Abstract
The transcriptional basis of vertebrate limb initiation, which is a well-studied system for the initiation of organogenesis, remains elusive. Specifically, involvement of the β-catenin pathway in limb initiation, as well as its role in hindlimb-specific transcriptional regulation, are under debate. Here, we show that the β-catenin pathway is active in the limb-forming area in mouse embryos. Furthermore, conditional inactivation of β-catenin as well as Islet1, a hindlimb-specific factor, in the lateral plate mesoderm results in a failure to induce hindlimb outgrowth. We further show that Islet1 is required for the nuclear accumulation of β-catenin and hence for activation of the β-catenin pathway, and that the β-catenin pathway maintains Islet1 expression. These two factors influence each other and function upstream of active proliferation of hindlimb progenitors in the lateral plate mesoderm and the expression of a common factor, Fgf10. Our data demonstrate that Islet1 and β-catenin regulate outgrowth and Fgf10-Fgf8 feedback loop formation during vertebrate hindlimb initiation. Our study identifies Islet1 as a hindlimb-specific transcriptional regulator of initiation, and clarifies the controversy regarding the requirement of β-catenin for limb initiation.
Keywords: Islet1, β-catenin, Mouse limb initiation, Fgf10
INTRODUCTION
During vertebrate development, a variety of progenitor cells are generated and multiple organs are constructed by coordinated proliferation and differentiation. A critical step during organogenesis is the initiation of organ development, where genetic mechanisms control growth and differentiation in a stereotypical morphogenetic process that results in a specific function and morphology for each organ. A thorough understanding of the processes and mechanisms that regulate organ initiation is, therefore, crucial for the study of developmental biology. The vertebrate limb has served as an ideal model system for understanding the mechanisms of organogenesis, including initiation, outgrowth, patterning and morphogenesis (Towers and Tickle, 2009; Zeller et al., 2009). It also provides a unique setting in which to study the development of the forelimb versus the hindlimb, which are two morphologically distinct but serially homologous organs (Duboc and Logan, 2011; Kawakami et al., 2003b; Logan, 2003).
During initiation of the vertebrate limb, cells in the lateral plate mesoderm (LPM) proliferate to initiate outgrowth, and, concomitantly, a signal emanating from the LPM acts on the overlying ectoderm (Capdevila and Izpisua Belmonte, 2001). It has been revealed that, both in the forelimb and hindlimb fields, fibroblast growth factor 10 (Fgf10) in the LPM activates the expression of another Fgf gene, Fgf8, in the surface ectoderm (Min et al., 1998; Ohuchi et al., 1997; Sekine et al., 1999), which then leads to the formation of a mesenchymal Fgf10-ectodermal Fgf8 feedback loop to maintain limb outgrowth. Although it is known that the Fgf10-Fgf8 feedback loop is an evolutionarily conserved molecular component that is essential for maintaining mesenchymal cell proliferation and limb outgrowth, two major questions remain with respect to vertebrate limb initiation. One is the involvement of the canonical Wnt/β-catenin pathway in this process, and the other is the difference between forelimb and hindlimb initiation.
Depending on the species, there remains some controversy as to the involvement of the canonical Wnt/β-catenin pathway. Our previous studies have demonstrated that Wnt/β-catenin signaling in the limb-forming region is an upstream regulator of Fgf10 in chick and zebrafish embryos (Kawakami et al., 2001; Ng et al., 2002). Wnt2b is expressed in the forelimb and pectoral fin-forming area in the chick and zebrafish, respectively, and Wnt8c is expressed in the hindlimb-forming area in chick embryos. These Wnts signal through the β-catenin-dependent pathway. Consistent with this, ectopic activation of the β-catenin pathway in the chick flank can induce ectopic Fgf10 expression and the formation of an extra limb. The reciprocal experiment, in which the Wnt/β-catenin pathway is downregulated, resulted in loss of limb and fin buds in the chick and zebrafish, respectively. In contrast to these studies, the expression of a Wnt ligand in the limb-forming area of mouse embryos has not been reported so far. Moreover, mice mutant in Lef/Tcf factors, which are transcription factors that mediate Wnt signaling by forming a complex with β-catenin, have not exhibited a lack of limb initiation (Galceran et al., 1999). Therefore, it remains to be clarified whether the β-catenin pathway regulates limb initiation in mouse embryos.
The issue of limb type-specific transcriptional regulation of initiation is complicated. Although the Fgf10-Fgf8 feedback loop operates on both the forelimb and hindlimb buds, it has become evident that transcriptional regulation upstream of active proliferation of LPM cells and mesenchymal Fgf10 expression is different in these two types of limbs. Tbx5, which encodes a T-box transcription factor, is exclusively expressed in the forelimb field, but not in the hindlimb field (Gibson-Brown et al., 1996). Tbx5–/– embryos show neither forelimb bud outgrowth nor Fgf10 expression in the forelimb field (Agarwal et al., 2003; Rallis et al., 2003). These reports have established Tbx5 as a crucial regulator of forelimb initiation. By contrast, the genetic mechanisms for hindlimb initiation remain elusive. Previous studies have reported that two transcription factor genes, Tbx4 and Pitx1, are exclusively expressed in the hindlimb field, but not in the forelimb field (Gibson-Brown et al., 1996; Szeto et al., 1999). However, gene-targeting experiments have revealed that neither is necessary for initiation of the hindlimb bud (Lanctot et al., 1999; Naiche and Papaioannou, 2003; Szeto et al., 1999). Tbx4-null embryos start both initial outgrowth to form the hindlimb bud and Fgf10 expression in the hindlimb-forming region. However, Fgf10 expression in the hindlimb bud is not maintained in the absence of Tbx4. These studies established that Tbx4 is not required for induction but for maintenance of Fgf10 expression (Naiche and Papaioannou, 2003). Pitx1–/– mice have small, but well-patterned hindlimbs. These reports highlight our lack of knowledge on the transcriptional regulation of hindlimb-specific initiation, as compared with the role of Tbx5 in forelimb initiation (summarized in Table S1 in the supplementary material). A recent report has demonstrated that Islet1, a LIM-homeodomain transcription factor gene, is transiently expressed in the hindlimb field, but not the forelimb field (Yang et al., 2006). Moreover, lineage analysis has shown that Islet1-expressing cells contribute to a large part of the hindlimb mesenchyme. These analyses make Islet1 a strong candidate to be a hindlimb-specific transcriptional regulator. However, Islet1-null embryos arrest by E9.5, prior to the initiation of the hindlimb bud in mice (Pfaff et al., 1996). Thus, the role of Islet1 in the development of the hindlimb remains elusive.
In order to clarify these two major issues in mouse limb initiation, we carried out conditional inactivation studies on the Ctnnb1 (which encodes β-catenin) and Islet1 genes in the LPM. Both resulted in loss of the hindlimb bud, demonstrating that the β-catenin pathway and Islet1 are required for initiation of the hindlimb. We show that both the β-catenin pathway and Islet1 lie upstream of active proliferation and expression of Fgf10 in the mesenchyme of the hindlimb-forming region. Furthermore, our analysis demonstrates that Islet1 function is required for the nuclear accumulation of β-catenin and for activation of the β-catenin-dependent pathway. β-catenin is not required for initiation of, but rather only for maintenance of Islet1 expression. Our study has clarified a controversy regarding the requirement of the β-catenin pathway and identified Islet1 as a factor regulating hindlimb initiation, thus clarifying our understanding of an important process during vertebrate limb initiation.
MATERIALS AND METHODS
Mouse lines
The Ctnnb1flox/flox line (Ctnnb1tm2Kem) was purchased from the Jackson Laboratory. Islet1flox/flox (Song et al., 2009; Sun et al., 2008), T-cre (Perantoni et al., 2005) and Hoxb6Cre (Lowe et al., 2000) lines have been reported. Fgf10 mutant mice (Min et al., 1998; Sahara and O'Leary, 2009) and the BATgal reporter line (Maretto et al., 2003) have been described.
Imaging analysis
β-catenin and laminin immunostaining were performed using anti-β-catenin (BD Bioscience, #610154) and anti-laminin (Sigma, L9393) in combination with Alexa 488 anti-mouse IgG and Alexa 568 anti-rabbit IgG (Invitrogen) with a standard protocol. Islet1 immunostaining was with rabbit anti-Islet1 antibody (1/1000) (Song et al., 2009) or monoclonal 39.4D5 (Developmental Studies Hybridoma Bank, final working concentration 4.5 μg/ml). Detection of β-galactosidase immunoreactivity in BATgal embryos was by rabbit anti-β-galactosidase antibody (MP Biomedicals, #55976, 1/5000). TUNEL analysis and phospho-histone H3 (pHis3) analysis were performed using rat anti-pHis3 (Sigma, H6409) and Cy3-labeled anti-rat IgG (Jackson ImmunoResearch) in combination with the In Situ Cell Death Detection Kit (Roche) according to the manufacturer's instruction. Fluorescent imaging analysis was performed with Leica TCS SP5 and TCS SPE microscopes. The graphs were constructed with the Leica SP5 software, and the quantification of the TUNEL/pHis3 analysis was with MetaMorph software. Scanning electron microscopy analysis was as previously described (Kawakami et al., 2006) using a Jeol JSM-6390LV microscope.
Statistical analysis
For statistical significance, sections from embryos of the same genotype were stained by immunofluorescence and DAPI and expressed as the percentage of immunopositive cells among DAPI-positive cells for each embryo. From each embryo, multiple sections were examined, and the number of embryos for each analysis was three or more. Statistical significance was examined by independent t-test.
In situ hybridization
Whole-mount and section in situ hybridization were performed following standard protocols (Bluske et al., 2009; Kawakami et al., 2009; Wilkinson, 1992).
RESULTS
Activation of the β-catenin pathway during limb initiation in mice
β-catenin is a crucial component of canonical Wnt signaling. Whereas β-catenin is abundant in the plasma membrane, its intracellular level is kept low. Cytosolic β-catenin is constitutively subjected to degradation, leading to low levels in the cytosol and nuclei. Upon stimulation of cells with a Wnt ligand, the degradation machinery is inhibited, and cytosolic β-catenin accumulates, leading to its translocation into the nucleus. This results in the formation of a complex with transcription factors such as Lef1 and Tcf, leading to the transcriptional activation of downstream genes (Nusse, 2005). Because nuclear accumulation of β-catenin is a hallmark of pathway activation, in order to clarify whether the β-catenin-dependent pathway is involved in mouse limb initiation, we first investigated the accumulation of nuclear β-catenin by fluorescent immunostaining and confocal imaging. In the prospective hindlimb field at E9.5, just prior to hindlimb outgrowth, when Fgf10 expression in the hindlimb field becomes evident (see Fig. S1 in the supplementary material), we observed two types of cells in the LPM with respect to nuclear β-catenin levels as revealed by fluorescence intensity. Whereas both types of cells showed high levels of β-catenin in the membrane, one clearly exhibited lower levels of nuclear β-catenin, with an elongated morphology (Fig. 1A,C,C′). By contrast, the other exhibited high levels of nuclear β-catenin and a relatively rounded morphology (Fig. 1A,B,B′), and constituted ∼24% of the total mesenchymal cells in the LPM at the hindlimb field at E9.5. These two types of cells are consistently observed in multiple sections from multiple embryos (Fig. 1D-F), indicating that the LPM is a heterogeneous tissue. These two types of cells are also observed in the forelimb field at E9.0-10.0 (the stage when forelimb buds initiate outgrowth), but are barely detected after E10.5 (data not shown). The levels of nuclear β-catenin suggest that stabilization, and hence activation, of the β-catenin pathway is temporally regulated in the LPM.
Fig. 1.
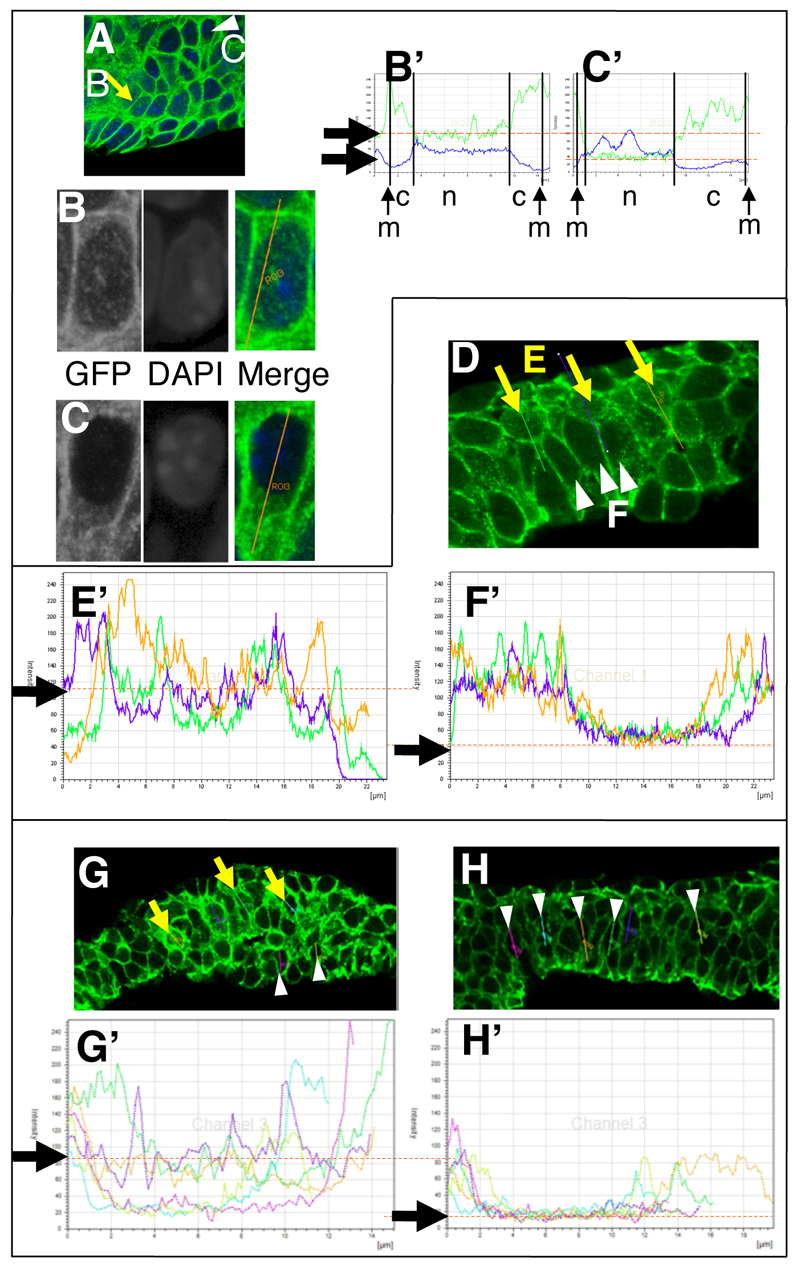
Accumulation of nuclear β-catenin in the lateral plate mesoderm (LPM) of the mouse embryo. (A) Fluorescent imaging analysis showing two types of cells with different levels of nuclear β-catenin in the LPM. (B,C) Higher magnification of the cells indicated by a yellow arrow (B) and white arrowhead (C) in A. GFP and DAPI images are shown in black and white. Merged images with GFP (green) and DAPI (blue) are also shown. (B′,C′) Fluorescence intensity analysis of B and C, with β-catenin signal in green and DAPI signal in blue. Higher DAPI signal indicates the nucleus, and the difference in the levels of nuclear β-catenin is indicated by orange dashed lines and arrows. The y-axis represents the fluorescence intensity. c, m and n indicate cytosol, membrane and nucleus, respectively. (D-F′) A typical image of E9.5 hindlimb field with β-catenin staining (D). Yellow arrows (E) and white arrowheads (F) represent cells with high and low nuclear β-catenin, respectively. Large numbers of these cells are detected simultaneously, and clearly have different levels of nuclear β-catenin as illustrated in E′ and F′. (G-H′) Accumulation of nuclear β-catenin is a characteristic of the limb-forming area. Fluorescent images of β-catenin in the hindlimb-forming area and interlimb-forming area of the E9.5 mouse embryo. The level of β-catenin is shown in G′ and H′ for the cells indicated by yellow arrows and white arrowheads, respectively. Dashed orange lines indicate differences in the nuclear β-catenin levels between the two types of cell.
Next, in order to clarify whether the nuclear localization of β-catenin is associated with limb initiation, we examined the LPM at the interlimb area of E9.0, E9.5 and E10.0 embryos using the same approach. In contrast to the LPM at the hindlimb-forming area, we have not detected any cells with high levels of nuclear β-catenin out of 3581 cells counted in the LPM from 32 sections (compare Fig. 1G,G′,H,H′), indicating that nuclear accumulation of β-catenin is spatially linked to limb initiation in the mouse embryo. These data suggest that the β-catenin pathway is active in the limb-forming area in the LPM at the time of limb outgrowth initiation.
To confirm the activation of the β-catenin pathway in the limb field in mouse embryos, we utilized the BATgal reporter mouse line. This line has been used to visualize β-catenin-dependent pathway activity by lacZ staining (Maretto et al., 2003). In addition to an intense signal in the tail bud, dorsal neural tube and somites, we also detected patchy signals in the hindlimb-forming area (see Fig. S2E in the supplementary material). This confirms activation of the β-catenin pathway in the limb-forming area.
Next, we examined the role of mesenchymal β-catenin by conditionally inactivating β-catenin, as β-catenin-null embryos exhibit severe gastrulation defects (Haegel et al., 1995). Inactivation of β-catenin as early as E7.5 using the T-cre line causes severe truncation of the body posterior to the heart level (Dunty et al., 2008) (see also Fig. S3 in the supplementary material), precluding it from being used in the analysis of limb initiation. Instead, we used the Hoxb6Cre line, in which Cre is activated in the LPM after E8.5 (Lowe et al., 2000). Since Cre activation takes place completely in the hindlimb-forming field, but only in the posterior half of the forelimb-forming field, we focused our analysis on hindlimb development. In order to confirm elimination of β-catenin we utilized both immunofluorescence confocal imaging and the BATgal reporter line, as they were used to demonstrate activation of the β-catenin pathway in the limb-forming area. In both cases, we observed significant reduction of mesenchymal β-catenin activity in the hindlimb field at E9.5, the stage just prior to hindlimb outgrowth, demonstrating an efficient inactivation of β-catenin in a mesoderm-specific manner (see Fig. S2A-F in the supplementary material) (hereafter, the Hoxb6Cre; Ctnnb1flox/flox embryos are referred to as Ctnnb1 cKO). The outgrowth of the hindlimb bud is evident at E10.0 in control littermate embryos (Hoxb6Cre; Ctnnb1+/flox), whereas the Ctnnb1 cKO embryos showed no hindlimb outgrowth in seven embryos examined by scanning electron microscopy between E9.5 and E10.0 (Fig. 2A,B).
Fig. 2.
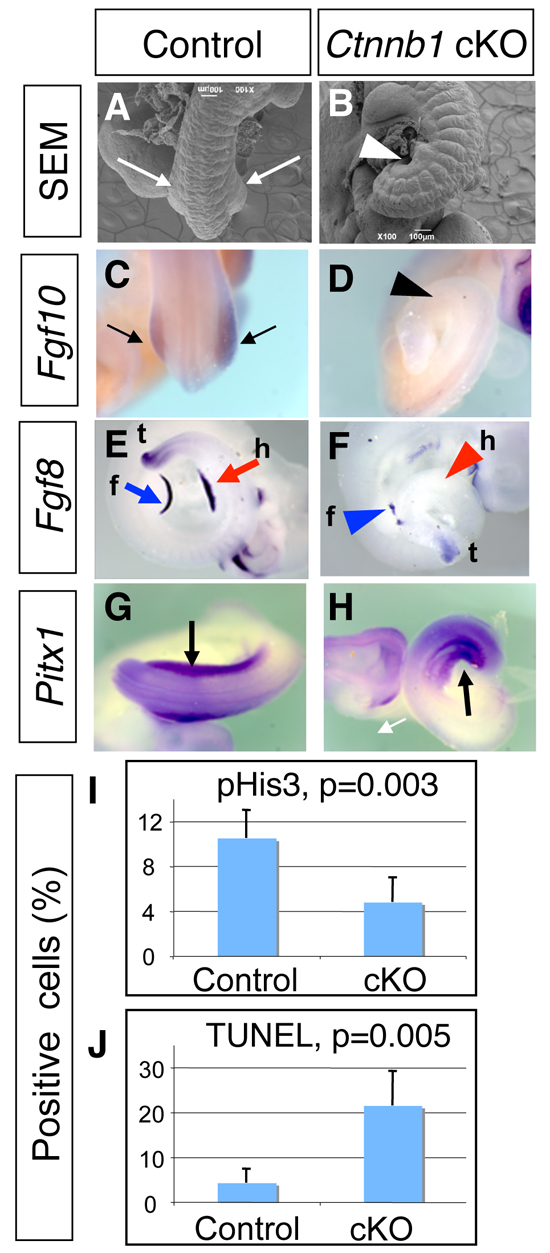
β-catenin is required for limb initiation in the mouse embryo. (A-H) Scanning electron microscopy analysis (A,B) and whole-mount mRNA expression analysis (C-H) of Ctnnb1 cKO embryos (B,D,F,H) and control littermate embryos (A,C,E,G). Hindlimb buds are visible in E10.0 control embryos (A, arrows), but are not formed in the Ctnnb1 cKO embryo (B, arrowhead). Fgf10 expression is detected in the hindlimb bud in the control embryo at E10.0 (C, arrows), but not in the Ctnnb1 cKO embryo (D, arrowhead). Fgf8 expression is detected in the forelimb (blue arrow) and hindlimb (red arrow) buds in the E10.0 control embryo (E). In the Ctnnb1 cKO embryo, Fgf8 expression is not detected in the hindlimb area (red arrowhead), and weak, discontinuous Fgf8 expression is detected in the forelimb bud (blue arrowhead). Pitx1 expression in the hindlimb-forming area is detected in both control (G, arrow) and Ctnnb1 cKO (H, arrow) embryos at E9.5. f, forelimb bud; h, hindlimb bud or hindlimb area; t, tail bud. (I,J) Quantitative analysis of multiple sections shows significant reduction of proliferation (I) and increased cell death (J) in the Ctnnb1 cKO embryo. Error bars indicate s.d.
Additionally, no Fgf10 expression in the LPM and no Fgf8 expression in the surface ectoderm were observed in the cKO embryos (Fig. 2C-F). These results demonstrate that the mesenchymal β-catenin pathway lies upstream of Fgf10 for hindlimb initiation. Unlike the complete loss of the hindlimb bud, we observed small forelimb buds in the Ctnnb1 cKO embryos (see Fig. S2G,H in the supplementary material). This could be because Hoxb6Cre-mediated inactivation takes place only in the posterior half of the forelimb region in the LPM, and the cells that escaped from Ctnnb1 gene inactivation contributed to proliferation and Fgf10 expression, leading to the formation of small forelimb buds. A similar phenotype has been observed in Lef1–/–; Tcf1–/– mutant embryos (Galceran et al., 1999). The Lef/Tcf family of factors form complexes with β-catenin and activate target gene expression. The smaller limb buds, but not the complete lack of limb buds, in Lef1–/–; Tcf1–/– embryos might be due to a functional redundancy with other transcription factors, such a Tcf3 and Tcf4. The Ctnnb1 cKO embryos die by E11.5, consistent with the fact that β-catenin activity is essential for the development of many other organs (Grigoryan et al., 2008). These reports concur with our observations and support the requirement of β-catenin in vertebrate limb initiation.
Our previous report on the differential expression of Wnt ligand genes in chick embryos suggested that the Wnt/β-catenin pathway might also have a role in the development of the differences between these limbs, namely forelimb versus hindlimb identity. Since Ctnnb1 cKO embryos develop no hindlimb, we examined this possibility by expression of the hindlimb-specific gene Pitx1, the expression of which starts prior to hindlimb bud outgrowth. Pitx1 has been demonstrated to be required and sufficient to determine hindlimb-specific characteristics in mice (DeLaurier et al., 2006; Lanctot et al., 1999; Szeto et al., 1999). Contrary to previous predictions, we observed normal expression of Pitx1 in Ctnnb1 cKO as compared with control embryos (Fig. 2G,H), suggesting that β-catenin activity is unlikely to be required for the genetic program that drives limb type specificity (Duboc and Logan, 2011; Kawakami et al., 2003b; Logan, 2003).
In the Ctnnb1 cKO embryos, we found increased cell death and reduced proliferation of cells in the LPM (Fig. 2I,J). Compared with littermate control embryos (n=4 embryos), both proliferation and cell death showed significant alterations in the Ctnnb1 cKO embryos (n=4 embryos), suggesting that β-catenin serves as a survival factor as well as a proliferation factor in LPM cells. Given that we detected normal expression of Pitx1, neither a general failure to activate genes in the LPM nor a simple loss of cells by apoptosis is a likely cause of the lack of hindlimb outgrowth. Rather, these observations suggest that a β-catenin-dependent genetic program is specifically required for hindlimb initiation as an upstream regulator of Fgf10, independent from serving as a cell survival factor in the LPM. This is further supported by our observation that neither cell proliferation nor cell death is changed in Fgf10–/– embryos (see Fig. S4 in the supplementary material). This suggests that these alterations in Ctnnb1 cKO embryos are not due to loss of Fgf10 expression in the LPM and are likely to be independent of the Ctnnb1-Fgf10 genetic cascade. Although the Fgf10-Fgf8 feedback loop is known to be required for cell survival in the developing limb bud (Sun et al., 2002), β-catenin-dependent cell proliferation and cell survival in the LPM seem to be regulated independently of FGF activity.
Taken together, our results demonstrate that β-catenin is required for the initiation of hindlimb bud outgrowth in mouse embryos, upstream of the Fgf10-Fgf8 feedback loop.
Islet1 regulates hindlimb-specific limb initiation
The limb initiation process seems to have a strong link to forelimb and hindlimb specificity. Tbx5 is specifically expressed in the forelimb field, and Tbx5–/– embryos fail to induce forelimb bud outgrowth and Fgf10 expression (Agarwal et al., 2003). Therefore, Tbx5 is required for forelimb initiation; however, the transcriptional regulation required for hindlimb initiation is still unclear. Tbx4 and Pitx1, two genes previously identified as expressed exclusively in the hindlimb field, are not required for initiation of hindlimb bud outgrowth or initiation of Fgf10 expression in the LPM (Lanctot et al., 1999; Naiche and Papaioannou, 2003; Szeto et al., 1999). Thus, our understanding of the transcriptional regulation of limb-type-specific initiation is limited to Tbx5 in the forelimb, whereas hindlimb-specific transcriptional regulation remains elusive.
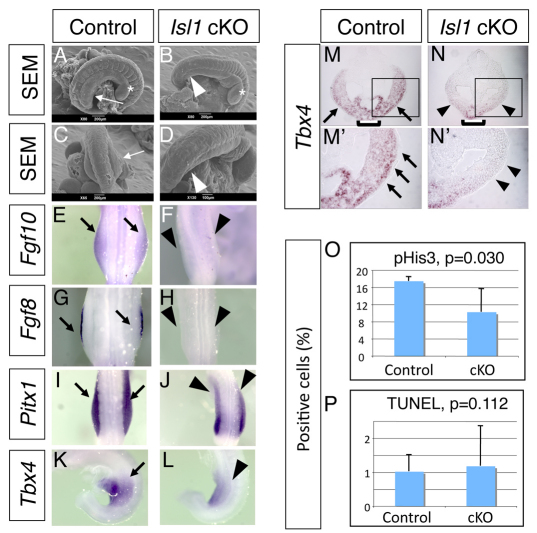
We have previously performed several screenings based on the expression pattern in chick embryos (Kawakami et al., 2004; Kawakami et al., 2003a; Trelles et al., 2002), by which we isolated Islet1 as a gene that is specifically expressed in the hindlimb field and not in the forelimb field (see Fig. S5A in the supplementary material). Although this same expression pattern has been reported in mouse embryos (Yang et al., 2006), its functional significance during limb development is unclear owing to the lethality of Islet1–/– embryos by E9.5 (Pfaff et al., 1996). As such, we inactivated Islet1 using an Islet1 conditional allele (Du et al., 2009; Song et al., 2009). Since Islet1 expression in the posterior body starts as early as E8.5 (see Fig. S5A in the supplementary material), we used the T-cre line, which activates Cre in the broad region of the mesoendoderm at E7.5 (Dunty et al., 2008; Yang et al., 2006). This strategy allowed us to successfully delete Islet1 from the LPM (see Fig. S5B in the supplementary material). T-cre; Islet1–/flox (hereafter referred to as Islet1 cKO) embryos exhibited no hindlimb outgrowth at E10.0, whereas forelimbs developed normally (Fig. 3A-D). Although we did not obtain Islet1 cKO embryos beyond E11.5, this result demonstrates a requirement of Islet1 for initiating hindlimb outgrowth. The Islet1 cKO embryo showed no Fgf10 expression in the LPM and no Fgf8 expression in the surface ectoderm, indicating that Islet1 acts upstream of the Fgf10-Fgf8 feedback loop (Fig. 3E-H). Thus, these data demonstrate that Islet1 is specifically required for initiation of the hindlimb, upstream of the Fgf10-Fgf8 feedback loop.
Fig. 3.
Islet1 is required for hindlimb initiation in the mouse embryo. (A-L) Scanning electron microscopy analysis (A-D) and whole-mount mRNA expression analysis (E-L) of Islet1 cKO embryos (B,D,F,H,J,L) and control littermate embryos (A,C,E,G,I,K). Hindlimb buds are visible in E10.0 control embryos (A,C, arrows), but are not formed in the Islet1 cKO embryo (B,D, arrowheads). Asterisks indicate similarly developed forelimb buds in both embryos. Expression of Fgf10 (E,F) and Fgf8 (G,H) is detected in the control embryo (E,G, arrows), but not in the Islet1 cKO embryo (F,H, arrowheads). Pitx1 expression in the hindlimb-forming area (I, arrows) is reduced in the Islet1 cKO embryo (J, arrowheads). (K) Lateral view of normal Tbx4 expression (arrow) in the control embryo. (L) Tbx4 expression in the LPM is reduced in the Islet1 cKO embryo (arrowhead). (M-N′) In situ hybridization of Tbx4 on cross-sections at the hindlimb-forming region at E9.5. M′ and N′ are magnifications of the boxed areas in M and N. Tbx4 is strongly expressed in the LPM (arrow) and the ventral body wall (bracket) in control embryos (M,M′). Tbx4 expression in the LPM is significantly downregulated (arrowhead), whereas the signal in the ventral body wall is clearly detectable (bracket) in the Islet1 cKO embryo. (O,P) Quantitative analysis of multiple sections shows significant reduction of proliferation (O) but no alteration of cell death (P) in the Islet1 cKO embryo. Error bars indicate s.d.
Islet1 regulates expression of Tbx4 but not Pitx1 in the LPM
Our findings compare and contrast two different regulators upstream of limb outgrowth and expression of Fgf10 – Islet1, a hindlimb-specific transcription factor, and Tbx5, a forelimb-specific transcription factor (Agarwal et al., 2003) – in the induction of hindlimb and forelimb bud outgrowth, respectively. Since the entire hindlimb bud is lost, it is not possible to examine other roles of Islet1 in hindlimb specification by morphological analyses. Therefore, we examined the expression of the hindlimb field-specific genes Pitx1 and Tbx4 in order to assess a possible role of Islet1 in the specification of limb identity. Genetic analysis has suggested that Tbx4 expression is driven by the hindlimb-specific program, although its role in hindlimb specificity remains controversial (Minguillon et al., 2005; Naiche and Papaioannou, 2007; Ouimette et al., 2010). We observed a slightly weaker signal for Pitx1 expression and significant downregulation of Tbx4 in the LPM (Fig. 3I-N′). Since the slight downregulation of Pitx1 could be due to a loss of cells in the hindlimb field, we examined proliferation and cell death. We found that Islet1 cKO embryos exhibit a significant reduction in the proliferation of cells in the LPM (n=5 embryos, Fig. 3O), as compared with control embryos (n=4 embryos), which could, at least in part, contribute to the absence of a hindlimb. However, unlike Ctnnb1 cKO embryos, we did not observe alterations in cell death (Fig. 3P). Such reduced proliferation might also contribute to the slightly weaker Pitx1 expression. By contrast, Tbx4 expression in the LPM is severely downregulated, although it is not completely abolished, as compared with persisting expression in the ventral body wall (Fig. 3M-N′). These data suggest that Islet1 is one of factors regulating Tbx4 expression in the hindlimb field, and other factors might participate in the regulation of Tbx4 expression. Although examination of hindlimb-specific morphology in the Islet1 cKO was not possible, these results indicate that Islet1 function is unlikely to be linked to determining hindlimb-specific characteristics in hindlimb progenitors before the formation of the hindlimb bud. Taken together, our analysis has identified Islet1 as a missing piece in the transcriptional regulation of hindlimb-specific initiation in the vertebrate embryo.
Relationship between the β-catenin pathway and Islet1
The fact that the losses of β-catenin and Islet1 led to the loss of hindlimb initiation raised the possibility that the β-catenin pathway and Islet1 might interact functionally to control a common process for limb initiation. In order to address this, we first examined whether nuclear β-catenin and Islet1 colocalize by performing a double immunostaining for β-catenin and Islet1 in sections prepared from the hindlimb-forming area of wild-type mouse embryos at E9.5. Among 16 sections from five embryos (total 2371 cells), we observed that, except for just two cells, nuclear β-catenin and Islet1 do not colocalize (Fig. 4A-C′). This suggests that the β-catenin pathway and Islet1 act on different populations in the hindlimb-forming area.
Fig. 4.
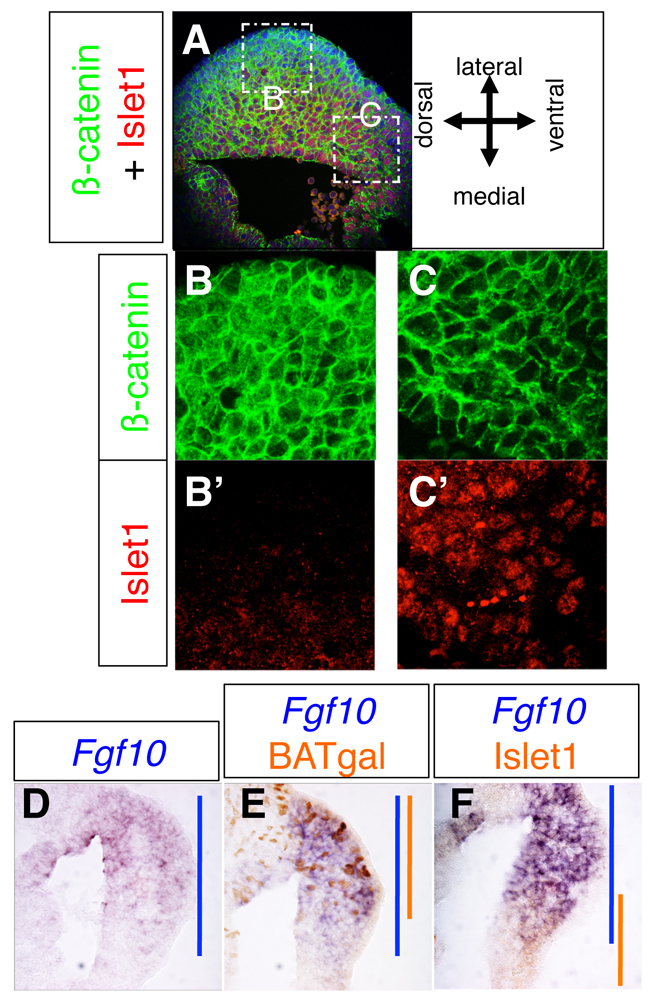
Spatial relationship between Islet1-expressing cells and nuclear β-catenin-positive cells during hindlimb initiation. (A-C′) A typical fluorescent image of β-catenin (green) and Islet1 (red) immunoreactivity on a section prepared from the hindlimb-forming area of an E9.5 wild-type mouse embryo. Insets B and C in A are shown in B,B′ and C,C′ at higher magnification. Nuclear β-catenin is predominantly detected in the lateral area (B,B′), whereas Islet1 is detected in the medial-ventral area (C,C′). (D-F) Spatial relationship between Fgf10 transcripts, β-catenin activity (BATgal) and Islet1 immunoreactivity. (D) Fgf10 mRNA distribution in a cross-section prepared from the middle region of the hindlimb-forming area at E9.5. (E) Spatial relationship between Fgf10 and β-catenin activity in a cross-section prepared from the middle region of hindlimb-forming area at E9.5. Fgf10 mRNA, purple; patchy BATgal signal, orange. The blue and orange bars represent the Fgf10 and BATgal expression domains, respectively, along the dorsal-ventral axis. (F) Spatial relationship between Fgf10 and Islet1 in a cross-section prepared from the middle region of hindlimb-forming area at E9.5. Fgf10 mRNA, purple; Islet1 immunoreactivity, orange. Blue and orange bars represent the expression domains of Fgf10 and Islet1, respectively, along the dorsal-ventral axis.
Because Fgf10 expression is completely lost in both Ctnnb1 cKO and Islet1 cKO hindlimb fields (Fig. 2C,D, Fig. 3E,F), we next examined which of the Islet1-positive cells or which cells with active β-catenin (BATgal-positive cells) overlap with Fgf10 expression. Fgf10 transcripts were detected in the broad region of the LPM, with stronger expression on the dorsal side (Fig. 4D). We observed that BATgal and Fgf10 signals overlapped on the dorsal side (Fig. 4E), whereas Islet1 immunoreactivity overlapped with Fgf10 signal on the ventral side (Fig. 4F). These results demonstrate that cells with active β-catenin and cells with Islet1, which constitute two spatially exclusive populations, overlap with Fgf10 expression.
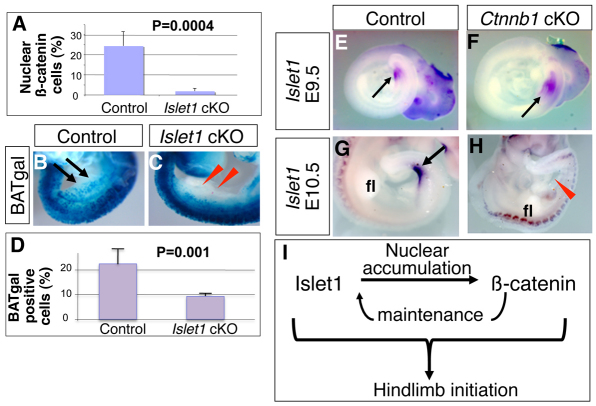
The mutually exclusive localization of Islet1 and nuclear β-catenin does not rule out the possibility that the β-catenin pathway and Islet1 affect each other functionally. To clarify this, we examined whether the loss of Islet1 affects β-catenin pathway activity, and vice versa. In the hindlimb field of Islet1 cKO embryos, only 1.8% of cells were nuclear β-catenin positive, whereas 24.3% of cells were nuclear β-catenin positive in control embryos (Fig. 5A). The hindlimb area of the Islet1 cKO embryos is almost indistinguishable from the interlimb area, where no nuclear β-catenin was detected (Fig. 1H). We further examined this using the BATgal reporter mice. The BATgal reporter signal in the hindlimb field is significantly downregulated in Islet1 cKO embryos as compared with control littermates at E9.5 (Fig. 5B,C). Examining six sections from two control embryos showed that 22.0% of cells in the LPM were BATgal positive, whereas 8.9% of cells in the LPM in six sections from two Islet1 cKO embryos were BATgal positive (Fig. 5D). These results suggest that Islet1 functions in the nuclear accumulation of β-catenin and activation of the β-catenin pathway.
Fig. 5.
Functional interaction between Islet1 and β-catenin during hindlimb initiation. (A) The percentage of cells with nuclear β-catenin is reduced in the LPM of Islet1 cKO mouse embryos at E9.5. (B,C) BATgal reporter activity in the hindlimb-forming area is reduced in the Islet1 cKO embryo (C, red arrowheads), compared with a littermate control embryo (B, black arrows) at E9.5. (D) Quantitative analysis of BATgal-positive cells in the LPM. The percentage of cells with BATgal signal is reduced in the LPM of Islet1 cKO embryos at E9.5. Error bars indicate s.d. (E-H) β-catenin activity is not required for expression of Islet1, but rather is involved in the maintenance of Islet1 expression. Islet1 is expressed similarly in control (E) and Ctnnb1 cKO (F) embryos at E9.5 (arrow indicates expression). Islet1 expression in the control embryo is detected in the very posterior proximal region of the hindlimb bud (G, arrow), but is not detected in the Ctnnb1 cKO embryo (H, red arrowhead) at E10.5. fl, forelimb bud. (I) Summary of the analysis. Islet1 functions in the nuclear accumulation of β-catenin, which in turn acts to maintain Islet1 expression. Both Islet1 and the β-catenin-dependent pathway lie upstream of hindlimb outgrowth and Fgf10 expression in the LPM for hindlimb initiation in vertebrates. For more details, see Discussion.
In the reciprocal experimental setting, we observed Islet1 expression in Ctnnb1 cKO embryos at E9.5, suggesting that the β-catenin function is not required for Islet1 expression (Fig. 5E,F), but rather that β-catenin seems to be required for the maintenance of Islet1 expression or for the survival of Islet1-expressing cells. This is because of the downregulation of Islet1 in the ventral-posterior region of the E10.5 embryo, as compared with control littermates (Fig. 5G,H).
Our analyses have shown that two genetic systems, involving Islet1 and β-catenin, regulate hindlimb initiation in the mouse embryo as upstream regulators of LPM cell proliferation and Fgf10 expression. Moreover, these two players functionally interact – Islet1 in the nuclear accumulation of β-catenin, and the β-catenin-dependent pathway in maintaining Islet1 expression (Fig. 5I).
DISCUSSION
In this study, we have shown that β-catenin and Islet1 regulate hindlimb initiation. Both genes are required for initiating outgrowth as well as for inducing Fgf10 expression, which provides the signal from the LPM to activate Fgf8 in the ectoderm, initiating the Fgf10-Fgf8 feedback loop for maintaining limb bud outgrowth.
β-catenin is required for hindlimb initiation
It has been controversial whether the β-catenin-dependent pathway is required for mouse limb initiation. Our previous studies have revealed that the β-catenin pathway regulates Tbx5 in the forelimb field, which in turn upregulates Fgf10 expression, during forelimb bud initiation in chick and zebrafish embryos (Kawakami et al., 2001; Ng et al., 2002). Our studies have also demonstrated that the β-catenin pathway regulates hindlimb initiation in the chick (Kawakami et al., 2001). This scenario has been uncertain in mammals because, thus far, no Wnt ligand knockout mice have shown defects in limb initiation (Grigoryan et al., 2008). Similarly, no Wnt ligand gene expression has been reported in the limb-forming area in mouse embryos. Several possibilities might account for these discrepancies, including the difficulty in detecting specific Wnt ligands at the low levels at which they are expressed, the limited sensitivity of our detection system, the short time window of expression, and rescue by functional redundancy of multiple ligands. Nonetheless, the possible involvement of the Wnt/β-catenin pathway in limb initiation has remained an open question. We have focused our study on β-catenin, given that its nuclear accumulation is the hallmark of activation of the signaling pathway, using confocal imaging analysis to directly assess the level of nuclear β-catenin and examine the activation status of the pathway. We also utilized a BATgal reporter line, which expresses lacZ under the control of a multimerized Lef/Tcf element, the activation of which depends on β-catenin (Maretto et al., 2003). Such a reporter line has proven useful for visualizing specific signaling activity in a variety of studies, and the confocal imaging and genetic analyses support each other in demonstrating that the β-catenin pathway is active in the limb-forming area.
In this study, we have revealed that β-catenin is required for mouse hindlimb initiation, upstream of active proliferation and expression of Fgf10 in the LPM, consistent with our previous studies in chick and zebrafish. In the absence of β-catenin, we observed increased cell death and reduced proliferation in the LPM. Since β-catenin is a multi-functional factor, it might interact with many factors to regulate cell survival and proliferation (Min et al., 1998; Sekine et al., 1999). The increased cell death and reduced proliferation are likely to be independent from the function of Fgf10 (Min et al., 1998; Sekine et al., 1999) because Fgf10–/– embryos exhibit neither increased cell death nor reduced proliferation (see Fig. S4 in the supplementary material). Although nuclear β-catenin-positive cells constitute ∼24% of LPM, the complete loss of Fgf10 expression and the lack of hindlimb bud outgrowth strongly suggest that the β-catenin pathway regulates Fgf10 as a genetically upstream factor for limb initiation, and, in addition, that it controls cell proliferation and survival. Besides the role of β-catenin-dependent signaling discussed here, we should also consider another possible scenario: a role of β-catenin in cell adhesion, given that only ∼24% of LPM cells are nuclear β-catenin-positive with a salt-and-pepper pattern. In support of this possibility, a previous study in chick embryos has shown that cell-cell adhesion is increased during limb bud formation (Heintzelman et al., 1978). Although the role of cell-cell adhesion in limb initiation in the mouse embryo is still to be investigated (Wada, 2011), both functions – in cell-cell adhesion and in the genetic pathway – might contribute during the β-catenin-dependent limb initiation process.
We speculate that β-catenin might also be required in the forelimb field. Our current genetic tool limits our experiments to the hindlimb field; very early inactivation of β-catenin mediated by T-cre causes a lack of mesoderm generation (Dunty et al., 2008), leading to severe truncation of the posterior body structure, which precludes analysis of β-catenin loss during limb development (see Fig. S3 in the supplementary material). Interestingly, Hoxb6Cre can recombine in cells in the posterior half of the forelimb field (Lowe et al., 2000), and Hoxb6Cre-mediated inactivation of Ctnnb1 resulted in a small forelimb bud compared with control littermates (see Fig. S2G,H in the supplementary material). This is likely to be due to limited inactivation of β-catenin only in the posterior region of the forelimb field, and cells that escaped inactivation would survive and proliferate and turn on Fgf10, leading to the formation of a small forelimb bud. Although we do not exclude the possibility of the involvement of an unidentified factor in the forelimb initiation process, our analysis has revealed that the β-catenin-dependent pathway is active in the limb-forming area, and that it functions upstream of cell proliferation and Fgf10 expression in the LPM during hindlimb initiation in the mouse.
Islet1, a hindlimb field-specific factor, is required for hindlimb initiation upstream of β-catenin activation
Although induction of Fgf10 is required for establishing the Fgf10-Fgf8 feedback loop to maintain mesenchymal proliferation for both forelimb and hindlimb development, the transcriptional programs that regulate Fgf10 expression are different in these two limbs. Tbx5 has been shown to be the transcription factor specifically regulating forelimb initiation in mice (Agarwal et al., 2003); however, hindlimb-specific transcriptional regulation has been elusive. Our analysis has revealed that Islet1, a new player in this field, specifically regulates initiation of the hindlimb, and has clarified the difference between the two limb-type-specific transcriptional regulation programs. Based on expression in the LPM before hindlimb outgrowth, as well as on an immediate decline in its expression after hindlimb bud outgrowth (see Fig. S5A in the supplementary material), Islet1 is likely to be acting only in the early stage of hindlimb development, similar to how Tbx5 is required only in the early stage of forelimb development (Hasson et al., 2007).
Interestingly, these two factors belong to different gene families, namely the T-box transcription factors (Tbx5) and LIM-homeodomain factors (Islet1). This leads to two possible scenarios. One is that Fgf10 contains multiple regulatory elements for expression in the limb-forming region. In this scenario, Tbx5 and Islet1 regulate different DNA sequences, yet they both genetically lie upstream of Fgf10 during the limb initiation process. Our observation that Fgf10 expression overlaps with both Islet1-positive cells and nuclear β-catenin-positive cells, which are located in a mutually exclusive manner in the hindlimb-forming region, also supports this scenario. Even in the hindlimb-forming region, Islet1 and β-catenin might regulate Fgf10 in different cell populations. Such a scenario adds another layer of complexity to the regulation of Fgf10 expression. Currently, it is not clear whether this is the case, as enhancer element(s) for Fgf10 expression in the forelimb and hindlimb fields are unknown.
Our data also provide a second scenario. During hindlimb initiation, Islet1 function is required for nuclear localization of β-catenin, and Ctnnb1 cKO showed no hindlimb outgrowth, suggesting that β-catenin mediates Islet1 function in limb initiation. Then, the β-catenin-dependent pathway regulates Fgf10 expression. In this second scenario, Tbx5 might have a similar role to Islet1 in the nuclear accumulation of β-catenin. Such a model would explain the requirement for limb-type-specific transcription factors, such as Islet1 and Tbx5, as well as the β-catenin-dependent pathway for limb initiation. These models need to be studied in the future.
Relationship between Islet1 and β-catenin
Our data demonstrate that both Islet1 and β-catenin are required for hindlimb initiation; however, it is still to be elucidated how Islet1 and β-catenin interact to initiate the hindlimb bud, given that they exhibit largely exclusive distributions in the hindlimb field. Both factors act in nuclei, Islet1 as a DNA-binding transcription factor and β-catenin as a transcriptional co-activator, and thus it is likely that their interaction is non-cell-autonomous. The finding that Islet1 is required for nuclear β-catenin accumulation and β-catenin activity (visualized by BATgal reporter, Fig. 5) suggests that Islet1 might stimulate production of an unknown secreted factor(s), which triggers activation of the β-catenin pathway. If Wnt-dependent mechanisms are involved, Islet1 may be regulating multiple Wnt ligands in the posterior hindlimb field to affect the β-catenin pathway, although specific Wnt ligands involved in this process are still unknown. Alternatively, Islet1-dependent factor(s) might be necessary to set up responsiveness to Wnt or other factors to trigger β-catenin activation. Similarly, β-catenin-dependent production of an unknown factor(s) might act on the maintenance of Islet1 expression. Investigation into these possibilities will be an important step towards a more detailed understanding of the mechanisms of limb initiation.
In conclusion, our data demonstrate that Islet1-dependent nuclear accumulation of β-catenin lies upstream of the initiation of hindlimb outgrowth and Fgf10 expression in the LPM of the hindlimb field in mouse embryos. Our analyses have clarified a hitherto controversial role for β-catenin in the vertebrate limb initiation process. Furthermore, we have uncovered transcriptional regulation of hindlimb initiation by Islet1, which contrasts with that of the forelimb by Tbx5, and unveiled distinct transcriptional control in different limb types in vertebrates.
Supplementary Material
Acknowledgments
We thank Dr Michael Kuehn for the Hoxb6Cre line; Dr Rolf Kemler for making the Ctnnb1flox/flox line available to the research community; Dr Thomas Neufeld for access to his microscope in the Developmental Biology Center at the University of Minnesota; and the B&H platform of the CMRB for their excellent technical help. The 39.4D5 antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242, USA.
Footnotes
Funding
This work was supported by the Minnesota Medical Foundation (3962-9211-09) and American Cancer Society Institutional Research Grant (IRG-58-001-52-IRG04) to Y.K., NINDS (5R37NS037116) and HHMI to S.P., the National Institutes of Health (R01 NS049357) to Y.N., and CIBER, MICINN, Fundacion Cellex, the G. Harold and Leila Y. Mathers Charitable Foundation, The Leona M. and Harry B. Helmsley Charitable Trust and Sanofi-Aventis to J.C.I.B. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.065359/-/DC1
References
- Agarwal P., Wylie J. N., Galceran J., Arkhitko O., Li C., Deng C., Grosschedl R., Bruneau B. G. (2003). Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130, 623-633 [DOI] [PubMed] [Google Scholar]
- Bluske K. K., Kawakami Y., Koyano-Nakagawa N., Nakagawa Y. (2009). Differential activity of Wnt/beta-catenin signaling in the embryonic mouse thalamus. Dev. Dyn. 238, 3297-3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J., Izpisua Belmonte J. C. (2001). Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell Dev. Biol. 17, 87-132 [DOI] [PubMed] [Google Scholar]
- DeLaurier A., Schweitzer R., Logan M. (2006). Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev. Biol. 299, 22-34 [DOI] [PubMed] [Google Scholar]
- Du A., Hunter C. S., Murray J., Noble D., Cai C. L., Evans S. M., Stein R., May C. L. (2009). Islet-1 is required for the maturation, proliferation, and survival of the endocrine pancreas. Diabetes 58, 2059-2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V., Logan M. P. (2011). Regulation of limb bud initiation and limb-type morphology. Dev. Dyn. 240, 1017-1027 [DOI] [PubMed] [Google Scholar]
- Dunty W. C., Jr, Biris K. K., Chalamalasetty R. B., Taketo M. M., Lewandoski M., Yamaguchi T. P. (2008). Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development 135, 85-94 [DOI] [PubMed] [Google Scholar]
- Galceran J., Farinas I., Depew M. J., Clevers H., Grosschedl R. (1999). Wnt3a–/– like phenotype and limb deficiency in Lef1(–/–)Tcf1(–/–) mice. Genes Dev. 13, 709-717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson-Brown J. J., Agulnik S. I., Chapman D. L., Alexiou M., Garvey N., Silver L. M., Papaioannou V. E. (1996). Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech. Dev. 56, 93-101 [DOI] [PubMed] [Google Scholar]
- Grigoryan T., Wend P., Klaus A., Birchmeier W. (2008). Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 22, 2308-2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegel H., Larue L., Ohsugi M., Fedorov L., Herrenknecht K., Kemler R. (1995). Lack of beta-catenin affects mouse development at gastrulation. Development 121, 3529-3537 [DOI] [PubMed] [Google Scholar]
- Hasson P., Del Buono J., Logan M. P. (2007). Tbx5 is dispensable for forelimb outgrowth. Development 134, 85-92 [DOI] [PubMed] [Google Scholar]
- Heintzelman K. F., Phillips H. M., Davis G. S. (1978). Liquid-tissue behavior and differential cohesiveness during chick limb budding. J. Embryol. Exp. Morphol. 47, 1-15 [PubMed] [Google Scholar]
- Kawakami Y., Capdevila J., Buscher D., Itoh T., Rodriguez Esteban C., Izpisua Belmonte J. C. (2001). WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell 104, 891-900 [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez-Leon J., Koth C. M., Buscher D., Itoh T., Raya A., Ng J. K., Esteban C. R., Takahashi S., Henrique D., et al. (2003a). MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat. Cell Biol. 5, 513-519 [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Tsukui T., Ng J. K., Izpisua Belmonte J. C. (2003b). Insights into the molecular basis of vertebrate forelimb and hindlimb identity. In Patterning in Vertebrate Development (ed. Tickle C.), pp. 198-213 Oxford: Oxford University Press; [Google Scholar]
- Kawakami Y., Esteban C. R., Matsui T., Rodriguez-Leon J., Kato S., Belmonte J. C. (2004). Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development 131, 4763-4774 [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez Esteban C., Raya M., Kawakami H., Marti M., Dubova I., Izpisua Belmonte J. C. (2006). Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 20, 3232-3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Uchiyama Y., Rodriguez Esteban C., Inenaga T., Koyano-Nakagawa N., Kawakami H., Marti M., Kmita M., Monaghan-Nichols P., Nishinakamura R., et al. (2009). Sall genes regulate region-specific morphogenesis in the mouse limb by modulating Hox activities. Development 136, 585-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C., Moreau A., Chamberland M., Tremblay M. L., Drouin J. (1999). Hindlimb patterning and mandible development require the Ptx1 gene. Development 126, 1805-1810 [DOI] [PubMed] [Google Scholar]
- Logan M. (2003). Finger or toe: the molecular basis of limb identity. Development 130, 6401-6410 [DOI] [PubMed] [Google Scholar]
- Lowe L. A., Yamada S., Kuehn M. R. (2000). HoxB6-Cre transgenic mice express Cre recombinase in extra-embryonic mesoderm, in lateral plate and limb mesoderm and at the midbrain/hindbrain junction. Genesis 26, 118-120 [DOI] [PubMed] [Google Scholar]
- Maretto S., Cordenonsi M., Dupont S., Braghetta P., Broccoli V., Hassan A. B., Volpin D., Bressan G. M., Piccolo S. (2003). Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. USA 100, 3299-3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H., Danilenko D. M., Scully S. A., Bolon B., Ring B. D., Tarpley J. E., DeRose M., Simonet W. S. (1998). Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 12, 3156-3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguillon C., Del Buono J., Logan M. P. (2005). Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev. Cell 8, 75-84 [DOI] [PubMed] [Google Scholar]
- Naiche L. A., Papaioannou V. E. (2003). Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 130, 2681-2693 [DOI] [PubMed] [Google Scholar]
- Naiche L. A., Papaioannou V. E. (2007). Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development 134, 93-103 [DOI] [PubMed] [Google Scholar]
- Ng J. K., Kawakami Y., Buscher D., Raya A., Itoh T., Koth C. M., Rodriguez Esteban C., Rodriguez-Leon J., Garrity D. M., Fishman M. C., et al. (2002). The limb identity gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development 129, 5161-5170 [DOI] [PubMed] [Google Scholar]
- Nusse R. (2005). Wnt signaling in disease and in development. Cell Res. 15, 28-32 [DOI] [PubMed] [Google Scholar]
- Ohuchi H., Nakagawa T., Yamamoto A., Araga A., Ohata T., Ishimaru Y., Yoshioka H., Kuwana T., Nohno T., Yamasaki M., et al. (1997). The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development 124, 2235-2244 [DOI] [PubMed] [Google Scholar]
- Ouimette J.-F., Jolin M. L., L’honore A., Gifuni A., Drouin J. (2010). Divergent transcriptional activities determine limb identity. Nat. Commun. 1, 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perantoni A. O., Timofeeva O., Naillat F., Richman C., Pajni-Underwood S., Wilson C., Vainio S., Dove L. F., Lewandoski M. (2005). Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 132, 3859-3871 [DOI] [PubMed] [Google Scholar]
- Pfaff S. L., Mendelsohn M., Stewart C. L., Edlund T., Jessell T. M. (1996). Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 84, 309-320 [DOI] [PubMed] [Google Scholar]
- Rallis C., Bruneau B. G., Del Buono J., Seidman C. E., Seidman J. G., Nissim S., Tabin C. J., Logan M. P. (2003). Tbx5 is required for forelimb bud formation and continued outgrowth. Development 130, 2741-2751 [DOI] [PubMed] [Google Scholar]
- Sahara S., O’Leary D. D. (2009). Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron 63, 48-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K., Ohuchi H., Fujiwara M., Yamasaki M., Yoshizawa T., Sato T., Yagishita N., Matsui D., Koga Y., Itoh N., et al. (1999). Fgf10 is essential for limb and lung formation. Nat. Genet. 21, 138-141 [DOI] [PubMed] [Google Scholar]
- Song M. R., Sun Y., Bryson A., Gill G. N., Evans S. M., Pfaff S. L. (2009). Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development 136, 2923-2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Mariani F. V., Martin G. R. (2002). Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature 418, 501-508 [DOI] [PubMed] [Google Scholar]
- Sun Y., Dykes I. M., Liang X., Eng S. R., Evans S. M., Turner E. E. (2008). A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat. Neurosci. 11, 1283-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto D. P., Rodriguez-Esteban C., Ryan A. K., O’Connell S. M., Liu F., Kioussi C., Gleiberman A. S., Izpisua-Belmonte J. C., Rosenfeld M. G. (1999). Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 13, 484-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers M., Tickle C. (2009). Growing models of vertebrate limb development. Development 136, 179-190 [DOI] [PubMed] [Google Scholar]
- Trelles R. D., Leon J. R., Kawakami Y., Simoes S., Belmonte J. C. (2002). Expression of the chick vascular endothelial growth factor D gene during limb development. Mech. Dev. 116, 239-242 [DOI] [PubMed] [Google Scholar]
- Wada N. (2011). Spatiotemporal changes in cell adhesiveness during vertebrate limb morphogenesis. Dev. Dyn. 240, 969-978 [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G. (1992). Whole mount in situ hybridization of vertebrate embryos. In In Situ Hybridization: a Practical Approach (ed. Wilkinson D. G.), pp. 75-83 Oxford, UK: IRL Press; [Google Scholar]
- Yang L., Cai C. L., Lin L., Qyang Y., Chung C., Monteiro R. M., Mummery C. L., Fishman G. I., Cogen A., Evans S. (2006). Isl1Cre reveals a common Bmp pathway in heart and limb development. Development 133, 1575-1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R., Lopez-Rios J., Zuniga A. (2009). Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat. Rev. Genet. 10, 845-858 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.