Abstract
Aim
to monitor microbial shifts during dental biofilm re-development
Methods
Supra and subgingival plaque samples were taken separately from 28 teeth in 38 healthy and 17 periodontitis subjects at baseline and immediately after tooth cleaning. Samples were taken again from 7 teeth in randomly selected quadrants during 1, 2, 4 and 7 days of no oral hygiene. Samples were analyzed using checkerboard DNA-DNA hybridization. Species counts were averaged within subjects at each time point. Significant differences in counts between healthy and periodontitis subjects were sought using the Mann-Whitney test.
Results
Total supra and subgingival counts were significantly higher in periodontitis on entry and reached or exceeded baseline values after day 2. Supragingival counts of Veillonella parvula, Fusobacterium nucleatum ss vincentii and Neisseria mucosa increased from 2 to 7 days. Subgingival counts were greater for Actinomyces, green and orange complex species. Significant differences between groups in supragingival counts occurred for 17 of 41 species at entry, 0 at day 7; for subgingival plaque these values were 39/41 taxa at entry, 17/41 at day 7.
Conclusions
Supragingival plaque re-development was similar in periodontitis and health, but subgingival species recolonization was more marked in periodontitis.
Keywords: oral bacteria, periodontal, health, periodontitis, biofilms, supragingival, subgingival
Introduction
Periodontal diseases are associated with bacterial species present in biofilms that colonize dental surfaces. Most information that describes the microbial composition of dental biofilms is based on studies of samples from mature dental plaque. Light and electron microscopy studies (Listgarten 1976, Listgarten 1999, Listgarten et al. 1975), in vitro adhesion and co-aggregation models (Gibbons & Nygaard M. 1970, Gibbons et al. 1988, Gibbons et al. 1991, Kolenbrander & London 1993, Kolenbrander et al. 1993, Stromberg & Boren 1992) and in vitro continuous culture studies (Stoodley et al. 1999, Bernimoulin 2003) have been helpful in describing possible changes that might occur in species composition during biofilm formation. However, few studies have actually examined the shifts in microbial species that occur during in vivo supra- or subgingival plaque development. The body of knowledge describing the microbial shifts that occur in microbial species during in vivo dental plaque development or re-development appears to result in large part from 5 studies that examined supragingival plaque samples taken from subjects who refrained from brushing during the period of plaque re-growth (Ritz 1967, Socransky et al. 1977, Zee et al. 1996b, Ramberg et al. 2003, Zee et al. 1996a, Li et al. 2004). Although informative, those studies did not evaluate subgingival plaque development. Further, they assessed a limited number of samples, bacterial taxa and subjects and did not evaluate subjects with periodontitis. Thus, the purpose of the present investigation was to compare changes in species' levels during early dental biofilm development in periodontally healthy and chronic periodontitis subjects who refrained from oral hygiene. The hypotheses to be tested were: 1) plaque development would be more rapid in periodontitis subjects than in periodontally healthy subjects; 2) the shift in levels of individual taxa over time would differ from periodontally healthy to periodontally diseased subjects; 3) the re-growth of specific periodontal pathogens such as Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola would be slower than the re-growth of other species.
Material and Methods
Subject population
The subject population consisted of 38 periodontally healthy and 17 chronic periodontitis subjects. The inclusion and exclusion criteria were as follows:
Inclusion criteria for healthy subjects
> 20 years of age, > 24 teeth, no pocket depth or attachment level measurements > 3 mm, < 20% of sites with overt gingival redness and/or bleeding on probing and willingness and ability to sign informed consent.
Inclusion criteria for periodontitis subjects
> 20 years of age, > 20 teeth, > 8 teeth with pocket depth and/or attachment level > 4 mm and willingness and ability to sign informed consent.
Exclusion criteria
Pregnancy or nursing, periodontal or antibiotic therapy in the previous 3 months, any systemic condition which might have influenced the course of periodontal disease or treatment (e.g. diabetes, AIDS), any systemic condition which required antibiotic coverage for routine periodontal procedures (e.g. heart conditions, joint replacements), any soft tissue lesions (e.g. leukoplakia, lichen planus) and current smokers.
Attempts were made to recruit approximately equal numbers of males and females. In addition, subjects of any racial/ethnic group were accepted for the study. All subjects were recruited at The Forsyth Institute. The study was approved by The Forsyth Institute Institutional Review Board and all subjects signed informed consent prior to entering the study. The baseline clinical characteristics of the subjects in the 2 groups are shown in Table 1.
Table 1.
Mean clinical parameters (± SD) of subject groups at baseline.
| Healthy | Periodontitis | p (Mann Whitney) | |
|---|---|---|---|
| N subjects | 38 | 17 | |
| Age | 32.32 ± 9.34 | 44.88 ± 11.94 | p < 0.001 |
| Number of missing teeth | 0.87 ± 1.55 | 2.18 ± 2.40 | p < 0.05 |
| % males | 39 | 35 | NS |
| % past smokers | 29 | 29 | NS |
| Pocket depth (mm) | 1.94 ± 0.30 | 2.70 ± 0.29 | p < 0.001 |
| Attachment level (mm) | 1.50 ±0.63 | 2.98 ± 1.17 | p < 0.001 |
| Plaque Index | 1.22 ±0.74 | 1.58 ± 0.73 | NS |
| % of sites with: | |||
| Gingival Redness | 47.69 ± 30.71 | 62.63 ± 34.75 | p < 0.001 |
| BOP | 7.33 ± 6.85 | 27.18 ± 12.79 | p < 0.001 |
| Suppuration | 0.00 ± 0.00 | 0.16 ± 0.38 | p < 0.01 |
| N White | 28 | 10 | |
| N African American | 1 | 4 | |
| N Asian | 6 | 2 | |
| N Other | 3 | 1 |
3 White and 2 Other subjects in the Healthy group were Hispanic, while 2 White subjects in the Periodontitis group were Hispanic
Monitoring and treatment protocols
Clinical monitoring
After determination of suitability and obtaining informed consent, subjects entered the study. All subjects were clinically monitored at entry. Clinical measurements were taken at 6 sites per tooth (mesiobuccal, buccal, distobuccal, distolingual, lingual, and mesiolingual) on all teeth excluding third molars (a maximum of 168 sites per subject) as previously described (Haffajee et al. 1983). The clinical parameters were measured in the following order: 1) Gingival Index (Loe & Silness 1963); 2) Plaque Index (Turesky et al. 1970); 3) Pocket Depth (mm); 4) Attachment level (mm); 5) Bleeding on probing (0 or 1); 6) Suppuration (0 or 1)
Pocket depth and attachment level measurements were made to the nearest mm using a North Carolina periodontal probe. Pocket depth and attachment level measurements were measured twice and the average of the pair of measurements was used for analysis. All clinical data were recorded on data sheets and entered into a computer using a prompted data entry program. The first set of supra and subgingival plaque samples were taken prior to the clinical measurements. Samples were taken by the same calibrated examiner at all sampling visits for a given subject.
Scaling and root planing or dental prophylaxis
At the entry visit, after the initial monitoring and sampling, all periodontitis subjects received full mouth scaling and root planning (SRP) at a single visit, using manual curettes and ultrasonic devices, followed by polishing and flossing. Periodontally healthy subjects received a dental prophylaxis using rubber cup and paste, followed by dental flossing. After the initial prophylaxis or SRP, subjects refrained from oral hygiene procedures for 7 days.
Microbiological sample taking and enumeration of organisms
Individual supra and subgingival plaque samples were taken separately from the mesio-buccal aspect of up to 28 teeth in each subject at entry and immediately after tooth cleaning. Thus, up to 28 samples per subject were taken at 2 visits (baseline and immediately after SRP) from 2 locations (supragingival and subgingival) for a total of up to 6160 samples (55 subjects × 28 teeth × 2 visits × 2 locations). Quadrants in each subject were randomly assigned to be sampled at the 1, 2, 4 and 7 day time points. Seven supra and separately 7 subgingival samples were taken at those time points providing up to an additional 3080 samples (55 subjects × 7 teeth × 4 visits × 2 locations).
Supragingival plaque samples were taken separately from each tooth using individual sterile Gracey curettes and evaluated for their content of 41 subgingival species using checkerboard DNA-DNA hybridization (Socransky et al. 1994, Socransky et al. 2004). After removal of any remaining supragingival plaque, subgingival plaque samples were taken separately from each tooth and evaluated as described above. Each sample was placed in individual tubes containing 0.15 ml TE (10 mM Tris-HCL, 0.1 mM EDTA, pH 7.6). 0.15 ml of freshly-prepared 0.5 M NaOH was added. The samples were boiled for 5 min and neutralized using 0.8 ml 5 M ammonium acetate and placed into the extended slots of a Minislot (Immunetics, Cambridge MA) and then concentrated onto a positively charged nylon membrane (Roche, Indianapolis, IN) by vacuum and fixed to the membrane by exposure to ultraviolet light followed by baking at 120°C for 20 min. The counts of the 41 species in each sample were determined using checkerboard DNA-DNA hybridization (Socransky et al. 1994, Socransky et al. 2004).
Data Evaluation
Clinical parameters including plaque index, gingival index, % of sites with bleeding on probing and suppuration as well as mean pocket depth and attachment level were computed for each subject, averaged within subjects and then averaged across subjects in the 2 groups at entry. Differences in clinical parameters between groups were sought using the Mann Whitney test.
Mean microbial counts of 41 test species in individual supragingival and subgingival biofilm samples were assessed separately. The counts for each species were averaged within each subject at each time point and then averaged across subjects in the 2 clinical groups separately. Up to 28 samples were averaged per subject pre tooth-cleaning and post tooth-cleaning, and 7 samples at days 1, 2, 4 and 7. Significance of differences over time (pre-cleaning, post-cleaning, and days 1, 2, 4 and 7) for each species were determined using the Friedman test. Pairwise comparisons between the post-cleaning and days 1, 2, 4 and 7 were determined individually using the Wilcoxon signed ranks test in the periodontally healthy and periodontitis groups separately for both the supra and the subgingival biofilm samples. Differences between clinical groups for each species at each time point were sought using the Mann-Whitney test. The mean values for each species at each time point in the 2 clinical groups were depicted graphically as “microbial profiles” ordered according to the microbial complexes described previously (Socransky et al. 1998).
Results
Changes in supragingival microbial species counts during biofilm re-development
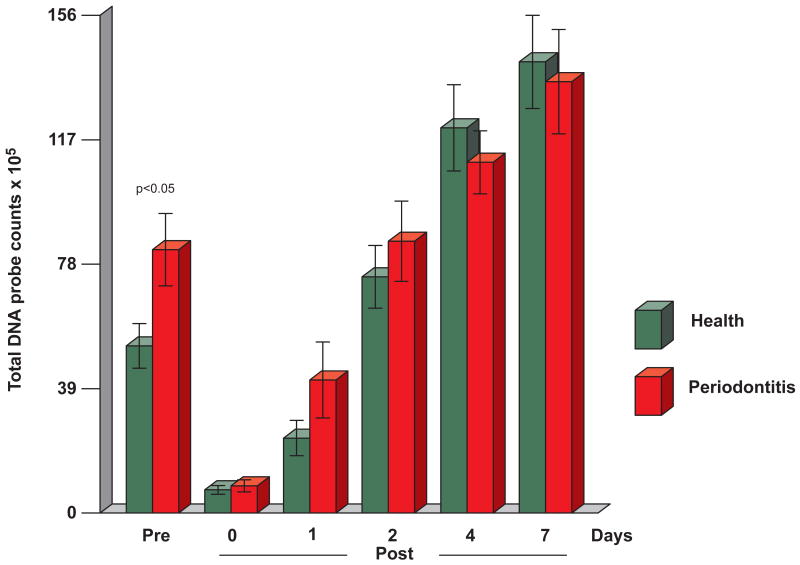
The changes in mean total DNA probe counts of the supragingival plaque samples obtained from periodontitis and periodontally healthy subjects are presented in Fig. 1. There was a significant difference in mean total DNA probe counts at baseline between periodontitis and periodontally healthy subjects. Mean (× 105, ± SEM) total supragingival DNA probe counts were 50.2±6.6, and 84.1±12.4 in healthy and periodontitis subjects respectively (p < 0.05). Mean total DNA probe counts exceeded baseline levels at day two in both periodontally healthy subjects and periodontitis subjects and a steady increase in total counts was observed until day 7. Differences between clinical groups in mean total microbial counts immediately after prophylaxis and at 1, 2, 4 and 7 days were not statistically significant. Interestingly, mean total counts in healthy subjects were greater than in periodontitis subjects at days 4 and 7. Cumulative plots of mean counts of the individual species in periodontally healthy and diseased subjects were remarkably similar other than higher total mean species counts in the periodontitis group at baseline (Fig. 2). Most species surpassed entry levels by day 2. A sharp increase in mean counts of species was observed in periodontally healthy subjects from 1 - 4 days and in periodontally diseased subjects from immediately post-cleaning to 2 days. Mean increases in species counts in both groups slowed thereafter.
Fig. 1.
Mean total DNA probe counts (×105, ± SEM) of supragingival biofilm samples taken at entry (pre-cleaning), immediately post-cleaning and after 1, 2, 4 and 7 days of biofilm development from 38 periodontally healthy subjects and 17 subjects with periodontitis. Total counts from each sample site were averaged in each subject at each time point and then averaged across subjects at each time point separately in each clinical group. Subjects did not perform oral hygiene procedures during the 7 days of the study. Significance of differences between clinical groups at each time point was determined using the Mann Whitney test. Mean total DNA probe counts differed significantly over time in each group (Friedman test, p < 0.001). The bars represent mean total DNA probe counts and the whiskers the SEM at each time point.
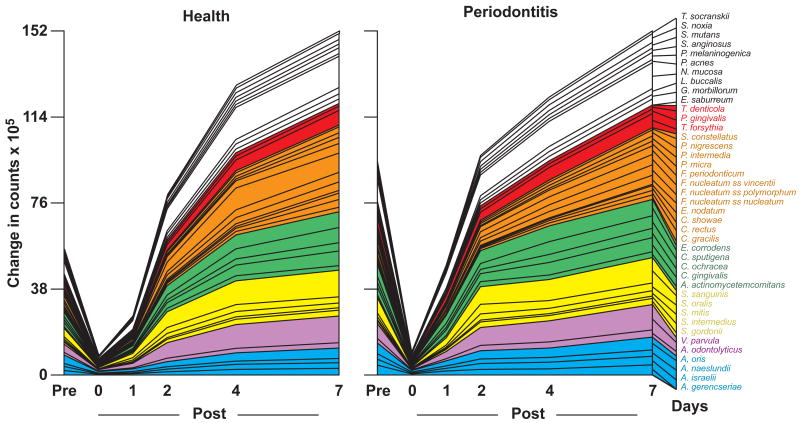
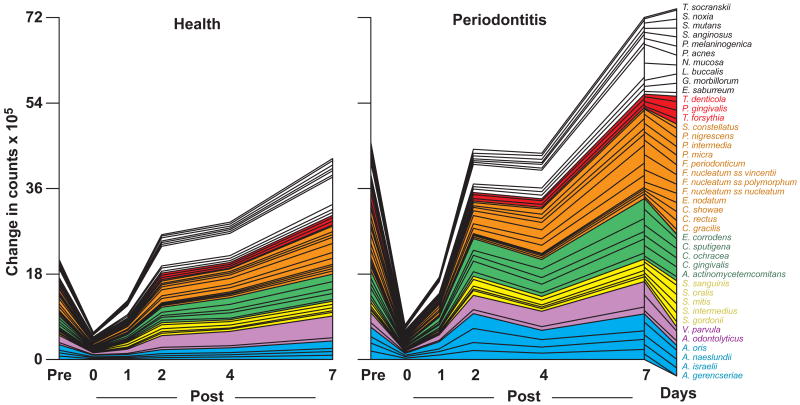
Fig. 2.
Cumulative mean counts (× 105) of 41 bacterial species in supragingival samples taken from 38 periodontally healthy and 17 subjects with periodontitis prior to professional removal of the dental biofilms, immediately after cleaning, and after 1, 2, 4 and 7 days of re-development. The subjects refrained from oral hygiene procedures for the 7 day test period. Samples were removed from the mesio-buccal aspect of each tooth (excluding third molars) pre-cleaning and immediately post-cleaning. In addition, supragingival samples were obtained from up to 7 teeth in randomly selected quadrants at 1, 2, 4 and 7 days after tooth cleaning. All samples were individually analyzed for their content of 41 taxa using checkerboard DNA-DNA hybridization. Species counts in the samples were averaged within each subject at each time point and then averaged across subjects in the 2 clinical groups. The plots present the cumulative mean values at each time point in each clinical group. The species were ordered and color-coded according to previously described microbial complexes (Socransky et al. 1998).
Mean counts (×105) for 41 individual bacterial species in supragingival plaque samples at different time points in the periodontally healthy and diseased subjects are shown in Fig. 3. There was a significant difference over time in mean counts of all the test species, from pre-therapy to 7 days (Friedman test, p<0.001). In addition, there was a significant difference over time for all species from the post-cleaning samples to 7 days (Friedman test, p<0.001), in both clinical groups.
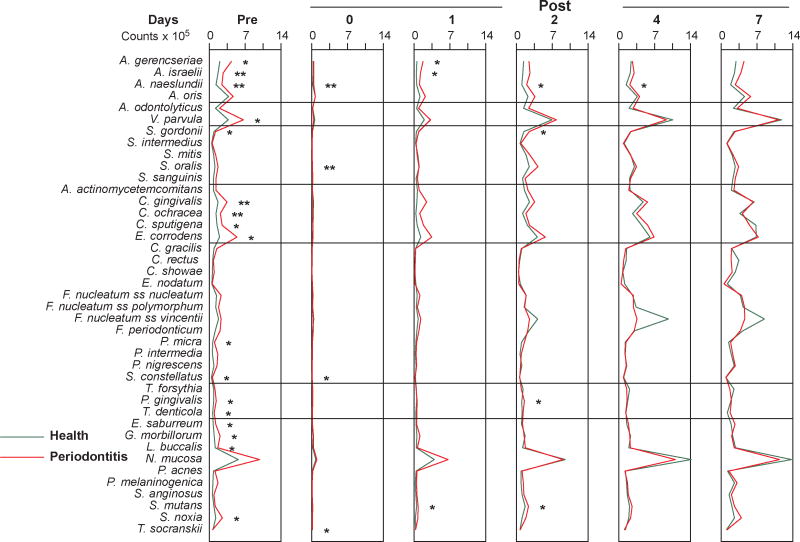
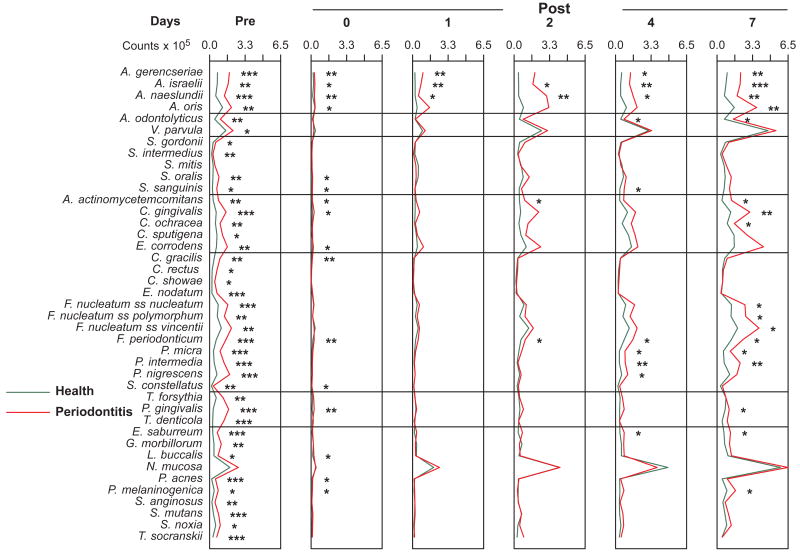
Fig. 3.
Mean counts (×105) of 41 bacterial species in supragingival samples taken from 38 periodontally healthy and 17 subjects with periodontitis prior to professional removal of the dental biofilms, immediately after cleaning, and after 1, 2, 4 and 7 days of biofilm re-development. The subjects refrained from oral hygiene procedures for the 7 day test period. Supragingival biofilm samples were removed from the mesio-buccal aspect of all teeth (excluding third molars) pre-cleaning and immediately post-cleaning. Supragingival samples were also obtained from up to 7 teeth in randomly selected quadrants at 1, 2, 4 and 7 days after tooth cleaning. All samples were individually analyzed for their content of 41 taxa using checkerboard DNA-DNA hybridization. For analysis, species counts in the samples were averaged within each subject at each time point and then averaged across subjects in the 2 clinical groups. Significance of differences in mean species counts between groups at each time point was determined using the Mann Whitney test; * p < 0.05, ** p < 0.01, *** p < 0.001. The species were ordered according to previously described microbial complexes (Socransky et al. 1998).
Pairwise comparisons between post-cleaning samples and each of the time points (i.e., 1, 2, 4, 7 days) indicated significant differences for all species when comparing the post-cleaning samples with those taken at day 2, day 4 or day 7 (Wilcoxon signed ranks test, p<0.001) in both clinical groups. When the post cleaning samples were compared with the samples collected at day 1 for the periodontally healthy subjects, A. gerencseriae, A. israelii, A. odontolyticus, C. sputigena, E. nodatum F. nucleatum ss polymorphum, F. nucleatum ss vincentii, F. periodonticum, P. nigrescens, T.denticola and T. socranskii did not differ significantly (p>0.05). For the periodontitis group, C. rectus, E. nodatum, P. intermedia and S. constellatus did not differ significantly from the post-cleaning visit to 1 day (Wilcoxon signed ranks test, p>0.05). At entry, counts of 17/41 taxa were significantly elevated in the periodontitis group, but few significant differences were observed during the observation period and none were present at day 7. Mean counts of individual species were quite similar post-prophylaxis and at 1, 2, 4 and 7 days in both groups. On day 1, “early colonizers” could be observed, including Veillonella parvula, Neisseria mucosa and A. oris in both groups. At the same time point, the periodontitis group exhibited somewhat increased counts of Capnocytophaga gingivalis, Capnocytophaga sputigena and Eikenella corrodens. On day 2, the increase observed at day 1 became more evident. Further, counts of Fusobacterium nucleatum ss vincentii (particularly in the periodontitis group) increased in both groups. On days 4 and 7, Actinomyces species reached entry levels; and the trends observed earlier became even more pronounced.
Changes in subgingival microbial species counts during biofilm re-development
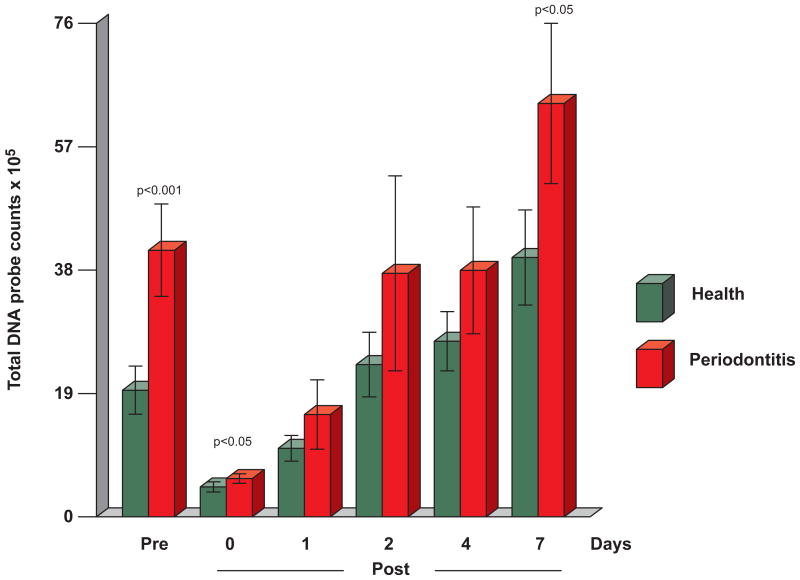
Mean (× 105, ± SEM) total subgingival DNA probe counts in healthy and periodontitis subjects respectively were 19.4±3.7, and 40.9±7.1 (p < 0.001) on entry and 4.6±0.8 and 5.9±0.7 (p < 0.05) immediately post-cleaning (Fig. 4). Although mean total DNA probe counts were consistently lower in samples from the periodontally healthy subjects than the periodontitis subjects at subsequent time points, the differences were statistically significant only at 7 days (39.8±7.3 vs 63.4±12.3, p< 0.05). In the periodontally healthy group, mean total counts surpassed entry levels at day 2 and exhibited gradual increases until day 7. In the periodontitis patients, a slower increase was observed, as levels seemed to plateau at days 2 and 4 and increase at day 7, when they surpassed entry levels.
Fig. 4.
Mean total DNA probe counts (×105, ± SEM) of subgingival biofilm samples taken pre-cleaning, immediately post-cleaning and after 1, 2, 4 and 7 days of biofilm development. Computation of counts and data analysis was performed as described in Fig. 1.
Cumulative mean species counts (Fig. 5) showed a rapid increase in individual mean species counts particularly for purple, green and orange complex taxa as well as N. mucosa in periodontally healthy subjects. In this group, mean total species counts reached or exceeded pre-therapy levels by 2 days. In the periodontitis group, this occurred only at day 7 (Figs. 4 and 5). These increases were more marked between days 1 and 2 and between days 4 and 7, suggesting that there were 2 waves of colonization. There was a sharp increase in counts of all taxa from day 1 to 2, then a plateau or decrease for a number of species, followed by a sharp increase in the counts of all taxa, except for Actinomyces naeslundii, Actinomyces gerencseriae and Actinomyces israelii.
Fig. 5.
Cumulative mean counts (× 105) of 41 bacterial species in subgingival samples taken from 38 periodontally healthy and 17 subjects with periodontitis prior to professional removal of the dental biofilms, immediately after cleaning, and after 1, 2, 4 and 7 days of re-development. Calculation of species counts and description of the plot were as described in Fig. 2.
Mean counts of the 41 enumerated bacterial species in subgingival plaque samples at the different time points in the periodontally healthy and diseased subjects are shown in Fig. 6. There was a significant difference over time in mean counts of all test species in both clinical groups, from pre-therapy to 7 days (Friedman test, p<0.001) and from the post-cleaning samples to 7 days (Friedman test, p<0.001). Pairwise comparisons between post-cleaning samples and each time point (i.e., 1, 2, 4 and 7 days) were performed. Multiple species did not differ significantly in the subgingival samples from the periodontally healthy group when the post-cleaning samples were compared with the samples from day 1. Species that did not differ significantly included A. gerencseriae, A. israelii, A. naeslundii, A. odontolyticus,, C. gingivalis, C. gracilis, E. nodatum, F. nucleatum ss nucleatum, P. intermedia, P. nigrescens, T. forsythia, T. denticola, E. saburreum, S. mutans, S. noxia and T. socranskii (Wilcoxon signed ranks test, p>0.05). For the periodontitis group, the only significant differences occurred for V. parvula and S. mitis (p<0.05). Most species in both clinical groups differed significantly from the post-cleaning samples to the day 2 samples. The exceptions were A. israelii in the periodontally healthy group and C. gracilis, C. rectus, E. nodatum, T. forsythia, T. denticola, P. melaninogenica and T. socranskii in the periodontitis group (p>0.05). From the post-cleaning samples to day 4, the mean counts of both E. nodatum and P. gingivalis did not differed significantly in the periodontitis subjects. The same comparison in the periodontally healthy group revealed that the mean counts of all species were significantly different. Pairwise comparisons between post cleaning samples and day 7 samples indicated significant changes for all species tested in both clinical groups (p<0.001)
Fig. 6.
Mean counts (× 105) of 41 bacterial species in subgingival samples taken from 38 periodontally healthy and 17 subjects with periodontitis prior to professional removal of the dental biofilms, immediately after cleaning, and after 1, 2, 4 and 7 days of biofilm re-development. Computation of counts and data analysis was performed as described in Fig. 3.
Mean counts of 39/41 species were significantly higher in the periodontitis than in the periodontally healthy subjects at entry. Mean counts of Actinomyces species were significantly higher in samples from the periodontitis group than the periodontally healthy group before and at most time points post-cleaning (Fig. 6). Interestingly, even at “0”days, i.e., immediately after cleaning, the test species could be quantified and levels of 16/41 taxa were significantly higher in the periodontitis group. At day 1, the most prominent taxa were V. parvula and N. mucosa in both groups, and A. oris in the periodontitis group. These trends became more evident on day 2, when counts of yellow and green complex species became elevated, as well as Fusobacterium nucleatum ss nucleatum and F. nucleatum ss. vincentii, in both groups. The major differences observed between groups resided in the higher levels of Actinomyces species, Capnocytophaga ochracea, C. gingivalis and E. corrodens in the periodontitis group.
By day 7, all taxa surpassed entry levels in both groups and 17/41 taxa exhibited significantly higher mean counts in samples from the periodontitis than the periodontally healthy subjects. Notably increased over time in both clinical groups were the mean counts of V. parvula, C. gingivalis, E. corrodens, Fusobacterium nucleatum subspecies, Prevotella melaninogenica and N. mucosa. The significant differences detected in mean counts between groups included Actinomyces species, green and orange complexes, as well as P. gingivalis. These increases started at day 2 and were clear at 7, when they were more evident in the periodontitis than in the periodontally healthy group. Red complex species were reduced immediately after cleaning in both groups and remained at low levels until day 7, when counts of P. gingivalis increased slightly in the periodontitis group, at day 7, although they remained at lower mean levels than at entry.
Discussion
The purpose of the present investigation was to examine the microbial shifts during the re-colonization of supra and subgingival tooth surfaces after professional tooth cleaning in the absence of oral hygiene. Since plaque re-colonization might have been affected by sampling, quadrants were randomly assigned to be sampled at the 1, 2, 4 and 7 day test points. For this reason, re-development could be determined only once per tooth, and only 7 of 28 teeth were sampled at each time point in each subject.
Development of supragingival biofilms
The significantly higher baseline mean bacterial counts of supragingival bacteria in periodontitis subjects than in periodontally healthy subjects (50.2±6.6 and 84.1±12.4 ×105) was similar to the results reported previously (Ximenez-Fyvie et al. 2000) (72.1±11.1 and 132.7±17.5 ×105, respectively) and it was in accord with the notion that more plaque would be present on coronal surfaces of teeth in subjects with periodontitis than in periodontally healthy individuals. The prophylaxis and SRP procedures could not completely eliminate the test species. Quantifiable levels of the taxa assessed could be observed at day “0”. This finding is in accord with results from previous studies.(Ramberg et al. 2003, Li et al. 2004) and suggest that cleaning measures can be further improved.
Our data demonstrated that re-colonization of species on the supragingival surfaces was quite similar in subjects with periodontitis or periodontal health. As demonstrated in previous investigations, (Ramberg et al. 2003, Simonsson et al. 1987b, Simonsson et al. 1987a), total supragingival counts reached pre-cleaning levels in 2 days, indicating that recolonization is an extremely rapid process. Certain species appeared to flourish during the 7 day recolonization phase in both clinical groups in supragingival biofilm. These included V. parvula, C. gingivalis, C. sputigena, E. corrodens, N. mucosa, and F. nucleatum ss vincentii (particularly in the periodontitis group) (Fig. 3). The increases in these species is in accord with previous reports (Ritz 1967, Zee et al. 1996a, Ramberg et al. 2003). Li et al. (2004) also showed similar results in a study that examined the very early stages of supragingival plaque development in 15 periodontally healthy subjects using the checkerboard DNA-DNA hybridization technique. Supragingival plaque samples were taken at 0, 2, 4, and 6 hr after cleaning. In that study, it was demonstrated that even though there was selectivity in the initial adhesion of different species, some increased rapidly after adhesion. Actinomyces oris was observed to increase from 0 to 2 hr but remained essentially constant to 6 hr. In our study, Actinomyces levels surpassed baseline values at day 2 and showed a modest increase at the remaining time points, which was also in accord with Ramberg et al. (2003). Li et al. (2004) observed that the major early increases that were observed during biofilm development were for Streptococcus mitis and Streptococcus oralis, which increased markedly up to 6 hr. Other species remained at negligible or low levels during the 6 hr time period. S. mitis and S. oralis also increased at the early time points of the present study. However, other taxa increased at the earliest post-cleaning time points in the present study. Major early increases occurred at Day 1 in the counts of V. parvula and N. mucosa, in both clinical groups. The major increase in Neisseria species that was observed in the present investigation has also been observed using cultural techniques (Ritz 1967). The difference in results from the Li et al. (2004) study may have been due to time of sampling (24 rather than 6 hours) and sample location (mesial rather than facial surfaces of the teeth).
Development of subgingival biofilms
The significantly higher baseline mean bacterial counts of subgingival bacteria in periodontitis subjects than in periodontally healthy subjects (19.4±3.7, and 40.9±7.1 ×105) was in accord with results reported by Sharawy et al. (1966)(16 mg and 84 mg wet weight) and Ximenez-Fyvie et al. (2000) (22.1±6.6 and 100.3±18.4 ×105). This was likely due to the greater number of deeper periodontal pockets in the periodontitis subjects than in the periodontally healthy subjects.
Both supra and subgingival samples exhibited a marked increase in biofilm species counts from 1 to 2 days; a plateau in counts between two and four days; and then a sharp rise in mean species counts between 4 and 7 days. It is not clear why there appeared to be these two phases in subgingival biofilm re-development. It might be speculated that the 2 phases represented a change in the environment brought about by the species that increased in the first wave. The proliferation of certain taxa might have led to changes in the levels of oxygen, availability of nutrients or pH, which might no longer be conducive to the growth of those species, but facilitated the increase of others This pattern suggests the occurrence of autogenic microbial species succession (Socransky & Haffajee 2005).
While redevelopment of biofilms was quite similar in periodontitis and periodontally healthy individuals for the supragingival plaque samples, there were quite marked differences in the redevelopment of subgingival biofilms. As expected, at baseline, the mean total DNA probe counts and counts of 39 of 41 species were significantly higher in the subgingival samples obtained from periodontitis subjects than in the samples obtained from periodontally healthy subjects. Unlike the supragingival environment, the re-development of biofilm in the subgingival area was more rapid in the periodontitis than in the periodontally healthy subjects. Notable was the modest increase in Actinomyces species particularly in the subjects with periodontitis. In addition, green and orange complex species such as C. gingivalis, E. corrodens, Fusobacterium subspecies and Prevotella intermedia increased more profoundly in the periodontitis than in the periodontally healthy subjects. This difference could be observed as early as day 2. To a lesser extent, counts of P. gingivalis also showed an increase over time in this group.
Differences in the development of supra- and subgingival biofilms
The present investigation showed that Streptococcus (yellow complex) species returned to baseline levels by 2 days in periodontitis and healthy subjects in both supra- and subgingival biofilms. In supragingival samples, their levels showed a modest increase until day 7. In subgingival biofilms these showed a plateau and a decrease in periodontally healthy and periodontitis subjects, respectively, from day 2 to 4, followed by an increase at day 7. The early increase may have been due to the re-adsorption of detached species in the oral cavity after cleaning procedures (Li et al. 2004).
The data of the present investigation also showed that, initial colonization appeared to involve members of the yellow, green, and purple complexes along with Actinomyces species (Figs. 3 and 6). Counts of Actinomyces species were close to pre-cleaning levels at day 7 in supragingival, whereas they increased to about or slightly above entry levels in subgingival plaque in both clinical groups. These findings are similar to those reported by Ramberg et al. (2003) but in contrast with those reported by Zee et al. (1996a). The latter investigators used predominant cultivable microbiota techniques to examine 1 pooled plaque sample from each of 5 “rapid” plaque-forming subjects and 6 “slow” plaque-forming subjects at 1, 2, 7 and 14 days. They indicated that Actinomyces species rose from a mean of 10 and 5% in “slow” and “rapid” groups respectively to 30 and 15% at 14 days. They also showed that Capnocytophaga species were low in proportion at 1 day but increased at 3 to 14 days. This was in accord with the findings of the present investigation in which C. gingivalis, C. ochracea and C. sputigena were low in counts at day 1 but increased significantly in levels, exceeding entry levels at day 4.
Even though the total DNA probe counts of subgingival biofilm samples exceeded baseline values at day 2 in both healthy and periodontitis subjects, there were striking differences between health and periodontitis in the patterns of recolonization. In periodontally healthy subjects, the counts of most species increased slowly over time, reaching or exceeding baseline values by 2 days. The reason for the difference between health and disease is not known, but one might speculate that the greater gingival crevice fluid (GCF) flow in the periodontitis subjects may have favored subgingival biofilm development. Alternatively, there may have been a greater reservoir of bacterial cells that were missed in the scaling of deep pockets or remaining in reservoirs within host cells or dentinal tubules.
The findings presented in this manuscript should be seen in light of the limitations of the study. The necessity of taking samples from randomly selected quadrants at days 1, 2, 4 and 7 prevented following biofilm changes at individual teeth. The 7 day duration of the study did not allow us to follow biofilm changes to a microbial composition that was similar to that of mature plaque. Also, the lack of a comparison group that continued self performed oral hygiene prevented determination of the effect of oral hygiene procedures on microbial shifts during re-colonization.
In summary, the hypothesis that biofilm redevelopment would be more rapid in periodontitis than in periodontally healthy subjects was rejected for supragingival biofilms but could not be rejected for subgingival biofilm re-development. The hypothesis that the shift in levels of individual taxa over time would differ from periodontally healthy to periodontally diseased subjects could not be rejected in that Actinomyces, green and orange complex species returned much more rapidly in subgingival biofilm samples from periodontitis than periodontally healthy individuals. The hypothesis that the re-growth of specific periodontal pathogens such as P. gingivalis, T. forsythia and T. denticola would be slower than the re-growth of other species was not rejected. Finally, the re-development of oral biofilm on dental tissues was so rapid that total DNA probe counts of many species exceeded baseline values at day 2.
Clinical Relevance.
Scientific Rationale: The study of dental biofilm re-development in the absence of oral hygiene in periodontal health and disease can show the dynamics of colonization by pathogens and host-compatible bacteria.
Principal findings: Supra and subgingival counts reached or exceeded baseline values after day 2. Differences between groups in supragingival counts occurred for 17/41 species at entry, 0 at day 7, and in subgingival plaque for 39/41 taxa at entry and 17/41 at day 7. Subgingival counts were greater for Actinomyces, green and orange complex species.
Practical implications: Microbial shifts during biofilm re-development can indicate critical periods for prevention and intervention.
Acknowledgments
Source of Funding Statement: Supported by NIDCR grants DE14368 and T32-DE-07327 (F.R.T.).
Footnotes
Conflicts of Interest: The authors declare that they have no conflict of interests.
References
- Bernimoulin JP. Recent concepts in plaque formation. Journal of Clinical Periodontogy. 2003;30 5:7–9. doi: 10.1034/j.1600-051x.30.s5.3.x. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Hay DI, Cisar JO, Clark WB. Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infection and Immunity. 1988;56:2990–2993. doi: 10.1128/iai.56.11.2990-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Hay DI, Schlesinger DH. Delineation of a segment of adsorbed salivary acidic proline-rich proteins which promotes adhesion of Streptococcus gordonii to apatitic surfaces. Infection and Immunity. 1991;59:2948–2954. doi: 10.1128/iai.59.9.2948-2954.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Nygaard M. Interbacterial aggregation of plaque bacteria. Archives of Oral Biology. 1970;15:1397–1400. doi: 10.1016/0003-9969(70)90031-2. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS, Goodson JM. Comparison of different data analyses for detecting changes in attachment level. Journal of Clinical Periodontology. 1983;10:298–310. doi: 10.1111/j.1600-051x.1983.tb01278.x. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Ganeshkumar N, Cassels FJ, Hughes CV. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 1993;7:406–413. doi: 10.1096/fasebj.7.5.8462782. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. Journal of Bacteriology. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Helmerhorst EJ, Leone CW, Troxler RF, Yaskell T, Haffajee AD, Socransky SS, Oppenheim FG. Identification of early microbial colonizers in human dental biofilm. Journal of Applied Microbiology. 2004;97:1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- Listgarten MA. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. Journal of Periodontology. 1976;47:1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- Listgarten MA. Formation of dental plaque and other oral biofilms. In: Nweman H, Wilson M, editors. Dental plaque revisited. Cardiff: Bioline; 1999. pp. 187–210. [Google Scholar]
- Listgarten MA, Mayo H, Tremblay R. Development of dental plaque on epoxy resin crowns in man. A light and electron microscopic study. Journal of Periodontology. 1975;46:10–26. doi: 10.1902/jop.1975.46.1.10. [DOI] [PubMed] [Google Scholar]
- Loe H, Silness J. Periodontal disease in pregnancy. I. Prevalence and Severity. Acta Odontologica Scandinavica. 1963;21:533–551. doi: 10.3109/00016356309011240. [DOI] [PubMed] [Google Scholar]
- Ramberg P, Sekino S, Uzel NG, Socransky S, Lindhe J. Bacterial colonization during de novo plaque formation. Journal of Clinical Periodontology. 2003;30:990–995. doi: 10.1034/j.1600-051x.2003.00419.x. [DOI] [PubMed] [Google Scholar]
- Ritz HL. Microbial population shifts in developing human dental plaque. Archives of Oral Biology. 1967;12:1561–1568. doi: 10.1016/0003-9969(67)90190-2. [DOI] [PubMed] [Google Scholar]
- Sharawy AM, Sabharwal K, Socransky SS, Lobene RR. A quantitative study of plaque and calculus formation in normal and periodontally involved mouths. Journal of Periodontology. 1966;37:495–501. doi: 10.1902/jop.1966.37.6.495. [DOI] [PubMed] [Google Scholar]
- Simonsson T, Edwardsson S, Glantz PO. Biophysical and microbiologic studies of “heavy” and “light” plaque formers. Scandinavian Journal of Dental Research. 1987a;95:43–48. [PubMed] [Google Scholar]
- Simonsson T, Ronstrom A, Rundegren J, Birkhed D. Rate of plaque formation--some clinical and biochemical characteristics of “heavy” and “light” plaque formers. Scandinavian Journal of Dental Research. 1987b;95:97–103. doi: 10.1111/j.1600-0722.1987.tb01814.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontology 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. Journal of Clinical Periodontology. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Smith C, Martin L, Haffajee JA, Uzel NG, Goodson JM. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiology and Immunology. 2004;19:352–362. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Manganiello AD, Propas D, Oram V, van Houte J. Bacteriological studies of developing supragingival dental plaque. Journal of Periodontal Research. 1977;12:90–106. doi: 10.1111/j.1600-0765.1977.tb00112.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin A. “Checkerboard” DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- Stoodley P, Boyle JD, Lappin-Scott HM. Biofilm structure and behaviour; influence of hydrodynamics and nutrients. In: Newman HN, Wilson M, editors. Dental Plaque Revisited. Cardiff: Bioline; 1999. pp. 63–72. [Google Scholar]
- Stromberg N, Boren T. Actinomyces tissue specificity may depend on differences in receptor specificity for GalNAc beta-containing glycoconjugates. I. Infection and Immunity. 1992;60:3268–3267. doi: 10.1128/iai.60.8.3268-3277.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turesky S, Gilmore ND, Glickman I. Reduced plaque formation by the chloromethyl analogue of victamine C. Journal of Periodontology. 1970;41:41–43. doi: 10.1902/jop.1970.41.41.41. [DOI] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. Journal of Clinical Periodontology. 2000;27:648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- Zee KY, Samaranayake LP, Attstrom R. Predominant cultivable supragingival plaque in Chinese “rapid” and “slow” plaque formers. Journal of Clinical Periodontology. 1996a;23:1025–1031. doi: 10.1111/j.1600-051x.1996.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Zee KY, Samaranayake LP, Attstrom R, Davies WI. Predominant cultivable microflora of supragingival dental plaque in Chinese individuals. Archives of Oral Biology. 1996b;41:647–653. doi: 10.1016/s0003-9969(96)00065-9. [DOI] [PubMed] [Google Scholar]