Abstract
Previous work suggests that gibberellins (GAs) play an important role in early seed development. To more fully understand the roles of GAs throughout seed development, tissue-specific transcription profiles of GA metabolism genes and quantitative profiles of key GAs were determined in pea (Pisum sativum) seeds during the seed-filling development period (8–20 d after anthesis [DAA]). These profiles were correlated with seed photoassimilate acquisition and storage as well as morphological development. Seed coat growth (8–12 DAA) and the subsequent dramatic expansion of branched parenchyma cells were correlated with both transcript abundance of GA biosynthesis genes and the concentration of the growth effector GA, GA1. These results suggest GA1 involvement in determining the rate of seed coat growth and sink strength. The endosperm’s PsGA20ox transcript abundance and the concentration of GA20 increased markedly as the endosperm reached its maximum volume (12 DAA), thus providing ample GA20 substrate for the GA 3-oxidases present in both the embryo and seed coat. Furthermore, PsGA3ox transcript profiles and trends in GA1 levels in embryos at 10 to 16 DAA and also in embryo axes at 18 DAA suggest localized GA1-induced growth in these tissues. A shift from synthesis of GA1 to that of GA8 occurred after 18 DAA in the embryo axis, suggesting that deactivation of GA1 to GA8 is a likely mechanism to limit embryo axis growth and allow embryo maturation to proceed. We hypothesize that GA biosynthesis and catabolism are tightly regulated to bring about the unique developmental events that occur during seed growth, development, and maturation.
GAs serve a vital role in coordinating growth and development throughout the life cycle of a plant, including seed development (Davies, 2004). In pea (Pisum sativum), KO (at the LH locus) encodes for ent-kaurene oxidase, which catalyzes a very early step in the GA biosynthesis pathway (Davidson et al., 2004). In the lh-2 mutation, there are very reduced GA levels in developing seeds, increased seed abortion, and much smaller seeds at maturity than in wild-type plants (Swain et al., 1993). Furthermore, fertilization of lh-2 ovaries with wild-type pollen partially reversed these seed mutant phenotypes. Taken together, this suggests that GAs play an important role in early pea seed development (Swain et al., 1993, 1995). However, it has been generally assumed that GAs are not required for seed development beyond these early stages (Garcia-Martinez et al., 1987; Swain et al., 1995), as the concentration of growth-active GAs in whole pea seed extracts decreases from a peak around 6 d after anthesis (DAA) to minimal or nondetectable levels by 12 DAA (Swain et al., 1993; Rodrigo et al., 1997), with nondetectable levels at seed maturity (Sponsel, 1983; Swain et al., 1993; Ayele et al., 2006). Additionally, developing pea seeds exposed to GA biosynthesis inhibitors can still produce viable mature seeds, albeit with reduced seed weight, germination, and shoot and root length (Garcia-Martinez et al., 1987). Thus, the possibility that tissue-specific regulation of GA biosynthesis occurs for unique developmental outcomes at later stages of seed development (10–20 DAA) has received minimal attention (Weber et al., 2005).
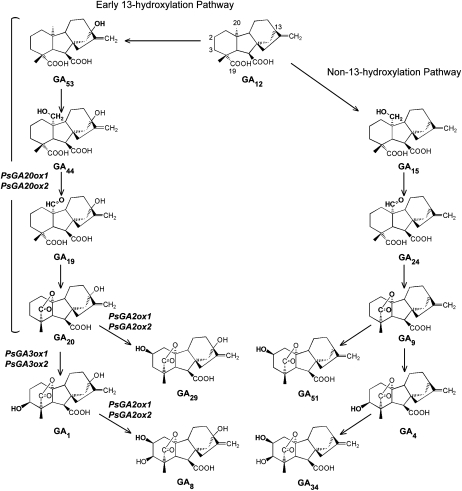
GAs have an ent-gibberellane structure, and β-hydroxylation at C-3 “activates” the GA molecule (e.g. GA1 is formed from the growth-inactive precursor, GA20; Fig. 1). In contrast, C-2 β-hydroxylation deactivates biologically active GA1 (C-2 β-hydroxylation can also prevent the conversion of a number of precursor GAs to bioactive forms). Bioactive GA levels are determined primarily through regulation at the third stage of the GA biosynthesis pathway (Fleet et al., 2003; Ayele et al., 2006; Fig. 1). In pea seeds, there are two GA biosynthesis pathways, one where C-13 remains nonhydroxylated or is hydroxylated much later and the other where C-13 is hydroxylated early (i.e. the sequential oxidation of GA12 [non-13-hydroxylated] or GA53 [13-hydroxylated] to bioactive GAs; Fig. 1). Following the formation of GA12 or GA53, the multifunctional enzyme GA 20-oxidase, which is encoded by two known genes in pea, PsGA20ox1 (García-Martínez et al., 1997) and PsGA20ox2 (Lester et al., 1996), catalyzes the sequential oxidation and loss of C-20 and the formation of a lactone ring (producing GA20 from GA53; Fig. 1). Subsequently, GA20 can be 3β-hydroxylated at C-3 to growth-active GA1 by two GA 3-oxidases encoded by PsGA3ox1 (Lester et al., 1997) and PsGA3ox2 (Weston et al., 2008). The GA 2-oxidase enzymes catalyze the 2β-hydroxylation of both GA1 and GA20, producing the biologically inactive metabolites GA8 and GA29, respectively (Lester et al., 1999; Martin et al., 1999). While the Escherichia coli heterologous expression products of the two known pea GA 2-oxidase genes are capable of catalyzing this reaction on either substrate, PsGA2ox1 has an approximately equal substrate affinity for both GA20 and GA1, whereas PsGA2ox2 has a much higher affinity for bioactive GA1 (Lester et al., 1999).
Figure 1.
The third stage of GA biosynthesis: the non-13-hydroxylation and early-13-hydroxylation GA biosynthesis pathways. Genes encoding for enzymes involved in this pathway in pea are indicated in italics. GA20 could also be synthesized from GA9 by 13-hydroxylation (not shown in the pathways here).
The differential regulation of the various PsGA20ox, PsGA3ox, and PsGA2ox genes during pea seed development as assessed from whole seed samples is evident from transcript-profiling experiments (Ozga et al., 2009). Consistent with the hypothesis that GAs play an important role in early pea seed development, transcript levels of PsGA3ox1 (which are responsible for the production of biologically active 3β-hydroxylated GAs) peak in the seeds immediately following fertilization (0 DAA; Ozga et al., 2003) and also during early seed development (approximately 4–7 DAA; Ozga et al., 2003, 2009). However, a third peak in seed PsGA3ox1 transcript levels, one with a greater magnitude, was observed from 10 to 14 DAA (Ozga et al., 2003, 2009), at a time when it is generally assumed that GAs are not required for seed development (Swain et al., 1995). Furthermore, there was no simple relationship between transcript abundance of the late-stage GA biosynthesis genes and GA concentration profiles from whole seed extracts across several stages of seed development (Ozga et al., 2009). Indeed, growth and development within the seed is dramatically different in the maternal tissue (seed coat) and the tissues derived from fertilization events (endosperm and embryo; Marinos, 1970). Each of these three seed tissues has distinct functions throughout development. The seed coat, made up of several discernible layers, provides a protective environment as well as serving as the conduit to provide sugars and other phloem-derived nutrients to the developing embryo (Van Dongen et al., 2003). The endosperm of pea is noncellular and transient (Marinos, 1970), and it also serves as a nutrient tissue for the developing embryo. The embryo itself is made up of two distinct tissues, the cotyledons and the embryo axis. The cotyledons differentiate into nutrient storage organs and the embryo axis into a miniature plant with a shoot and root meristem, which will progress into quiescence as the seed matures. It seems highly likely that tissue-specific regulation of GA biosynthesis will occur for unique developmental outcomes within each tissue of the seed, events that will not be reflected in whole seed analysis. Indeed, this is the case during very early pea seed development (4 DAA), when GA metabolism and bioactive GA profiles vary in the endosperm/embryo tissue relative to that seen in the seed coat (Rodrigo et al., 1997).
To clarify the tissue-specific nature of GA biosynthesis as the seed transitions from being predominantly seed coat tissue to predominantly embryo tissue (from 8 to 20 DAA), the transcription profiles of key GA biosynthesis and catabolism genes as well as metabolite profiles (concentrations) of key GAs were investigated in the seed coat, endosperm, and embryo (and in cotyledons and embryo axes) of pea at specific time points. The assessment of photoassimilate acquisition and storage, as well as seed coat and cotyledon morphological development, was also employed to correlate the physiology of the developing seed with its hormone biosynthesis and metabolism. Since the small seed size of model dicot plants such as Arabidopsis (Arabidopsis thaliana) and Medicago truncatula generally preclude studies of this nature, the developmental model proposed in this study for GA biosynthesis and catabolism during pea seed development can serve as a general dicot model for further understanding of the role of GAs in seed development.
RESULTS AND DISCUSSION
Seed Development in Pea
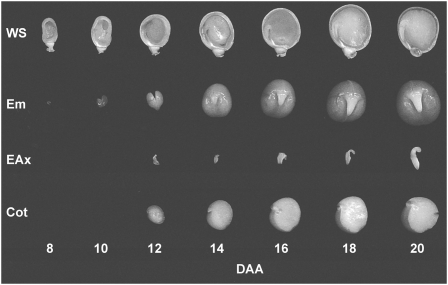
The seed coat, embryo, and endosperm tissues undergo a variety of changes in physiology and structure as development progresses toward a viable, mature seed (Fig. 2). During the developmental period studied, the seed transitions from mainly consisting of seed coat tissue with liquid endosperm to mainly embryo tissue with no liquid endosperm. It is likely that extensive coordination must be maintained through tissue-to-tissue signaling in order for normal seed development to occur. Seed coat growth (Fig. 2) and fresh weight (Fig. 3A) mainly occurred between 8 and 12 DAA (Figs. 2 and 3). At this time, endosperm volume increased markedly, reaching maximum volume at 12 DAA, before being rapidly absorbed by the developing embryo between 12 and 14 DAA (Figs. 2 and 3B). By 14 to 16 DAA, the majority of the seed tissue’s fresh weight was embryo, consisting of rapidly growing cotyledons and embryo axis (Fig. 3).
Figure 2.
Representative pea seed tissues over 20 d of development. Bisected whole seeds (WS) and embryos (Em) from 8 to 20 DAA are shown, as are embryo axes (EAx) and cotyledons (Cot) from 12 to 20 DAA.
Figure 3.
Development of seed tissues. A, Whole seed, embryo, and seed coat fresh weight (fwt) between 6 and 20 DAA. B, Embryo, embryo axis (inset), and cotyledon fresh weight and endosperm volume (per seed) between 8 and 20 DAA. Data are expressed as means ± se. In some cases, se bars are not visible as they are obscured by the symbols. Sample size varies by time point: n = 53 to 228 for whole seeds (except at 8 DAA, where n = 8), n = 43 to 106 for embryos, n = 32 to 110 for seed coats (except at 8 DAA, where n = 11), n = 22 to 109 for embryo axis tissue (except at 14 DAA, where n = 8), n = 21 to 107 for cotyledons (except at 14 DAA, where n = 9), and n = 10 to 28 for endosperm tissue.
Morphology and Physiology of Developing Seed Coats
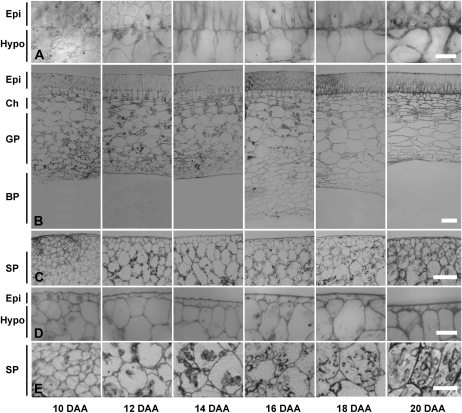
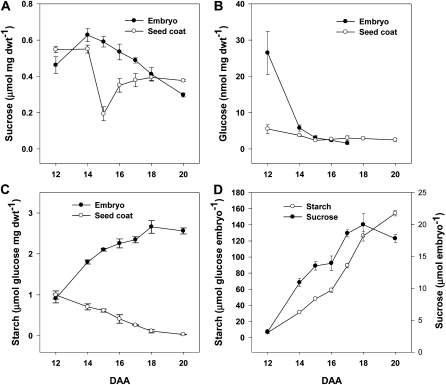
The morphology and function of the seed coat changes throughout its development; therefore, understanding the morphological state and function of the tissue temporally will aid in making meaningful predictions of the potential roles of GAs during seed coat growth and development. The seed coat of pea consists of several discernible layers that, depending on developmental stage, can include an epidermis subtended by a single layer of hypodermal cells, followed by multiple layers of chlorenchyma, ground parenchyma, and branched parenchyma cells (Fig. 4, A and B). Each of these seed coat cell types has its own developmental pattern. Similar to that reported by Van Dongen et al. (2003), differentiation of the epidermal cells into a palisade of macrosclerids was observed in the seed coat from 10 to 16 DAA (Fig. 4, A and B). Cell wall thickening was observed in the epidermis, particularly from 16 DAA onward (Fig. 4B). Organization of the hypodermis also occurred, as cells developed from roughly circular (in cross-section) hypodermal cells at 10 DAA to hourglass-shaped hypodermal cells by 14 DAA (Fig. 4A). Chlorenchyma cells, which immediately subtend the hypodermis, maintained a relatively constant size from 10 to 16 DAA, then expanded from 16 to 18 DAA (Figs. 4B and 5A). The cells of the ground parenchyma layer increased in cross-sectional area from 10 to 14 DAA (Figs. 4B and 5A). This ground parenchyma cell expansion phase was followed by a pause in cell cross-sectional area growth between 14 and 16 DAA. A marked increase in cell expansion then occurred from 16 to 18 DAA (Fig. 5A). The increase in the cross-sectional area of the ground parenchyma/chlorenchyma cells by 18 DAA may act to compensate for the loss of the peripheral ground parenchyma cells (closest to the embryo) following mechanical compaction by the expanding embryo (Figs. 4B and 5A). Symplasmic unloading of phloem nutrients that are ultimately destined for the embryo occurs in the ground parenchyma and chlorenchyma cell layers of the pea seed coat (Van Dongen et al., 2003). Starch also transiently accumulates in the plastids of these cell layers (Fig. 4B; Rochat and Boutin, 1992). At 12 DAA, seed coat starch levels were high and equivalent to starch levels (on a dry weight basis) found in the embryo (Fig. 6C). As seed development proceeded, both the seed coat starch levels (Fig. 6C) and the prevalence of starch-containing plastids in the ground parenchyma and chlorenchyma cell layers (Fig. 4B) decreased to minimal levels by 20 DAA. Coincidentally, embryo starch levels increased (Fig. 6C). Studies using the pea rb mutant (mutation in ADP-Glc pyrophosphorylase; Rochat et al., 1995; Lloyd et al., 1996) suggest that this transient pool of seed coat starch is required for maximal embryo growth and final seed size.
Figure 4.
Morphology of seed coat (A and B) and cotyledon (C–E) tissues throughout development. Left to right: 10, 12, 14, 16, 18, and 20 DAA. Magnification bars are 100 μm (B and C), 20 μm (A and D), and 50 μm (E; storage parenchyma cells from C at higher magnification). Images of 10-, 14-, and 18-DAA seed coats and 10-, 12-, 16-, and 20-DAA cotyledons are representative of two independent samples. Images of 12- and 16-DAA seed coats and 14- and 18-DAA cotyledons are representative of three independent samples. Image of the 20-DAA seed coat is representative of four independent samples. Epi, Epidermis; Hypo, hypodermis; Ch, chlorenchyma; GP, ground parenchyma; BP, branch parenchyma; SP, storage parenchyma.
Figure 5.
Average cross-sectional area of seed coat (A) and cotyledon (B) cells between 10 and 20 DAA. Subsamples (average number of cells per subsample = 98 cells) were obtained from two biological replicates, and data are presented as means of biological replicates ± se. In some cases, error bars are concealed by data points. Embryo expansion crushes the branch parenchyma after 16 DAA and the ground parenchyma after 18 DAA, so data at these times are not presented.
Figure 6.
A to C, Suc (A), Glc (B), and starch (C) levels expressed on a per mg dry weight (dwt) basis in the seed coats and embryos over seed development (12–20 DAA). D, Embryo starch and Suc levels expressed on a per embryo basis over seed development (12–20 DAA).
The innermost layer of the seed coat is made up of several layers of branched parenchyma cells. While ground parenchyma cell size was relatively constant between 14 and 16 DAA, the branched parenchyma cells substantially increased in size during this period (Figs. 4B and 5A). The branched parenchyma cells were irregular in appearance, with extensive intercellular spaces present from 10 to 14 DAA (Fig. 4B). This morphology was also confirmed by assessing the wide variation in branched parenchyma cell shape within the biological replicates (data not shown). These intercellular spaces are both air and liquid filled (Van Dongen et al., 2003), and the branched parenchyma cells make extensive contacts with the liquid endosperm prior to absorption of the latter. The dramatic expansion of the branched parenchyma cell layer between 14 and 16 DAA marks one of the most distinctive morphological changes within the seed coat (Fig. 4B). The presence of this expanded branched parenchyma cell layer (from 16 to 18 DAA) was transient, and as seed coat development proceeded, complete compression of the branch parenchyma cells by the expanding embryo occurred (Fig. 4B). The cell wall remnants from the compression of this seed coat cell type (and inner ground parenchyma cells) form a boundary layer between the intact seed coat’s ground parenchyma cells and the cotyledons (Van Dongen et al., 2003).
GA Gene Expression and GA Levels in Developing Seed Coats
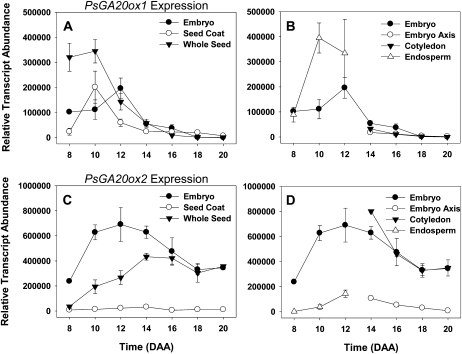
Both PsGA20ox1 and PsGA20ox2 encode enzymes responsible for catalyzing the three reactions that convert GA53 to GA20. PsGA20ox1 transcript abundance in the seed coat (Fig. 7A) peaked at 10 DAA, followed by a 3-fold increase in the level of seed coat GA20 by 12 DAA (6-fold increase per seed; Table I; Supplemental Table S1). The endosperm could be an additional source for seed coat-localized GA20 at this time (see endosperm section below). A second peak in GA20 concentration was detected at 16 DAA, prior to the rapid expansion of ground parenchyma and chlorenchyma cells in the seed coat at 18 DAA (Fig. 5A). GA20ox2 transcript abundance was minimal in the seed coat from 8 to 20 DAA (Fig. 7C).
Figure 7.
Expression of PsGA20ox1 and PsGA20ox2 genes (their enzyme products are responsible for catalyzing the reactions that convert GA53 to GA20) in seed tissues from 8 to 20 DAA. Data are expressed as means ± se; n = 2 to 5 independent samples, except for 8-DAA embryo (n = 1) and 14-DAA seed coat (n = 6). Error bars are present at each point but may be obscured by symbols if the se is too small. All samples are normalized to allow comparison between tissues for the same gene.
Table I. Abundance of 13-hydroxylated GAs, GA20, GA29, GA1, and GA8, and the non-13-hydroxylated GA, GA9, in developing seed tissues from 10 DAA to 20 to 21 DAA.
Endogenous GA4 and GA3 were not detected, but their internal standards were recovered in all samples. Values are means of two independent samples ± se; if no se is given, n = 1. na, Not available; nd, not detected (endogenous GA not detected, but internal standard recovered).
| Tissue | GA20 | GA29 | GA1 | GA8 | GA9 |
| ng g−1 fresh wt | |||||
| Embryo | |||||
| 10 DAA | 184.94 | 16.54 | 0.37 | 4.45 | na |
| 12 DAA | 386.6 ± 73.34 | 53.06 ± 2.61 | 0.13 ± 0.01 | 2.08 ± 0.03 | 113.74 |
| 14 DAA | 121.80 ± 9.91 | 64.30 ± 3.88 | 0.13 ± 0.03 | 2.03 ± 0.14 | 131.27 ± 21.23 |
| 16 DAA | 297.08 ± 0.32 | 83.65 ± 3.93 | 0.26 ± 0.04 | 2.27 ± 0.04 | 73.90 ± 5.13 |
| 18 DAA | 89.21 ± 8.79 | 75.39 ± 4.46 | nd | 1.18 ± 0.01 | 2.68 ± 0.81 |
| Seed coat | |||||
| 10 DAA | 5.00 ± 0.18 | 9.66 ± 0.16 | 1.57 ± 0.44 | 8.35 ± 0.41 | nd |
| 12 DAA | 17.05 ± 1.97 | 127.33 ± 26.25 | 0.15 ± 0.03 | 2.85 ± 0.36 | nd |
| 14 DAA | 12.65 ± 1.97 | 124.77 ± 6.82 | 0.21 ± 0.04 | 1.15 ± 0.06 | nd |
| 16 DAA | 41.95 ± 4.39 | 113.54 ± 6.02 | nd | 1.20 ± 0.03 | 0.79 ± 0.17 |
| 18 DAA | 27.79 ± 8.15 | 147.01 ± 6.37 | nd | 1.34 | nd |
| Embryo axis | |||||
| 18 DAA | 1,256.79 | 233.62 | 1.03 | 11.69 | 4.49 |
| 20–21 DAA | 372.98 ± 10.9 | 273.92 ± 21.18 | nd | 4.5 ± 1.5 | na |
| Cotyledon | |||||
| 18 DAA | 163.00 ± 38.27 | 205.83 ± 5.86 | 0.07 ± 0.07 | 1.30 ± 0.30 | 11.09 ± 0.36 |
| ng mL−1 | |||||
| Endosperm | |||||
| 10 DAA | 3.43 ± 0.57 | 0.66 ± 0.26 | 0.06 ± 0.05 | 0.38 ± 0.09 | nd |
| 12 DAA | 190.7 ± 21.22 | 18.34 ± 4.92 | nd | 0.24 ± 0.06 | 3.58 ± 0.05 |
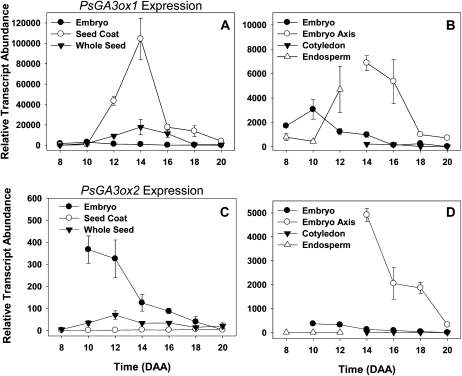
The enzyme products of the PsGA3ox1 and PsGA3ox2 genes are responsible for catalyzing the 3β-hydroxylation of GA20 to growth-active GA1 and also GA9 to GA4. Levels of PsGA3ox1 transcript were low in the seed coat at 8 DAA but began to increase by 10 DAA (5.5-fold; Fig. 8A). Based on the relatively low mRNA levels of the two PsGA2ox catabolic genes from 8 to 10 DAA (Fig. 9, A and C) and the increasing levels of PsGA20ox1, our data suggest that a moderate flux through the GA biosynthesis pathway from precursor GAs to growth-active GA1 occurs at this developmental stage. Consistent with these GA biosynthesis pathway transcription profiles, GA1 at 1.6 ng g−1 fresh weight was detected in the seed coat at 10 DAA (Table I). The presence of GA1 in the seed coat at this developmental stage may promote the substantial increase in seed coat fresh weight that we observed up to 12 DAA (Fig. 3A). Furthermore, since GA1 in general is considered to elicit its response in the tissue where it is synthesized (Yamaguchi, 2008), it is possible that GA1 synthesis occurs within different seed coat tissues at different developmental stages to bring about stage-specific growth and development of these tissues. Nakayama et al. (2002) reported that immunohistochemical staining with an anti-GA A1-methyl ester antiserum showed that GA1 and/or GA3 were localized around starch grains in the seed coats of young developing seeds of morning glory (Pharbitis nil). The localization of GA1 and/or GA3 in the seed coats was coincident with increased expression and colocalization of PnAmy1 (a GA-inducible α-amylase gene), suggesting the participation of a GA-inducible α-amylase in starch grain digestion. It is possible that the GA1 detected in pea seed coats at 10 DAA may be mainly localized in the ground parenchyma cells, where it stimulates increases in ground parenchyma cell expansion (Fig. 5A) and also initiates GA-inducible α-amylase starch grain digestion (since starch levels decrease from 12 to 20 DAA in this cell layer; Figs. 4B and 6C). Rodrigo et al. (1997) found GA1, GA3, and a small amount of GA4 in the seed coats of 4-DAA cv Alaska seeds, and all can be considered as growth-effector GAs. However, neither endogenous GA4 nor GA3 was detected in our study in any of the seed coat samples, even though the internal standards ([2H2]GA4 and [2H2]GA3) were detected (Table I). GA4’s immediate precursor, GA9, occurred at minimal to nondetectable levels in the seed coat tissue over the entire developmental period studied (Table I).
Figure 8.
Expression of PsGA3ox1 and PsGA3ox2 genes (their enzyme products are responsible for catalyzing the conversion of GA20 to bioactive GA1) in seed tissues from 8 to 20 DAA. Data are expressed as means ± se; n = 2 to 5 independent samples, except for 8-DAA embryo (n = 1) and 14-DAA seed coat (n = 6). Error bars are present at each point but may be obscured by symbols if the se is too small. All samples are normalized to allow comparison between tissues for the same gene.
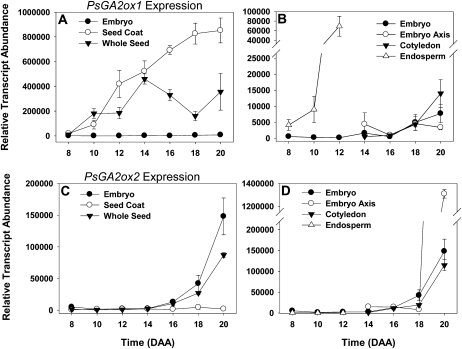
Figure 9.
Expression of PsGA2ox1 and PsGA2ox2 genes (which encode for enzymes that catalyze the deactivation of GA1 to GA8 and GA20 to GA29) in seed tissues from 8 to 20 DAA. Data are expressed as means ± se; n = 2 to 5 independent samples, except for 8-DAA embryo (n = 1) and 14-DAA seed coat (n = 6). Error bars are present at each point but may be obscured by symbols if the se is too small. All samples are normalized to allow comparison between tissues for the same gene.
As seed coat development progressed, the transcript abundance of PsGA3ox1 increased markedly in the seed coat from 10 to 12 DAA and peaked at 14 DAA (Fig. 8A). This peak in seed coat PsGA3ox1 transcript abundance preceded the dramatic expansion of branched parenchyma cells between 14 and 16 DAA (Figs. 4B and 5A). It is possible that the seed coat-produced GA1 detected at 14 DAA (Table I) was tissue specific, mainly stimulating the rapid cell expansion of branched parenchyma cells between 14 and 16 DAA. In contrast, GA1 at 10 DAA could be mainly localized within the ground parenchyma cell layer, where it would be able to stimulate the initiation of GA-inducible α-amylase starch grain digestion and cell expansion in this seed coat layer. It should be noted that the expansion of the branched parenchyma layer occurred immediately following the complete absorption of endosperm by the developing embryo (Figs. 3B and 4B), thereby bringing the developing embryo into complete contact with the seed coat (contact point; 16 DAA; Fig. 2). The expansion of the branched parenchyma layer was also temporally correlated with a rapid decrease in seed coat Suc levels (Fig. 6A). Data from fluorescent tracer experiments suggest that symplasmic transport of Suc occurs from the phloem into the chlorenchyma and ground parenchyma cell layers but not into the branched parenchyma cell layer of the pea seed coat (Van Dongen et al., 2003). Furthermore, work by Weber et al. (1995) suggests that the innermost seed coat parenchyma cell layer of Vicia faba (homologous to the branched parenchyma layer in pea seed coats) hydrolyzes, by means of its apoplastic cell wall invertase activity, phloem-derived Suc into Glc and Fru, destined for the endospermal cavity. These workers suggest that the cleavage of Suc acts to increase seed sink strength by (1) increasing the concentration gradient of Suc between the apoplastic space in the inner layers of seed coat and the seed coat parenchyma cells, which contain the symplasmically unloaded phloem-derived Suc, and (2) lowering the water potential in the apoplast due to the accumulation of hexoses (Weber et al., 1995). The rapid decrease in seed coat Suc levels that we observed (Fig. 6A) from 14 to 16 DAA is likely due to the uptake of Suc and/or Suc-derived hexoses from the apoplast by the branched parenchyma cells and the embryo. This would support the expansion of branched parenchyma cells as well as further growth of the embryo. It is also possible that an increase in seed coat invertase activity may have occurred concurrently with the expansion of the branched parenchyma cell layer. However, a concomitant increase in Glc levels (Fig. 6B) was not observed with the transient drop in Suc levels within the seed coat. Possible reasons for this lack of correlation could include a rapid uptake of both Suc and hexoses by the expanding cells of the branched parenchyma layer and the embryo. Finally, the expansion of the branched parenchyma cell layer was very transient. By 18 DAA, this layer was substantially reduced, and by 20 DAA, it was completely crushed by the expanding embryo (Fig. 4B). It should be noted that our use of a tall pea cultivar that contains the wild-type PsGA3ox1 gene (LE), the very transient nature of the expanded branched parenchyma layer, and the fact that this layer is easily detached from the remainder of the seed coat during tissue sectioning may all be factors contributing to the absence of previous reports of the expansion of the branched parenchyma layer subsequent to endosperm absorption by the developing pea embryo.
Both PsGA2ox1 and PsGA2ox2 encode for catabolic enzymes capable of catalyzing the reactions that convert GA20 to GA29 and/or GA1 to GA8. As noted above, the GA 2-oxidase encoded for by PsGA2ox1 can use both GA20 and GA1 as substrates, while the GA 2-oxidase produced by PsGA2ox2 is primarily used in the inactivation of bioactive GA1 to GA8 (Lester et al., 1999). Transcript abundance of seed coat PsGA2ox1 increased steadily over seed coat development (from 8 to 20 DAA; Fig. 9A), suggesting increased GA20 and/or GA1 catabolism. In contrast, PsGA2ox2 was expressed only minimally in the seed coat from 8 to 20 DAA (Fig. 9C). Consistent with the PsGA2ox1 transcription profile, the levels of the 2β-hydroxylated catabolite, GA29, increased 13-fold from 10 to 12 DAA and remained at elevated levels through 18 DAA (Table I). Very similar trends were seen for the GA8-to-GA1 ratios in the seed coat (e.g. a ratio of 5:1 at 10 DAA, increasing to 19:1 at 12 DAA; Table I). From 12 to 14 DAA, the concentration of GA1 in the seed coat remained low but detectable. GA1 was not detected at 16 to 18 DAA, although GA8 was present. These data suggest that the levels of GA1 are being maintained in the developing seed coat by a GA 2-oxidase encoded by PsGA2ox1 (i.e. both GA20 and GA1 are being catabolized to maintain levels of GA1 during most of the period of rapid seed coat growth). The substantial metabolism of [13C,3H]GA20 to [13C,3H]GA29 in SLN but not sln pea seed coats (at 20 DAA, sln contains a null mutation in PsGA2ox1; Ross et al., 1995; Lester et al., 1999) is also consistent with this interpretation.
The relationship between transcript abundance of the late-stage GA biosynthesis and catabolism genes and GA concentration profiles in pea seed tissues is consistent in most instances with the assumption that mRNA levels are indicative of protein levels and enzyme activity. This assumption is supported by previously published data obtained using pea ovary (Ozga et al., 1992, 2003, 2009; van Huizen et al., 1995, 1997) and pea stem tissues (O’Neill and Ross, 2002). Therefore, we believe that mRNA levels can be used to aid in meaningful predictions of the potential roles of GAs during pea seed growth and development. The changes in seed coat transcription profiles of the GA biosynthesis and catabolism genes, and the concentration profiles of key GAs, also show correlations with seed coat photoassimilate acquisition and storage changes that may imply that GAs are involved in determining both the growth rate and sink strength of the seed coat.
GA Gene Expression and GA Levels in the Endosperm
The endosperm of pea is noncellular (Marinos, 1970), and the endospermic cytoplasm contains multiple endoplasmic reticulum membranes, Golgi apparatus, ribosomes, mitochondria, and nuclei (Marinos, 1970; Melkus et al., 2009). Marinos (1970) notes that the pea endospermic cytoplasm occupies only a small fraction of the total volume of the “liquid endosperm” (cytoplasm plus vacuolar solutes). After fertilization, the endosperm increases in volume until approximately 12 DAA; at this point, the endosperm and the embryo have reached the same mass equivalent (Fig. 3B). Melkus et al. (2009) found that the pea endosperm Suc concentration peaked (at 250 mm) when the embryo and endosperm reached the same mass equivalent, with the Suc concentration remaining constant thereafter. Subsequently, the endosperm is absorbed by the rapidly growing embryo in the seeds within the span of 2 d (by 14 DAA; Fig. 3B).
Transcript abundance of both of the PsGA20ox genes increased markedly in the endosperm as it reached maximum volume (Figs. 3B and 7, B and D). Concomitantly, GA20 increased 56-fold (10–12 DAA; Table I). At 12 DAA, on a per seed basis (Supplemental Table S1), GA20 levels were 4.3-fold higher in the endosperm than in the seed coat, and approximately half that was found in the embryo. Interestingly, GA9, the final non-13-hydroxylated product of GA 20-oxidase activity, was not detectable at 10 DAA but was found in the endosperm at 12 DAA (Table I). An increase in endosperm PsGA3ox1 transcript abundance was also observed (Fig. 8B), although little to no GA1, no GA4 or GA3, and only a small amount of GA8 were detected from 10 to 12 DAA (see endosperm; Table I). If the increased PsGA3ox1 transcript abundance from 10 to 12 DAA did lead to increased GA 3-oxidase protein and enzyme activity, the very low levels of both GA1 and GA8 (GA8 is the immediate catabolite of GA1) in 12-DAA endosperm may be indicative of high GA1 and GA8 catabolism and/or transport from the endosperm. The GA catabolic gene, PsGA2ox1, did increase markedly in the endosperm from 10 to 12 DAA (Fig. 9B), and that could be indicative of high GA1 catabolism to GA8. Alternatively, it is possible that posttranscriptional regulation of PsGA3ox1 or posttranslational regulation of the GA 3-oxidase occurred, leading to a reduced level of GA 3β-hydroxylase activity in the 10- to12-DAA endosperm. Furthermore, the increase in PsGA2ox1 transcript (Fig. 9B; 17-fold from 8 to 12 DAA) was concurrent with a 28-fold increase in endosperm GA29 levels (10 to 12 DAA; Table I). We should note that the GA profile in our 10- to 12-DAA pea endosperm differs appreciably from that reported by Rodrigo et al. (1997) for the endosperm of 4-DAA cv Alaska pea seeds, where GA4 and GA3 were the main GAs. Rodrigo et al. (1997) also reported that 4-DAA endosperm produced only non-13-hydroxylated GA metabolites when fed [14C]GA12. The differences in GA profiles observed at 4 DAA (Rodrigo et al., 1997) and 10 to 12 DAA (this study) are not due to different genetic backgrounds (both studies used cv Alaska). Furthermore, Rodrigo et al. (1997) found high levels of GA3 in whole seeds from 4 to 8 DAA but only minimal levels of GA3 in 10-DAA whole seeds and no detectable GA3 in 12-DAA whole seeds. The rapid decrease of GA3 to nondetectable levels in seeds as the endosperm reaches its maximal volume (about 12 DAA) is consistent with data in this study and suggests that GA biosynthesis in the endosperm is developmentally regulated and that the GA biosynthesis pathway utilized early in development (4 DAA) differs markedly from that found later (10–12 DAA). Overall, the dramatic increase in GA20 levels (56-fold) as the endosperm reaches its maximum volume (12 DAA) suggests that this high GA20 concentration may act as a signal for tissue expansion (i.e. by providing a large pool of substrate for GA 3-oxidases localized in both embryo and seed coat tissues). It is this tissue expansion that ultimately leads to rapid absorption of the endosperm.
Morphology and Physiology of Developing Embryos
Embryo growth (as measured by fresh weight changes) and embryo size increased rapidly from 8 to 20 DAA (Figs. 2 and 3). The endosperm surrounds the growing embryo until 14 DAA, at which time the endosperm is completely absorbed by the embryo as the embryo expands to fill the seed cavity (Figs. 2 and 3B). The epidermal cell layer of the cotyledons had a relatively higher proportion of cells in prophase or metaphase (as indicated by condensed nuclei) at 10 DAA (Fig. 4D) compared with later stages, which is suggestive of relatively rapid cell division at 10 DAA. Marinos (1970) and Van Dongen et al. (2003) reported that the formation of invaginations of the cell walls of the cotyledonary epidermis occurred prior to endosperm absorption using transmission electron microscopy and cryo-scanning electron microscopy, respectively. The invagination of these epidermal cells produces cotyledon transfer cells, and these transfer cells are proposed to be important for nutrient uptake from the endosperm cavity (Borisjuk et al., 2002). However, the resolution afforded by light microscopy in our study did not permit the observation of these structures.
The single layer of cells immediately subtending the cotyledonary epidermis became organized into an ordered hypodermis between 10 and 14 DAA (Fig. 4, C and D), an event previously documented by Bain and Mercer (1966). While the cotyledonary storage parenchyma cells increased markedly in size (Figs. 4C and 5B) concomitant with both starch (Fig. 6C) and protein (Flinn and Pate, 1968) accumulation from 14 to 20 DAA, the hypodermal cells grew less, and there was a noticeable lack of storage vacuoles in the hypodermal cell layer relative to the subtending storage parenchyma (Fig. 4, D and E). Speculatively, the cotyledonary hypodermal cells may be part of the nutrient transport pathway leading from the epidermis to the storage parenchyma cells of the cotyledons.
GA Gene Expression and GA Levels in Developing Embryos
Transcript levels of both of the PsGA20ox genes were high in the embryo during early seed development (8–12 DAA; Fig. 7, A and C). While PsGA20ox2 mRNA abundance remained at elevated levels until 20 DAA (Fig. 7C), PsGA20ox1 transcript levels peaked at 12 DAA and then decreased to very low levels by 18 to 20 DAA (Fig. 7A). Thus, relatively high levels of GA20 occurred in the embryo from 10 to 18 DAA, peaking at 12 DAA, a period when transcript levels of both embryo PsGA20ox1 and PsGA20ox2 genes were most abundant (Table I). High concentrations of GA20 were also observed in the endosperm at 12 DAA, implying that the endosperm is a possible additional source for embryo-localized GA20. The pH of pea endosperm is approximately 5.5 (Murray, 1980). In this slightly acid environment, a greater proportion of the GA20 pool would have the carboxylic acid moiety protonated, reducing the polarity of the molecule and facilitating passive movement across the membranes of cells of both the cotyledons and seed coats. If GA20 is sequestered within the cotyledonary cells, a concentration gradient that supports the movement of GA20 into this tissue is possible. GA9 (which can also be synthesized by GA 20-oxidase) was detected in the embryo, with relatively high levels occurring from 12 to 16 DAA (Table I). Frydman et al. (1974) also identified both GA20 and GA9 in developing pea seeds. Although no method of internal standardization was used to estimate recovery losses in the Frydman et al. (1974) study, the general trends in the quantitative profiles of GA20 and GA9 over seed development observed in their study were consistent with that observed in this study. Interestingly, Sponsel and MacMillan (1977) found that pea seeds at a similar developmental stage (from 200 to 500 mg fresh weight) were capable of converting [3H]GA9 to [3H]GA20, a conversion also seen for Salix using [2H]GA9 (Junttila et al., 1992). Since detectable levels of GA4 were not observed (Table I), it seems reasonable to postulate that GA9 is C-13 hydroxylated to GA20 in the LE pea embryo at this stage of development. By 18 DAA, GA9 levels had markedly decreased relative to levels of GA20, suggesting that GA20 is the preferred GA for storage in the cotyledons as the seed matures. Upon germination, indirect evidence suggests that GA20 is transported from the cotyledons to the embryo axis, where GA1 biosynthesis occurs, thereby promoting the rapid growth of the developing seedling (Ross et al., 1993; Ayele, 2006).
Transcript abundance of both PsGA3ox genes, whose enzyme products are capable of converting GA20 to growth-active GA1, was elevated in the young embryo (at 10 DAA; Fig. 8, B and C), and GA1 was also detected in the embryo at this time (Table I). After 12 DAA, both PsGA3ox1 (Fig. 8B) and PsGA3ox2 (Fig. 8C) transcript abundance decreased in the embryo. Coincident with the initial decrease in PsGA3ox1 transcript abundance (between 10 and 12 DAA), embryo GA1 concentration decreased 2.8-fold by 12 DAA. GA1 remained detectable at low levels until 16 DAA, after which it could not be detected (Table I). Higher levels of both GA1 and GA8 and lower levels of GA29 at 10 DAA (Table I) suggest that a greater flux to GA1 biosynthesis is occurring in the embryo as a whole at 10 DAA than during subsequent developmental stages (12–20 DAA). Much earlier in seed development (4 DAA), when the embryo is likely at or near the globular stage and is attached to the suspensor (Marinos, 1970), Rodrigo et al. (1997) detected only small amounts of GA3 and GA20 in the embryo tissue. This is further evidence that GA biosynthesis in the embryo is developmentally regulated and that older embryos, well beyond the early developmental stages, do produce GA1. It is likely that GA1 is required for stimulating embryo growth throughout the growth-specific phases of seed development.
Transcript levels of the embryo-derived PsGA2ox1 and PsGA2ox2 catabolism genes were relatively low from 8 to 16 DAA, then they increased (11- to 16-fold) from 16 to 20 DAA (Fig. 9, B–D). Embryo GA29 levels were relatively low at 10 DAA, increased 3-fold by 12 DAA, and then remained stable through 18 DAA (Table I). On a per seed basis (Supplemental Table S1) from 10 to 14 DAA, GA29 was higher in the seed coat than in the embryo. At 16 to 18 DAA, GA29 levels were approximately equal in the seed coat and embryo. By 21 DAA, Sponsel (1983) found that GA29 abundance in the pea embryo (mainly localized in the cotyledons) was greater than that of the seed coat, and it remained so from 24 DAA to seed maturity (cv Progress No. 9). Feeding studies with labeled GA20 suggested that in the storage phase of Progress No. 9 pea seeds, embryo-derived GA29 can also be transported to the seed coat for catabolism to GA29-catabolite (Sponsel, 1983). Overall, these data suggest that catabolism of GA20 increases in the embryo as the seed matures.
Embryo Axis- and Cotyledon-Localized GA Biosynthesis and Catabolism
The embryo axis and cotyledons of the embryo execute vastly different developmental programs throughout seed development. Nutrient storage is accomplished primarily by the cotyledons, and storage-phase cotyledon development is directed primarily to the biosynthesis and storage of starches and proteins. The large size difference between the cotyledons and the embryo axis that make up the embryo (Fig. 2) results in the underrepresentation of embryo axis metabolite and transcript levels in whole embryo samples. Therefore, the determination of GA biosynthesis gene transcript levels and quantitative GA profiles in the cotyledons and embryo axes separately should lead to a better understanding of GA metabolism in these developmentally diverse tissues.
The majority of the embryo-derived PsGA20ox2 mRNA was present in the cotyledons relative to that found in the embryo axis in 14- to 20-DAA seeds (Fig. 7D). Although PsGA20ox2 transcript abundance was high in the cotyledons and the abundance of both PsGA20ox genes was lower in the embryo axis from 14 to 20 DAA (Fig. 7, B and D), GA20 concentrations were 7.7-fold greater in the embryo axis than in the cotyledons at 18 DAA (Table I). Levels of PsGA2ox1 transcript (Fig. 9B) and the immediate catabolic product of GA20, GA29 (Table I), were similar in both the cotyledons and the embryo axis during this developmental period. However, if transport of GA29 from the embryo to the seed coat occurs at this stage of seed development (observed in storage-phase seeds by Sponsel [1983]) and if it is specific to the cotyledonary tissue of the embryo, it is possible that higher cotyledon catabolism of GA20 to GA29 leads to lower GA20 levels in the cotyledons than that in the embryo axis. Alternatively, since GA20 is considered to be the major mobile GA in pea stem tissue (Proebsting et al., 1992), it is possible that in addition to in situ GA20 synthesis in the embryo axis, GA20 might also be transported from the cotyledons to the embryo axis. In general, GA transport is considered to be passive, moving from cell to cell, or through the vascular system (Hoad, 1995). Since the vascular system in the cotyledons does not differentiate from procambium until after imbibition of the mature seed (Smith and Flinn, 1967), potential GA transport to the embryo axis would be nonvascular.
Within the embryo, transcript abundance of both PsGA3ox1 and PsGA3ox2 was markedly higher in the embryo axis at 14 DAA (the earliest point in time at which embryo axes could be reliably obtained using our methods) relative to their abundance in the cotyledons (Fig. 8, B and D). Subsequently, embryo axis PsGA3ox1 and PsGA3ox2 transcript levels decreased to low levels by 20 DAA. Consistent with these PsGA3ox transcript profiles, GA1 was present in the embryo axis at 18 DAA (Table I), a time when the embryo axis is still actively growing (Fig. 3B). Transcript abundance of both of these genes was minimal in the cotyledons from 14 to 20 DAA, and minimal to no GA1 was detected in the 18-DAA cotyledons (Table I).
From 18 to 20 DAA, embryo axis PsGA2ox2 transcript abundance increased dramatically (Fig. 9D; 166-fold). This occurred coincidentally with decreases in embryo axis PsGA3ox transcript abundance (Fig. 8, B and D) and GA1 levels (nondetectable by 20–21 DAA; Table I). This strongly implies a shift from GA biosynthesis (to the growth-active GA1) to catabolic GA metabolism (GA1 deactivation) in the embryo axis and is likely an important mechanism for limiting embryo axis growth and allowing embryo maturation to proceed. The elevated embryo axis PsGA2ox2 transcript abundance that we observed at 20 DAA is likely maintained in the embryo axis until seed maturity, as Ayele et al. (2006) reported high levels of PsGA2ox2 transcript in the embryo axes at 12 h after seed imbibition. A marked reduction in embryo axis PsGA2ox2 transcript abundance was observed by 24 h after imbibition, an event that may be necessary for seed germination (observed at 48 h after imbibition; Ayele et al., 2006).
CONCLUSION
There are tissue-specific developmental patterns in the seed coat, embryo, embryo axis, and endosperm tissues that we conclude are likely in part regulated by tissue-specific developmental regulation of GA1 biosynthesis and catabolism (Fig. 10; Table I). Although we cannot exclude the possibility that other GA20ox, GA3ox, and GA2ox genes occur within the pea genome that may be expressed in seed tissues, the expression patterns of the known pea GA20ox, GA3ox, and GA2ox genes assessed in this study correlate well, in general, with the tissue-specific levels of 3-deoxy GA precursors, bioactive GA1, and the 2β-hydroxylated deactivated GA metabolites as development proceeds from a seed that is mainly seed coat tissue with liquid endosperm to one that is mainly embryo tissue with no liquid endosperm. Because our results correlate GA gene expression and GA levels with seed morphology and photoassimilate acquisition and storage during the mid stages of seed development, further research to test these correlations and determine causal relationships will be needed to confirm specific roles for GAs in these developmental processes. Important tools for further investigation include the use of both GA biosynthesis inhibitors and pea lines that are isogenic for GA biosynthesis and catabolism gene mutations. These should include PsGA3ox1 (LE, le), PsGA2ox1 (SLN, sln), PsKAO1 (LH, lh), and PsCPS1 (LS, ls).
Figure 10.
The major changes in GA biosynthesis and catabolism gene expression profiles of developing seed coats, embryos, cotyledons, and embryo axes of pea. BP, Branch parenchyma; GP, ground parenchyma.
In summary, in the seed coat, GA biosynthesis and catabolism gene transcript profiles and quantitative changes in GA concentrations imply that there is a GA1-stimulated increase in seed coat fresh weight up to 12 DAA, which is driven mainly through an increase in ground parenchyma cell expansion. Concomitantly, starch grain digestion (observed from 12 to 20 DAA in the ground parenchyma layer) may be initiated by GA1-induced α-amylase during this time. A peak in PsGA3ox1 transcript abundance precedes the dramatic expansion of the branched parenchyma cells, suggesting that seed coat-produced GA1 is involved in the stimulation of rapid branched parenchyma cell expansion. A steady increase in transcript abundance of PsGA2ox1 during seed coat development, along with higher levels of the 2β-hydroxylated catabolite of GA20, GA29, and a high ratio of GA8 to GA1, suggest an elevated GA20 and GA1 catabolism as the development of the seed coat progresses. Overall, these GA pathway data, when considered together with changes that occur in photoassimilate acquisition and storage as well as in morphological development, indicate that anabolic and catabolic GA metabolism may play an important role in determining growth rate and sink strength of the seed coat.
In the endosperm, the dramatic increase in GA20 levels (56-fold) as the endosperm reached its maximum volume (12 DAA), likely driven by elevated endosperm PsGA20ox transcript levels, suggests that high GA20 concentration may act as a signal for tissue expansion by providing a pool of substrate for the GA 3-oxidases localized in both embryo and seed coat tissues. It is this tissue expansion that ultimately leads to rapid absorption of the endosperm. Embryo growth was rapid from 8 to 20 DAA, and high transcript levels of PsGA20ox genes, particularly PsGA20ox2, dominated the embryo’s GA biosynthesis transcript profile. This transcript profile was associated with high levels of embryo GA20, the immediate precursor of GA1. PsGA20ox2 transcripts were present in much greater abundance in the cotyledons than the embryo axis, and this is consistent with a primary role for this PsGA20ox homolog in seed storage tissues. Transcript abundance of embryo PsGA3ox1 and PsGA3ox2 genes was highest earlier in the developmental profile (8–12 DAA), and growth-active GA1 continued to be present in embryos at 10 to 16 DAA, well beyond the early developmental stages. The relatively high PsGA3ox1 and PsGA3ox2 transcript levels in the embryo axes and low levels in the cotyledons at 14 to 18 DAA, along with the detection of GA1 in 18-DAA embryo axes (with minimal to no GA1 detected in 18-DAA cotyledons), suggest that GA1-stimulated growth and development processes are focused in the embryo axis in the maturing embryo. Finally, there is a shift from anabolic growth-active GA1 biosynthesis to a pattern of catabolic GA deactivation in the embryo axis as the seed continues to mature. This is a likely mechanism for limiting embryo axis growth and allowing embryo maturation to proceed.
MATERIALS AND METHODS
Plant Material
Seeds of pea (Pisum sativum I3 ’Alaska‘ type) were planted at an approximate depth of 2.5 cm in 3-L plastic pots (three seeds per pot) in 1:1 Sunshine No. 4 potting mix (Sun Gro Horticulture) and sand. Plants were grown at the University of Alberta in a climate-controlled growth chamber with a 16-h-light/8-h-dark photoperiod (19°C/17°C) with an average photon flux density of 383.5 μE m−2 s−2 (measured with a LI-188 photometer [Li-Cor Biosciences]). Flowers were tagged at anthesis, and seeds were harvested at selected stages as identified by date of anthesis and, where appropriate, pericarp length and width. Seeds were harvested directly onto ice and, if required, dissected immediately and then stored at −80°C. Seeds were harvested whole or dissected into seed coat, endosperm, and embryo at 8, 10, and 12 DAA. At 14, 16, 18, and 20 DAA, seeds were dissected into seed coat and embryo or seed coat, cotyledons, and embryo axis. When harvested, endosperm was removed from the seeds using a micropipette and immediately frozen on dry ice. Endosperm samples were centrifuged briefly before RNA or metabolite extraction to remove any contaminating cellular tissue. Seed coats and embryos at 8, 10, 12, and 14 DAA were transferred into distilled water in microfuge tubes or 20-mL scintillation vials on ice and further washed three times with distilled water to remove any remaining endosperm prior to freezing at −80°C.
Carbohydrate Analysis
Carbohydrate analysis was performed at the University of Alberta. Frozen seed coats and embryos were lyophilized and ground to a fine powder in liquid nitrogen using a mortar and pestle. Ground samples (20–42 mg dry weight) were extracted with 8 mL of 80% ethanol. The extract was placed at room temperature for 1 h, then immersed in a 75°C to 80°C water bath for 5 min, followed by centrifugation at 4,200 rpm for 15 min. The supernatant was removed, and the pellet was reextracted with 8 mL of 80% aqueous ethanol and reprocessed as described above. After removal of the supernatant, the pellet was stored at −20°C prior to starch analysis.
For determination of the soluble sugars, the supernatants were pooled and dried under vacuum, and the residue was resuspended in hexane (4 mL), followed by 4 mL of water for organic solvent partitioning to remove chlorophyll and lipids. The samples were partitioned three to five times (4 mL each) with hexane, and the aqueous phase was dried under vacuum. Dried samples were solubilized in 0.5 to 4 mL of water. For Glc determination, a 50-μL aliquot of the sample was brought up to 150 μL with 0.1 m acetic acid buffer (pH 4.6) prior to measuring Glc levels using a Glc oxidase/peroxidase (GOPOD) assay (Megazyme International).
For Suc determination, a 50-μL sample aliquot was incubated with 100 μL of invertase (1,500 units mL−1 invertase in 0.1 m acetic acid buffer, pH 4.6) at room temperature for 20 min to cleave Suc to Glc and Fru. Glc levels were determined after invertase cleavage using the GOPOD assay. For each sample, Suc was determined by subtracting the value obtained for Glc in the no-invertase-treatment subsample from that obtained in the invertase-treated subsample.
Total starch was determined based on the amyloglucosidase/α-amylase method (Megazyme). α-Amylase (3 mL, 300 units mL−1) was added to the pellet, and the sample was vortexed and incubated in boiling water for 6 min. After incubation, 100 μL of amyloglucosidase (3,250 units mL−1) and 4 mL of 200 mm sodium acetate buffer (pH 4.5) were added to each sample. The samples were vortexed and then incubated for 30 min at 50°C. After incubation, the volume of each sample was adjusted to 12 or 24 mL with water.
For the GOPOD assay, 1.5 mL of GOPOD solution (minimally 12,000 units L−1 Glc oxidase and 650 units L−1 peroxidase; Megazyme) was added to 50 μL of sample solution and incubated for 20 min in a 50°C water bath. Subsequently, the absorbance of the samples was measured at 510 nm using a Spectra MAX190 spectrophotometer (Molecular Devices) in a 96-well plate format and compared to a Glc standard curve made from a dilution series of 1 g L−1 Glc stock. The GOPOD assay was run in duplicate for each sample, and the average of the duplicates was used for the sample value.
RNA Isolation and Processing
RNA isolation, processing, and quantitation were performed at the University of Alberta. Tissues were ground in liquid nitrogen, and subsamples of 20 to 300 mg fresh weight were removed for total RNA isolation using a guanidinium thiocyanate-phenol-chloroform extraction (Ozga et al., 2003). After extraction with either TRIzol (Invitrogen) or TRI reagent (Ambion) and centrifugation at 4°C in a benchtop centrifuge to remove cellular debris, a phase partition separation using chloroform (0.2 mL mL−1 Tri reagent) was performed, and the organic phase was discarded. RNA was precipitated from the aqueous phase with isopropanol (0.25 mL mL−1 Tri reagent) and a high-salt solution (1.2 m sodium citrate and 0.8 m NaCl) to remove polysaccharides. The RNA pellet was resuspended, and RNA was precipitated with 8 m aqueous LiCl. The RNA pellet was again resuspended, and a final precipitation with 3 m sodium acetate (pH 5.2; final concentration of 96.77 mm) and 100% ethanol (final concentration of 64.5% [v/v]) was performed. The RNA was pelleted and washed twice with 70% aqueous ethanol and then resuspended and treated with DNase (DNA-free kit; Ambion). Diethyl pyrocarbonate-treated water was utilized throughout this procedure to reduce RNase contamination. RNA concentration was quantified by measurement at A260, and RNA purity was estimated with A260/A280 and A260/A230 ratios. RNA samples were diluted to 25 ng μL−1 and aliquoted to 96-well plates in a sterile laminar flow hood to reduce contamination.
Quantitative Reverse Transcription-PCR
Transcript quantification was performed on a model 7700 sequence detector (Applied Biosystems) except for PsGA3ox2 quantification, where a StepOnePlus system (Applied Biosystems) was used. Reverse transcription and quantification of PsGA3ox1, PsGA3ox2, PsGA2ox1, PsGA2ox2, PsGA20ox1, and PsGA20ox2 were performed using TaqMan One-Step RT-PCR Master Mix (Applied Biosystems; final concentration of 1×) and 200 ng of DNase-treated total RNA (final concentration of 8 ng μL−1) in duplicate at a final volume of 25 μL per well. The final concentration of forward and reverse primers was 300 nm each, and the final concentration of the probe was 100 nm. Reverse transcription was carried out for 30 min at 48°C. DNA polymerase antibody was denatured at 95°C for 10 min. Quantification was carried out for 40 cycles of the following program: denaturation at 95°C for 15 s, primer annealing and extension at 60°C for 1 min. Probes were labeled at the 5′ end with 6-carboxyfluorescein and at the 3′ end with the minor groove binder quencher (Applied Biosystems).
As a loading control, 18S rRNA was quantified from 3 ng of DNase-treated total RNA generated from a single dilution of the original 8 ng μL−1 stock (final concentration of 120 pg μL−1) of each sample using the same master mix, primer and probe concentrations, and thermocycling conditions. Given the wide variety of tissues and large developmental time spans used in this study, many commonly used control genes (including actin and ubiquitin) are not expressed at consistent levels and thus are not suitable. A mixture of primers containing 3′ hydroxyl and C6NH2 chain terminators in a 1:9 ratio was used to quantify 18S transcript levels. The addition of competitive primers allows a larger amount of template to be used while maintaining an acceptable reaction profile, effectively decreasing the variation that would be introduced during the serial dilution of RNA samples. The 18S probe was labeled at the 5′ end with VIC and at the 3′ end with the 6-carboxy-tetramethyl-rhodamine quencher (Applied Biosystems). RNA templates and reaction components were aliquoted to 96-well plates in a sterile laminar flow hood, and all tools and the hood itself were washed regularly with RNase Zap (Ambion) to reduce RNase contamination. The mean 18S rRNA threshold cycle (Ct) value (18.899) was calculated by averaging the Ct values from all samples (n = 672). Any sample that substantially deviated from the mean plate Ct mean was removed from the data set. The coefficient of variation of the 18S rRNA Ct among the remaining samples (n = 640) was low (3%); therefore, the target amplicon mRNA values were not normalized to the 18S signal (Livak and Schmittgen, 2001; Ozga et al., 2003, 2009).
Probes and primers used in this study are described in Supplemental Table S2. Probes and primers for PsGA3ox1 and 18S rRNA were designed by Ozga et al. (2003). Probes and primers for PsGA2ox1, PsGA2ox2, PsGA20ox1, and PsGA20ox2 were designed by Ayele et al. (2006). Probe and primers for PsGA3ox2 were designed by Ozga et al. (2009). Transcript levels were calculated using the ΔCt method (Livak and Schmittgen, 2001) using the following formula, where X is an arbitrary value equal to or greater than the highest assayed Ct value and E is the reaction efficiency for the amplicon in question: transcript abundance = (1 + E)(X − Ct).
Reaction efficiency was calculated by diluting a single RNA sample over several log concentrations (typically from 400–500 to 0.05–0.08 ng per reaction) and running quantitative reverse transcription (qRT)-PCR as previously described. Data were plotted on a semilog graph of Ct and log(input RNA), and a linear regression was calculated (Pfaffl, 2006). If the r2 value was sufficiently high, the slope of this equation was then used to calculate reaction efficiency (E; as a percentage) with the following formula: E = (10[−1/slope] − 1) × 100.
qRT-PCR Reaction Efficiency
qRT-PCR reaction efficiency for each amplicon was calculated (Supplemental Table S3) in order to allow more accurate comparison of transcript levels between samples, as the assumption of 100% reaction efficiency in the calculations for relative transcript abundance may inflate differences in gene expression if actual reaction efficiency is less than 100%. The calculated regressions explained the majority of the variation in Ct value (r2 ≥ 0.990), and reactions proceeded with relatively high efficiency.
Hormone Extraction
Plant tissues were harvested and dissected at the University of Alberta as described above, and frozen tissues were freeze dried prior to transport to the University of Calgary for hormone extraction and quantification. Liquid endosperm samples were centrifuged briefly after harvesting to remove any contaminating cellular materials and stored at −80°C prior to transport on dry ice to the University of Calgary. Solid tissues were ground with liquid nitrogen and washed sea sand. Ground tissue and liquid endosperm samples were extracted with 80% (v/v) aqueous methanol (MeOH). Internal standards were added to the solvent as follows: 20 to 40 ng each of [2H2]GA1, [2H2]GA3, [2H2]GA4, [2H2]GA8, [2H2]GA9, [2H2]GA20, and [2H2]GA29 (deuterated GAs were obtained from Prof. L.N. Mander, Research School of Chemistry, Australian National University). The 80% MeOH extract was then filtered through a No. 2 Whatman filter (55 mm; Whatman International) and purified with a C18 preparative column consisting of a syringe barrel filled with 3 g of C18 (Waters) preparative reverse-phase material (Koshioka et al., 1983). The 80% aqueous MeOH eluate from this column was dried in vacuo at 35°C.
HPLC
The dried sample was dissolved in 1 mL of 10% aqueous MeOH with 1% acetic acid and injected into the HPLC column using the method described by Koshioka et al. (1983). The HPLC apparatus (Waters) consisted of two pumps (model M-45), an automated gradient controller (model 680), and a Rheodyne injector (model 7125). The solvent reservoir for pump A was filled with 10% MeOH in 1% acetic acid (water:MeOH:acetic acid = 89:10:1 [v/v]), while pump B was 100% MeOH. A reverse-phase C18 Radial-PAK μ-Bondapak column (10 cm × 8 mm) was used with a manually implemented 10% to 73% gradient program at a flow rate of 2 mL min−1 as follows: 0 to 10 min (pump A, 100%; pump B, 0%), 10 to 50 min (pump A, 30%; pump B, 70%), 50 to 80 min (pump A, 0%; pump B, 100%), and 80 to 90 min (pump A, 100%; pump B, 0%).
The HPLC fractions (4 mL) were dried in vacuo at 35°C. Fractions from the C18 HPLC, which were expected (based on retention times, min 9–32) to contain GA1, GA3, GA8, GA20, and GA29 were transferred with MeOH to 2-mL glass vials and dried in vacuo. Later fractions (min 33–44; GA4 and GA9) from the C18 HPLC were grouped using MeOH and again dried in vacuo, then subjected to Nucleosil N(CH3)2 HPLC (column, 5 μm, 250 mm × 4.6 mm i.d.), which was isocratically eluted with 0.1% HOAc in MeOH at 1.2 mL min−1 (Jacobsen et al., 2002). Three-minute fractions from the N(CH3)2 HPLC were taken to dryness in vacuo. Subsequently, fractions that would contain GA4 and GA9 (min 16–42) were transferred with MeOH to 2-mL glass vials and dried in vacuo.
Gas Chromatography and Mass Spectrometry
Samples were methylated by ethereal CH2N2 for 20 min and then trimethylsilylated with N,O-bis(trimethylsilyl)trifluoroacetamide and 1% trimethylchlorosilane (Hedden, 1987; Gaskin and MacMillan, 1991). The identification and quantification of plant hormones was carried out using a gas chromatograph connected to a mass spectrometer using the selected ion monitoring mode. The derivatized sample was injected into a capillary column installed in an Agilent 6890 gas chromatograph with a capillary direct interface to an Agilent 5973 mass selective detector. The dimensions of the capillary column were 0.25-μm film thickness, 0.25-mm internal diameter, and 30-m length (DB-1701, model J&W122-0732; J&W Scientific). The gas chromatograph temperature program was as follows: 1 min at 60°C, followed by an increase to 240°C at a rate of 25°C min−1 and an increase at 5°C min−1 to 280°C, where it remained constant for 15 min before returning to 60°C. The interface temperature was maintained at 280°C. The dwell time was 100 ms, and data were processed using HP G1034C MS ChemStation Software.
Comparisons of gas chromatography retention times of both the GAs and [2H2]GAs and the relative intensities of molecular ion (M+) pairs were used to identify endogenous GAs. Relative intensities of at least two other characteristic mass-to-charge ratio ion pairs for each endogenous GA and its deuterated standard were also compared. All stable isotope-labeled internal standards were added at the extraction stage with appropriate purification and chromatography being accomplished (see above) prior to gas chromatography-mass spectrometry-selected ion monitoring. Quantification was accomplished by reference to the stable isotope-labeled internal standard using equations for isotope dilution analysis adapted by D.W. Pearce (Jacobsen et al., 2002) from Gaskin and MacMillan (1991).
Histology
Seed coat tissues for histology studies were obtained by making two parallel cuts from top to bottom (attachment site of the funiculus to the pericarp) of the seed approximately 0.5 to 1 mm apart, as indicated in Supplemental Figure S1. Embryo samples were obtained from the regions directly underneath these sections, and excess material from the inside of the cotyledons was trimmed to promote efficient fixation and resin infiltration. The final dimensions of the seed coat samples were 0.5 to 1 mm wide by 5 to 6 mm long (thickness of the entire seed coat). Final dimensions of embryo samples were 0.5 to 1 mm wide, 5 to 6 mm long, and 2 to 3 mm thick. Sections were taken from both right and left halves of the seed, and embryo samples were obtained from regions away from the cotyledon-cotyledon interface (Supplemental Fig. S1).
Samples were fixed in a solution of aqueous 0.2% (v/v) gluteraldehyde, 3% (v/v) paraformaldehyde, 2 mm CaCl2, 10 mm Suc, and 25 mm PIPES at pH 7. Fixation was carried out overnight under vacuum to increase infiltration, then at atmospheric pressure for 2 d. Prior to embedding, tissues were washed three times for 10-min intervals with 25 mm PIPES and then dehydrated using a graded ethanol series of 30%, 50%, and 70% (v/v) aqueous ethanol for 15 min each. The 70% aqueous ethanol was replaced, and tissues were stored for several days. Further dehydration was then carried out with 80%, 96%, and 100% ethanol (twice) at 15-min intervals, then with 50% ethanol:50% (v/v) propylene oxide, then with 100% propylene oxide before embedding. Tissues were embedded in Spurr’s epoxy resin (the hard resin protocol; Spurr, 1969) and cured at 70°C for 4 h and then at 60°C for 3 d. Blocks were trimmed with blades and fine-toothed saws to a maximum surface area of approximately 2 mm2. Seed coat samples were sliced to 1-μm-thick sections and cotyledon samples to 2-μm-thick sections with a Reichert-Jung Ultracut E ultramicrotome using glass knives made by hand with an LKB Broma Knifemaker II. Sections were transferred from the water-filled collection boat to glass slides, stretched briefly with a JBS heat pen, and dried on a 60°C slide warmer. Slides were stained with 0.05% (w/v) toluidine blue in water for 3 min at room temperature and then were briefly washed with distilled water to remove excess stain. After drying at room temperature, slides were observed with a Zeiss Primostar light microscope, and micrographs were taken with a Photometrics CoolSNAP CF camera. Image editing was performed with Adobe Photoshop. All scale bars were generated by calibration to a stage micrometer, not using field-of-view calculations. Cell area was estimated by manually tracing cells from each tissue type and stage using MetaMorph software (version 7.0r4; Molecular Devices). The average subsample size for cell area calculations was 98 cells per layer and time point, examined in at least two biological replicates.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Orientation of histological sections.
Supplemental Table S1. GA quantitation in ng seed1.
Supplemental Table S2. qRT-PCR primers and probes.
Supplemental Table S3. qRT-PCR reaction efficiency.
Supplementary Material
References
- Ayele BT. (2006) Gibberellin biosynthesis during germination and young seedling growth of pea. PhD thesis. University of Alberta, Edmonton, Canada: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayele BT, Ozga JA, Kurepin LV, Reinecke DM. (2006) Developmental and embryo axis regulation of gibberellin biosynthesis during germination and young seedling growth of pea. Plant Physiol 142: 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JM, Mercer FV. (1966) Subcellular organization of the developing cotyledons of Pisum sativum L. Aust J Biol Sci 19: 49–67 [Google Scholar]
- Borisjuk L, Walenta S, Rolletschek H, Mueller-Klieser W, Wobus U, Weber H. (2002) Spatial analysis of plant metabolism: sucrose imaging within Vicia faba cotyledons reveals specific developmental patterns. Plant J 29: 521–530 [DOI] [PubMed] [Google Scholar]
- Davidson SE, Smith JJ, Helliwell CA, Poole AT, Reid JB. (2004) The pea gene LH encodes ent-kaurene oxidase. Plant Physiol 134: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. (2004) The plant hormones: their nature, occurrence, and functions. Davies PJ, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Springer, Dordrecht, The Netherlands, pp 1–15 [Google Scholar]
- Fleet CM, Yamaguchi S, Hanada A, Kawaide H, David CJ, Kamiya Y, Sun TP. (2003) Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol 132: 830–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn AM, Pate JS. (1968) Biochemical and physiological changes during maturation of fruit of the field pea (Pisum arvense L.). Ann Bot (Lond) 32: 479–495 [Google Scholar]
- Frydman VM, Gaskin P, MacMillan J. (1974) Qualitative and quantitative analyses of gibberellins throughout seed maturation in Pisum sativum cv. Progress No. 9. Planta 118: 123–132 [DOI] [PubMed] [Google Scholar]
- García-Martínez JL, López-Diaz I, Sánchez-Beltrán MJ, Phillips AL, Ward DA, Gaskin P, Hedden P. (1997) Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol 33: 1073–1084 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Sponsel VM, Gaskin P. (1987) Gibberellins in developing fruits of Pisum sativum cv. Alaska: studies on their role in pod growth and seed development. Planta 170: 130–137 [DOI] [PubMed] [Google Scholar]
- Gaskin P, MacMillan J. (1991) GC-MS of the Gibberellins and Related Compounds: Methodology and a Library of Spectra. University of Bristol (Cantock’s Enterprises), Bristol, UK [Google Scholar]
- Hedden P. (1987) Gibberellins. Rivier L, Crozier A, eds, Principles and Practice of Plant Hormone Analysis, Vol 1. Academic Press, London, pp 9–110 [Google Scholar]
- Hoad GV. (1995) Transport of hormones in the phloem of higher plants. Plant Growth Regul 16: 173–182 [Google Scholar]
- Jacobsen JV, Pearce DW, Poole AT, Pharis RP, Mander LN. (2002) Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol Plant 115: 428–441 [DOI] [PubMed] [Google Scholar]
- Junttila O, Jensen E, Pearce D, Pharis RP. (1992) Stimulation of shoot elongation in Salix by gibberellin A9: is activity dependent upon hydroxylation to GA1 via GA20? Physiol Plant 84: 113–120 [Google Scholar]
- Koshioka M, Takeno K, Beall FD, Pharis RP. (1983) Purification and separation of plant gibberellins from their precursors and glucosyl conjugates. Plant Physiol 73: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Ait-Ali T, Martin DN, Reid JB. (1996) A gibberellin 20-oxidase cDNA (accession no. U58830) from pea seed (PGR 96-050). Plant Physiol 111: 13538756508 [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. (1997) Mendel’s stem length gene (Le) encodes a gibberellin 3β-hydroxylase. Plant Cell 9: 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. (1999) Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J 19: 65–73 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lloyd JR, Wang TL, Hedley CL. (1996) An analysis of seed development in P. sativum. XIX. Effect of mutant alleles at the r and rb loci on starch grain size and on the content and composition of starch in developing pea seeds. J Exp Bot 47: 171–180 [Google Scholar]
- Marinos N. (1970) Embryogenesis of the pea (Pisum sativum). I. The cytological environment of the developing embryo. Protoplasma 70: 261–279 [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. (1999) The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol 121: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkus G, Rolletschek H, Radchuk R, Fuchs J, Rutten T, Wobus U, Altmann T, Jakob P, Borisjuk L. (2009) The metabolic role of the legume endosperm: a noninvasive imaging study. Plant Physiol 151: 1139–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DR. (1980) Functional significance of acid phosphatase distribution during embryo development in Pisum sativum L. Ann Bot (Lond) 45: 273–281 [Google Scholar]
- Nakayama A, Park S, Zheng-Jun X, Nakajima M, Yamaguchi I. (2002) Immunohistochemistry of active gibberellins and gibberellin-inducible α-amylase in developing seeds of morning glory. Plant Physiol 129: 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill DP, Ross JJ. (2002) Auxin regulation of the gibberellin pathway in pea. Plant Physiol 130: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Brenner ML, Reinecke DM. (1992) Seed effects on gibberellin metabolism in pea pericarp. Plant Physiol 100: 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM, Ayele BT, Ngo P, Nadeau CD, Wickramarathna AD. (2009) Developmental and hormonal regulation of gibberellin biosynthesis and catabolism in pea fruit. Plant Physiol 150: 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Yu J, Reinecke DM. (2003) Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiol 131: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. (2006) Relative quantification. Dorak T, ed, Real-Time PCR. Taylor & Francis Group, New York, pp 63–82 [Google Scholar]
- Proebsting WM, Hedden P, Lewis MJ, Croker SJ, Proebsting LN. (1992) Gibberellin concentration and transport in genetic lines of pea: effects of grafting. Plant Physiol 100: 1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat C, Boutin JP. (1992) Temporary storage compounds and sucrose-starch metabolism in seed coats during pea seed development (Pisum sativum). Physiol Plant 85: 567–572 [Google Scholar]
- Rochat C, Wuilleme S, Boutin JP, Hedley CL. (1995) A mutation at the rb gene, lowering ADPGPPase activity, affects storage product metabolism of pea seed coats. J Exp Bot 46: 415–421 [Google Scholar]
- Rodrigo MJ, García-Martínez JL, Santes CM, Gaskin P, Hedden P. (1997) The role of gibberellins A1 and A3 in fruit growth of Pisum sativum L. and the identification of gibberellins A4 and A7 in young seeds. Planta 201: 446–455 [Google Scholar]
- Ross JJ, Reid JB, Swain SM. (1993) Control of stem elongation by gibberellin A1: evidence from genetic studies including the slender mutant sln. Aust J Plant Physiol 20: 585–599 [Google Scholar]
- Ross JJ, Reid JB, Swain SM, Hasan O, Poole AT, Hedden P, Willis CL. (1995) Genetic regulation of gibberellin deactivation in Pisum. Plant J 7: 513–523 [Google Scholar]
- Smith DL, Flinn AM. (1967) Histology and histochemistry of the cotyledons of Pisum arvense L. during germination. Planta 74: 72–85 [DOI] [PubMed] [Google Scholar]
- Sponsel V. (1983) The localization, metabolism and biological activity of gibberellins in maturing and germinating seeds of Pisum sativum cv. Progress No. 9. Planta 159: 454–468 [DOI] [PubMed] [Google Scholar]
- Sponsel V, MacMillan J. (1977) Further studies on the metabolism of gibberellins (GAs) A9, A20 and A29 in immature seeds of Pisum sativum cv. Progress No. 9. Planta 135: 129–136 [DOI] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Swain SM, Reid JB, Ross JJ. (1993) Seed development in Pisum: the lhi allele reduces gibberellin levels in developing seeds, and increases seed abortion. Planta 191: 482–488 [Google Scholar]
- Swain SM, Ross JJ, Reid JB, Kamiya Y. (1995) Gibberellins and pea seed development: expression of the lhi, ls and le5839 mutations. Planta 195: 426–433 [Google Scholar]
- Van Dongen JT, Ammerlaan AM, Wouterlood M, Van Aelst AC, Borstlap AC. (2003) Structure of the developing pea seed coat and the post-phloem transport pathway of nutrients. Ann Bot (Lond) 91: 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM. (1997) Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol 115: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM, Twitchin B, Mander LN. (1995) Seed and 4-chloroindole-3-acetic acid regulation of gibberellin metabolism in pea pericarp. Plant Physiol 109: 1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Buchner P, Wobus U. (1995) Seed coat-associated invertases of fava bean control both unloading and storage functions: cloning of cDNAs and cell type-specific expression. Plant Cell 7: 1835–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 253–279 [DOI] [PubMed] [Google Scholar]
- Weston DE, Elliott RC, Lester DR, Rameau C, Reid JB, Murfet IC, Ross JJ. (2008) The pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiol 147: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.