Abstract
Mutations in tripartite motif protein 32 (TRIM32) are responsible for several hereditary disorders that include limb girdle muscular dystrophy type 2H (LGMD2H), sarcotubular myopathy (STM) and Bardet Biedl syndrome. Most LGMD2H mutations in TRIM32 are clustered in the NHL β-propeller domain at the C-terminus and are predicted to interfere with homodimerization. To get insight into TRIM32's role in the pathogenesis of LGMD2H and to create an accurate model of disease, we have generated a knock-in mouse (T32KI) carrying the c.1465G > A (p.D489N) mutation in murine Trim32 corresponding to the human LGMD2H/STM pathogenic mutation c.1459G > A (p.D487N). Our data indicate that T32KI mice have both a myopathic and a neurogenic phenotype, very similar to the one described in the Trim32-null mice that we created previously. Analysis of Trim32 gene expression in T32KI mice revealed normal mRNA levels, but a severe reduction in mutant TRIM32 (D489N) at the protein level. Our results suggest that the D489N pathogenic mutation destabilizes the protein, leading to its degradation, and results in the same mild myopathic and neurogenic phenotype as that found in Trim32-null mice. Thus, one potential mechanism of LGMD2H might be destabilization of mutated TRIM32 protein leading to a null phenotype.
INTRODUCTION
Tripartite motif protein 32 (TRIM32) is an E3 ubiquitin ligase (1,2) consisting of three domains common for tripartite motif (TRIM) family members, including RING-finger, B-box and coiled-coil regions (3). In addition, its C-terminus forms a β-propeller NHL domain [an element defined by amino acid sequence homologies to regions of Ncl-1, HT2A and Lin-41 proteins (4)], which is involved in protein homodimerization (5). Being ubiquitously expressed, TRIM32 has been shown to interact with myosin (1) and Argonaute-1 (6); it can ubiquitinate multiple substrates, including actin (1), dysbindin (7), PIASy (8), Abl-2 (9) and c-Myc (6) and is involved in diverse pathological processes ranging from cancer to muscular dystrophy.
Currently, seven mutations in the Trim32 gene have been linked to limb girdle muscular dystrophy type 2H (LGMD2H). The LGMDs are hereditary muscle-wasting disorders involving muscles of the pelvis and shoulder girdle. LGMD2H is a mild autosomal recessive muscular dystrophy with highly variable phenotypes encompassing a spectrum of patients that range from asymptomatic to wheelchair bound. Mutations in Trim32 responsible for LGMD2H include two missense mutations [c.1459G>A (p.D487N) (10), c.1180G>A (p.R394H) (5)], one codon deletion [c.1761–1763delGAT (p.D588 del) (5)], three frameshift mutations [c.1559delC (p.T520TfsX13) (5), c.1753–1766dup (p.I590LfsX38) (11), c.1560delC (p.C521VfsX13) (12)] and one intragenic deletion that removes the entire open reading frame [del 30 586 bp + insert 2 bp (12)]. The first-described LGMD2H missense mutation (p.D487N) (10) also causes sarcotubular myopathy (STM), an allelic disorder that is characterized by a more severe muscular dystrophy phenotype than LGMD2H, suggesting these disparate clinical phenotypes are on the same disease spectrum (13). Interestingly, six of the LGMD2H mutations are clustered in the conserved C-terminal NHL β-propeller domain of TRIM32. Using molecular modeling, it has been predicted that at least some of these mutations in the C-terminus might cause conformational changes that could impact protein–protein interactions and homodimerization (5) and therefore impair the normal biological function of TRIM32.
Not only do mutations in TRIM32 cause muscular dystrophy, but they also result in a disorder called Bardet Biedl syndrome (BBS) (14). BBS is a complex and genetically heterogeneous disorder involving retinal dystrophy, obesity, kidney abnormalities, and polydactyly. Remarkably, no muscular dystrophy symptoms have been reported for BBS patients. In contrast to LGMD2H mutations, the p.P130S mutation causing BBS type 11 is located in a different region of TRIM32 called the B-box zinc-finger domain, which is a region that may recognize DNA, RNA, protein, and lipid substrates. It is not understood why different mutations in one gene can result in such clinically diverse phenotypes; therefore, use of mouse models expressing the various mutant genes will be very informative.
Previously, we created genetically modified mice lacking TRIM32 [AKA, Trim32 knock-out mouse (T32KO)] and observed both myopathic and neurogenic phenotypes caused by TRIM32 deficiency (15). The muscles of this mouse model demonstrated myopathic features similar to those found in patients with the muscular disorders LGMD2H and STM. These features included an increased number of fibers with multiple centrally located nuclei, fiber splitting, abnormal fiber diameter variability, targetoid fibers lacking succinic dehydrogenase (SDH) or nicotinamide adenine dinucleotide (NADH) staining, a dilated sarcotubular system with abnormal accumulation of membranous structures and z-line streaming. In addition, these studies revealed a high level of TRIM32 expression in normal mouse brain compared with skeletal muscle. Intriguingly, Trim32-null mice showed a decrease in the concentration of neurofilament proteins in the brain and a reduced motoraxon diameter. These axonal changes were accompanied by a shift toward a slower motor unit type resulting in an elevated type I slow myosin isotype and a concomitant reduction in the type II fast myosin in T32KO soleus muscle. These data suggest that muscular dystrophy due to the disruption of the Trim32 gene involves both myopathic and neurogenic characteristics. In accordance with our findings, neurogenic features were also evident in LGMD2H patients, where a slight dominance of type I muscle fibers, decreased motor and sensory nerve conduction velocities and myopathic and neurogenic electromyography abnormalities in the leg muscles were observed (5,12).
In an effort to better understand the role of TRIM32 in muscle as well as pathogenic mechanisms occurring in LGMD2H, we have generated a Trim32 knock-in mouse (T32KI) carrying the common LGMD2H/STM mutation c.1465G > A (p.D489N) in murine TRIM32 corresponding to human LGMD2H/STM pathogenic mutation c.1459G > A (p.D487N). The data from this study show that, like in the T32KO mice, muscles of T32KI mice have myopathic and neurogenic features. Furthermore, analysis of Trim32 gene expression in T32KI mice demonstrated a dramatic reduction in mutated TRIM32 (D489N) at the protein level. These studies suggest that the D489N mutation destabilizes the TRIM32 protein, resulting in loss of TRIM32 and muscle pathology similar to the myopathy described in the Trim32-null mouse.
RESULTS
T32KI mice have a mild myopathic phenotype
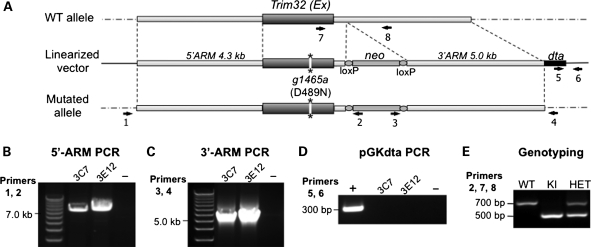
We successfully generated a knock-in mouse carrying the pathogenic D489N mutation (c.1465G > A) at the Trim32 locus using the targeting vector pGKneolox2DTA, which was electroporated into mouse embryonic stem (ES) cells (Fig. 1). Two clones, which underwent homologous recombination and contained the c.1465G > A mutation at the Trim32 locus (verified by sequencing), were used to generate a knock-in mouse.
Figure 1.
Generation of the T32KI (D489N) mouse. (A) Genomic structure of Trim32 and the construct that was electroporated to ES cells to elicit homologous recombination to introduce the D489N mutation (mutation is denoted with asterisks). The presence of the neo-cassette and dta gene aids in selection and genotyping. Primers used for genotyping and selection of positive ES cell clones are indicated by black numbered arrows. (B) PCR for the 5′-arm (using primers 1 and 2) of genomic DNA isolated from electroporated ES cells as template. Two positive clones are shown (3C7 and 3E12). (C) PCR for the 3′-arm (using primers 3 and 4) of genomic DNA isolated from electroporated ES cells as template. (D) PCR of diptheria toxin gene (primers 5 and 6) that is contained in the plasmid but that should be excluded after correct homologous recombination. Both positive clones do not contain this region of plasmid DNA, as expected. (E) T32KI mouse genotyping strategy. Genotyping PCR results in production of a 500 bp band in KI, amplified from the allele with the integrated neo-cassette (primers 2 and 7), and 700 bp band from the WT allele (primers 7 and 8). PCR from HET containing both alleles (primers 2, 7 and 8) yields both bands.
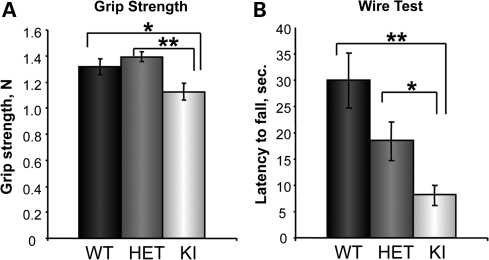
Several muscle strength tests demonstrated that muscle performance in T32KI animals was decreased compared with wild-type (WT) littermates (Fig. 2). The average grip strength peak tension was reduced in T32KI animals compared with WT or heterozygote (HET) littermates (Fig. 2A). T32KI mice were less successful on the wire test, exhibiting a shorter hang time, compared with WT or HET littermates (Fig. 2B). Thus, the T32KI mouse is weaker than its WT counterpart, similar to the T32KO mouse generated previously (15).
Figure 2.
T32KI muscle performance. (A) Forelimb grip strength of KI (n= 9), WT (n= 9) and HET (n= 18) animals (age 6–9 months) was measured using a grip strength meter and data expressed as percent of WT value ±SEM. Grip strength of T32KI mice was reduced by 14%, compared with WT or HET littermates (** –P< 0.0008, * –P< 0.05). (B) Wire hang test performance. Latency to fall was compared within genotypes as the average time in seconds ±SEM, n= 18–22, age 6–9 months. Latency to fall in T32KI animals was 3.7 times less, compared with WT littermates, and 1.9 times less, compared with HET (** –P<0.0005, * –P<0.03). Difference between WT and HET animals was not statistically significant (P= 0.08).
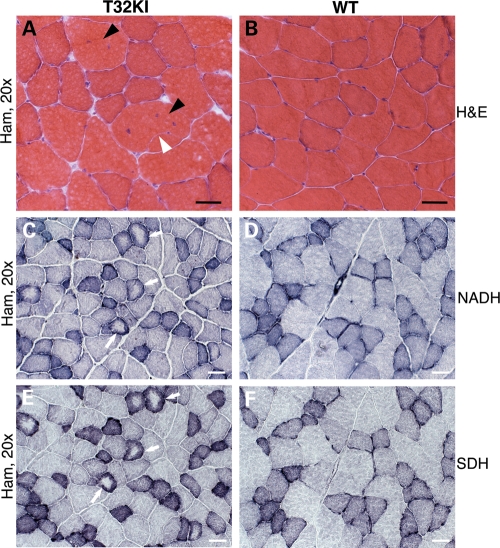
Histological assessment of muscle cross-sections by light microscopy revealed mild dystrophic changes in T32KI muscles (Fig. 3). Centrally located nuclei and fiber splitting were observed (Fig. 3A). NADH and SDH staining revealed targetoid fibers (Fig. 3C and E). These findings suggest defects in sarcoplasmic reticulum and mitochondrial oxidative enzymes in T32KI muscles. Taken together, these results suggest a mild myopathic phenotype in T32KI mice very similar to the one seen in T32KO (15).
Figure 3.
Histology of T32KI muscles. (A–F) Micrographs of cross-sections of hamstring muscles of 12-month-old animals. (A and B) H&E staining of T32KI (A) and WT (B) muscle shows internalized nuclei (black arrowheads) and fiber splitting (white arrowhead). (C and D) NADH-TR and (E and F) SDH staining. (C and E) Serial sections of T32KI muscles; (D and F) serial sections of WT muscles. Arrows point to fibers with central pallor, indicating defects in both mitochondrial and SR staining. Scale bar is 50 μm.
Myosin isoform shift
Previously, we demonstrated that the T32KO phenotype has both myogenic and neurogenic features (15). T32KO mice had a reduction in the concentration of neurofilament proteins and in accordance with the role for neurofilaments in controlling radial growth of axons (16–18), T32KO mice demonstrated reduced motor axon diameters. Axonal diameter dictates the rate of neuronal firing and consequently affects the motor unit type: axons with larger diameters innervate type II (fast) muscle fibers, whereas smaller axons innervate type I (slow) muscle fibers (19). In agreement with their smaller axonal diameter, T32KO mice showed a slight predominance of slow myosin type I isoform (15).
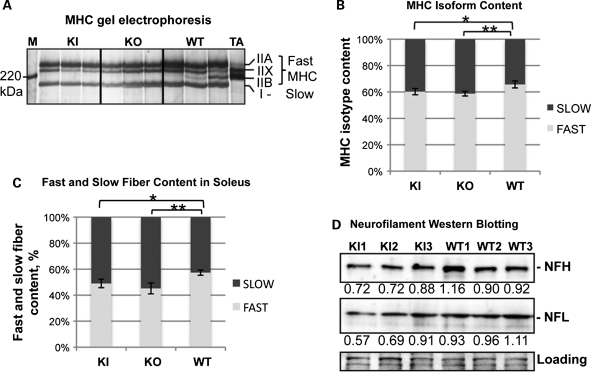
T32KI mice were examined to determine whether the same shift in myosin isoform expression that we observed in the T32KO could also be detected in the T32KI. Myosin heavy chain (MHC) gel electrophoresis was carried out and revealed a shift in the MHC isoform distribution in T32KI soleus muscles toward a slower phenotype (Fig. 4A and B). Slow MHC type I was increased by 5.5% in T32KI soleus muscle compared with the WT. Moreover, immunohistochemical staining of soleus muscle with anti-slow MHC antibody also demonstrated a statistically significant increase in the percent of slow fibers in the T32KI (Fig. 4C).
Figure 4.
Slow type I myosin isoform predominance in T32KI. (A) MHC isoform gel electrophoresis of soleus muscle lysates from 12-month-old animals. Positions of individual MHC isoforms are indicated. M—benchmark protein ladder (Invitrogen, Carlsbad, CA, USA). TA—tibialis anterior muscle extract was loaded to ensure position of fast MHC isoforms type IIX and IIB, since this muscle does not contain any slow type I MHC. (B) Quantification of the gels shown in (A). The fraction corresponding to individual MHC isoforms was calculated as a percent of total MHC content ±SEM in each sample. This accounted for the differences in loading. Amount of MHC type II (fast) is decreased and amount of MHC type I (slow) is increased in T32KO and T32KI soleus muscles by ∼7 and 5.5% correspondingly, compared with the WT (** –P< 0.003, * –P< 0.015). (C) Immunohistochemical analysis of soleus muscles using anti-slow MHC-specific antibody revealed increased amount of slow (type I) fibers in T32KO soleus by 12.1% (n= 8) and in T32KI soleus by 8.3% (n= 6), compared with the WT (n= 13). (D) Neurofilament protein concentrations are reduced in T32KI brains by ∼25%. Western blots of T32KI brain lysates using NFH, and NFL-specific antibodies. Ponceau S staining is shown for loading control. Numbers indicate densitometry data of relative amount of protein in the corresponding bands.
Because the T32KO mouse showed a reduction in neurofilament (NF) protein concentration, we asked whether a similar reduction was observed in T32KI brains. Western blotting of brain lysates from T32KI mice was carried out and probed for neurofilament proteins using isoform specific antibodies. Heavy (NFH) and light (NFL) neurofilament proteins were reduced by ∼25% in T32KI, compared with the WT (Fig. 4D). Therefore, the T32KI reproduces both the myogenic and the neurogenic components of the T32KO phenotype.
Trim32 levels of expression
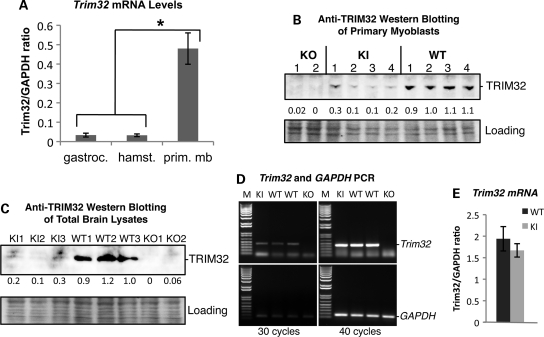
TRIM32 is a ubiquitously expressed protein; however, its mRNA expression in skeletal muscles is 100 times lower than in the brain, making detection of the protein in skeletal muscle very challenging (15). In spite of its low expression in muscle, the primary pathogenic feature of the C-terminal LGMD2H mutations is skeletal muscle weakness and no mental retardation. To gain insight into Trim32's expression in muscle, we performed real-time PCR of primary myoblasts versus skeletal muscle tissue. This analysis revealed that the level of Trim32 mRNA expression is ∼16 times higher in myoblasts than in skeletal muscle tissue (Fig. 5A). Thus, myoblasts appear to be the main site of expression of Trim32 in skeletal muscle. Not surprisingly, anti-TRIM32 western blot analysis readily detected TRIM32 protein in primary myoblasts derived from WT mice. However, in T32KI myoblasts, the level of mutated TRIM32 (D489N) was greatly reduced (Fig. 5B). Western blotting of total brain lysates using anti-TRIM32 antibody also revealed nearly complete loss of TRIM32 protein expression in T32KI samples, compared with the WT level (Fig. 5C).
Figure 5.
Trim32 levels of expression. (A) Trim32 mRNA was isolated from WT primary myoblast cells and WT skeletal muscle tissue (gastrocnemius and hamstring group of muscles). Real-time PCR revealed that the concentration of Trim32 mRNA is ∼16 times higher in myoblasts than in skeletal muscles. (B and C) Anti-TRIM32 western blot analysis of primary myoblast lysates (B) derived from WT, T32KI and T32KO mice and total brain lysates (C) indicates severe reduction in mutated TRIM32 protein in T32KI cultured myoblasts and T32KI brains, compared with WT levels. Ponceau S staining is shown as a loading control. Numbers under the blots indicate densitometry data of relative amount of TRIM32 in the corresponding bands. (D) RT–PCR of Trim32 mRNA expression in brain (upper panels). GAPDH PCR (lower panels) is shown as a control for RNA/cDNA preparation. Left panels, 30 PCR amplification cycles; right panels, 40 cycles. T32KO lacks Trim32 mRNA, whereas T32KI has the same level of Trim32 mRNA expression as WT animals. (E) Real-time PCR demonstrates similar levels of Trim32 mRNA in T32KI and WT primary myoblasts.
To verify that the reduction in TRIM32 concentration in the T32KI was not due to mRNA distortion, we performed several rounds of reverse transcription polymerase chain reaction (RT–PCR) using a varying number of cycles (Fig. 5D). As expected, no Trim32 mRNA was detected in the T32KO, whereas the same level of Trim32 mRNA expression could be detected in the T32KI as in WT animals. Real-time PCR verifies equal level of Trim32 mRNA in T32KI and WT primary myoblasts (Fig. 5E). Therefore, the Trim32 mutation c.1465G > A (p.D489N) leads to the loss of TRIM32 at the protein level without affecting its mRNA expression. This dramatic reduction in TRIM32 protein results in a myopathic phenotype in mice, similar to muscular defects observed in Trim32-null animals. These studies identify the mechanism by which the common Trim32 missense mutation leads to a muscular dystrophy.
DISCUSSION
In the present investigation, genetically modified mice were created that express the common LGMD2H/STM mutation in Trim32. Analysis of the T32KI phenotype and expression levels of D489N-mutated TRIM32 in mouse tissues suggest that the D489N pathogenic mutation destabilizes the protein leading to its insufficiency. As a result of TRIM32 loss, T32KI mice reproduce the same mild myopathic phenotype with a neurogenic component as that found in Trim32-null mice.
Consistent with our finding of mutated TRIM32 protein loss, Albor et al. (8) reported a very low basal level of TRIM32 in fibroblasts isolated from LGMD2H patients carrying the homozygous D487N mutation (corresponding to D489N in the mouse protein), compared with levels of TRIM32 in healthy donor fibroblasts. The authors hypothesized that the D487N mutation reduced TRIM32 protein stability, which resulted in a failure of mutated TRIM32 to accumulate in the cell in vivo. Moreover, it has been shown recently by Borg et al. (12) that truncated TRIM32 was not detected in muscle lysates from patients harboring a C-terminal frameshift mutation c.1560delC (p.Cys521ValfsX13). Compound HET patients with a heterozygous intragenic deletion that removes the entire Trim32 open reading frame on one allele, and a c.1560delC mutation on the other, lack TRIM32 protein completely. Given that the entire Trim32 open reading frame is contained within a single exon, it is unlikely that the c.1560delC mutation would lead to a nonsense-mediated mRNA decay, since premature termination codons residing within the last exon do not normally trigger nonsense-mediated decay (20). In support of impaired protein turnover of the mutated protein rather than mRNA distortion as the mechanism of TRIM32 loss, our data also demonstrate normal mRNA expression for c.1465G > A (p.D489N) mutated Trim32, indistinguishable from the WT Trim32 levels (Fig. 5D and E). It is noteworthy that most mutated forms of TRIM32 (with mutations in the NHL domain) can be heterologously and stably expressed in different systems (1,5,7,8). This ability to express the mutant protein in vitro may be explained by the high expression levels achieved from strong, constitutively active promoters in such systems.
The majority of the LGMD2H/STM mutations is clustered in the highly conserved C-terminal NHL domain of TRIM32 and may cause conformational changes in the protein (5). Such conformational changes often result in decreased protein stability. Genetic diseases caused by missense mutations that lead to instability and loss of the encoded proteins are not uncommon (21,22). For example, the D1424N substitution in non-muscular myosin II (MYH9) responsible for the phenotypes in May-Hegglin anomaly/Fechtner syndrome causes the MYH9 protein to become highly unstable (23). Interestingly, in both cases (mutation D487N in TRIM32 and D1424N in MYH9), a charge change occurs due to the substitution of negatively charged aspartic acid to uncharged asparagine, which could disturb the electrostatic intramolecular interactions.
NHL domains are conserved motifs forming six-bladed β-propeller structures of antiparallel β-sheets (24,25) found in many pro- and eukaryotic proteins. Aspartic acid at position 487 (human TRIM32) or 489 (mouse TRIM32) is one of the most highly conserved residues in these domains; the majority of the over 100 known NHL motifs have an aspartate, several have a glutamate and one has a lysine, at this position (4,10). Non-conservative substitution of this residue most likely will affect the structure and stability of the mutated protein. Dokholyan Group's Eris webserver (26) allows for the estimation of the effect of a single amino acid mutation on the thermodynamic stability of the protein with a known structure. The folding-free energy difference (▵▵G) is calculated so that it results in a positive value when the mutated protein is less stable than the WT protein (27). We used the known X-ray crystal structures of two NHL-repeat β-propeller domains from Protein Data Bank (PDB) for ▵▵G calculation by Eris server: PDB #1Q7F (Brain tumor Brat-2 NHL domain) and PDB #1RWI (extracellular domain of Mycobacterium tuberculosis receptor Ser/Thr protein kinase PKND). Mutations E > N in 1Q7F structure and D > N in 1RWI at positions corresponding to D489 in mouse TRIM32 were predicted to have positive ▵▵G of 3.13 and 2.07 kcal/mol, respectively, suggesting that the substitution of these positively charged, highly conserved residues to neutral asparagines will result in reduced protein stability.
Therefore, accumulating evidence suggests that mutations in a highly conserved C-terminal NHL domain of TRIM32 most likely disrupt the structure of the β-propeller. Such misfolding might affect its interactions with stabilizing partners or destabilize the protein directly resulting in its loss. It is also possible that the genetic background can affect the stability of the mutated protein. LGMD2H/STM is a disease with extremely variable phenotypes and the ages of onset including range of the patients from asymptomatic to wheelchair-bound. The same point mutation in TRIM32 (D487N) leads to diverse phenotypes with different severity even among the members of the same family. Therefore, genetic heterogeneity may play an important role in the severity of LGMD2H symptoms. Genetic background variations that contribute to protein degradation or instability (for, example, more active proteasomal degradation machinery or reduced chaperone activity) will likely result in more severe phenotypes in such LGMD2H patients. In conclusion, one of the potential mechanisms of LGMD2H might be insufficiency of E3-ubiquitin ligase TRIM32 due to protein instability caused by pathogenic mutations.
MATERIALS AND METHODS
All experimental protocols and use of animals were conducted in accordance with the National Institute of Health Guide for Care and Use of Laboratory Animals and approved by the UCLA Institutional Animal Care and Use Committee.
Generation of Trim32 knock-in mice
T32KI mice were generated using targeting vector pGKneolox2DTA containing dephteria toxin gene (dta) and neomycin cassette (neo) for selection. Mutation c.1465G > A (p.D489N) was introduced in the Trim32 locus by site-directed mutagenesis (Stratagene, La Jolla, CA, USA). 5′-arm genomic Trim32 DNA (6.9 kb) including the entire single Trim32 exon carrying mutation (c.1465G > A) and 3′-arm (5 kb) containing 3′-untranslated genomic Trim32 DNA was cloned into the pGK vector (Fig. 1A). Electroporation of the ES cells with pGK vector carrying the mutated Trim32 locus resulted in generation of four clones, which underwent homologous recombination with the targeting vector (Fig. 1A). Positive clones were selected by extensive PCR (Fig. 1B–D; see PCR and genotyping section for primer details). These clones contained the c.1465G>A mutation at the Trim32 locus as verified by sequencing. Two positive clones of ES cells were injected into blastocysts. Original founder mice were 129 SvEvBrd × C57 BL/6 chimeras. Germline transmission was enabled by backcrossing these high-grade chimeras to BALB/cJ WT animals. HETs from this cross were interbred to produce knock-in (KI) and WT homozygotes (genotyping is shown on Fig. 1E; see PCR and genotyping section for primer details). All analyses were performed on interbred mice on a mixed 129 SvEvBrd × C57 BL/6J × BALB/cJ background.
PCR and genotyping
Real-time PCR was performed as described previously (15).
PCR analysis for selection of ES cell clones with correct recombination was carried out using genomic DNA isolated from ES cell clones (Fig. 1B–D). Primers for the 5′-arm PCR: (1) fwd 5′-GATAAGTCTCAACAACATCAAAACC; (2) rev 5′-GGTTCCGGATCCACTAGTTCTAGAGC. Primers for the 3′-arm PCR: (3) fwd 5′-CGACCTGCAGCCCAAGCTAGCTTATCG; (4) rev 5′-GCAGCCTCACTGTGCCTCATATGGTC. Primers for diptheria toxin gene PCR: (5) fwd 5′-GATGATGTTGTTGATTCTTCTAAATC; (6) rev 5′-GACTTAAACCTAACTCTTTCTTAATAG.
Genotyping was performed using genomic tail DNA (Fig. 1E). Genotyping primers: (2) rev 5′-GGTTCCGGATCCACTAGTTCTAGAGC; (7) fwd 5′-GGTGGCTACAGCGTCCTTATTCG; (8) rev 5′-CATCCTCTGGCACATCTTCAATG.
Grip strength and wire hang tests
Grip tests were performed using a grip strength meter Chatillon DFIS2 (AMETEK, Sellersville, PA, USA). For the wire hang test, the mouse was placed on a suspended wire that was 2 ft from the floor. The latency to fall off the wire in seconds was scored in five trials.
Muscle histology and immunohistochemistry
Muscles were dissected from the mice, placed in O.C.T. Compound (Sakura Finetek, Torrance, CA, USA) and frozen in isopentane cooled in liquid nitrogen. Frozen sections were cut at 10 µm and kept frozen until use. Histology of the tissue was assessed by staining with hematoxylin and eosin (H&E). Enzyme histochemical stains were also performed and included NADH-tetrazolium reductase (NADH-TR) and SDH.
To assess fast and slow fiber content in soleus muscle, anti-slow MHC antibody 1:50 (Leica Microsystems, Buffalo Grove, IL, USA) was used in immunohistochemical staining.
All images were acquired using an Axio Imager M1 microscope with software Axio Vision 4.8 (Carl Zeiss MicroImaging, LLC, Thornwood, NY, USA).
Tissue extract preparation and western blot analysis
For western blot analysis, muscles and brains were homogenized in reducing sample buffer [80 mm Tris, pH 6.8, 0.1m dithiothreitol, 2% SDS and 10% glycerol with protease inhibitor cocktail [Sigma-Aldrich, St Louis, MO, USA)] using a Dounce homogenizer. Forty micrograms of total protein per lane were loaded on polyacrylamide gel followed by transfer to nitrocellulose membrane.
Antibodies used for western blotting included: anti-NFL and anti-NFH (Millipore, Billerica, MA, USA). Secondary antibodies conjugated with horseradish peroxidase were from Sigma-Aldrich. Specific signals were developed using ChemiGlow substrate (Alpha Innotech, San Leandro, CA, USA). Images of the blots were acquired using FlourChem FC2 Imager (Alpha Innotech). Densitometry was performed using AlphaEase FC Software Version 6.0.2 (Alpha Innotech).
MHC isoform gel electrophoresis
The procedure was previously described (28) with modifications (29). Gels were stained with silver staining kit (Bio-Rad, Hercules, CA, USA).
Antibodies
We generated specific rabbit affinity-purified anti-peptide antibody against TRIM32 as described previously by Locke et al. (7).
Primary myoblast culture
Myogenic cells were isolated as previously described (30,31).
Statistical analysis
Statistical analysis of all data was carried out by Student's t-test. Differences were considered statistically significant if the P-value was <0.05. Error bars on all graphs are represented by standard errors of means (SEM).
FUNDING
This work was supported by funding from the National Institute of Arthritis, Musculoskeletal and Skin Diseases (RO1 AR052693, RO1 AR/NS48177), and funding from NIAMS for a P30 Muscular Dystrophy Core Center (P30AR057230-01) and the Muscular Dystrophy Association.
ACKNOWLEDGEMENTS
We thank Ms Jane Wen, Mr Leonel Martinez and Ms Julia Overman for excellent technical support and Dr Irina Kramerova for helpful discussions and reading of the manuscript. The authors are grateful to the UTSW transgenic facility.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kudryashova E., Kudryashov D., Kramerova I., Spencer M.J. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 2005;354:413–424. doi: 10.1016/j.jmb.2005.09.068. doi:10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 2.Horn E.J., Albor A., Liu Y., El-Hizawi S., Vanderbeek G.E., Babcock M., Bowden G.T., Hennings H., Lozano G., Weinberg W.C., et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25:157–167. doi: 10.1093/carcin/bgh003. doi:10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- 3.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. doi:10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slack F.J., Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem. Sci. 1998;23:474–475. doi: 10.1016/s0968-0004(98)01299-7. doi:10.1016/S0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 5.Saccone V., Palmieri M., Passamano L., Piluso G., Meroni G., Politano L., Nigro V. Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum. Mutat. 2008;29:240–247. doi: 10.1002/humu.20633. doi:10.1002/humu.20633. [DOI] [PubMed] [Google Scholar]
- 6.Schwamborn J.C., Berezikov E., Knoblich J.A. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. doi:10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locke M., Tinsley C.L., Benson M.A., Blake D.J. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 2009;18:2344–2358. doi: 10.1093/hmg/ddp167. doi:10.1093/hmg/ddp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albor A., El-Hizawi S., Horn E.J., Laederich M., Frosk P., Wrogemann K., Kulesz-Martin M. The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkappaB. J. Biol. Chem. 2006;281:25850–25866. doi: 10.1074/jbc.M601655200. doi:10.1074/jbc.M601655200. [DOI] [PubMed] [Google Scholar]
- 9.Kano S., Miyajima N., Fukuda S., Hatakeyama S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008;68:5572–5580. doi: 10.1158/0008-5472.CAN-07-6231. doi:10.1158/0008-5472.CAN-07-6231. [DOI] [PubMed] [Google Scholar]
- 10.Frosk P., Weiler T., Nylen E., Sudha T., Greenberg C.R., Morgan K., Fujiwara T.M., Wrogemann K. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am. J. Hum. Genet. 2002;70:663–672. doi: 10.1086/339083. doi:10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cossee M., Lagier-Tourenne C., Seguela C., Mohr M., Leturcq F., Gundesli H., Chelly J., Tranchant C., Koenig M., Mandel J.L. Use of SNP array analysis to identify a novel TRIM32 mutation in limb-girdle muscular dystrophy type 2H. Neuromuscul. Disord. 2009;19:255–260. doi: 10.1016/j.nmd.2009.02.003. doi:10.1016/j.nmd.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Borg K., Stucka R., Locke M., Melin E., Ahlberg G., Klutzny U., Hagen M., Huebner A., Lochmuller H., Wrogemann K., et al. Intragenic deletion of TRIM32 in compound heterozygotes with sarcotubular myopathy/LGMD2H. Hum. Mutat. 2009;30:E831–E844. doi: 10.1002/humu.21063. doi:10.1002/humu.21063. [DOI] [PubMed] [Google Scholar]
- 13.Schoser B.G., Frosk P., Engel A.G., Klutzny U., Lochmuller H., Wrogemann K. Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann. Neurol. 2005;57:591–595. doi: 10.1002/ana.20441. doi:10.1002/ana.20441. [DOI] [PubMed] [Google Scholar]
- 14.Chiang A.P., Beck J.S., Yen H.J., Tayeh M.K., Scheetz T.E., Swiderski R.E., Nishimura D.Y., Braun T.A., Kim K.Y., Huang J., et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc. Natl Acad. Sci. USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. doi:10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudryashova E., Wu J., Havton L.A., Spencer M.J. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum. Mol. Genet. 2009;18:1353–1367. doi: 10.1093/hmg/ddp036. doi:10.1093/hmg/ddp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Q., Couillard-Despres S., Julien J.P. Delayed maturation of regenerating myelinated axons in mice lacking neurofilaments. Exp. Neurol. 1997;148:299–316. doi: 10.1006/exnr.1997.6654. doi:10.1006/exnr.1997.6654. [DOI] [PubMed] [Google Scholar]
- 17.Elder G.A., Friedrich V.L., Jr., Bosco P., Kang C., Gourov A., Tu P.H., Lee V.M., Lazzarini R.A. Absence of the mid-sized neurofilament subunit decreases axonal calibers, levels of light neurofilament (NF-L), and neurofilament content. J. Cell Biol. 1998;141:727–739. doi: 10.1083/jcb.141.3.727. doi:10.1083/jcb.141.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lariviere R.C., Julien J.P. Functions of intermediate filaments in neuronal development and disease. J. Neurobiol. 2004;58:131–148. doi: 10.1002/neu.10270. doi:10.1002/neu.10270. [DOI] [PubMed] [Google Scholar]
- 19.Cullheim S., Kellerth J.O. A morphological study of the axons and recurrent axon collaterals of cat alpha-motoneurones supplying different functional types of muscle unit. J. Physiol. 1978;281:301–313. doi: 10.1113/jphysiol.1978.sp012423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng J., Belgrader P., Zhou X., Maquat L.E. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pakula A.A., Sauer R.T. Genetic analysis of protein stability and function. Annu. Rev. Genet. 1989;23:289–310. doi: 10.1146/annurev.ge.23.120189.001445. doi:10.1146/annurev.ge.23.120189.001445. [DOI] [PubMed] [Google Scholar]
- 22.Waters P.J. Degradation of mutant proteins, underlying ‘loss of function’ phenotypes, plays a major role in genetic disease. Curr. Issues Mol. Biol. 2001;3:57–65. [PubMed] [Google Scholar]
- 23.Deutsch S., Rideau A., Bochaton-Piallat M.L., Merla G., Geinoz A., Gabbiani G., Schwede T., Matthes T., Antonarakis S.E., Beris P. Asp1424Asn MYH9 mutation results in an unstable protein responsible for the phenotypes in May-Hegglin anomaly/Fechtner syndrome. Blood. 2003;102:529–534. doi: 10.1182/blood-2002-09-2783. doi:10.1182/blood-2002-09-2783. [DOI] [PubMed] [Google Scholar]
- 24.Edwards T.A., Wilkinson B.D., Wharton R.P., Aggarwal A.K. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 2003;17:2508–2513. doi: 10.1101/gad.1119403. doi:10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Good M.C., Greenstein A.E., Young T.A., Ng H.L., Alber T. Sensor domain of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknD, forms a highly symmetric beta propeller. J. Mol. Biol. 2004;339:459–469. doi: 10.1016/j.jmb.2004.03.063. doi:10.1016/j.jmb.2004.03.063. [DOI] [PubMed] [Google Scholar]
- 26.Yin S., Ding F., Dokholyan N.V. Eris: an automated estimator of protein stability. Nat. Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. doi:10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 27.Gilis D., Rooman M. Predicting protein stability changes upon mutation using database-derived potentials: solvent accessibility determines the importance of local versus non-local interactions along the sequence. J. Mol. Biol. 1997;272:276–290. doi: 10.1006/jmbi.1997.1237. doi:10.1006/jmbi.1997.1237. [DOI] [PubMed] [Google Scholar]
- 28.Talmadge R.J., Roy R.R. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. J. Appl. Physiol. 1993;75:2337–2340. doi: 10.1152/jappl.1993.75.5.2337. [DOI] [PubMed] [Google Scholar]
- 29.Kohn T.A., Myburgh K.H. Electrophoretic separation of human skeletal muscle myosin heavy chain isoforms: the importance of reducing agents. J. Physiol. Sci. 2006;56:355–360. doi: 10.2170/physiolsci.RP007706. doi:10.2170/physiolsci.RP007706. [DOI] [PubMed] [Google Scholar]
- 30.Horsley V., Jansen K.M., Mills S.T., Pavlath G.K. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. doi:10.1016/S0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- 31.Rando T.A., Blau H.M. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. doi:10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]