Abstract
Despite a rapidly-growing scientific and clinical brain imaging literature based on functional MRI using blood oxygenation level dependent (BOLD)1 signals, it remains controversial if BOLD signals in a particular region can be caused by activation of local excitatory neurons2. This difficult question is central to the interpretation and utility of BOLD, with major significance for fMRI studies in basic research and clinical applications3. Using a novel integrated technology unifying optogenetic4–13 control of inputs with high-field fMRI signal readouts, we show here that specific stimulation of local CaMKIIα-expressing excitatory neurons, either in neocortex or thalamus, elicits positive BOLD signals at the stimulus location with classical kinetics. We also show that optogenetic fMRI (ofMRI) allows visualization of the causal effects of specific cell types defined not only by genetic identity and cell body location, but also by axonal projection target. Finally, we show that ofMRI within the living and intact mammalian brain reveals BOLD signals in downstream targets distant from the stimulus, indicating that this approach can be used to map the global effects of controlling a local cell population. In this respect, unlike both conventional fMRI studies based on correlations14 and fMRI with electrical stimulation which will also directly drive afferent and nearby axons, this ofMRI approach provides causal information about the global circuits recruited by defined local neuronal activity patterns. Together these findings provide an empirical foundation for the widely-employed fMRI BOLD signal, and the features of ofMRI define a potent tool that may be suitable for functional circuit analysis as well as global phenotyping of dysfunctional circuitry.
Blood oxygenation level-dependent functional magnetic resonance imaging (BOLD fMRI)1 is a widely used technology for non-invasive whole brain imaging. BOLD signals reflect complex and incompletely understood changes in cerebral blood flow (CBF), cerebral blood volume (CBV), and cerebral metabolic rate of oxygen consumption (CMRO2) following neuronal activity2,15. Candidate circuit elements for triggering various kinds of BOLD signals include excitatory neurons, mixed neuronal populations, astroglia, and axonal tracts or fibers of passage16,17. Importantly, it is not clear which kinds of activity are capable of triggering BOLD responses, placing limitations on interpretation for both clinical and scientific applications. For example, it is sometimes assumed that positive BOLD signals can be triggered by increased activity of local excitatory neurons, but this remains to be shown empirically, a challenge which seriously confounds fMRI interpretation18,19. Moreover, the use of MRI-compatible electrodes for local stimulation, while of pioneering significance, will nevertheless drive all local excitatory, inhibitory, and modulatory cell types, as well as antidromically drive non-local cells that happen to have axons within the stimulated region, thereby confounding functional circuit mapping using BOLD. We sought to address these challenges by integrating high-field fMRI output with optogenetic stimulation4–13, in which single-component microbial light-activated transmembrane conductance regulators are introduced into specifically targeted cell types and circuit elements7,8 using cell type-specific promoters to allow millisecond-scale targeted activity modulation in vivo9–11.
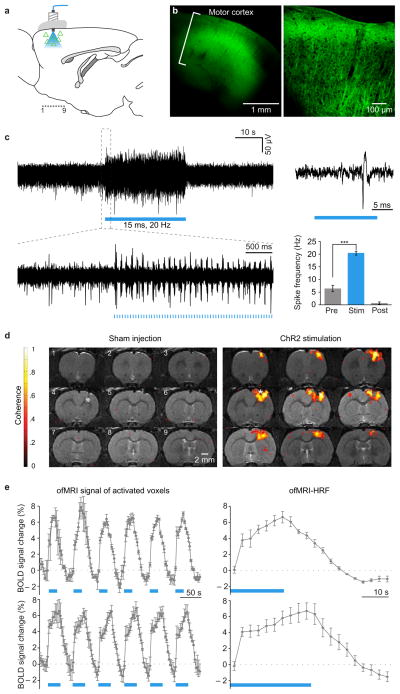
In adult rats, primary motor cortex (M1) was injected with the adeno-associated viral vector AAV5-CaMKIIα::ChR2(H134R)-EYFP to drive expression of a channelrhodopsin (ChR2) specifically in CaMKIIα-expressing principal cortical neurons, but not in GABAergic or glial cells7,9–12; here the cortical virus injection site was also employed as the optical stimulation site for BOLD and electrophysiological functional studies (Fig 1a). To minimize susceptibility artifact during MRI scanning, the implanted cannula, optical fiber and accessories were custom-fabricated from MR-compatible materials. Confocal imaging (Fig. 1b) and optrode recording (simultaneous optical stimulation and electrical recording) under 1.3–1.5% isoflurane anaesthesia20 (Fig. 1c) were conducted to validate the expression and functionality, respectively, of ChR2-EYFP under these conditions; in line with previous optogenetic studies11, 473 nm light pulses at 20 Hz (15 ms pulse width) delivered through the optical fiber were found to reliably drive local neuronal firing (Fig. 1c) in vivo.
Figure 1. ofMRI: optogenetic excitation of CaMKIIα neocortical cells drives local positive BOLD.
a, Transduced cells (triangles), light, and locations (1..9) of coronal slices. b, Confocal images of ChR2-EYFP expression in M1 (left); higher magnification (right). c, Optrode recordings during 473nm optical stimulation (20 Hz/15 ms pulsewidth; blue); spiking is significantly elevated (two-sample t-test; p<0.001; n=3). d, BOLD activation observed with AAV5-CaMKIIα::ChR2-EYFP but not with saline injection (p<0.001; asterisk: optical stimulation). e, Left, ofMRI hemodynamic response (averaged across activated voxels in motor cortex) during 20s (n=3) and 30s (n=8) optical stimuli. Right, Mean over stimulus repetitions; baseline: mean pre-stimulation signal.
To assess fMRI signals, we acquired 0.5 mm coronal slices centered on M1, >10 days after virus injection (Fig. 1d). Intubated animals were placed on a custom-designed MRI-compatible cradle with a stereotaxic frame and ventilated with 1.3–1.5% isoflurane. A 3.5 cm-diameter custom-designed transmit/receive single-loop surface coil was apposed to the cranium and a long 300 μm diameter optical fiber inserted through the implanted cannula; in this configuration, the cradle with the animal was placed into the iso-center of the magnet while the laser diode itself was maintained outside the 5 Gauss perimeter. In order to minimize systemic physiological confounds, the ventilation volume, frequency, endtidal CO2, and rectal temperature levels were carefully maintained at narrow levels known to produce most robust and reproducible BOLD signals in anesthetized animals (3.0–3.5 cc/stroke, 50–60 stroke/min, 3.5%, 34–38°C)20. fMRI scans were performed at 7.0 Tesla (T) field strength using conventional GRE-BOLD fMRI and passband b-SSFP fMRI21,22 (Supplementary Material). Both pulse sequences were designed to have 3.5 × 3.5 cm2 in-plane FOV, 0.5 × 0.5 × 0.5 mm3 spatial resolution and 3 s temporal resolution. GRE-BOLD fMRI was designed to be a 2D, multi-slice, gradient-echo (GRE) sequence with 4-interleave spiral readout23; 750 ms TR and 12 ms TE resulting in 23 slices covering 1.15 cm slice direction volume. This specific design allowed large-volume mapping of the brain during optogenetic control with high temporal resolution.
473 nm light pulses at 20 Hz (15 ms pulse width) were delivered to targeted CaMKIIα-expressing principal neurons, and in response, robust optically-evoked BOLD signals were observed in cortical gray matter at the virus injection and optical stimulation site, while in control animals (injected with saline instead of opsin-AAV) no detectable BOLD signal could be elicited (Fig. 1). Stimulus-synchronized BOLD hemodynamic responses from activated M1 voxels are displayed in Fig. 1d, and mean optogenetic fMRI hemodynamic response functions (ofMRI-HRF) in Fig. 1e. Evoked BOLD was dominated by positive signals while driving these excitatory CaMKIIα-positive cells; in contrast, optically driving inhibitory parvalbumin-positive cells10, which may have unique connectivity with local neuronal circuitry or vasculature, additionally gave rise to a zone of negative BOLD, consistent with the GABAergic phenotype, surrounding the local positive BOLD signal (Supplementary Fig. 4). Strikingly, the BOLD dynamics observed by optically driving the defined CaMKIIα principal cell population embedded within the mixed M1 cell population precisely matched dynamics of conventional stimulus-evoked BOLD-fMRI24,25. In particular, the ofMRI-HRF signal onset occurred after 3 seconds but within 6 seconds of stimulus onset; likewise offset was reflected by a drop in BOLD signal contrast beginning within 6 seconds and returning to baseline in ~20 seconds after optical stimulation (Fig. 1e; upper panel: n=3, lower panel: n=8). Finally, the pronounced post-stimulus undershoot observed during systemic somatosensory stimulation in humans26 and animals24 was preserved in ofMRI-HRFs as well (Fig. 1e). All of these dynamic properties derived from driving a defined, specific (Methods; Supplementary Figure 1a) cell population correspond closely to prior measurements on conventional sensory-evoked BOLD.
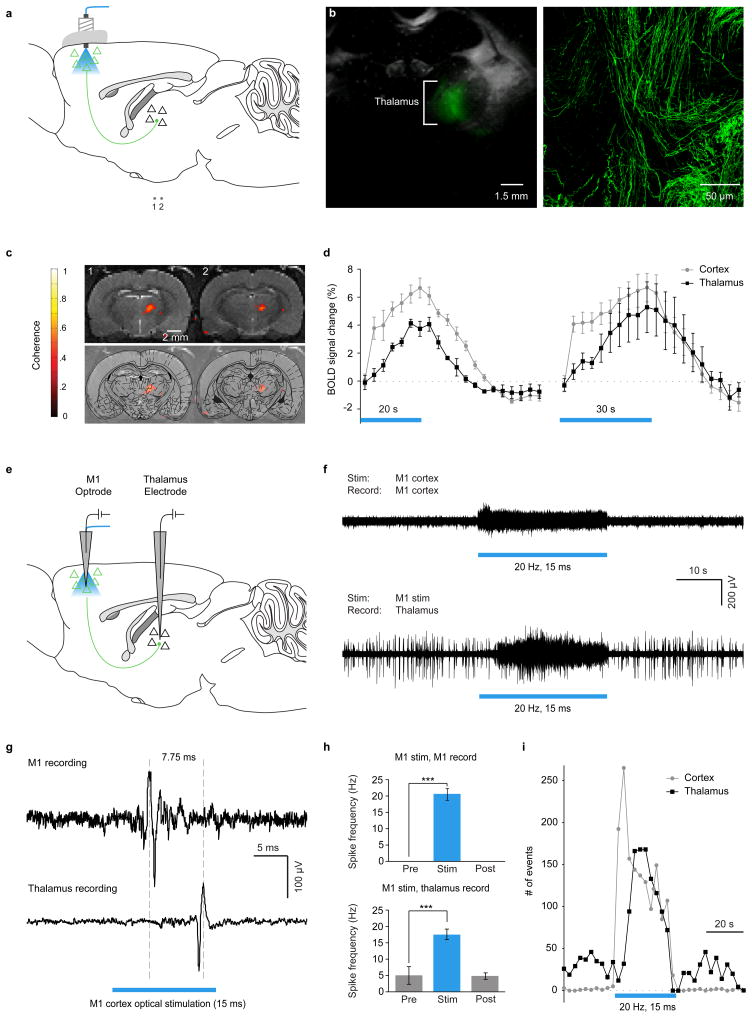
In order to study macrocircuit properties of the brain using optogenetic fMRI, it will be important to assess feasibility of monitoring long-range activity in synaptically connected brain areas. MRI-compatible electrodes for local stimulation represent a major advance but in addition to driving all local excitatory, inhibitory, and modulatory cell types, will also antidromically drive non-local cells that happen to have axons within the stimulated region, posing a challenge for functional mapping using BOLD. We therefore employed high-resolution fMRI slices capturing thalamic nuclei (coronal slices shown in Fig. 2a) to monitor downstream responses during optical stimulation of M1 cortical neurons. Figure 2b illustrates the observed specific expression of ChR2(H134R)-EYFP in cortico-thalamic axonal projection fibers while thalamic cell bodies revealed no ChR2 expression, as expected from the cortical injection protocol (Supplementary Fig. 5). Local optical stimulation was then delivered to cortex during fMRI, to determine if unidirectionally triggered BOLD responses could be observed and measured (this method eliminates the antidromic drive confound from which electrical stimulation suffers, thereby allowing true global causal connectivity mapping). Figures 2c and 2d summarize the thalamic ofMRI-HRFs; robust thalamic BOLD signals in response to M1 stimulation were observed, but with properties quite distinct from the intracortical CaMKIIα + response described above. A markedly reduced initial rise and slope for onset kinetics of positive-BOLD downstream thalamic recruitment was observed (Fig. 2d, black traces; local cortical BOLD signals shown for comparison, gray traces; cortical BOLD activation is shown in Fig. 1).
Figure 2. Nonlocal mapping of causal role of cells defined by location and genetic identity.
a, AAV5-CaMKIIα::ChR2-EYFP injection and optical stimulation in M1. Slices in (c): “1” and “2”. b, Fluorescence/bright-field: ChR2-EYFP in thalamus (left); confocal image reveals expression limited to axons. c, Thalamic ofMRI during M1 optical stimulation (top); superimposed on Paxinos atlas (bottom). d, ofMRI-HRF summary. e, M1 optrode and thalamic electrode. f, Thalamic spiking follows M1 optical stimulation; delay consistent with BOLD. g, Typical M1 and thalamus spikes with M1 optical excitation. h, M1 and thalamus spiking summary (two-sample t-test; p<0.001; n=5). i, Spike-frequency time histograms.
Given the unusual kinetics, we sought to determine if this delayed thalamic BOLD response would be discrepant with local thalamic electrical activity, assessed with simultaneous optrode stimulating/recording in motor cortex and electrode recoding in thalamus (Fig. 2e). However, a strikingly similar pattern was observed with direct recording in thalamus, including a commensurate delay in spike-rate increase for thalamic neurons compared to cortical neurons during cortical optogenetic drive (Fig. 2f), further supporting the tight correspondence between positive BOLD and local neuronal excitation2,27,28. Additional characterization revealed that after this ~5 second delay presumably related to network properties, successfully evoked spikes recorded in thalamus reliably followed cortical spikes by several milliseconds, as expected (Fig. 2g). Summary data on mean spike rates is presented in Figure 2h, and on spike rate dynamics in Figure 2i; further details on the passband bSSFP-fMRI21,22 method we developed for small animal imaging with more robust whole-brain mapping capability than traditional BOLD are presented in the Supplementary Material (Supplementary Figure 3).
Since true functional outputs of genetically defined neurons in a brain region can be globally mapped with ofMRI (Fig. 2), it is conceivable that additional levels of specificity could also be achieved. For example, M1 excitatory pyramidal neurons form a genetically- and anatomically-defined class of cell, but within this class are cells that each project to different areas of the brain or spinal cord and therefore have fundamentally distinct roles. Genetic tools may not advance far enough to separate all of these different cell classes, pointing to the need for other promoter-independent targeting methods13. But ofMRI raises the current possibility of globally mapping the casual roles of these cells, accessing them by means of connection topology—i.e. by the conformation of their functional projection patterns in the brain. We therefore sought to test this possibility by selectively driving the M1 CaMKIIα-expressing cells that project to thalamus.
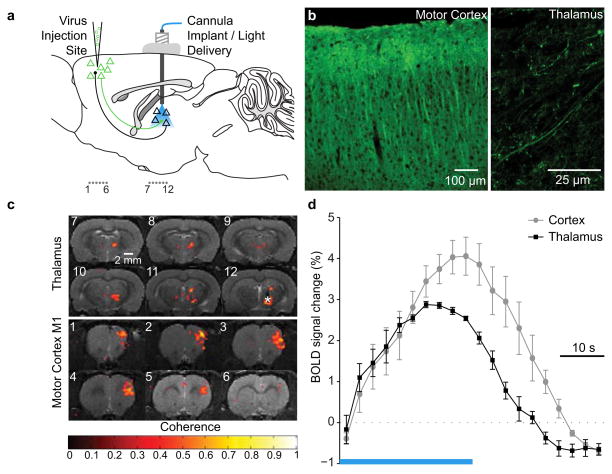
An optical fiber was stereotactically placed in thalamus of animals that had received M1 cortical viral injections (Fig. 3a); posthoc validation of ChR2 expression (Fig. 3b) confirmed ChR2–YFP in cortical neurons and in cortico-thalamic projection fibers. ChR2 readily triggers spikes in illuminated photosensitive axons, that both drive local synaptic output and back-propagate throughout the axon, to the soma of the stimulated cell; note that unlike the case with electrical stimulation, specificity is maintained for driving the targeted (photosensitive) axons, and therefore this configuration in principle allows ofMRI mapping during selective control of the M1 cortical cells that project to thalamus. Indeed robust BOLD signals were observed both locally in thalamus (Fig. 3c: coronal slices 7–12) and also in M1 (Fig. 3d: coronal slices 1–6), consistent with the anticipated recruitment of the topologically targeted cells both locally and distally. These data demonstrate that ChR2-expressing axonal fiber stimulation alone is sufficient to elicit BOLD responses in remote areas, and illustrate the feasibility for in vivo mapping of the global impact of cells defined not only by anatomical location and genetic identity, but also by connection topology.
Figure 3. Control of cells defined by location, genetic identity, and wiring during ofMRI.
a, M1 injection of AAV5-CaMKIIα::ChR2-EYFP and optical stimulation of thalamus. Coronal slices shown in (c) marked as “1..6” and “7..12”. b, ChR2 expression pattern confirming expression in cortical neurons (left) and cortico-thalamic projections (right; see also Supplementary Fig. 5). c, BOLD ofMRI data obtained in thalamus (above) and cortex (below). d, ofMRI-HRF for cortical (gray) and thalamic (black) BOLD signals elicited by optical stimulation of cortico-thalamic fibers in thalamus. Both ofMRI-HRFs ramp slowly by comparison with intracortical results in Figure 1.
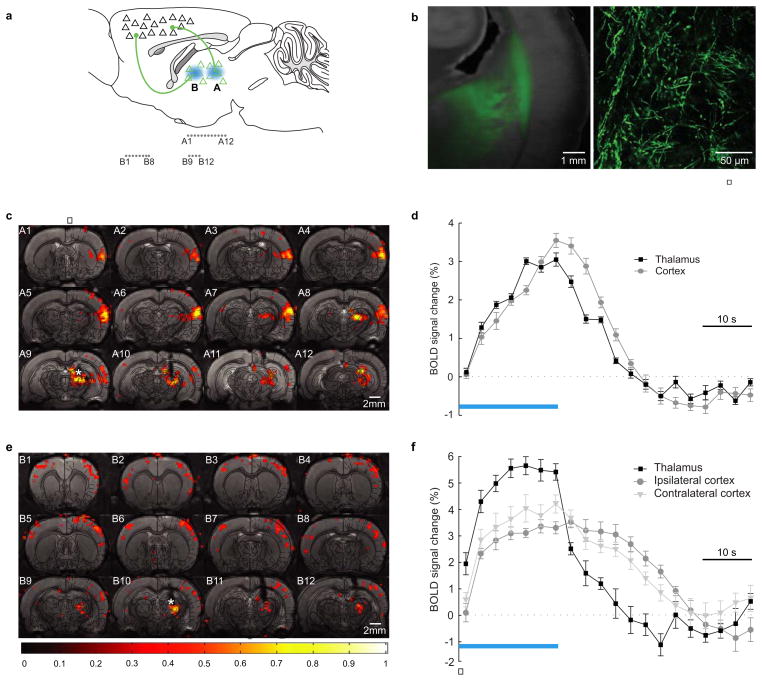
We further explored the global mapping capabilities of ofMRI. It has been suggested that thalamic projections to motor cortex may be more likely than those to sensory cortex, to involve both ipsilateral and contralateral pathways, since in many cases motor control and planning must involve bilateral coordination29. This principle is challenging to assess at the functional level, since electrode-based stimulation will drive antidromic as well as orthodromic projections, and hence may mistakenly report robust cortico-thalamic rather than thalamocortical projections. We therefore sought to globally map functional connectivity arising from initial drive of anterior or posterior thalamic nucleus projections, employing ofMRI. After injecting CaMKIIα::ChR2 into thalamus (Fig. 4), we found that optical stimulation of posterior thalamic nuclei resulted in a strong BOLD response, both at the site of stimulation as expected and in the posterior ipsilateral somatosensory cortex (S2) (Fig. 4a–d). Optically stimulating excitatory cell bodies and fibers in the more anterior thalamic nuclei resulted in BOLD response at the site of stimulation and also significant ipsilateral and contralateral cortical BOLD responses (Fig. 4e,f), consistent with the proposed bilaterality of anterior thalamocortical nuclei involvement in motor control and coordination30.
Figure 4. Recruitment of bilateral cortices by anterior thalamus.
a, Thalamic injection of AAV5-CaMKIIα::ChR2-EYFP and posterior/anterior optical stimulation. Coronal slices marked “A1...A12” and “B1...B12”. b, Fluorescence overlaid onto bright-field (left) and confocal image (right) illustrating transduction in thalamus (left) and cortical projections in internal and external capsule (right). c, Posterior thalamus stimulation-evoked ofMRI signal in ipsilateral thalamus and somatosensory cortex. d, ofMRI-HRFs. Excited volumes: 5.5±1.3mm3 (thalamus); 8.6±2.5mm3 (somatosensory cortex) (n=3). e, Anterior thalamus stimulation-evoked ofMRI signal in ipsilateral thalamus and bilateral motor cortex. f, ofMRI-HRFs. Excited volumes: 1.5mm3 (thalamus); 10.1mm3 (ipsilateral cortex); 3.7mm3 (contralateral cortex).
Together, these results illustrate the power of optogenetic fMRI in shedding light on the controversial identification of positive BOLD signals with increased local neuronal excitation, providing empirical underpinning for fMRI BOLD. We also find that the properties of integrated optogenetics and BOLD-fMRI (ofMRI) allow for global mapping of the causal connectivity of defined neurons in specific brain regions, fundamentally extending the capabilities of pharmacological or electrode-based methods (of course, contributions from additional cells and processes downstream of the defined optically-triggered population are expected and indeed represent an important aspect of this approach; it is, however, important to note that absence of a BOLD signal does not prove the absence of connectivity). Finally, we demonstrate that ofMRI allows causal connectivity mapping of cells defined not only genetically but also by circuit topology, or the conformation of their connections in vivo. Together, the ofMRI methods and findings reported here provide tools and approaches for further probing and defining the causal generation of BOLD signals; these results may accelerate the search for global circuit-disease endophenotypes, as well as the dynamical mapping and reverse engineering of intact neural circuitry.
METHODS SUMMARY
Virus-mediated opsin expression
The pAAV-CaMKIIα-hChR2(H134R)-EYFP plasmid was designed and constructed by standard methods and packaged as AAV5. Virus was stereotaxically injected and cannulas placed at the locations where optical stimulation was planned12. Concentrated virus was delivered using a 10μl syringe and 34-gauge needle; volume and flow rate (0.1μl/min) were controlled by injection pump. Maps and clones are available at optogenetics.org.
Optogenetic fMRI
Rodent subjects were connected to the optical fiber and ventilator (1.3% isofluorane), physiological monitoring systems, and radiofrequency coil, and placed in the MR-compatible stereotaxic frame. After subject placement in the scanner, blue (473 nm) light pulsed at 20 Hz (15 ms pulsewidth) was periodically applied through the optical fiber at 1 min intervals while repeated BOLD scans of large brain volumes21,22 were conducted.
In vivo recording and analysis
After ofMRI, simultaneous optical stimulation and electrical recording in living rodents was conducted using an optrode composed of an extracellular tungsten electrode (1 MΩ, ~125 μm) attached to an optical fiber (~200 μm) with the tip of the electrode deeper than the tip of the fiber to ensure illumination of the recorded neurons12.
Opsin Expression Validation and Immunohistochemistry
To validate specificity, sensitivity and spatial distribution of opsin expression, brain slices were prepared for optical microscopy and immunohistochemistry. 40 μm-thick coronal sections were cut on a freezing microtome and stored in cryoprotectant (25% glycerol, 30% ethylene glycol, in PBS) at 4ºC until processed for immunohistochemistry. Confocal fluorescence images were acquired on a scanning laser microscope using oil immersion objectives12.
Supplementary Material
Acknowledgments
J.H.L. is supported by NIH training grant 1K99EB008738. R.D. is supported by an NSF Graduate Research Fellowship. V.G. is supported by SGF and SIGF (Stanford Graduate Fellowships). We acknowledge Gary H. Glover, John M. Pauly and Dwight G. Nishimura for the generous support and advice, and Cholawat Pacharinsak for assistance with rat intubation. We would also like to thank the entire Deisseroth lab for discussions and support, and Vasiliy Karasev, Zhongnan Fang, and Cory Jones for help with histological quantification and fMRI data analysis. K.D. is supported by the Keck, Snyder, Woo, Yu, McKnight, and Coulter Foundations, as well as by CIRM, NIMH, NIDA, and the NIH Director’s Pioneer Award.
Footnotes
AUTHOR CONTRIBUTIONS
J.H.L., F.Z., R.D., V.G., and K.D. designed the experiments. D.K. provided information on the animal fMRI setup, J.H.L. developed the fMRI methods, and J.H.L. and R.D. conducted the fMRI experiments. R.D., V.G., and F.Z. conducted animal surgery and preparation. J.H.L., L.E.F., R.D. and V.G. conducted optrode recordings. R.D., V.G. and I.G. acquired the confocal microscope images. J.H.L., R.D. and I.G. performed histology, confocal imaging for quantification. C. R. prepared the viral vectors. J.H.L., R.D., V.G., F.Z., D.K. and K.D. prepared the figures and wrote the paper. K.D. supervised all aspects of the work.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Ogawa S, et al. Intrinsic signal changes accompanying sensory stimulation: Functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci USA. 1992;89:5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412 (6843):150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JD, Blum KI. Reward and decision. Neuron. 2002;36 (2):193–198. doi: 10.1016/s0896-6273(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 4.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8 (9):1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 5.Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3 (10):785–792. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 6.Deisseroth K, et al. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26 (41):10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446 (7136):633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 8.Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007;8 (8):577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- 9.Aravanis AM, et al. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4 (3):S143–156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- 10.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459 (7247):698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science (New York, NY) 2009;324 (5925):354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5 (3):439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141 (1):154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friston KJ. Functional and effective connectivity in neuroimaging: a synthesis. Human Brian Mapping. 1994;2:56–78. [Google Scholar]
- 15.Ugurbil K, et al. Functional mapping in the human brain using high magnetic fields. Philos Trans R Soc Lond B Biol Sci. 1999;354 (1387):1195–1213. doi: 10.1098/rstb.1999.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453 (7197):869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- 17.Douglas RJ, Martin KA. A functional microcircuit for cat visual cortex. J Physiol. 1991;440:735–769. doi: 10.1113/jphysiol.1991.sp018733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sirotin YB, Das A. Anticipatory haemodynamic signals in sensory cortex not predicted by local neuronal activity. Nature. 2009;457 (7228):475–479. doi: 10.1038/nature07664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cauli B, et al. Cortical GABA Interneurons in Neurovascular Coupling: Relays for Subcortical Vasoactive Pathways. Journal of Neuroscience. 2004;24 (41):8940–8949. doi: 10.1523/JNEUROSCI.3065-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masamoto K, Kim T, Fukuda M, Wang P, Kim SG. Relationship between neural, vascular, and BOLD signals in isoflurane-anesthetized rat somatosensory cortex. Cereb Cortex. 2007;17 (4):942–950. doi: 10.1093/cercor/bhl005. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, et al. Full-brain coverage and high-resolution imaging capabilities of passband b-SSFP fMRI at 3T. Magn Reson Med. 2008;59 (5):1099–1110. doi: 10.1002/mrm.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Hargreaves BA, Hu BS, Nishimura DG. Fast 3D imaging using variable-density spiral trajectories with applications to limb perfusion. Magn Reson Med. 2003;50 (6):1276–1285. doi: 10.1002/mrm.10644. [DOI] [PubMed] [Google Scholar]
- 23.Glover GH, Lee AT. Motion artifacts in fMRI: comparison of 2DFT with PR and spiral scan methods. Magn Reson Med. 1995;33 (5):624–635. doi: 10.1002/mrm.1910330507. [DOI] [PubMed] [Google Scholar]
- 24.Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: the balloon model. Magn Reson Med. 1998;39 (6):855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- 25.Boynton GM, Engel SA, Glover GH, Heeger DJ. Linear systems analysis of functional magnetic resonance imaging in human V1. J Neurosci. 1996;16 (13):4207–4221. doi: 10.1523/JNEUROSCI.16-13-04207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donahue MJ, et al. Theoretical and experimental investigation of the VASO contrast mechanism. Magn Reson Med. 2006;56 (6):1261–1273. doi: 10.1002/mrm.21072. [DOI] [PubMed] [Google Scholar]
- 27.Lauritzen M. Reading vascular changes in brain imaging: is dendritic calcium the key? Nat Rev Neurosci. 2005;6 (1):77–85. doi: 10.1038/nrn1589. [DOI] [PubMed] [Google Scholar]
- 28.Nir Y, Dinstein I, Malach R, Heeger DJ. BOLD and spiking activity. Nature neuroscience. 2008;11(5):523–524. doi: 10.1038/nn0508-523. author reply 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alloway KD, Olson ML, Smith JB. Contralateral corticothalamic projections from MI whisker cortex: potential route for modulating hemispheric interactions. The Journal of comparative neurology. 2008;510 (1):100–116. doi: 10.1002/cne.21782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuramoto E, et al. Two types of thalamocortical projections from the motor thalamic nuclei of the rat: a single neuron-tracing study using viral vectors. Cereb Cortex. 2009;19 (9):2065–2077. doi: 10.1093/cercor/bhn231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.