowest band represents the native Rubisco.
In both cyanobacteria and the green algae, the arrangement of the CCM components within the cell or chloroplast is critical to the functioning of the CCM. In a current model of the C. reinhardtii CCM (Moroney and Ynalvez, 2007; Spalding, 2008; Yamano et al., 2010), a higher than ambient concentration of CO2 inside the thylakoid lumen is predicted to be generated by CAH3. Since the thylakoid lumen is acidified in the light, most Ci in the lumen will be converted to CO2. This CO2 will either diffuse into the pyrenoid region to be fixed by Rubisco or diffuse back into the stroma. Since the stroma becomes more basic in the light, any CO2 that leaks out of the pyrenoid has a chance to be converted back to HCO3− by CAH6, thus reducing CO2 leakage. In cia6 cells, net Ci accumulation was reduced (Fig. 2C). This reduced accumulation could be due to a lower rate of Ci uptake or to a reduction in the ability of the cells to retain Ci, but we cannot discriminate between these two possibilities. It is clear that any disruption of the pyrenoid potentially disconnects the arrangement of these CCM components and reduces CO2 assimilation efficiency.
Recently, there has been increasing interest in the possible interaction between the LCIB/LCIC complex and the pyrenoid (Yamano et al., 2010). Two roles for such a localization have been proposed: a role in recapturing CO2 using stromal carbonic anhydrase (CAH6) and a role as a structural barrier in avoiding CO2 leakage from the pyrenoid. It would be intriguing to investigate how the LCIB/LCIC complex would localize in the cia6 mutant background, in which the pyrenoid is absent and Rubisco is no longer concentrated in this specialized region of the chloroplast. For example, would low CO2 still induce the migration of the LCIB/LCIC complex to the vicinity of the pyrenoid-like structure in the mutant cia6 cell? These results might shed light on whether the aggregation of the LCIB/LCIC complex needs the presence of pyrenoid or not, or whether the complex is actually associating with the thylakoid tubules or the pyrenoid.
Apparently, CIA6 is required for the formation of pyrenoid in C. reinhardtii; however, the exact function of CIA6 in the CCM or pyrenoid formation is not clear. Predicted to contain a SET domain by Pfam (Bateman et al., 2002), the CIA6 protein is likely to act as a Lys methyltransferase (Dillon et al., 2005; Qian and Zhou, 2006; Ng et al., 2007). In plants, other than modifying histones, SET domain-containing proteins were also found to methylate Lys-14 on Rubisco large subunit (Klein and Houtz, 1995; Ying et al., 1999; Trievel et al., 2002, 2003). Since in the cia6 mutant cells the Rubisco-containing pyrenoid is not well developed, it could be speculated that one possible function of CIA6 could be acting as the C. reinhardtii Rubisco methyltransferase, modifying the conformation of this holoenzyme and in some way allowing it to organize or self-assemble into a pyrenoid. However, the Rubisco-methylating activity of CIA6 could not be found in vitro. It should be noted that, unlike the pea RbcL, with methylated Lys-14 on the N-terminal tail, the C. reinhardtii RbcL’s Lys-14 is unmethylated. Instead, crystal structure data from C. reinhardtii Rubisco indicated the presence of methyl-Cys-256 and methyl-Cys-369 (Taylor et al., 2001), with the first Cys buried at the interface of RbcL and RbcS and the latter Cys positioned on the external surface of RbcL. Notwithstanding the potential electrostatic problem of a Cys fitting in the narrow cleft of the SET domain’s catalytic site, neither Cys is thought to easily enter the catalytic channel of RLsMT, as compared with Lys-14 on the flexible N-terminal tail. On the other hand, by examining the amino acid sequences of CIA6 and its 13 homologs, it was found that the invariant Tyr residue on the SET domain N terminus is present, indicating the possible presence of the target Lys-binding site. However, in all 13 CIA6 homologs, the highly conserved catalytic site motif NHS could not be found, which could argue that these proteins may belong to a subclass of SET domain-containing proteins that do not have catalytic functions but emphasize structural roles in relation to Rubisco aggregation and pyrenoid formation. Alternatively, based on the fact that CIA6 homologs are only found in green algae and higher plants but not in cyanobacteria or the sequenced diatom genomes, it could be speculated that the function of CIA6 may not be limited to or directly linked to the formation of pyrenoid structure or CCM function.
A notable phenotype of cia6 C. reinhardtii cells was its increased chlorophyll content under photoautotrophic conditions (Fig. 8). This observation implies that the function of CIA6 might be related to chloroplast organization or nuclear/chloroplast coordination. Like the chloroplast, the pyrenoid reproduces by binary fission during cell division (Goodenough, 1970; Goodenough and Levine, 1970; Harris et al., 2009). The disruption of the pyrenoid might occur if chloroplast structure or division is not well coordinated in the CIA6-deficient cell. We are working at present on obtaining an Arabidopsis knockout line with a CIA6 homolog gene disrupted to see whether the chloroplast structure is affected in that plant. Given the fact that multiple small pyrenoids were sometimes observed in a single mutant chloroplast, the possibility that the mutant pyrenoid division problem led to the altered chlorophyll concentration could not be excluded.
In summary, CIA6 is a member of a new gene family found in green algae and in higher plants. When this gene is knocked out in C. reinhardtii, the cell has a disrupted pyrenoid and a dysfunctional CCM. When the wild-type gene is returned to cia6 mutant cells, the normal pyrenoid morphology and CCM function are restored. To our knowledge, this is the first report of a gene outside of the Rubisco structure genes that affects pyrenoid structure. The linkage of the pyrenoid structure and the CCM provides strong support for CCM models in which the localization of Rubisco to the pyrenoid is essential.
MATERIALS AND METHODS
Cell Cultures and Growth
Chlamydomonas reinhardtii culture conditions were similar to those used previously (Rawat and Moroney, 1991). The strain D66 (nit2−, cw15, mt+) was obtained from Rogene Schnell (University of Arkansas, Little Rock), and strain CC124 (nit1−, nit2−, mt−) was obtained from the C. reinhardtii Duke University Stock Center (http://www.chlamy.org/). TAP and minimal (without acetate) liquid media were prepared according to Sueoka (1960). TAP plates and minimal medium plates were prepared by adding 1.2% (w/v) agar. Cell cultures were started from inoculating C. reinhardtii colonies from TAP plates into 100 mL of TAP liquid medium and grown with continuous shaking and light (100 μmol m−2 s−1) until the culture reached early log phase. TAP-grown cultures were harvested and washed with minimal liquid medium twice and then split into two flasks containing minimal medium and bubbled with high CO2 (5% [v/v] CO2 in air) until it reached a cell density of about 2 to 3 × 106 cells mL−1. To induce CCM, one flask was switched from high-CO2 to low-CO2 (0.01% [v/v] CO2 in air) bubbling for 1 d or otherwise as indicated; the other flask was kept on high CO2 as the control.
C. reinhardtii Mutagenesis and Electroporation
Strain D66 was mutagenized by transformation with a linearized pSP124s plasmid (Lumbreras et al., 1998) using the electroporation procedure described by Shimogawara et al. (1998) with modifications described by Colombo et al. (2002). Transformants were first selected on TAP plates containing the antibiotic zeocin (7.5 μg mL−1; Invitrogen). Antibiotic-resistant strains were then screened for a CCM-deficient phenotype in a low-CO2 chamber (0.01% [v/v] CO2 in air) in light (100 μmol m−2 s−1).
Nucleic Acid and Protein-Blot Analysis
Total DNA was isolated according to Newman et al. (1990). Southern-blot analysis was carried out following the guidelines of Sambrook et al. (2001). Briefly, restriction enzyme-digested DNA (2 μg in each lane) was loaded and separated on a 0.8% (w/v) agarose gel and blotted onto a nylon membrane (Schleicher & Schull). [32P]dCTP-labeled probes were prepared using a random primer procedure. For quantitative RT-PCR analysis, RNA was first extracted using Trizol reagent following the guidelines provided (Invitrogen). Contaminating DNA was removed by DNase treatment (Roche Applied Science) and was further cleaned up using an RNeasy kit (Qiagen). Total RNA was used for cDNA synthesis following the manufacturer’s instructions (Roche Applied Science) using poly(dT) primers. Synthesized cDNA was then subjected to quantitative RT-PCR analysis using SYBR Green Premix (Takara) using an ABI 7000 Real Time PCR System (Applied Biosystem).
Identification of Flanking Regions and Genetic Linkage Analysis
Adaptor-mediated PCR was used to identify the DNA flanking the pSP124s insertion using a modified version of the Genome Walker Kit (Clontech). Homology searches were performed using the BLAST server (Altschul et al., 1997) and the Joint Genome Institute C. reinhardtii database version 4.0 site (Merchant et al., 2007). Genetic crosses and tetrad analysis were performed as described previously (Sears et al., 1980; Moroney et al., 1986; Harris et al., 2009). Briefly, cia6 (mt+) and CC124 (mt−) cell cultures were subjected to nitrogen starvation in high light overnight and combined for mating for 1 h. Aliquots (0.3 mL) were plated on nitrogen-minus minimal medium containing 4% nitrogen-depleted agar and stored in the dark for zygote maturation for 10 d. The zygotes on the maturation plates were transferred to 1.2% agar TAP medium plates incubated overnight for meiotic germination. Tetrad dissections were then carried out, and linkage was determined by association of the bleomycin resistance gene with those progeny that grew poorly on low CO2 levels.
Photosynthesis Assays
Cultures were started heterotrophically in 100 mL of TAP medium. After reaching the log phase, the cultures were then transferred to 1 L of minimal medium and bubbled with 5% CO2 until they reached a cell density of about 3 × 106 cells mL−1. The cultures were then subjected to CCM induction for 12 h at 100 μE m−2 s−1. The affinity for external Ci (K0.5[DIC]) was estimated as described by Pollock and Colman (2001). Briefly, cells containing 100 μg of chlorophyll were suspended in CO2-free HEPES buffer (pH 7.3) that was previously bubbled with N2. The cell suspension was transferred to the electrode chamber (Rank Brothers) and was allowed to deplete any endogenous DIC until no net oxygen exchange was observed. The DIC concentrations of the medium were controlled by injecting NaHCO3 solution into the chamber. The light intensity for photosynthesis assay was adjusted at 300 μE m−2 s−1. The K0.5[DIC] value is calculated as the DIC concentration required for half-maximal rates of oxygen evolution (Badger, 1985).
Complementation
Complementation of the mutant strain cia6 was achieved by transforming cia6 cells with the entire CIA6 genomic region spanning the entire gene using the glass bead method (Kindle, 1990). Briefly, cells were grown to 3 × 106 cells mL−1 in 100 mL of TAP before transferring to 1 L of minimal medium bubbled with air until the cell density reached 2 × 106 cells mL−1. Transformation was started by mixing 0.3 mL of cell suspension (3 × 108 cells mL−1), 300 mg of sterilized glass beads, 5 μg of DNA, and 0.1 mL of 20% polyethylene glycol. The mixture was then agitated at top speed in a 15-mL tube for 15 s. After beads settled to the bottom, the cell suspension was plated onto minimal medium plates and maintained in a low-CO2 chamber (0.007% [v/v] CO2 in air). Colonies that were able to grow heterotrophically under low-CO2 conditions were examined using light microscopy to screen for the strains with a pyrenoid. RNA was then extracted from putative complemented strains to confirm the reappearance of the full-length CIA6 mRNA.
Immunolocalization Studies Using Electron Microscopy
The immunolocalization procedure was performed as described previously (Mitra et al., 2004). Briefly, about 2 mL of cell suspension was fixed with an equal volume of 1% OsO4, 2% formaldehyde, 0.5% glutaraldehyde, and 0.1 mm sodium cacodylate buffer (pH 7.2) for 30 min. The solution was then extracted into a 10-mL syringe with a Swinney filter holder fitted with a 13-mm-diameter, 5-μm-pore polycarbonate filter and fixed for another 30 min, followed by 15-min washes with 0.1 m cacodylate buffer containing 0.02 m Gly five times. Materials were rinsed with distilled water and stained with 0.5% uranyl acetate in the dark for 30 min. After this, excess cells were rinsed and the samples were dehydrated using an ethyl alcohol series. Samples were then infiltrated and embedded in LR White resin (Electron Microscopy Sciences). Embedded samples were sectioned with a DuPont Sorvall microtome to 70 nm. TEM sections were mounted on collodion-coated nickel grids.
The immunocytochemical procedure was similar to the method of Borkhsenious et al. (1998) with some modifications. Sections were pretreated with 2% sodium-metaperiodate (Sigma) for 15 min to remove any residual glutaraldehyde, then blocked two times for 30 min in blocking solution (2% bovine serum albumin and 0.1% Tween 20 in phosphate-buffered saline). The sections were incubated for 90 min with Rubisco antibody (1:50) or with the preimmune serum with the same dilution. The grids were transferred to a 1:50 dilution of Protein A-gold label (20 nm; Sigma) for 1 h. Antibodies were all diluted in blocking solution. Finally, the sections were rinsed with distilled water and photographed by TEM.
The fraction of Rubisco in the pyrenoid and in the chloroplast stroma was measured as described previously (Borkhsenious et al., 1998). Here, the pyrenoid area is excluded from the total chloroplast stroma. The immunogold particle density in the pyrenoid (Dp; no. μm−2) was first calculated by dividing the total number of immunogold particles in the pyrenoid by the area of pyrenoid. Similarly, the immunogold particle density in the chloroplast stroma (Ds; no. μm−2) was calculated by dividing the total number of immunogold particles in the chloroplast stroma by the total area of chloroplast stroma. The total immunogold particles in the pyrenoid or stroma were then calculated by multiplying the immunogold density (D) averaged from 25 TEM thin sections and the average volume of each compartment (Lacoste-Royal and Gibbs, 1987). The fraction of Rubisco in the pyrenoid (Fractionp) was further calculated by dividing the total particles in the pyrenoid by the total number of particles from pyrenoid and chloroplast stroma. The final equation is as follows: Fractionp = 100 × [Dp × 2.4/(Dp × 2.4 + Ds × 35.6)], where D = number of particles per area.
Other Methods
The CO2 concentration in the growth chambers was measured using an infrared gas analyzer (Analytical Development Co.). Protein concentration was determined using Bradford 10× Dye Reagent (Bio-Rad) with bovine serum albumin as standard. The growth curve experiments were standardized based on chlorophyll mass (1.9 μg mL−1) and approximately the same cell density. Chlorophyll content was measured as the total content from chlorophyll a plus chlorophyll b (Arnon, 1949; Holden, 1976). Chlorophyll was extracted by 100% methanol and was quantified spectrophotometrically (Arnon, 1949) and calculated according to Holden (1976). Cell density values were determined by direct counting in a hemacytometer chamber (Hausser Scientific). For pyrenoid staining and observation using light microscopy, C. reinhardtii cells were stained with 0.05% bromphenol blue in 0.1% HgCl2 (Kuchitsu et al., 1988).
Sequence data from this article can be found in the GenBank data library under accession number JF288753.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A higher chlorophyll content per cell was observed in the mutant cia6.
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR. (1985) Photosynthetic oxygen exchange. Annu Rev Plant Physiol 36: 27–53 [Google Scholar]
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD. (1998) The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can J Bot 76: 1052–1071 [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bold H, Wynne M. (1985) Introduction to the Algae, Ed 2 Prentice-Hall, Englewood, NJ [Google Scholar]
- Borkhsenious ON, Mason CB, Moroney JV. (1998) The intracellular localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii Plant Physiol 116: 1585–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo SL, Pollock SV, Eger KA, Godfrey AC, Adams JE, Mason CB, Moroney JV. (2002) Use of the bleomycin resistance gene to generate tagged insertional mutants of Chlamydomonas reinhardtii that require elevated CO2 for optimal growth. Funct Plant Biol 29: 231–241 [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. (2005) The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Wang Y, Spalding MH. (2009) Thylakoid lumen carbonic anhydrase (CAH3) mutation suppresses air-dier phenotype of LCIB mutant in Chlamydomonas reinhardtii. Plant Physiol 149: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genkov T, Meyer M, Griffiths H, Spreitzer RJ. (2010) Functional hybrid Rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas. J Biol Chem 285: 19833–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Beardall J, Raven JA. (2005) CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol 56: 99–131 [DOI] [PubMed] [Google Scholar]
- Goodenough UW. (1970) Chloroplast division and pyrenoid formation in Chlamydomonas reinhardtii. J Phycol 6: 1–6 [Google Scholar]
- Goodenough UW, Levine RP. (1970) Chloroplast structure and function in ac-20, a mutant strain of Chlamydomonas reinhardtii. 3. Chloroplast ribosomes and membrane organization. J Cell Biol 44: 547–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths D. (1970) The pyrenoid. Bot Rev 36: 29–58 [Google Scholar]
- Gutknecht J, Bisson MA, Tosteson FC. (1977) Diffusion of carbon dioxide through lipid bilayer membranes: effects of carbonic anhydrase, bicarbonate, and unstirred layers. J Gen Physiol 69: 779–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH, Stern DB, Witman G. (2009) The Chlamydomonas Sourcebook, Ed 2 Academic Press, Amsterdam [Google Scholar]
- Hemsley A, Poole I. (2004) The Evolution of Plant Physiology: From Whole Plants to Ecosystems. Elsevier Academic Press, Amsterdam [Google Scholar]
- Henk M, Rawat M, Hugghins S, Lavigne L, Ramazanov Z, Mason C, Moroney J. (1995) Pyrenoid morphology in Rubisco and CO2 concentrating mutants of Chlamydomonas reinhardtii. Mathis P, , Proceedings of the Xth International Photosynthesis Congress, Vol V Springer, Montpellier, France, pp 595–598 [Google Scholar]
- Holden M. (1976) Chlorophylls. Goodwin TW, , Chemistry and Biochemistry of Plant Pigments, Ed 2, Vol 2 Academic Press, London, pp 461–488 [Google Scholar]
- Houtz RL, Poneleit L, Jones SB, Royer M, Stults JT. (1992) Posttranslational modifications in the amino-terminal region of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase from several plant species. Plant Physiol 98: 1170–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Takeda H. (1995) Species-specific differences of pyrenoids in Chlorella (Chlorophyta). J Phycol 31: 813–818 [Google Scholar]
- Inwood W, Yoshihara C, Zalpuri R, Kim KS, Kustu S. (2008) The ultrastructure of a Chlamydomonas reinhardtii mutant strain lacking phytoene synthase resembles that of a colorless alga. Mol Plant 1: 925–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson J, Clarke AK, Chen ZY, Hugghins SY, Park YI, Husic HD, Moroney JV, Samuelsson G. (1998) A novel alpha-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J 17: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Houtz RL. (1995) Cloning and developmental expression of pea ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit N-methyltransferase. Plant Mol Biol 27: 249–261 [DOI] [PubMed] [Google Scholar]
- Kuchitsu K, Tsuzuki M, Miyachi S. (1988) Characterization of the pyrenoid isolated from unicellular green-alga Chlamydomonas reinhardtii: particulate form of Rubisco protein. Protoplasma 144: 17–24 [Google Scholar]
- Lacoste-Royal G, Gibbs SP. (1987) Immunocytochemical localization of ribulose-1,5-bisphosphate carboxylase in the pyrenoid and thylakoid region of the chloroplast of Chlamydomonas reinhardtii. Plant Physiol 83: 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Stevens DR, Purton S. (1998) Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J 14: 441–447 [Google Scholar]
- McKay RML, Gibbs SP, Vaughn KC. (1991) RuBisCo activase is present in the pyrenoid of green algae. Protoplasma 162: 38–45 [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M, Lato SM, Ynalvez RA, Xiao Y, Moroney JV. (2004) Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 135: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M, Mason CB, Xiao Y, Ynalvez RA, Lato SM, Moroney JV. (2005) The carbonic anhydrase gene families of Chlamydomonas reinhardtii. Can J Bot 83: 780–795 [Google Scholar]
- Miura K, Yamano T, Yoshioka S, Kohinata T, Inoue Y, Taniguchi F, Asamizu E, Nakamura Y, Tabata S, Yamato KT, et al. (2004) Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol 135: 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Abe T, Tsuzuki M, Fujiwara S, Sato N, Hirata A, Sonoike K, Nozaki H. (1999) Role of pyrenoids in CO2 concentrating mechanism: comparative morphology, physiology and molecular phylogenetic analysis of closely related strains of Chlamydomonas and Chloromonas (Volvocales). Planta 208: 365–372 [Google Scholar]
- Morita E, Kuroiwa H, Kuroiwa T, Nozaki H. (1997) High localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in the pyrenoids of Chlamydomonas reinhardtii (Chlorophyta), as revealed by cryofixation and immunogold electron microscopy. J Phycol 33: 68–72 [Google Scholar]
- Moroney JV, Husic HD, Tolbert NE, Kitayama M, Manuel LJ, Togasaki RK. (1989) Isolation and characterization of a mutant of Chlamydomonas reinhardtii deficient in the CO2 concentrating mechanism. Plant Physiol 89: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Tolbert NE, Sears BB. (1986) Complementation analysis of the inorganic carbon concentrating mechanism of Chlamydomonas reinhardtii. Mol Gen Genet 204: 199–203 [Google Scholar]
- Moroney JV, Ynalvez RA. (2007) Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot Cell 6: 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SM, Boynton JE, Gillham NW, Randolph-Anderson BL, Johnson AM, Harris EH. (1990) Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics 126: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC. (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769: 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock SV, Colman B. (2001) The inhibition of the carbon concentrating mechanism of the green alga Chlorella saccharophila by acetazolamide. Physiol Plant 111: 527–532 [DOI] [PubMed] [Google Scholar]
- Pollock SV, Colombo SL, Prout DL, Jr, Godfrey AC, Moroney JV. (2003) Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low-CO2 atmosphere. Plant Physiol 133: 1854–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR. (1989) Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype: evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiol 91: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM. (2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461 [DOI] [PubMed] [Google Scholar]
- Pronina NA, Semenenko VE. (1992) Carbonic anhydrase activity and fatty-acid composition of photosystem deficient and high CO2 required mutants of Chlamydomonas reinhardtii. Photosynth Res 34: 201 [Google Scholar]
- Qian C, Zhou MM. (2006) SET domain protein lysine methyltransferases: structure, specificity and catalysis. Cell Mol Life Sci 63: 2755–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazanov Z, Rawat M, Henk MC, Mason CB, Matthews SW, Moroney JV. (1994) The induction of the CO2-concentrating mechanism is correlated with the formation of the starch sheath around the pyrenoid of Chlamydomonas reinhardtii. Planta 195: 210–216 [Google Scholar]
- Raunser S, Magnani R, Huang Z, Houtz RL, Trievel RC, Penczek PA, Walz T. (2009) Rubisco in complex with Rubisco large subunit methyltransferase. Proc Natl Acad Sci USA 106: 3160–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat M, Henk MC, Lavigne LL, Moroney JV. (1996) Chlamydomonas reinhardtii mutants without ribulose-1,5-bisphosphate carboxylase-oxygenase lack a detectable pyrenoid. Planta 198: 263–270 [Google Scholar]
- Rawat M, Moroney JV. (1991) Partial characterization of a new isoenzyme of carbonic anhydrase isolated from Chlamydomonas reinhardtii. J Biol Chem 266: 9719–9723 [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Russell DW, Fritsch EF. (2001) Molecular Cloning: A Laboratory Manual, Ed 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schnell RA, Lefebvre PA. (1993) Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Reinhold L, Kaplan A. (1995) Low activation state of ribulose-1,5-bisphosphate carboxylase/oxygenase in carboxysome-defective Synechococcus mutants. Plant Physiol 108: 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears BB, Boynton JE, Gillham NW. (1980) The effect of gametogenesis regimes on the chloroplast genetic system of Chlamydomonas reinhardtii. Genetics 96: 95–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H. (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraiwa Y, Miyachi S. (1983) Factors controlling induction of carbonic anhydrase and efficiency of photosynthesis in Chlorella vulgaris llh cells. Plant Cell Physiol 24: 919–923 [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23: 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH. (2008) Microalgal carbon-dioxide-concentrating mechanisms: Chlamydomonas inorganic carbon transporters. J Exp Bot 59: 1463–1473 [DOI] [PubMed] [Google Scholar]
- Spalding MH, Spreitzer RJ, Ogren WL. (1983) Carbonic anhydrase-deficient mutant of Chlamydomonas reinhardtii requires elevated carbon dioxide concentration for photoautotrophic growth. Plant Physiol 73: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ, Goldschmidt-Clermont M, Rahire M, Rochaix J-D. (1985) Nonsense mutations in the Chlamydomonas chloroplast gene that codes for the large subunit of ribulosebisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA 82: 5460–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. (1960) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 46: 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Taylor TC, Backlund A, Bjorhall K, Spreitzer RJ, Andersson I. (2001) First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii. J Biol Chem 276: 48159–48164 [DOI] [PubMed] [Google Scholar]
- Trievel RC, Beach BM, Dirk LMA, Houtz RL, Hurley JH. (2002) Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111: 91–103 [DOI] [PubMed] [Google Scholar]
- Trievel RC, Flynn EM, Houtz RL, Hurley JH. (2003) Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat Struct Biol 10: 545–552 [DOI] [PubMed] [Google Scholar]
- Vaughn KC, Campbell EO, Hasegawa J, Owen HA, Renzaglia KS. (1990) The pyrenoid is the site of ribulose 1,5-bisphosphate carboxylase/oxygenase accumulation in the hornwort (Bryophyta: Anthocerotae) chloroplast. Protoplasma 156: 117–129 [Google Scholar]
- Wang Y, Spalding MH. (2006) An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 103: 10110–10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Lee J-H, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272 [DOI] [PubMed] [Google Scholar]
- Yamano T, Tsujikawa T, Hatano K, Ozawa S, Takahashi Y, Fukuzawa H. (2010) Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol 51: 1453–1468 [DOI] [PubMed] [Google Scholar]
- Ying Z, Mulligan RM, Janney N, Houtz RL. (1999) Rubisco small and large subunit N-methyltransferases: bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J Biol Chem 274: 36750–36756 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.