Abstract
Familial hypertrophic cardiomyopathy (FHC) is an inherited disorder affecting roughly one in 500 people. Its hallmark is abnormal thickening of the ventricular wall, leading to serious complications that include heart failure and sudden cardiac death. Treatment is complicated by variation in the severity, symptoms and risks for sudden death within the patient population. Nearly all of the genetic lesions associated with FHC occur in genes encoding sarcomeric proteins, indicating that defects in cardiac muscle contraction underlie the condition. Detailed biophysical data are increasingly available for computational analyses that could be used to predict heart phenotypes based on genotype. These models must integrate the dynamic processes occurring in cardiac cells with properties of myocardial tissue, heart geometry and haemodynamic load in order to predict strain and stress in the ventricular walls and overall pump function. Recent advances have increased the biophysical detail in these models at the myofilament level, which will allow properties of FHC-linked mutant proteins to be accurately represented in simulations of whole heart function. The short-term impact of these models will be detailed descriptions of contractile dysfunction and altered myocardial strain patterns at the earliest stages of the disease—predictions that could be validated in genetically modified animals. Long term, these multi-scale models have the potential to improve clinical management of FHC through genotype-based risk stratification and personalized therapy.
Keywords: hypertrophic cardiomyopathy, multi-scale, cardiac muscle
1. Introduction
Efforts to model cardiac function in silico are a prototype for interdisciplinary science, combining techniques from the fields of engineering, computer science, medical imaging, molecular biology, biophysics and physiology among others. Computational models are being applied at many different biological scales to further understanding of the heart in normal and diseased conditions, with the prospect of greatly accelerating progress on both fronts (reviewed in [1]). The goal of this review is to assess the progress and potential of current experimental and computational techniques towards unravelling the relationship between genotype and phenotype in a specific class of cardiomyopathy-causing mutations.
Unexplained hypertrophic cardiomyopathy (HCM) is present in one out of every 500 adults in the USA [2], and in the majority of cases it can be traced to genetic factors [3]. The inherited form of the disease, known as familial hypertrophic cardiomyopathy (FHC), is associated with increased left ventricular wall thickness, myocardial fibrosis, myocyte disarray and increased risk of sudden cardiac death. No cure exists for the condition, and treatments to alleviate symptoms are limited. The use of implantable cardioverter–defibrillators against the threat of sudden cardiac death has successfully reduced mortality among FHC patients, as have surgical procedures that remove excess myocardium from the intraventricular septum [4]. Still, these patients will require long-term management of remaining disease complications.
Genetic linkage studies, the first appearing two decades ago [5], have identified mutations to sarcomeric genes as the primary cause of FHC [6]. These discoveries have answered some basic questions about FHC, but have also raised new ones that are the subject of intensive research efforts [3]. The extent and pattern of hypertrophy, risk for sudden cardiac death, age of symptom onset and overall prognosis are highly variable in the patient population. This phenotypic diversity seems generally explained by the large number (greater than 500) of individual mutations documented in the current medical literature [3], and suggests that predictions of disease phenotypes may be possible for specific genotypes. However, the identification of a substantial number of apparently asymptomatic, gene-positive individuals poses a new challenge to that idea and indicates that phenotypes of advanced FHC are sensitive to multiple factors [7]. In the light of these findings, Tardiff [8, p. 765] has proposed in a recent review that ‘a renewed focus on the most proximal events in both the molecular and clinical pathogenesis of [FHC] will be necessary to achieve the central goal of using genotype information to manage affected patients’.
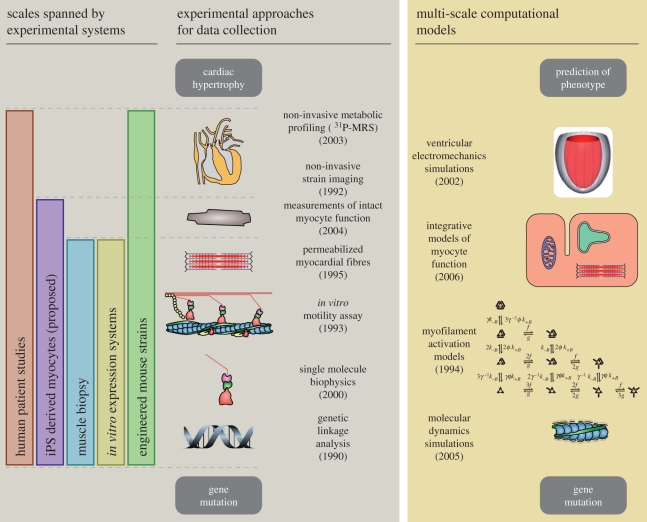
The implicit hypothesis in current FHC research is that hypertrophy, regardless of its advanced form, is the result of altered acute function at the level of the cardiac sarcomere. Experimental and computational tools exist at present that together could be used to predict this kind of proximal, pre-hypertrophic phenotype based on the properties of mutant proteins (figure 1). When applied to animal models of FHC, bridging genotype and phenotype in this way would enable much more detailed descriptions of the disease process, and the generation of focused, testable hypotheses. Further on, these tools could provide a quantitative means of stratifying risk among patients and assist in clinical decision-making.
Figure 1.
Diagram of key experimental methods and potential computational tools for studying familial hypertrophic cardiomyopathy (FHC). Approximate dates show when each experimental approach was first applied to FHC research. Multi-scale computational approaches have not yet been applied to the study of FHC, but many of the necessary tools to do so have emerged in recent years (dates reflect the publication of applicable modelling techniques). [9] P-MRS refers to phosphorus-31 magnetic resonance spectroscopy [10].
In the meantime, many groups continue to pursue FHC research using more traditional approaches (see [6,8,11] for recent reviews). These include genome-wide association studies in humans to identify new mutations, mouse lines engineered to have FHC-linked mutations and in vitro studies of altered sarcomeric proteins. A new approach being explored at present is to create induced pluripotent stem cells from human somatic tissues that can then be differentiated into heart cells [12]. Myocytes derived from patients harbouring FHC mutations could then be used for functional cell-scale assays or as a means of obtaining mutant proteins for molecular studies. Genetically engineered mice have the advantage of allowing systematic molecular, structural and functional studies at several scales, but, in practice, the integration of these data is usually done qualitatively. Furthermore, secondary effects such as the development of heart failure, multi-genic interactions or epigenetic factors can confound phenotypes. As multi-scale models achieve their full potential, we anticipate that they will strengthen current and emerging approaches by integrating data quantitatively and assisting in efforts to account for secondary factors.
2. Sarcomeric proteins
Approximately 70 per cent of inherited forms of HCM can be linked to genes encoding proteins found in the sarcomere [3]. The sarcomeres of cardiac muscle cells are responsible for producing the contractile force, and are composed of two overlapping arrays of protein filaments: thick filaments, containing the motor protein myosin, and thin filaments, consisting of polymerized actin decorated with the regulatory proteins troponin and tropomyosin (figure 2). Contraction is initiated when Ca2+ binds to the troponin complex, triggering a series of allosteric signalling events that move tropomyosin on the surface of the actin filament to expose binding sites for myosin heads [13]. Myosin cyclically interacts with actin in a process that converts energy in the form of adenosine triphosphate (ATP) into mechanical work, sliding thick and thin filaments past each other and causing the muscle to shorten.
Figure 2.
Schematic of the cardiac sarcomere and main protein constituents. The sarcomere consists of interdigitated arrays of thick and thin filaments. The expanded view labels nine principal components: myosin heavy chain (MHC), essential light chain (ELC), regulatory light chain (RLC), myosin binding protein C (MyBP-C), tropomyosin (Tm), troponin I (TnI), troponin T (TnT), troponin C (TnC) and actin. Cardiomyopathy-linked mutations have been identified in the genes encoding each of these nine proteins, which represent the most common causes of inherited HCM [6]. At rest, force is inhibited by Tm, which blocks the myosin binding site on actin (left most myosin molecule; note that the second myosin head in each case is omitted for clarity). Force production is initiated in the sarcomere when Ca2+ binds to a low-affinity site on TnC (labelled), triggering a shift in the Tm position to expose binding sites (myosin in the centre). Once attached, myosin releases energy obtained from ATP hydrolysis to rotate its lever arm, distending its tether to the thick filament and producing force. Evidence suggests that disease-linked mutations alter the way this system produces force in response to Ca2+.
The first mutations definitively linked with human FHC were found in the gene MYH7, which encodes β-myosin heavy chain (MHC). β-MHC mutations remain the most common cause of inherited HCM [6]. MHC is the largest of the three subunits that form the myosin molecule, and contains domains responsible for actin binding and nucleotide hydrolysis. The other two subunits of myosin, essential light chain (ELC) and regulatory light chain (RLC), associate with and stabilize an α-helix of MHC known as the lever arm. Motion of the lever arm is responsible for force production by myosin [14]. Relatively rare but well-documented FHC mutations are found in the human genes MYL2 and MYL3 encoding ELC and RLC, respectively, suggesting that these proteins play an important functional role [11]. Mutations to the gene MYBPC3, which encodes the thick filament protein myosin binding protein C (MyBP-C), are the second most frequent among individuals with FHC [11]. While the ability of MyBP-C to modulate sarcomere function is well established, the structural and functional details of its regulatory activity have not been fully determined.
Nearly all other known FHC-causing mutations occur in thin filament proteins (see [8] for a review). These include cardiac actin (ACTC), which forms the thin filament and its myosin binding sites. Tropomyosin sterically blocks these binding sites during relaxation, and mutations to the gene for its α isoform (TPM1) are linked to FHC. Cardiac troponin T (TNNT2) anchors the other troponin subunits to tropomyosin, and is thought to stabilize end-to-end overlap of adjacent tropomyosins. Cardiac troponin I (TNNI3) participates directly in Ca2+-dependent regulation of contraction by binding to actin in a way that prevents the movement of tropomyosin [13]. When Ca2+ binds to troponin C (TNNC1), transfer of the inhibitory domains of troponin I from actin to the N-terminal domain of troponin C allows movement of tropomyosin and the formation of actin–myosin crossbridges.
Studies examining the properties and functional consequences of mutant sarcomeric proteins are numerous, and have added substantially to our understanding of genotype–phenotype connections in FHC (see Tardiff [8] and Harris et al. [11] for detailed reviews). Studies on purified proteins having point mutations seen in human FHC patients have been performed, but FHC genes have also been cloned and used to create transgenic and gene-targeted animal models (primarily mice). The resulting data range from measurements on the activity of single molecules (e.g. [15]) to measurements of cardiac function in vivo [16].
Perhaps the most frequently used experiment is to measure the Ca2+ sensitivity of force in skinned myocardial preparations containing mutant proteins (see Bai et al. [17] for a recent example). In these experiments, tissue is treated with a detergent to compromise cell membranes, enabling the Ca2+ concentration around the myofilaments to be set directly by the bathing solution. The myocardial sample is attached between a force transducer and a motor-controlled lever, and steady-state force produced by the preparation can be measured at varying Ca2+ concentrations. The force–Ca2+ relation obtained in this way is typically parametrized by fitting points with the Hill equation,
 |
where Fmax is the force at saturating Ca2+ concentration,  is the Ca2+ concentration at half-maximal force (often referred to as the ‘Ca2+ sensitivity’) and nH is the Hill coefficient.
is the Ca2+ concentration at half-maximal force (often referred to as the ‘Ca2+ sensitivity’) and nH is the Hill coefficient.
A trend emerging from the many steady-state force–Ca2+ relations measured in the presence of mutant sarcomeric proteins is that mutations linked to HCM tend to increase Ca2+ sensitivity of the myofilaments, while the small (but significant) number of mutations linked to dilated cardiomyopathy (DCM) tend to decrease it [8,18]. This result is significant, but the ability of Ca2+ sensitivity to predict phenotype seems limited in that the magnitude of the sensitivity change from baseline does not appear to predict disease severity. Moreover, a single parameter cannot describe the multi-dimensional phenotypic space of FHC, which includes varied patterns of hypertrophy and distinct risks for arrhythmia for different mutations.
Even in a case where Ca2+ sensitivity measurements agree with the general HCM/DCM paradigm, the genotype–phenotype connection is only partially achieved. In such a case, the initial challenge to relate a specific mutation to the type of cardiac remodelling becomes one of relating the mutation to Ca2+ sensitivity. Knowing the amino acid sequence of normal and mutant proteins should provide important clues, and some progress is being made in this regard through the use of molecular dynamics (MD) simulations (see Ertz-Berger et al. [19] and Lorenz & Holmes [20]). MD uses protein structure data to predict the motion of atoms within molecules on picosecond time scales. In one case, MD has been used to predict motion of a critical region of the troponin T (TnT) molecule in the presence of FHC-linked mutations R92W and R92L [19]. Simulations showed that both mutations tended to destabilize helical structures in the protein, increasing flexibility of the molecule relative to the wild-type sequence. At the same time, the degree of flexibility was different between the two mutations. This may explain differences in severity of hypertrophy that are seen between R92W and R92L in both humans and transgenic mice with these mutations.
The advantage of MD simulations is that they enable direct functional predictions based on single amino acid substitutions, but they are limited by time scale and the feasibility of simulating molecular systems large enough to make direct predictions of Ca2+-activated force. Even indirect predictions are difficult owing to remaining questions about the molecular mechanisms of myofilament function; exactly how flexibility of a single region in TnT might change Ca2+ sensitivity is not currently known, for instance. Techniques for MD will undoubtedly improve in the future, and could ultimately overcome these limitations. In the meantime, it may be possible to bridge the gap between molecular behaviour and Ca2+ sensitivity using integrative models of myofilament activation.
3. Integrative myofilament function
Integrative models of myofilament function predict Ca2+ activation on the basis of known structural interactions and conformational states of myofilament proteins (recent examples include [21–23]). While clearly lacking in the molecular detail achievable through MD simulations, these models have the advantage of being able to reproduce functional measurements made in cardiac muscle preparations. In general, these models represent Ca2+ binding to troponin C, activation of the troponin–tropomyosin regulatory switch and crossbridge cycling, with interactions among these processes that are suggested by experimental evidence. Integrative myofilament models could be used to analyse force–Ca2+ relations and other properties in muscle-containing FHC mutant proteins, pointing towards one or more of the simplified processes they represent as the main functional change(s) induced by the mutation. This has the potential of focusing atomistic simulations towards specific structures and interactions among myofilament proteins.
Improvements in these types of myofilament models in recent years have centred around representation of cooperative activation [24] by Ca2+. Cooperativity describes the steep, sigmoidal relation observed between Ca2+ concentration and contractile force under steady-state conditions (represented by the parameter nH in the Hill equation). The molecular origin of striated muscle cooperativity is widely believed to be end-to-end interactions arising between adjacent tropomyosin molecules on the actin thin filament [25,26]. According to the steric blocking model of muscle regulation [13], tropomyosin blocks myosin binding to actin under low Ca2+ conditions. Interactions among adjacent tropomyosins are thought to couple myosin binding sites such that they tend to be exposed in an ‘all-or-nothing’ fashion consistent with steeply cooperative behaviour.
A number of theoretical and computational models have been developed to describe these and other putative mechanisms of myofilament cooperativity under steady-state conditions [27–32]. Others have been formulated for the purpose of predicting myofilament activation during the transient changes in [Ca2+] that occur in the beating heart [9,21,33]. Ideally, these models would be capable of translating measurements of myofilament Ca2+ sensitivity and cooperativity from skinned muscle preparations into their true effects under physiological conditions. In spite of the variety and the number of published myofilament models, this exciting prospect remains largely unrealized.
Our own recent work has led to the formulation of a Markov model of myofilament activation that can simultaneously reproduce the steady-state and dynamic force–Ca2+ relations [23]. The model is based on the three states of the cardiac thin filament regulatory unit originally proposed by McKillop & Geeves [28] and later verified by structural data [34]. When transitions between the three states are considered as dependent on the states of neighbouring regulatory units (through tropomyosin interactions), the system exhibits both steady-state and dynamic aspects of cooperative activation. The model was validated against data collected in skinned cardiac muscle under a variety of conditions, including the addition of NEM-S1 (a soluble myosin sub-fragment that binds actin with high affinity), elevated inorganic phosphate concentration and shortened thin filament length, among others.
The ability of this model to reproduce experiments that use molecular-scale perturbations supports its use in analysing steady-state force–Ca2+ relations in the presence of FHC-linked mutations. A simplified example of this is shown in figure 3a, where the model is used to reproduce a leftward shift and loss of cooperativity in the force–Ca2+ relation seen in the FHC-linked tropomyosin mutation E180G [17]. Both changes were achieved simultaneously by lowering a single parameter, the free energy change associated with nearest-neighbour interactions between tropomyosins. Full details of the model, including parameters, are presented in the electronic supplementary material that accompanies this paper. This cursory observation implicates increased flexibility of tropomyosin or effects on end-to-end tropomyosin interactions as the functional consequence of E180G, rather than a simple alteration in thin filament state equilibrium [17]. Insights such as these could lead to focused MD or coarse-grained molecular model studies with direct tie-ins to higher level function.
Figure 3.
Myofilament models can bridge the gap between steady-state and dynamic force–Ca2+ relations. (a) Qualitative changes in the steady-state force–pCa relation owing to the Tm mutation E180G [17], including an increase in Ca2+ sensitivity and decrease in cooperativity (steepness of the curve) were recapitulated in a myofilament model [23] by reducing the amount of nearest-neighbour Tm coupling (control and E180G Tm are solid and dashed lines, respectively). This parameter change is indicative of either increased Tm flexibility or destabilization of end-to-end binding. Each curve has been normalized by maximum tension to emphasize the difference in Ca2+ sensitivity. (b) The model can also be used to predict the effects of the E180G Tm mutation on twitch dynamics. Using the same parameters as in (b), twitches were elicited in response to an idealized Ca2+ transient (not shown). The simulation suggests that the mutation increases diastolic tension and slows rates of both contraction and relaxation, consistent with known phenotypes [17].
The Markov model of thin filament activation, fitted to skinned muscle data, could also be used to predict the impact of mutant proteins on function in vivo. The model was shown to reproduce behaviour of intact muscle, correctly predicting twitch force in response to measured Ca2+ transients. As an illustration, an isometric twitch was predicted here using the same parameter set that reproduced steady-state Ca2+-activated behaviour for the muscle containing the E180G tropomyosin mutant (figure 3b). The predicted effect, based on the steady-state force–Ca2+ relation, is the prolongation of twitch tension and slower relaxation in consequence of the E180G tropomyosin mutation. One further advantage of the Markov model is computational tractability, meaning that the model can be upwardly integrated into models of cells and even the whole heart without the loss of biophysical detail.
4. Ventricular myocytes
During each heartbeat, the signal to contract reaches cardiac muscle cells in the form of an electrical impulse. At the level of the cell, this electrical excitation causes rapid release of Ca2+ ions into the cytosol from intracellular stores to activate contraction in the sarcomeres. This process from electrical stimulation to force production is known as cardiac excitation–contraction coupling or simply EC coupling [35].
Several cellular structures and a host of related proteins are involved in EC coupling in ventricular myocytes [35]. Ion channels and their accessory proteins at the cell membrane are responsible for detecting and propagating the transient changes in membrane potential that initiate contraction. Changes in membrane potential open L-type Ca2+ channels, which triggers Ca2+ release from the sarcoplasmic reticulum (SR) in a process known as Ca2+-induced Ca2+ release (CICR). Membrane-bound proteins in the SR are responsible for Ca2+ re-uptake from the cytosol, which lowers Ca2+ and causes contraction to end.
It could be argued that the entire process of force generation by the myofilaments forms part of EC coupling. Traditionally, EC coupling has been considered complete at the point Ca2+ binds to troponin C (TnC), but two observations suggest that this view neglects potentially important mechanisms in the behaviour of cardiac muscle. The first is that the Ca2+ affinity of TnC is increased nearly 10-fold by myosin binding to actin [36]. This means that, during a twitch, the capacity of TnC as a buffer of cytosolic Ca2+ cannot be considered constant. A second general observation is that, in numerous cases, modification of myofilament proteins alters the overall Ca2+ sensitivity (and presumably Ca2+ buffering) of the sarcomeres, whether through post-translational modification [37] or cardiomyopathy-linked mutations [17]. The abundance of TnC in the cytosol means that factors modifying myofilament Ca2+ sensitivity have the potential to influence EC coupling in the whole cell, one form of a phenomenon known as mechanoelectric feedback [38]. Hence, an understanding of the relationship between excitation and the force produced by contraction depends critically on the properties of myofilaments.
Integrative ventricular myocyte models that combine representations of electrophysiology, Ca2+ handling and myofilament contraction allow quantitative study of the complex interplay among these processes [39–41]. Computational models of myocyte electrophysiology have evolved over several decades and now include mechanistic descriptions of many ion channels and transporters. The most recent of these are being used to predict the effects of mutated channel proteins on cardiac action potentials [42]. Innovative mathematical approaches developed in the last decade have simultaneously improved the biophysical accuracy and the computational efficiency of Ca2+ handling and CICR models [43]. We recently used a canine EC coupling model containing this improved CICR representation coupled with the contraction model of Rice et al. [22] to investigate sources of electromechanical heterogeneity in endocardial, mid-myocardial and epicardial myocytes [39]. One prediction from that work was that differences in EC coupling alone could not explain more rapid contraction and relaxation rates in epicardial cells. Instead, an increase in the rate of crossbridge cycling consistent with elevated expression of the faster α-MHC isoform was needed to explain experimental measurements. This result was subsequently supported by the discovery of increased α-MHC expression in porcine ventricular epicardium that correlated with faster crossbridge kinetics in skinned epicardial cells [44].
Integrative models of myocyte electromechanics have great potential in the realm of FHC research for several reasons. Many FHC mutations are associated with high risk for lethal arrhythmias [8], and the myocyte is the simplest system in which sarcomeric mutations could exert an effect on electrical activity in the heart. Myocytes are also the simplest experimental preparation in which the effects of FHC mutations on twitch characteristics can be observed. Typically in these experiments, Ca2+ transients and unloaded cell shortening are measured in myocytes isolated from engineered mice expressing FHC-linked mutant proteins. Twitch characteristics gleaned from unloaded shortening measurements are frequently different from wild-type controls, but interpretation of twitch phenotypes based on genotype is complicated by the fact that the Ca2+ transients driving twitch can also be altered in these animals (e.g. the E22K mutation in MYL3 [45]). By combining experiments with quantitative model analysis, the effects of a mutated sarcomeric protein on twitch itself can be separated from those exerted by the Ca2+ transient. For example, our recent model of myofilament activation [23], when coupled with appropriate equations for length and velocity dependence of contraction [41], can be used to quantitatively link measured Ca2+ transients with cell shortening (figure 4; see the electronic supplementary material for additional details). Ca2+-handling differences could be accounted for by fitting model parameters to reproduce measured shortening in response to measured Ca2+ transients. Differences in the fitted contraction model parameters would reveal the nature of functional changes to myofilament activation caused by the mutant protein.
Figure 4.
Myofilament models are capable of reproducing contraction events in living myocytes. (a) A measured Ca2+ transient was used as input to a myofilament activation model coupled with equations representing an internal elastic load to simulated cell shortening. (b) Free parameters were adjusted in the model until predicted shortening (dotted trace) matched the experimentally measured response (solid trace, 0.2% relative error). Rat ventricular myocyte at 25°C.
A biophysically detailed electromechanical myocyte model would allow even more integrative analysis to examine not only altered twitch, but also the basis for alterations to the Ca2+ transient. Ca2+ handling undergoes certain well-known adaptive changes in hypertrophy and heart failure that are not directly dependent on the myofilaments. However, acute changes in Ca2+ handling early in the disease process could be mediated by changes to Ca2+ buffering by TnC caused by mutant proteins. An integrative myocyte model that represents dynamic buffering of Ca2+ by myofilaments containing normal versus mutant proteins would be capable of assessing the feasibility of such a mechanism.
5. Transmural heterogeneity
Any influence that FHC mutations have on cardiac function takes place against the backdrop of naturally occurring spatial variation in expression and composition of proteins in the sarcomere (and elsewhere) that modify EC coupling properties of individual cells [46–48]. Simulations suggest that regional differences in contractile behaviour critically affect mechanics and pump function of the heart [49], and this raises the possibility that FHC mutations could alter or otherwise interact with natural heterogeneities to produce disease pathology. Regional patterns in the morphology and duration of myocyte action potentials were among the first of many heterogeneous properties described in the past 20 years [50]. Since then, differences in ion channel current density [51], Ca2+ transients [48,52,53], myosin isoform expression [44,54–56], myofilament protein phosphorylation [57,58] and unloaded cell shortening [48,52] have been noted. In some cases, it has been possible to correlate variations at the molecular level to functional differences among myocytes isolated from the respective myocardial regions [44,46,59]; however, much remains to be learned about the practical implications of observed heterogeneities, even at the level of individual cells.
The same may be said of the role these heterogeneities play in the function of the whole heart. For instance, cells in the outer, epicardial region of the left ventricle tend to have shorter action potentials and Ca2+ transients, and contract more rapidly than those in the inner or endocardial region [48,52]. It has been proposed that this type of heterogeneity coordinates contraction in the heart [48,60]. The hypothesis is that faster contraction in epicardial cells would allow them to ‘catch up’ to endocardial cells, which are activated earlier during the cardiac cycle. However, a computational model predicts that heterogeneities affect ventricular deformation throughout most of systole, not just in its opening moments (figure 5) [49].
Figure 5.
A three-dimensional model can be used to predict the effects of cell-type distribution on ventricular mechanics and function. The simulation data rendered in this figure were generated during a previous study (see [49] for complete details). (a) This view of the model shows the ventricular geometry and orientation of cardiac fibres. (b) The model was used to predict left ventricular (LV) pressure waveforms during systole. Here, a physiological distribution of cell types (baseline) was compared with a hypothetical case in which the ventricle is composed entirely of mid-myocardial cells (all mid). Circles mark the opening and closing of the aortic valve. (c) The two cases also show differences in their patterns of ventricular wall strain, shown here on cross sections through the ventricular wall. These results suggest that accounting for normally occurring heterogeneity as a backdrop for the effects of sarcomeric mutations will be critical in correctly predicting early myocardial strain phenotypes. Blue, endocardial region; red, mid-myocardial region; green, epicardial region.
There is strong evidence that in some cases FHC mutations interfere directly with transmurally heterogeneous properties, which could be an intriguing source of phenotypic variation. For example, RLC contains a phosphorylation site that modulates contractile behaviour in heart muscle, and the degree of phosphorylation has been shown to be greater in the epicardial region than in the endocardium [57]. The FHC-linked E22K mutation in MYL3 is not phosphorylatable by myosin light chain kinase in vitro [61], which in vivo should attenuate the naturally occurring gradient. One potent rationale for developing multi-scale ventricular models of FHC is to predict the effects of such interactions on myocardial function.
6. Modelling and measurement of myocardial strain
Cardiac hypertrophy is observed clinically in response to hypertension or any other condition that places increased load on the heart. HCM is the default diagnosis for patients presenting with increased left ventricular wall thickness, but without obvious causative factors such as hypertension [6]. When mutant sarcomeric proteins were identified as the main cause of inherited HCM, it made logical sense to many as alterations to the contractile apparatus of the heart would be expected to result in changes in mechanical loading and therefore trigger hypertrophy. In recent years, non-invasive imaging studies have shown that alterations to normal patterns of myocardial strain coincide with or even precede remodelling of heart tissue in various pathologies [62,63]. These results offer a possible explanation for the phenotypic diversity in patterns of ventricular hypertrophy seen among FHC patients. Each mutation, acting on a background of spatially heterogeneous sarcomeric and cellular properties, has the potential of affecting myocardial strain in the left ventricle in different ways. Subtle differences in strain and loading could give rise to diverse remodelling phenotypes.
Multi-scale models of ventricular mechanics could be used to explore the connection between altered sarcomere behaviour and patterns of myocardial strain. A general approach would be to use data from mice expressing a mutant FHC protein to create a finite-element mesh of the left ventricle with realistic three-dimensional geometry and contractile behaviour. Properties of isolated myocardium from these mice could be studied and used to construct mechanistic myofilament and cell-level parameter sets. These in turn would be embedded in fully coupled electromechanical simulations of the left ventricle to predict global and regional function of the myocardium, which could be validated against in vivo strain measurements.
It may be particularly instructive to build and compare multi-scale models of hearts having either hypertrophic or DCM-linked sarcomeric mutations. Modelling hearts at an early age prior to anatomical changes could reveal patterns of myocardial strain unique to each type of ventricular remodelling. If validated, these models would offer a powerful tool for relating mutations to the mechanical cues that drive myocardial remodelling at the level of individual cells. Simulations of this kind could also suggest new imaging-based strategies that would allow clinicians to non-invasively detect the existence of underlying molecular pathologies.
Recent advances in magnetic resonance (MR) imaging and software have made the process of creating realistic meshes of left ventricular geometry more rapid and accessible, even in mice [64]. The orientation of cardiac fibres in the left ventricle is a critical determinant of ventricular mechanics and can be measured through diffusion tensor MR imaging [65] or traditional histological methods [66]. Regional strain in the myocardium for model validation can be acquired using either MR tagging with harmonic phase (HARP) tracking or echocardiographic strain imaging (speckle-tracking). It is critical to note that both MR tagging and speckle-tracking have recently been shown to be accurate enough to detect changes in regional strain in mice at time points that precede remodelling and changes in heart function [63,64]. This means that FHC mutant mice could be studied at a young age (less than eight weeks), prior to the occurrence of hypertrophy and the emergence of heart failure, in line with the paradigm proposed by Tardiff [8].
New methods for coupling cell-level models with models of action potential propagation and ventricular biomechanics have emerged in recent years. These advances are important as they allow experimental data obtained in reduced systems such as skinned fibres or isolated cardiac myocytes to be scaled up for predicting function at the level of the intact heart. An initial challenge in this area was numerical instability in simulations arising from coupling length-dependent contraction models with iterative solvers for the finite-element mechanics problem [67,68]. Naive coupling is accomplished through an operator splitting approach, wherein the system of differential and algebraic equations (DAEs) representing cellular electromechanics is solved separately from the partial differential equations (PDEs) that govern mechanical deformation of the three-dimensional ventricular mesh. Sarcomere length and velocity are computed from the mechanics PDEs and supplied to the cell-level DAEs at discrete time intervals throughout the simulation. Values of active tension computed by cell-level DAEs are passed to the mechanics PDEs at the same time. Instability results in this case because, as the iteration scheme deforms nodes in the mesh to balance forces, a constant value for active tension is assumed even though length dependence in the contractile model means that it should change for each iteration [67]. Taking extremely small time steps or re-solving the entire cell model, DAEs with each mechanics iteration can mitigate instability but are both computationally prohibitive. Instead, the so-called ‘update’ schemes have been employed in which simplified forms of the cell model are used to update or re-compute an approximate value for active tension with each iteration [49,67,68].
Physiological loads can be applied to ventricular models by coupling left ventricular volume and boundary conditions on the endocardial surface to lumped-parameter systems models of the circulation [69]. This has enabled simulations of regional cardiac function that are providing clinically relevant insights [70]. These simulations that mimic in vivo loading will allow accurate validation of regional myocardial strains against those measured in animal models. Haemodynamic parameters, which are frequently assessed in engineered mouse models of FHC [71], are also produced in these simulations and would provide an additional point of validation.
7. Limitations
Using a multi-scale modelling approach to study FHC mutations entails several challenges that will have to be addressed in implementation. Some limitations are common to the broader field of multi-scale heart modelling (see Clayton et al. [72] for a detailed review). For instance, many questions remain about the ability of cell models to reproduce behaviour of myocardial tissue. In most cases, functionally coupled cells are represented using continuum approximations, which neglect complex aspects of tissue microstructure such as voids and the presence of fibroblasts or other non-myocyte cells [72]. More work is needed to understand the implications of tissue heterogeneity at this scale. There is also evidence that models constructed from single cell data do not always extend well to conditions present in tissue. Cherry & Fenton [73] demonstrated that two published models of canine ventricular myocyte electrophysiology displayed substantially different spiral wave dynamics in two-dimensional tissue simulations, in spite of the fact that the models are based on data from the same species. Hence, cell-level models that are useful for studying one question may not be suitable for addressing others, particularly where crossing scales are concerned.
Other obstacles to multi-scale modelling in FHC are more specific to the disease. As in the human population, there can be large phenotypic variation in murine strains engineered to have human FHC mutations (e.g. [74]). Without a consistent pattern of ventricular hypertrophy, it would be harder to link changes in regional mechanics to localized areas of tissue remodelling. It may be necessary to construct mouse-specific models for strains with highly variable hypertrophic phenotypes, which would greatly increase the cost and effort required.
Studying the proximal events in FHC has been advocated in large part because later stages involve secondary responses that complicate interpretation of data and could be major sources of phenotypic variation [8]. While many confounding influences can be avoided by studying young animals, some compensatory pathways, such as those regulating blood pressure, are likely to be altered from the earliest stages of FHC. For instance, evidence of altered β-adrenergic signalling at the level of the myofilaments can be seen in transgenic mice expressing non-phosphorylatable myosin RLC [75]. If an FHC mutation indirectly alters phosphorylation or the isoform expression profile of other myofilament proteins, these changes will have to be considered in order to properly account for their influence on Ca2+-contraction dynamics [76].
Another potentially critical component of acute responses to mutant sarcomeric proteins is their impact on cell metabolism. The tendency of FHC-linked mutations to cause gains in function such as increased myofibrillar Ca2+ sensitivity and shortening velocity implicates inefficient use of ATP as a disease mechanism. While we have focused on mechanical stimuli as drivers of FHC in this review, other authors have suggested that altered energy metabolism is the central cause. This hypothesis originated in a study demonstrating lower phosphocreatine to ATP ratios in FHC patients that were independent of the specific gene mutation [10], and similar findings have been noted in engineered mouse lines (see [77] and references therein). Integrative models of ventricular myocytes that include mitochondrial bioenergetics have been developed (e.g. [78]), and could be used to investigate the metabolic consequences of altered myofilament function in multi-scale models.
These points reinforce the necessity for careful validation of modelling results at all scales and particularly in vivo if multi-scale models of FHC are to be useful. It is likely that the models, their scope, the volume and the type of data included and even experimental protocols used for validation will have to be optimized before multi-scale models can accurately predict time-varying patterns of myocardial strain or other properties that could be correlated with hypertrophy. Iteration between computational models and experiments may constitute a limitation of the approach, but it also represents a significant advantage since integrative analysis of this kind enables the completeness of hypotheses to be quantitatively assessed.
8. Conclusion
Multi-scale models of the heart have the potential to facilitate the next advances in understanding and treating FHC by providing a quantitative link between sarcomeric mutations and the conditions that precede maladaptive hypertrophy. The acute effects of mutant contractile proteins on function of the whole organ cannot be predicted in a useful way by intuition alone, considering the volume and complexity of information at intervening biological scales. On the other hand, multi-scale heart models are advancing to the point that they will soon be capable of predicting pre-hypertrophy strain patterns on the basis of genotype. These hypothesized ‘pre-hypertrophy phenotypes’ will allow those studying FHC mutations in mice to shift focus from the endpoint of hypertrophy to the more subtle mechanical changes that might be present at much earlier stages.
Acknowledgements
We thank Dr Kenneth S. Campbell for the use of laboratory equipment to measure myocyte function (figure 4), secured through NIH grant HL090749. This research was also supported in part by NIH grants 5P01HL46345 (Knowlton), P41RR08605 (Arzberger) and 1R01HL96544 (McCulloch).
References
- 1.Trayanova N. A. 2011. Whole-heart modeling: applications to cardiac electrophysiology and electromechanics. Circ. Res. 108, 113–128 10.1161/CIRCRESAHA.110.223610 (doi:10.1161/CIRCRESAHA.110.223610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maron B. J., Gardin J. M., Flack J. M., Gidding S. S., Kurosaki T. T., Bild D. E. 1995. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation 92, 785–789 [DOI] [PubMed] [Google Scholar]
- 3.Force T., et al. 2010. Research priorities in hypertrophic cardiomyopathy: report of a Working Group of the National Heart, Lung and Blood Institute. Circulation 122, 1130–1133 10.1161/CIRCULATIONAHA.110.950089 (doi:10.1161/CIRCULATIONAHA.110.950089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coats C. J., Elliott P. M. 2008. Current management of hypertrophic cardiomyopathy. Curr. Treat Options Cardiovasc. Med. 10, 496–504 10.1007/s11936-008-0042-9 (doi:10.1007/s11936-008-0042-9) [DOI] [PubMed] [Google Scholar]
- 5.Geisterfer-Lowrance A. A., Kass S., Tanigawa G., Vosberg H. P., Mckenna W., Seidman C. E., Seidman J. G. 1990. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell 62, 999–1006 10.1016/0092-8674(90)90274-I (doi:10.1016/0092-8674(90)90274-I) [DOI] [PubMed] [Google Scholar]
- 6.Seidman C. E., Seidman J. G. 2011. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: a personal history. Circ. Res. 108, 743–750 10.1161/CIRCRESAHA.110.223834 (doi:10.1161/CIRCRESAHA.110.223834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho C. Y., et al. 2010. Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N. Engl J. Med. 363, 552–563 10.1056/NEJMoa1002659 (doi:10.1056/NEJMoa1002659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tardiff J. C. 2011. Thin filament mutations: developing an integrative approach to a complex disorder. Circ. Res. 108, 765–782 10.1161/CIRCRESAHA.110.224170 (doi:10.1161/CIRCRESAHA.110.224170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landesberg A., Sideman S. 1994. Mechanical regulation of cardiac muscle by coupling calcium kinetics with cross-bridge cycling: a dynamic model. Am. J. Physiol. 267, H779–H795 [DOI] [PubMed] [Google Scholar]
- 10.Crilley J. G., et al. 2003. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J. Am. Coll. Cardiol. 41, 1776–1782 10.1016/S0735-1097(02)03009-7 (doi:10.1016/S0735-1097(02)03009-7) [DOI] [PubMed] [Google Scholar]
- 11.Harris S. P., Lyons R. G., Bezold K. L. 2011. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ. Res. 108, 751–764 10.1161/CIRCRESAHA.110.231670 (doi:10.1161/CIRCRESAHA.110.231670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beqqali A., Van Eldik W., Mummery C., Passier R. 2009. Human stem cells as a model for cardiac differentiation and disease. Cell. Mol. Life Sci. 66, 800–813 10.1007/s00018-009-8476-0 (doi:10.1007/s00018-009-8476-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon A. M., Homsher E., Regnier M. 2000. Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 14.Spudich J. A. 2001. The myosin swinging cross-bridge model. Nat. Rev. Mol. Cell Biol. 2, 387–392 10.1038/35073086 (doi:10.1038/35073086) [DOI] [PubMed] [Google Scholar]
- 15.Greenberg M. J., Watt J. D., Jones M., Kazmierczak K., Szczesna-Cordary D., Moore J. R. 2009. Regulatory light chain mutations associated with cardiomyopathy affect myosin mechanics and kinetics. J. Mol. Cell. Cardiol. 46, 108–115 10.1016/j.yjmcc.2008.09.126 (doi:10.1016/j.yjmcc.2008.09.126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsoutsman T., et al. 2008. Severe heart failure and early mortality in a double-mutation mouse model of familial hypertrophic cardiomyopathy. Circulation 117, 1820–1831 10.1161/CIRCULATIONAHA.107.755777 (doi:10.1161/CIRCULATIONAHA.107.755777) [DOI] [PubMed] [Google Scholar]
- 17.Bai F., Weis A., Takeda A. K., Chase P. B., Kawai M. 2011. Enhanced active cross-bridges during diastole: molecular pathogenesis of tropomyosin's HCM mutations. Biophys. J. 100, 1014–1023 10.1016/j.bpj.2011.01.001 (doi:10.1016/j.bpj.2011.01.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson P., Griffiths P. J., Watkins H., Redwood C. S. 2007. Dilated and hypertrophic cardiomyopathy mutations in troponin and alpha-tropomyosin have opposing effects on the calcium affinity of cardiac thin filaments. Circ. Res. 101, 1266–1273 10.1161/CIRCRESAHA.107.156380 (doi:10.1161/CIRCRESAHA.107.156380) [DOI] [PubMed] [Google Scholar]
- 19.Ertz-Berger B. R., He H., Dowell C., Factor S. M., Haim T. E., Nunez S., Schwartz S. D., Ingwall J. S., Tardiff J. C. 2005. Changes in the chemical and dynamic properties of cardiac troponin T cause discrete cardiomyopathies in transgenic mice. Proc. Natl Acad. Sci. USA 102, 18 219–18 224 10.1073/pnas.0509181102 (doi:10.1073/pnas.0509181102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenz M., Holmes K. C. 2010. The actin–myosin interface. Proc. Natl Acad. Sci. USA 107, 12 529–12 534 10.1073/pnas.1003604107 (doi:10.1073/pnas.1003604107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niederer S. A., Hunter P. J., Smith N. P. 2006. A quantitative analysis of cardiac myocyte relaxation: a simulation study. Biophys. J. 90, 1697–1722 10.1529/biophysj.105.069534 (doi:10.1529/biophysj.105.069534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice J. J., Wang F., Bers D. M., Tombe P. P. D. 2008. Approximate model of cooperative activation and crossbridge cycling in cardiac muscle using ordinary differential equations. Biophys. J. 95, 2368–2390 10.1529/biophysj.107.119487 (doi:10.1529/biophysj.107.119487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell S. G., Lionetti F. V., Campbell K. S., Mcculloch A. D. 2010. Coupling of adjacent tropomyosins enhances cross-bridge-mediated cooperative activation in a Markov model of the cardiac thin filament. Biophys. J. 98, 2254–2264 10.1016/j.bpj.2010.02.010 (doi:10.1016/j.bpj.2010.02.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bremel R. D., Weber A. 1972. Cooperation within actin filament in vertebrate skeletal muscle. Nat. New Biol. 238, 97–101 [DOI] [PubMed] [Google Scholar]
- 25.Mclachlan A. D., Stewart M. 1975. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J. Mol. Biol. 98, 293–304 10.1016/S0022-2836(75)80119-7 (doi:10.1016/S0022-2836(75)80119-7) [DOI] [PubMed] [Google Scholar]
- 26.Pan B. S., Gordon A. M., Luo Z. X. 1989. Removal of tropomyosin overlap modifies cooperative binding of myosin S-1 to reconstituted thin filaments of rabbit striated muscle. J. Biol. Chem. 264, 8495–8498 [PubMed] [Google Scholar]
- 27.Dobrunz L. E., Backx P. H., Yue D. T. 1995. Steady-state [Ca2+]i-force relationship in intact twitching cardiac muscle: direct evidence for modulation by isoproterenol and EMD 53998. Biophys. J. 69, 189–201 10.1016/S0006-3495(95)79889-7 (doi:10.1016/S0006-3495(95)79889-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKillop D. F., Geeves M. A. 1993. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys. J. 65, 693–701 10.1016/S0006-3495(93)81110-X (doi:10.1016/S0006-3495(93)81110-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice J. J., Stolovitzky G., Tu Y., De Tombe P. P. 2003. Ising model of cardiac thin filament activation with nearest-neighbor cooperative interactions. Biophys. J. 84, 897–909 10.1016/S0006-3495(03)74907-8 (doi:10.1016/S0006-3495(03)74907-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiner J. S., Solaro R. J. 1982. Activation of thin-filament-regulated muscle by calcium ion: considerations based on nearest-neighbor lattice statistics. Proc. Natl Acad. Sci. USA 79, 4637–4641 10.1073/pnas.79.15.4637 (doi:10.1073/pnas.79.15.4637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith D. A., Geeves M. A. 2003. Cooperative regulation of myosin-actin interactions by a continuous flexible chain II: actin-tropomyosin-troponin and regulation by calcium. Biophys. J. 84, 3168–3180 10.1016/S0006-3495(03)70041-1 (doi:10.1016/S0006-3495(03)70041-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou G., Phillips G. N. 1994. A cellular automaton model for the regulatory behavior of muscle thin filaments. Biophys. J. 67, 11–28 10.1016/S0006-3495(94)80475-8 (doi:10.1016/S0006-3495(94)80475-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice J. J., Winslow R. L., Hunter W. C. 1999. Comparison of putative cooperative mechanisms in cardiac muscle: length dependence and dynamic responses. Am. J. Physiol. 276, H1734–H1754 [DOI] [PubMed] [Google Scholar]
- 34.Vibert P., Craig R., Lehman W. 1997. Steric-model for activation of muscle thin filaments. J. Mol. Biol. 266, 8–14 10.1006/jmbi.1996.0800 (doi:10.1006/jmbi.1996.0800) [DOI] [PubMed] [Google Scholar]
- 35.Bers D. M. 2002. Cardiac excitation-contraction coupling. Nature 415, 198–205 10.1038/415198a (doi:10.1038/415198a) [DOI] [PubMed] [Google Scholar]
- 36.Davis J. P., Norman C., Kobayashi T., Solaro R. J., Swartz D. R., Tikunova S. B. 2007. Effects of thin and thick filament proteins on calcium binding and exchange with cardiac troponin C. Biophys. J. 92, 3195–3206 10.1529/biophysj.106.095406 (doi:10.1529/biophysj.106.095406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi T., Solaro R. J. 2005. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu. Rev. Physiol. 67, 39–67 10.1146/annurev.physiol.67.040403.114025 (doi:10.1146/annurev.physiol.67.040403.114025) [DOI] [PubMed] [Google Scholar]
- 38.Kohl P., Sachs F. 2001. Mechanoelectric feedback in cardiac cells. Phil. Trans. R. Soc. Lond. A 359, 1173–1185 10.1098/rsta.2001.0824 (doi:10.1098/rsta.2001.0824) [DOI] [Google Scholar]
- 39.Campbell S., Flaim S., Leem C., Mcculloch A. 2008. Mechanisms of transmurally-varying myocyte electromechanics in an integrated computational model. Phil. Trans. R. Soc. A 366, 3361–3380 10.1098/rsta.2008.0088 (doi:10.1098/rsta.2008.0088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niederer S. A., Smith N. P. 2007. A mathematical model of the slow force response to stretch in rat ventricular myocytes. Biophys. J. 92, 4030–4044 10.1529/biophysj.106.095463 (doi:10.1529/biophysj.106.095463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice J., Wang F., Bers D., De Tombe P. 2008. Approximate model of cooperative activation and crossbridge cycling in cardiac muscle using ordinary differential equations. Biophys. J. 95, 2368–2390 10.1529/biophysj.107.119487 (doi:10.1529/biophysj.107.119487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva J. R., Pan H., Wu D., Nekouzadeh A., Decker K. F., Cui J., Baker N. A., Sept D., Rudy Y. 2009. A multiscale model linking ion-channel molecular dynamics and electrostatics to the cardiac action potential. Proc. Natl Acad. Sci. USA 106, 11 102–11 106 10.1073/pnas.0904505106 (doi:10.1073/pnas.0904505106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenstein J., Hinch R., Winslow R. 2006. Mechanisms of excitation-contraction coupling in an integrative model of the cardiac ventricular myocyte. Biophys. J. 90, 77–91 10.1529/biophysj.105.065169 (doi:10.1529/biophysj.105.065169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stelzer J. E., Norman H. S., Chen P. P., Patel J. R., Moss R. L. 2008. Transmural variation in myosin heavy chain isoform expression modulates the timing of myocardial force generation in porcine left ventricle. J. Physiol. (Lond) 586, 5203–5214 10.1113/jphysiol.2008.160390 (doi:10.1113/jphysiol.2008.160390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szczesna-Cordary D., Jones M., Moore J. R., Watt J., Kerrick W. G. L., Xu Y., Wang Y., Wagg C., Lopaschuk G. D. 2007. Myosin regulatory light chain E22K mutation results in decreased cardiac intracellular calcium and force transients. FASEB J. 21, 3974–3985 10.1096/fj.07-8630com (doi:10.1096/fj.07-8630com) [DOI] [PubMed] [Google Scholar]
- 46.Wan X., Bryant S. M., Hart G. 2003. A topographical study of mechanical and electrical properties of single myocytes isolated from normal guinea-pig ventricular muscle. J. Anat. 202, 525–536 10.1046/j.1469-7580.2003.00187.x (doi:10.1046/j.1469-7580.2003.00187.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bryant S. M., Shipsey S. J., Hart G. 1999. Normal regional distribution of membrane current density in rat left ventricle is altered in catecholamine-induced hypertrophy. Cardiovasc. Res. 42, 391–401 10.1016/S0008-6363(99)00033-4 (doi:10.1016/S0008-6363(99)00033-4) [DOI] [PubMed] [Google Scholar]
- 48.Cordeiro J., Greene L., Heilmann C., Antzelevitch D., Antzelevitch C. 2004. Transmural heterogeneity of calcium activity and mechanical function in the canine left ventricle. Am. J. Physiol. Heart Circ. Physiol. 286, H1471–H1479 10.1152/ajpheart.00748.2003 (doi:10.1152/ajpheart.00748.2003) [DOI] [PubMed] [Google Scholar]
- 49.Campbell S. G., Howard E., Aguado-Sierra J., Coppola B. A., Omens J. H., Mulligan L. J., Mcculloch A. D., Kerckhoffs R. C. P. 2009. Effect of transmurally heterogeneous myocyte excitation-contraction coupling on canine left ventricular electromechanics. Exp. Physiol. 94, 541–552 10.1113/expphysiol.2008.044057 (doi:10.1113/expphysiol.2008.044057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antzelevitch C., Sicouri S., Litovsky S. H., Lukas A., Krishnan S. C., Di Diego J. M., Gintant G. A., Liu D. W. 1991. Heterogeneity within the ventricular wall. Electrophysiology and pharmacology of epicardial, endocardial, and M cells. Circ. Res. 69, 1427–1449 [DOI] [PubMed] [Google Scholar]
- 51.Rosati B., Grau F., Rodriguez S., Li H., Nerbonne J. M., Mckinnon D. 2003. Concordant expression of KChIP2 mRNA, protein and transient outward current throughout the canine ventricle. J. Physiol. (Lond) 548, 815–822 10.1113/jphysiol.2002.033704 (doi:10.1113/jphysiol.2002.033704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kondo R. P., Dederko D. A., Teutsch C., Chrast J., Catalucci D., Chien K. R., Giles W. R. 2006. Comparison of contraction and calcium handling between right and left ventricular myocytes from adult mouse heart: a role for repolarization waveform. J. Physiol. 571, 131–146 10.1113/jphysiol.2005.101428 (doi:10.1113/jphysiol.2005.101428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laurita K. R., Katra R., Wible B., Wan X., Koo M. H. 2003. Transmural heterogeneity of calcium handling in canine. Circ. Res. 92, 668–675 10.1161/01.RES.0000062468.25308.27 (doi:10.1161/01.RES.0000062468.25308.27) [DOI] [PubMed] [Google Scholar]
- 54.Bouvagnet P., Mairhofer H., Leger J. O., Puech P., Leger J. J. 1989. Distribution pattern of alpha and beta myosin in normal and diseased human ventricular myocardium. Basic Res. Cardiol. 84, 91–102 10.1007/BF01907006 (doi:10.1007/BF01907006) [DOI] [PubMed] [Google Scholar]
- 55.Carnes C. A., Geisbuhler T. P., Reiser P. J. 2004. Age-dependent changes in contraction and regional myocardial myosin heavy chain isoform expression in rats. J. Appl. Physiol. 97, 446–453 10.1152/japplphysiol.00439.2003 (doi:10.1152/japplphysiol.00439.2003) [DOI] [PubMed] [Google Scholar]
- 56.Dool J. S., Mak A. S., Friberg P., Wahlander H., Hawrylechko A., Adams M. A. 1995. Regional myosin heavy chain expression in volume and pressure overload induced cardiac hypertrophy. Acta Physiol. Scand. 155, 396–404 10.1111/j.1748-1716.1995.tb09989.x (doi:10.1111/j.1748-1716.1995.tb09989.x) [DOI] [PubMed] [Google Scholar]
- 57.Davis J. S., Hassanzadeh S., Winitsky S., Lin H., Satorius C., Vemuri R., Aletras A. H., Wen H., Epstein N. D. 2001. The overall pattern of cardiac contraction depends on a spatial gradient of myosin regulatory light chain phosphorylation. Cell 107, 631–641 10.1016/S0092-8674(01)00586-4 (doi:10.1016/S0092-8674(01)00586-4) [DOI] [PubMed] [Google Scholar]
- 58.Hidalgo C., Wu Y., Peng J., Siems W. F., Campbell K. B., Granzier H. 2006. Effect of diastolic pressure on MLC2v phosphorylation in the rat left ventricle. Arch. Biochem. Biophys. 456, 216–223 10.1016/j.abb.2006.06.026 (doi:10.1016/j.abb.2006.06.026) [DOI] [PubMed] [Google Scholar]
- 59.Flaim S. N., Giles W. R., Mcculloch A. D. 2006. Contributions of sustained INa and IKv43 to transmural heterogeneity of early repolarization and arrhythmogenesis in canine left ventricular myocytes. Am. J. Physiol. Heart Circ. Physiol. 291, H2617–H2629 10.1152/ajpheart.00350.2006 (doi:10.1152/ajpheart.00350.2006) [DOI] [PubMed] [Google Scholar]
- 60.Kerckhoffs R. C. P., Bovendeerd P. H. M., Kotte J. C. S., Prinzen F. W., Smits K., Arts T. 2003. Homogeneity of cardiac contraction despite physiological asynchrony of depolarization: a model study. Ann. Biomed. Eng. 31, 536–547 10.1114/1.1566447 (doi:10.1114/1.1566447) [DOI] [PubMed] [Google Scholar]
- 61.Szczesna D., Ghosh D., Li Q., Gomes A. V., Guzman G., Arana C., Zhi G., Stull J. T., Potter J. D. 2001. Familial hypertrophic cardiomyopathy mutations in the regulatory light chains of myosin affect their structure, Ca2+ binding, and phosphorylation. J. Biol. Chem. 276, 7086–7092 10.1074/jbc.M009823200 (doi:10.1074/jbc.M009823200) [DOI] [PubMed] [Google Scholar]
- 62.Rosen B. D., et al. 2005. Left ventricular concentric remodeling is associated with decreased global and regional systolic function: the Multi-Ethnic Study of Atherosclerosis. Circulation 112, 984–991 10.1161/CIRCULATIONAHA104.500488 (doi:10.1161/CIRCULATIONAHA104.500488) [DOI] [PubMed] [Google Scholar]
- 63.Bauer M., Cheng S., Jain M., Ngoy S., Theodoropoulos C., Trujillo A., Lin F. C., Liao R. 2011. Echocardiographic speckle-tracking-based strain imaging for rapid cardiovascular phenotyping in mice. Circ. Res. 108, 908–916 10.1161/CIRCRESAHA.110.239574 (doi:10.1161/CIRCRESAHA.110.239574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chuang J. S., Zemljic-Harpf A., Ross R. S., Frank L. R., Mcculloch A. D., Omens J. H. 2010. Determination of three-dimensional ventricular strain distributions in gene-targeted mice using tagged MRI. Magn. Reson. Med. 64, 1281–1288 10.1002/mrm.22547 (doi:10.1002/mrm.22547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strijkers G. J., Bouts A., Blankesteijn W. M., Peeters T. H. J. M., Vilanova A., Van Prooijen M. C., Sanders H. M. H. F., Heijman E., Nicolay K. 2009. Diffusion tensor imaging of left ventricular remodeling in response to myocardial infarction in the mouse. NMR Biomed. 22, 182–190 10.1002/nbm.1299 (doi:10.1002/nbm.1299) [DOI] [PubMed] [Google Scholar]
- 66.Vetter F. J., Mcculloch A. D. 1998. Three-dimensional analysis of regional cardiac function: a model of rabbit ventricular anatomy. Prog. Biophys. Mol. Biol. 69, 157–183 10.1016/S0079-6107(98)00006-6 (doi:10.1016/S0079-6107(98)00006-6) [DOI] [PubMed] [Google Scholar]
- 67.Niederer S. A., Smith N. P. 2008. An improved numerical method for strong coupling of excitation and contraction models in the heart. Prog. Biophys. Mol. Biol. 96, 90–111 10.1016/j.pbiomolbio.2007.08.001 (doi:10.1016/j.pbiomolbio.2007.08.001) [DOI] [PubMed] [Google Scholar]
- 68.Whiteley J. P., Bishop M. J., Gavaghan D. J. 2007. Soft tissue modelling of cardiac fibres for use in coupled mechano-electric simulations. Bull. Math. Biol. 69, 2199–2225 10.1007/s11538-007-9213-1 (doi:10.1007/s11538-007-9213-1) [DOI] [PubMed] [Google Scholar]
- 69.Kerckhoffs R. C. P., Neal M. L., Gu Q., Bassingthwaighte J. B., Omens J. H., Mcculloch A. D. 2007. Coupling of a 3D finite element model of cardiac ventricular mechanics to lumped systems models of the systemic and pulmonic circulation. Ann. Biomed. Eng. 35, 1–18 10.1007/s10439-006-9212-7 (doi:10.1007/s10439-006-9212-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kerckhoffs R. C., Omens J. H., Mcculloch A. D., Mulligan L. J. 2010. Ventricular dilation and electrical dyssynchrony synergistically increase regional mechanical nonuniformity but not mechanical dyssynchrony: a computational model. Circ. Heart Fail. 3, 528–536 10.1161/CIRCHEARTFAILURE.109.862144 (doi:10.1161/CIRCHEARTFAILURE.109.862144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palmer B. M., et al. 2004. Role of cardiac myosin binding protein C in sustaining left ventricular systolic stiffening. Circ. Res. 94, 1249–1255 10.1161/01.RES.0000126898.95550.31 (doi:10.1161/01.RES.0000126898.95550.31) [DOI] [PubMed] [Google Scholar]
- 72.Clayton R. H., et al. 2011. Models of cardiac tissue electrophysiology: progress, challenges and open questions. Prog. Biophys. Mol. Biol. 104, 22–48 10.1016/j.pbiomolbio.2010.05.008 (doi:10.1016/j.pbiomolbio.2010.05.008) [DOI] [PubMed] [Google Scholar]
- 73.Cherry E. M., Fenton F. H. 2007. A tale of two dogs: analyzing two models of canine ventricular electrophysiology. Am. J. Physiol. Heart Circ. Physiol. 292, H43–H55 10.1152/ajpheart.00955.2006 (doi:10.1152/ajpheart.00955.2006) [DOI] [PubMed] [Google Scholar]
- 74.Muthuchamy M., et al. 1999. Mouse model of a familial hypertrophic cardiomyopathy mutation in alpha-tropomyosin manifests cardiac dysfunction. Circ. Res. 85, 47–56 [DOI] [PubMed] [Google Scholar]
- 75.Scruggs S. B., Hinken A. C., Thawornkaiwong A., Robbins J., Walker L. A., De Tombe P. P., Geenen D. L., Buttrick P. M., Solaro R. J. 2009. Ablation of ventricular myosin regulatory light chain phosphorylation in mice causes cardiac dysfunction in situ and affects neighboring myofilament protein phosphorylation. J. Biol. Chem. 284, 5097–5106 10.1074/jbc.M807414200 (doi:10.1074/jbc.M807414200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Varian K. D., Janssen P. M. L. 2007. Frequency-dependent acceleration of relaxation involves decreased myofilament calcium sensitivity. Am. J. Physiol. Heart Circ. Physiol. 292, H2212–H2219 10.1152/ajpheart.00778.2006 (doi:10.1152/ajpheart.00778.2006) [DOI] [PubMed] [Google Scholar]
- 77.Ingwall J. S. 2009. Energy metabolism in heart failure and remodelling. Cardiovasc. Res. 81, 412–419 10.1093/cvr/cvn301 (doi:10.1093/cvr/cvn301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cortassa S., Aon M. A., O'Rourke B., Jacques R., Tseng H. J., Marban E., Winslow R. L. 2006. A computational model integrating electrophysiology, contraction, and mitochondrial bioenergetics in the ventricular myocyte. Biophys. J. 91, 1564–1589 10.1529/biophysj.105.076174 (doi:10.1529/biophysj.105.076174) [DOI] [PMC free article] [PubMed] [Google Scholar]