Abstract
Psoriasis is a chronic and complex inflammatory skin disease with lesions displaying dramatically altered mRNA expression profiles. However, much less is known about the expression of small RNAs. Here, we describe a comprehensive analysis of the normal and psoriatic skin miRNAome with next-generation sequencing in a large patient cohort. We generated 6.7 × 108 small RNA reads representing 717 known and 284 putative novel microRNAs (miRNAs). We also observed widespread expression of isomiRs and miRNA*s derived from known and novel miRNA loci, and a low frequency of miRNA editing in normal and psoriatic skin. The expression and processing of selected novel miRNAs were confirmed with qRT-PCR in skin and other human tissues or cell lines. Eighty known and 18 novel miRNAs were 2–42-fold differentially expressed in psoriatic skin. Of particular significance was the 2.7-fold upregulation of a validated novel miRNA derived from the antisense strand of the miR-203 locus, which plays a role in epithelial differentiation. Other differentially expressed miRNAs included hematopoietic-specific miRNAs such as miR-142-3p and miR-223/223*, and angiogenic miRNAs such as miR-21, miR-378, miR-100 and miR-31, which was the most highly upregulated miRNA in psoriatic skin. The functions of these miRNAs are consistent with the inflammatory and hyperproliferative phenotype of psoriatic lesions. In situ hybridization of differentially expressed miRNAs revealed stratified epidermal expression of an uncharacterized keratinocyte-derived miRNA, miR-135b, as well as the epidermal infiltration of the hematopoietic-specific miRNA, miR-142-3p, in psoriatic lesions. This study lays a critical framework for functional characterization of miRNAs in healthy and diseased skin.
INTRODUCTION
Psoriasis (PS) is a chronic, inflammatory skin disease that affects 2–3% of Caucasians, and is less common in other populations (1). In psoriatic lesions, hyperproliferation and defective terminal differentiation of keratinocytes impair barrier formation, infiltration of activated immune cells leads to inflammation and interactions between the two cell types perpetuate disease (2,3). Transcriptome analyses have revealed approximately 1300 protein-coding genes with altered expression in psoriatic skin (4,5). However, much less is known about the expression of non-coding RNAs (ncRNAs), such as microRNAs (miRNAs), in psoriatic skin.
miRNAs are a class of short, regulatory RNAs that play critical roles in human development and disease (6–9). They are transcribed as long stem-loop precursors, which undergo a number of processing steps resulting in the generation of a functional ∼22 nucleotide (nt) single-stranded miRNA. Most miRNA precursors are cleaved through a canonical pathway involving the RNase type III enzymes Drosha and Dicer (10). One key exception is some intronic miRNAs, termed miRtrons, for which spliceosome processing replaces the Drosha cleavage step (11,12). Following cleavage, a single-strand of the RNA duplex (miRNA strand) is incorporated into the RNA-induced silencing complex (RISC) and the unincorporated strand (miRNA* strand) is rapidly degraded (13,14).
Mature miRNAs function by directing RISC to target mRNA transcripts. Target recognition is largely dependent on reverse complementarity between the 5′ miRNA seed region, defined as nt 2–7, and the 3′UTR of the transcript (15,16). miRNA binding may result in translational repression or destabilization of the transcript, although it is still unclear which mechanism is dominant in metazoans (17–19). A single miRNA has the potential to regulate many different targets. Thus, spatial and temporal regulation of miRNAs is crucial to prevent unwanted miRNA–mRNA target interactions.
The first evidence of a role for miRNAs in skin differentiation came from the observation that epidermal specific knockouts of Dicer1 [and later Dgcr8 (DiGeorge syndrome critical region gene 8)] in the mouse embryo perturb development of the epidermis and epidermal appendages (20,21). In the case of PS, two independent miRNA microarray studies interrogating approximately 561 known miRNAs revealed 2- to 8-fold changes in the expression of 23 miRNAs (22,23). However, these few miRNA expression changes are inconsistent with the dramatic shift in cellular composition, differentiation and gene expression characteristic of psoriatic lesions. Hence, a more comprehensive profiling method is needed to advance our understanding of the role of miRNAs in PS. Recently, small RNA library construction coupled with next-generation sequencing (NGS) has been performed with RNA from human tissues and cell lines, providing insights into cancer and other human diseases (24–27).
Here, we describe miRNA expression profiling of 67 normal and psoriatic human skin samples with NGS of small RNAs. We detected mature miRNAs derived from 613 known miRNA precursor families and 284 putative novel miRNA loci in human skin. Digital gene expression analysis (DGE) revealed dramatic changes in global miRNA expression, reflecting defects in keratinocytes, immune cells and vasculature. We identified 80 known and 18 novel miRNAs that were differentially expressed by a fold change of two or more in involved or uninvolved psoriatic skin versus normal skin. We also observed widespread expression of isomiRs and miRNA*s derived from known and novel miRNA loci, and a low frequency of miRNA editing in both normal and psoriatic skin. This study greatly increases our understanding of miRNA expression in human skin under normal and diseased conditions and provides a critical foundation for functional characterization of miRNAs in human skin.
RESULTS
Small RNA sequencing
We obtained punch biopsies from the involved (PP) and uninvolved (PN) skin of PS patients and normal skin (NN) from healthy donors (Supplementary Material, Table S1). Such biopsies are predominantly composed of keratinocytes, but also contain cells derived from epidermal appendages, vasculature, dermal fibroblasts and skin homing immune cell populations. We extracted total RNA using a method that preserved small RNAs and constructed small RNA libraries from 20 NN, 23 PN and 24 PP biopsies. We independently sequenced each of the 67 libraries on the Illumina GAIIx platform, generating 1.1 billion raw and 670 million qualified reads. Raw sequencing data generated in this study have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/, last accessed 8-4-11, Series accession number GSE31037). Three technical replicates produced from a single PP skin biopsy were highly correlated in pairwise comparisons (r2 = 0.994, 0.990 and 0.989; Supplementary Material, Fig. S1). The lengths of the adapter-trimmed, qualified reads were normally distributed around an average length of 22 nt (Supplementary Material, Fig. S2). We mapped qualified reads to miRNA precursors, functional ncRNAs and build 37 (hg19) of the human genome. The number of reads and mapping proportions were roughly equivalent in the NN, PN and PP skin categories (Supplementary Material, Table S2).
Supplementary Material, Figure S3 outlines our mapping strategy and the distribution of read alignments in the cumulative data set (NN, PN and PP combined). Briefly, 305 million (46%) reads aligned to known miRNA precursors, with 99.9% of these overlapping the annotated, mature miRNA or miRNA* sequence ±3 nt. Another 187 million reads aligned to a variety of other functional ncRNAs such as small nucleolar RNAs, small cajal body-associated RNAs and transfer RNAs (data not shown). An additional 41 million reads aligned to other regions of the human reference sequence, including intergenic, intronic and less frequently, exonic regions. Novel miRNA predictions were derived from this pool of reads. The remaining 136 million reads that did not align perfectly to any of these databases were re-mapped to known mature miRNAs with relaxed stringency for the analysis of global miRNA editing patterns.
Known miRNAs in normal and psoriatic skin
We first characterized the expression of known miRNAs in NN, PN and PP skin by examining reads that aligned perfectly to mature miRNA sequences with a 3 nt extension at either end. This flexibility was introduced to allow the detection of various miRNA isoforms that differ at the 3′ or 5′ end because of imperfect enzymatic processing, termed isomiRs (Fig. 1; 24,28). Although 3′ heterogeneity between isomiRs is thought to be largely inconsequential, heterogeneity at the 5′ end is predicted to change the function of the miRNA by shifting the seed sequence. Consistent with this, 5′ heterogeneity was much less prevalent than 3′ heterogeneity, indicating that the vast majority of mature miRNAs derived from a given precursor strand have identical target profiles. The complete set of known miRNA alignments has been provided as Supplementary Material, File S1.
Figure 1.
Example of small RNA reads aligning to a known miRNA precursor (miR-1-1). The mature miR-1-1 sequence, as annotated in miRBase, is underlined. The minimum free energy secondary structure is denoted by dot-bracket notation, where ‘(’ and ‘)’ represent complementary bases and ‘.’ represent non-complementary bases. Numbers in the right column represent the abundance of digital read counts corresponding to each sequence.
A total of 717 mature miRNAs and 176 mature miRNA*s were represented by at least one read in the cumulative data set (Supplementary Material, Table S3). We also detected miRNA* production from several known miRNA loci that are currently lacking miRNA* annotations, including a well-characterized skin-expressed miRNA, miR-203 (Supplementary Material, File S1). The 10 miRNAs represented by the most reads in each skin category (NN, PN and PP) were largely overlapping and accounted for 70% of miRNA reads overall (Table 1). Notably, eight of the 10 most highly represented miRNAs in NN skin (let-7a/b/c/f, miR-143, miR-203, miR-21 and miR-24) overlapped with a set of 47 highly abundant miRNAs previously cloned and sequenced from murine skin (20). One such miRNA, miR-203, has been functionally characterized in skin morphogenesis as a repressor of ▵Np63 (p63 isoform with N-terminal truncation) in the suprabasal layers of the epidermis (29,30). The two highly represented miRNAs in human skin that were not detected by cloning and sequencing in murine skin, miR-26a and miR-451, have murine homologs. Thus, their absence may reflect species-specific differences in skin architecture. Alternatively, their high read counts in our data set may be due to intrinsic bias introduced by miRNA library preparation, resulting in an overestimation of abundance. All other highly expressed miRNAs in murine skin were also detected by NGS in human skin. Differential expression for these and other known miRNAs in NN, PN and PP skin is discussed below, following a description of novel miRNAs detected in skin.
Table 1.
Most abundant known miRNAs expressed in normal, uninvolved psoriatic and involved psoriatic skin
| Mature miRNA | NN |
PN |
PP |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of readsa | %miRNA readsb | Rank | Number of readsa | %miRNA readsb | Rank | Number of readsa | %miRNA readsb | Rank | |
| hsa-let-7a | 48 065 414 | 33.34 | 1 | 39 606 288 | 30.60 | 1 | 44 266 619 | 25.67 | 1 |
| hsa-let-7f | 15 566 363 | 10.80 | 2 | 13 566 250 | 10.48 | 2 | 19 113 977 | 11.08 | 2 |
| hsa-miR-143 | 11 265 038 | 7.81 | 3 | 11 234 955 | 8.68 | 3 | 12 681 332 | 7.35 | 4 |
| hsa-miR-203 | 4 821 268 | 3.34 | 4 | 5 378 508 | 4.16 | 4 | 9 866 015 | 5.72 | 5 |
| hsa-miR-451 | 3 743 389 | 2.60 | 5 | 3 464 570 | 2.68 | 6 | 4 089 856 | 2.37 | 7 |
| hsa-miR-21 | 3 739 479 | 2.59 | 6 | 3 975 815 | 3.07 | 5 | 18 345 268 | 10.64 | 3 |
| hsa-let-7b | 3 625 405 | 2.51 | 7 | 3 360 180 | 2.60 | 7 | 3 345 155 | 1.94 | 9 |
| hsa-miR-26a | 3 526 144 | 2.45 | 8 | 3 133 159 | 2.42 | 9 | 3 160 312 | 1.83 | 10 |
| hsa-miR-24 | 3 375 273 | 2.34 | 9 | 3 179 154 | 2.46 | 8 | 5 118 190 | 2.97 | 6 |
| hsa-let-7c | 2 370 013 | 1.64 | 10 | 1 873 647 | 1.45 | 10 | 1 678 735 | 0.97 | 21 |
| miR-378 | 1 195 818 | 0.83 | 24 | 1 579 574 | 1.22 | 12 | 3 974 309 | 2.30 | 8 |
| Total | 101 293 604 | 70.25 | — | 90 352 100 | 69.80 | — | 125 639 768 | 72.86 | — |
NN, normal skin; PN, uninvolved psoriatic skin; PP, involved psoriatic skin.
aReads that mapped to the annotated mature miRNA ± 3 nt in NN, PN or PP skin.
bAbundance expressed as the percentage of all reads that mapped to annotated mature miRNAs ± 3 nt in NN, PN or PP skin.
Features and genomic distribution of novel miRNAs
Over 1200 mature human miRNAs are registered in miRBase v16. However, as NGS methods improve the depth and quality of miRNA profiling, much debate remains about the true number of human miRNAs. We computationally predicted and prioritized novel miRNAs from small RNA reads that mapped to the human reference sequence on the basis of four criteria: (i) predicted RNA hairpin structure, (ii) presence of miRNA and miRNA* reads aligning to the hairpin stems, (iii) characteristic 3′ overhangs of the Dicer-cleaved miRNA/miRNA* duplex, and (iv) evidence of spliceosome processing for some predicted intronic novel miRNAs.
We identified 284 putative novel miRNA loci which produced 284 mature miRNAs and 227 cognate miRNA*s that were represented by at least one read in the cumulative data set (Supplementary Material, Table S4). The complete set of novel miRNA alignments has been provided as Supplementary Material, File S2. However, pending additional validation of loci associated with very few reads, the bulk of subsequent analyses focused on the 57 novel miRNA loci that were represented by at least four reads per library (268 total reads).
Compared with the known miRNAs expressed in skin, novel miRNAs were typically represented by fewer reads. For example, the most highly represented novel miRNA overall, novel #117, would have ranked in the 72nd percentile for known miRNA read count. Collectively, the top ten most highly represented mature novel miRNAs in each skin category accounted for <0.01% of miRNA reads overall (Table 2). In addition to these novel miRNAs, we validated 21 newly reported miRNA loci from other recent high-throughput sequencing studies as well as a previously described non-canonical miRNA processed from the ACA45 (small cajal-body-specific RNA 15) snoRNA (Supplementary Material, Table S5; 25,31–33).
Table 2.
Most abundant novel miRNAs expressed in normal, uninvolved psoriatic and involved psoriatic skin
| Identifier/location | Sequencea | NN |
PN |
PP |
|||
|---|---|---|---|---|---|---|---|
| Number of readsb | Rank | Number of readsb | Rank | Number of readsb | Rank | ||
| #117/chr14:104583678-104583927(−) | TTGAACTGTTAAGAACCACTGG | 1515 | 1 | 2036 | 2 | 5214 | 1 |
| #342/chr17:39673318-39673567(−) | CACCGACTCTGTCTCCTGCAG | 2816 | 2 | 2065 | 1 | 1199 | 4 |
| #31/chr17:6558659-6558908(−) | TTGGACAGAAAACACGCAGGA | 1174 | 3 | 1480 | 3 | 847 | 6 |
| #78/chr6:33665829-33666078(+) | TCAGGTGTGGAAACTGAGGCAG | 999 | 4 | 1191 | 4 | 2323 | 2 |
| #76/chr6:6169476-6169725(+) | TACAGATGCAGATTCTCTGACTTC | 869 | 5 | 676 | 7 | 697 | 8 |
| #102/chr9:135820997-135821246(+) | GTCGGTGCAAAAGTCATCACGGT | 683 | 6 | 817 | 6 | 554 | 9 |
| #254/chr3:32547709-32547958(−) | GCAAAAGTAATTGTGGTTTTTG | 659 | 7 | 613 | 8 | 762 | 7 |
| #72/chr5:150901536-150901785(−) | TCGGGCGCAAGAGCACTGCAGT | 648 | 8 | 927 | 5 | 1011 | 5 |
| #97/chr8:125834132-125834381(+) | TTAGCCAATTGTCCATCTTTAG | 403 | 9 | 542 | 9 | 429 | 11 |
| #13/chr1:243509367-243509616(+) | TCTGTGAGACCAAAGAACTACT | 332 | 10 | 302 | 14 | 469 | 10 |
| #6/chr1:82174775-82175024(+) | TAAAAGTAATTGTGGTATTTGC | 277 | 13 | 353 | 10 | 1414 | 3 |
| Total | — | 10 375 | — | 11 002 | — | 14 919 | — |
NN, normal skin; PN, uninvolved psoriatic skin; PP, involved psoriatic skin.
aMost abundant sequence derived from the novel miRNA precursor.
bReads that mapped to the most abundant mature miRNA ± 3 nt in NN, PN or PP skin.
Supplementary Material, Table S6 provides the genomic distribution of novel miRNAs with respect to intergenic regions, introns, 3′UTRs, 5′UTRs, exons and ncRNAs compared with the distribution of the known miRNAs we detected in skin. Novel miRNAs showed a higher frequency of intronic localization and a lower frequency of intergenic localization compared with known miRNAs. Of the 185 novel miRNAs that aligned to introns, 62 (34%) aligned to the 3′ end such that the 3′ end of the mature miRNA sequence was within 2 nt of the intron–exon boundary. Ten intronic novel miRNAs aligned to putative miRtrons that were <100 nt in length. Six novel miRNAs mapped uniquely to the antisense strand of known miRNA loci: novel #195/miR-33b, #117/miR-203, #220/miR-371, #273-1/miR-219-1, #273-2/miR-219-2 and #233/miR-1245. Detailed alignments for these novel antisense miRNAs have been provided as Supplementary Material, File S3.
Comparative genomic analysis reveals a recent origin for novel miRNAs
We used comparative genomics to assess conservation of the 57 most highly represented novel and recently described miRNA loci in skin. We classified miRNAs as conserved if both the seed sequence and local hairpin structure were maintained. Interestingly, 24/47 (51%) novel and 5/10 (50%) recently described miRNAs were human-specific. An additional 21/47 (45%) novel and 4/10 (40%) recently described miRNAs were conserved in Macaca mulatta but not Mus musculus, suggesting that they may be primate specific. Only three novel or recently described miRNAs were conserved further down the mammalian lineage: novel #116 (Loxodonta africana), novel #117 (Canis lupus familiaris) and ACA45 small RNA/novel #2456 (L. africana). However, these loci overlapped with other conserved genomic elements: miR-203 (antisense strand), TRAF3 (TNF receptor-associated factor 3) 3′UTR and ACA45. Thus, conservation of these three miRNAs is likely to be an indirect consequence of the conservation of overlapping genomic features. The overall lack of conservation of novel and recently described miRNAs suggests they may have human- or primate-specific functions. However, the possibility remains that some of these newly evolved miRNAs are transient evolutionary intermediates that have yet to develop important regulatory functions. Complete multi-species alignments have been provided as Supplementary Material, File S4.
Validation of novel miRNAs
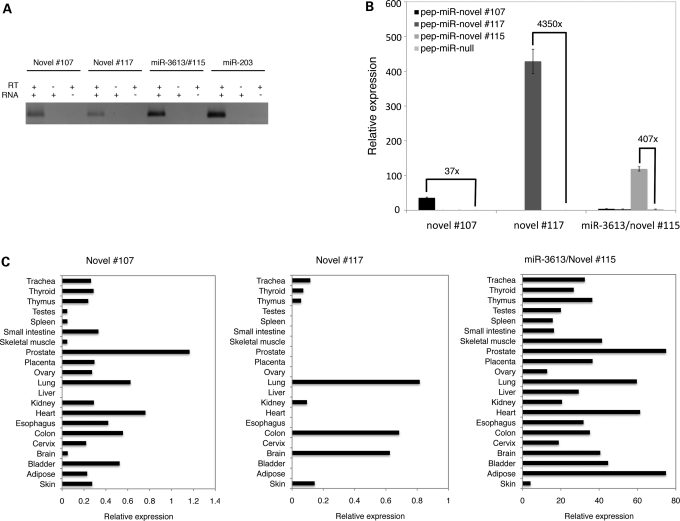
Experimental validation of novel miRNAs is important, particularly in the face of low abundance and poor conservation. We used stem-loop qRT-PCR to validate the expression and processing of two novel and one recently described miRNAs: novel #107, novel #117 and miR-3613/novel #115. We found that all three miRNAs yielded a PCR product of the same length as an abundant known miRNA, miR-203, in NN skin, indicating that these novel miRNAs are endogenously expressed in skin (Fig. 2A). To further confirm that the predicted novel miRNA precursors were recognized by the human miRNA-processing machinery, we ectopically expressed the novel #107, novel #117 and miR-3613/novel #115 precursors in HEK293 cells. Each over-expressed precursor yielded a mature miRNA product that was at a substantially higher level than that seen in wild-type cells, indicating that these novel miRNA precursors were indeed processed into mature miRNAs in human cells (Fig. 2B). Variability in the degree of over-expression from each novel miRNA construct probably reflects differences in precursor-processing efficiency.
Figure 2.
Experimental validation of novel miRNAs with qRT-PCR. (A) Endogenous expression of mature novel miRNAs in skin in the presence of RNA and reverse transcriptase, RNA only or reverse transcriptase only. (B) Levels of mature novel miRNAs following ectopic expression of novel miRNA loci with ‘pep-miR’ vectors in HEK293 cells (see Materials and Methods). Relative expression was calculated with respect to the endogenous snoRNA Z30 (see Materials and Methods). ‘x’ refers to the fold change in mature miRNA expression following ectopic expression of the precursor. (C) Endogenous expression levels of novel miRNAs in 21 human tissues. Relative expression was calculated with respect to the endogenous snoRNA Z30 (see Materials and Methods).
We next assessed whether these three novel miRNAs were skin-specific by performing qRT-PCR on RNA from 20 different human tissues. Each miRNA was expressed in other human tissues at highly variable levels, indicating that these miRNAs are not skin-specific and are subject to tissue-specific transcriptional or post-transcriptional regulation (Fig. 2C).
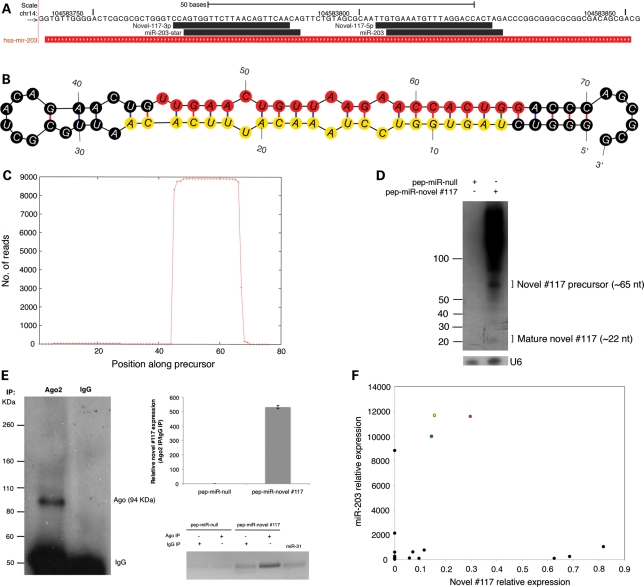
Novel #117 was of particular significance because it appeared to be an antisense miRNA derived from the miR-203 locus (Fig. 3A). The existence of #117 as a bona fide miRNA was confirmed by several lines of evidence. First, it displayed all the features of miRNA biogenesis, including hairpin formation, the presence of miRNA and miRNA* reads and 3′ overhangs on the highest likelihood miRNA/miRNA* duplex (Fig. 3B and C). Second, ectopic expression of the #117 locus in HEK293 cells (which do not endogenously express this miRNA) resulted in two distinct northern blot bands of ∼65 and ∼22 nt, a pattern that is consistent with the expected sizes of the precursor and mature miRNA species, respectively (Fig. 3D). Third, ectopically expressed #117 co-immunoprecipitated with argonaute 2 (Ago2), a core component of the RISC complex, in HEK293 cells, suggesting that the mature miRNA is functionally competent (Fig. 3E). Fourth, there was no correlation between the expression patterns of miR-203 and novel #117 in a panel of human tissues (Fig. 3F). Taken together, these results support the transcription and processing of two distinct miRNAs derived from the sense and antisense strands at this locus. Henceforth, we will refer to novel #117 as miR-203-AS. Notably, miR-203 and miR-203-AS do share sequence similarity, but the imperfect palindromic nature of the locus leads to the production of distinct sense and antisense products. Of particular importance is a single base difference in their seed regions, which would indicate that the two miRNAs have distinct target profiles. Because miR-203 is more abundant than miR-203-AS in all human tissues, and has long been recognized as a miRNA, we conclude that miR-203-AS is the minor product derived from this locus.
Figure 3.
Features of a validated antisense miRNA locus (miR-203/novel #117). (A) Alignment of the dominant, known and novel small RNA reads derived from the miR-203 locus. Forward strand sequence encoding miR-203 is indicated. The 3′ and 5′ arms of novel #117 correspond to the miRNA and miRNA* strands, respectively. (B) Predicted minimum free energy secondary structure of novel #117. Red circles represent the dominant mature miRNA sequence. Yellow circles represent the dominant mature miRNA* sequence. (C) Distribution of small RNA reads along the length of the novel #117 precursor. (D) Northern blot showing the accumulation of precursor and mature forms of novel #117 following ectopic expression of the #117 locus with a ‘pep-miR’ vector in HEK293 cells (see Materials and Methods). The endogenous U6 snRNA served as an input control. (E) Co-immunoprecipitation (IP) of ectopically expressed novel #117 with argonaute 2 (Ago2). Left, western blot showing specificity of the Ago2 antibody. Top right, relative abundance of novel #117 in Ago2 IP compared with IgG control as indicated by qRT-PCR. Bottom right, RT-PCR bands corresponding to Ago2 and IgG IPs quantified in the top right panel. (F) Correlation of miR-203 and novel #117 expression in NN (green circle), PN (yellow circle), PP skin (red circle) and other human tissues (black circles). Relative expression was calculated with respect to the endogenous snoRNA Z30 (see Materials and Methods).
Altered expression of known and novel miRNAs in psoriatic skin
We performed DGE analysis to compare the expression of known and novel miRNAs in NN, PN and PP skin. First, we normalized the number of miRNA reads to the total number of reads that mapped to the reference sequence in each skin category. Next, we implemented a detection threshold of 268 raw reads in the cumulative data set (an average of four reads per individual library), which was empirically adjusted to allow the lowest detection threshold while minimizing noise. This resulted in a set of 512 known, 13 recently described and 47 novel mature miRNAs and miRNA*s.
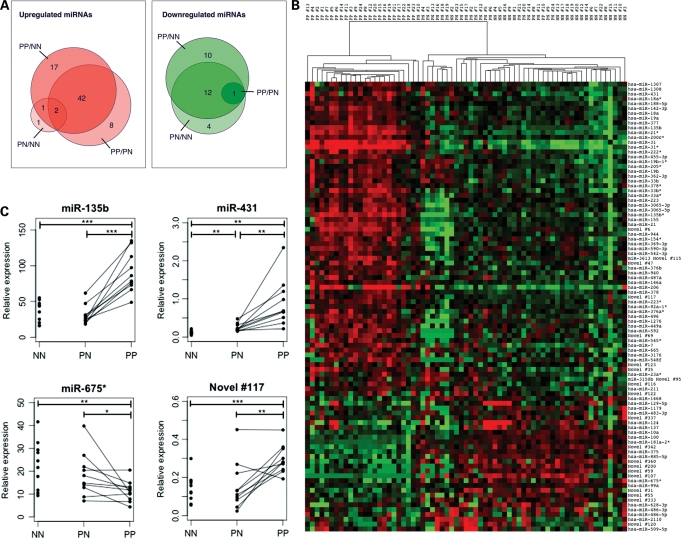
Ninety-eight miRNAs, including 18 novel miRNAs, were significantly differentially expressed with a ±2-fold change in at least one comparison: PP/NN, PP/PN or PN/NN (Bonferroni adjusted P < 0.05; Table 3, Supplementary Material, Table S7). Seventy-one miRNAs were upregulated, whereas 27 were downregulated. This is substantially greater than the number of miRNAs previously shown to be ±2-fold differentially expressed in psoriatic skin from two previous microarray analyses (22,23). The majority of miRNA expression changes occurred in PP skin compared with both PN and NN (Fig. 4A). Thus, clustering of samples based on the expression of the top 98 differentially expressed miRNAs completely distinguished PP from PN and NN skin (Fig. 4B). Only five miRNAs were ±2-fold differentially expressed between PN and NN skin: miR-31, miR-206, miR-509-5p, miR-675* and novel #122. These findings are consistent with mRNA expression changes that have been described in psoriatic skin, in that there are more upregulated than downregulated transcripts and relatively few changes between PN and NN skin (4,5). Many more miRNAs exhibited more modest fold changes in psoriatic skin. A complete list of miRNAs that were ±1.4-fold differentially expressed in at least one comparison has been provided as Supplementary Material, Table S8.
Table 3.
Top ten upregulated and downregulated miRNAs in involved psoriatic versus normal skin
| Mature miRNA | Fold changea |
Adjusted P-value | Total reads (raw) | ||
|---|---|---|---|---|---|
| PP/NN | PP/PN | PN/NN | |||
| hsa-miR-31 | 42.93 | 14.03 | 3.06 | 0 | 55 883 |
| hsa-miR-31* | 34.51 | 24.17 | 1.43 | 0 | 10 412 |
| hsa-miR-206 | 12.80 | 4.84 | 2.65 | 0 | 2156 |
| hsa-miR-21* | 8.80 | 6.08 | 1.45 | 0 | 8754 |
| hsa-miR-135b* | 8.00 | 7.55 | 1.06 | 5.7324E − 131 | 694 |
| hsa-miR-33b* | 5.70 | 5.20 | 1.10 | 5.87623E − 73 | 521 |
| hsa-miR-135b | 5.65 | 5.32 | 1.06 | 0 | 5362 |
| hsa-miR-222* | 5.59 | 3.35 | 1.67 | 0 | 3902 |
| hsa-miR-1268 | 4.45 | 3.03 | 1.47 | 0 | 11 154 |
| Novel #6 | 4.10 | 3.36 | 1.22 | 1.0861E − 185 | 2044 |
| Novel #107 | −8.07 | −6.06 | −1.33 | 2.72295E − 24 | 326 |
| hsa-miR-124 | −5.87 | −3.02 | −1.94 | 0 | 4518 |
| hsa-miR-675* | −4.44 | −1.99 | −2.23 | 4.98729E − 46 | 617 |
| Novel #360 | −4.16 | −3.77 | −1.10 | 3.62519E − 20 | 434 |
| Novel #200 | −3.79 | −3.75 | −1.01 | 7.61644E − 13 | 308 |
| Novel #342 | −2.94 | −2.04 | −1.43 | 5.0832E − 210 | 6080 |
| hsa-miR-483-3p | −2.93 | −1.61 | −1.82 | 1.48677E − 10 | 290 |
| hsa-miR-129-5p | −2.62 | −4.51 | 1.72 | 6.5151E − 170 | 2723 |
| hsa-miR-486-3p | −2.56 | −2.37 | −1.08 | 4.71991E − 56 | 2085 |
| hsa-miR-885-5p | −2.56 | −2.33 | −1.10 | 4.13693E − 22 | 875 |
NN, normal skin; PN, uninvolved psoriatic skin; PP, involved psoriatic skin.
aBased on normalized read counts in NN, PN and PP skin (reported in Supplementary Material, Table S7).
Figure 4.
Differential expression of known and novel miRNAs in normal, uninvolved psoriatic and involved psoriatic skin. (A) Venn diagram indicating the number of differentially expressed miRNAs that were ±2-fold differentially expressed in any of the three comparisons: PP/NN, PP/PN, PN/NN. (B) Heat map showing hierarchical clustering of skin samples on the basis of 98 differentially expressed miRNAs. (C) qRT-PCR levels of differentially expressed miR-135b, miR-431, miR-675* and novel #117/miR-203-AS in 10 NN, 10 PN and 10 PP patients. Lines indicate matched uninvolved and involved samples from the same patient (***P < 0.001, **P < 0.01,*P < 0.05). Relative expression was calculated with respect to the endogenous snoRNA Z30 (see Materials and Methods). NN, normal skin; PN, uninvolved psoriatic skin; PP, involved psoriatic skin.
miRNAs that were derived from the same precursor were frequently co-regulated. There were 66 differentially expressed miRNAs/miRNA*s for which a cognate miRNA*/miRNA also passed our detection threshold for DGE. Eighty percent of these cognate strands exhibited a similar expression trend in the DGE data set (±1.4-fold cut-off). One exception was miR-431/431*. miR-431 was upregulated in PN and PP skin compared with NN (3.2- and 3.9-fold, respectively), whereas miR-431* remained unchanged (1.2- and 1.0-fold). We subsequently validated this finding with stem-loop qRT-PCR (Fig. 4C, Supplementary Material, Fig. S4). Thus, a subset of differentially expressed miRNAs may exhibit altered strand selection in psoriatic skin.
We selected 13 differentially expressed miRNAs for independent validation with stem-loop qRT-PCR and confirmed that 8/10 known and 3/3 novel miRNAs were significantly differentially expressed in PP skin compared with PN or NN (Fig. 4C, Supplementary Material, Fig. S4). However, changes between NN and PN skin did not validate as well, presumably because the fold changes were typically smaller. Overall, qRT-PCR levels and normalized digital read counts for these 13 differentially expressed miRNAs were highly correlated (r2 = 0.979).
RNA in situ hybridization
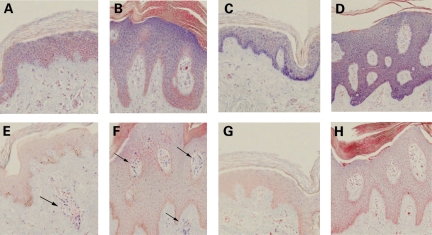
We analyzed the expression patterns of three selected miRNAs in PP and PN skin with RNA in situ hybridization: miR-135b (PP/NN = 5.6, PP/PN = 5.3); miR-205 (PP/NN = 1.56, PP/PN = 1.15); and miR-142-3p (PP/NN = 2.52, PP/PN = 2.95). Although miR-205 had not met our 2-fold threshold for differential expression in PS, its modest upregulation in PP skin coupled with its previously described role in the establishment of epithelial cell fate (34) pointed towards a role in PS pathogenesis.
We found that miR-135b and miR-205 were variably expressed in the stratified layers of the epidermis in PN and PP skin. miR-135b was expressed at higher levels in the suprabasal epidermis (Fig. 5A and B), which is similar to the expression pattern of miR-203, described elsewhere (22). In contrast, miR-205 was expressed at higher levels in the basal epidermis (Fig. 5C and D). These staining patterns are strongly suggestive of a role for miR-135b and miR-205 in keratinocyte differentiation. miR-142-3p displayed strong evidence of immune cell staining (Fig. 5E and F), which is consistent with the previously described expression of miR-142-3p in hematopoietic tissues (35). Interestingly, miR-142-3p exhibited staining to lumen-like structures within the dermis in PN skin (Fig. 5E), and staining to similar structures within the epidermis in PP skin (Fig. 5F), supporting a role for miR-142-3p in epidermal inflammation in psoriatic lesions.
Figure 5.
RNA in situ hybridization for skin-expressed miRNAs in uninvolved psoriatic and involved psoriatic skin sections. Expression of miR-135b in the (A) PN and (B) PP epidermis. Expression of miR-205 in (C) PN and (D) PP epidermis. Expression of miR-142-3p in (E) dermal immune cells (arrowheads) in PN skin and (F) dermal/epidermal immune cells (arrowheads) in PP skin. Scramble-miR background signal in (G) PN and (H) PP skin. PN, uninvolved psoriatic skin; PP, involved psoriatic skin.
miRNA editing
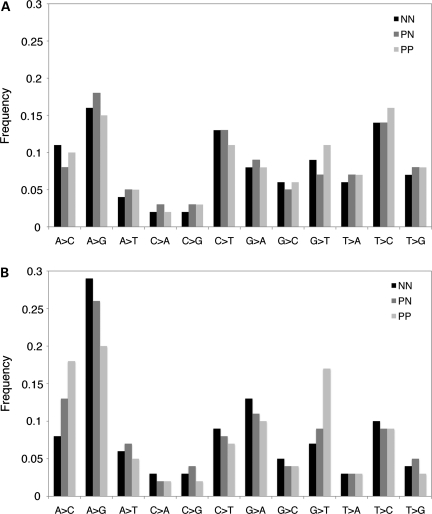
In order to analyze global miRNA editing patterns in NN, PN and PP skin, we examined reads that aligned to mature miRNAs with one mismatch. We first filtered out spurious editing events by removing mismatches with a low quality score or mismatches that were consistent with non-templated nucleotide additions at the 3′ terminus. The remaining single mismatch reads represented 1% of total miRNA reads, indicating that miRNA editing occurs at a low frequency. We next analyzed the frequencies of all possible single-base substitutions, with particular focus on the two forms of substitutional RNA editing that have been previously described in mammals: cytosine deamination (C → U conversions) and adenosine deamination (A → I conversions; 36,37). A → I and C → U conversions would appear as A → G and C → T mismatches in the sequencing data set, respectively. If there were an unbiased distribution of all possible substitutions, each would represent ∼8.3% of mismatched reads. However, we observed a clear over-representation of A → G and C → T substitutions, which accounted for 16.3 and 12.3% of single mismatch reads in the cumulative data set, respectively (P < 0.0001; Fig. 6A). A high relative frequency of T → C substitutions (14.7%) was also observed, although the underlying mechanism is unknown (Fig. 6A).
Figure 6.
Patterns of miRNA editing in normal, uninvolved psoriatic and involved psoriatic skin. Global frequencies of single base pair substitutions detected in (A) full-length miRNAs and (B) miRNA seed regions in NN, PN and PP skin. NN, normal skin; PN, uninvolved psoriatic skin; PP, involved psoriatic skin.
We next examined the frequency of single-base substitutions specifically in the miRNA seed region, which is the primary determinant of target recognition. We found that the frequency of A → G substitutions in the seed region was 8.7% higher than that in the full-length miRNA (P < 0.0001; Fig. 6B). In contrast, the frequency of C → T substitutions in the seed region was 4.3% lower than that in the full-length miRNA, restoring the frequency of C → T substitutions to background levels within the seed (Fig. 6B). Taken together, these results suggest that mature miRNAs are subject to cytosine and adenosine deamination, and that adenosine, but not cytosine, deamination in the miRNA seed region might be an important mechanism for modulating miRNA target interactions. We observed only minor differences in the frequencies of A → G and C → T substitutions between NN, PN and PP skin, but the possibility remains that a small set of individual miRNAs are subject to differential editing in psoriatic skin.
DISCUSSION
Deep sequencing of small RNAs has made it possible to comprehensively probe the miRNAome of normal and diseased human tissues. In the present study, we have leveraged this technology in skin to produce the largest small RNA data set from any human tissue to date. The depth of this data set allowed us to detect low abundance, novel and edited miRNAs with unparalleled sensitivity. We have shown extensive alterations to the psoriatic miRNAome, many of which have not been previously reported, including the differential expression of a novel antisense miRNA derived from the miR-203 locus. Overall, this work lays a critical foundation for future studies characterizing the role of miRNAs in skin development and disease.
Reliability of miRNA profiling with NGS
Small RNA sequencing may be subject to intrinsic bias introduced by mechanisms such as non-random adapter ligation or sequence-based differences in PCR efficiency (38). Such bias could result in skewing of absolute quantification of miRNAs, but should not affect comparative analysis of singular miRNAs. Despite this potential bias, we observed a close correlation between normalized digital read counts and qRT-PCR levels, suggesting that NGS of small RNAs is a reliable method for miRNA profiling. Furthermore, we observed strong concordance between the present study and a previous study analyzing miRNA expression in murine skin by traditional cloning and sequencing (20). Thus, we conclude that digital read count is an adequate proxy for miRNA abundance.
We also replicated several miRNA expression changes from two previous miRNA microarray studies which identified 23 miRNAs as ±2-fold differentially expressed in PS (22,23). Of these 23, we conclusively confirmed 6: miR-21, miR-31, miR-142-3p, miR-146a, miR-223 and miR-378 (±2-fold change in our study). We tentatively confirmed an additional seven: miR-17-5p, miR-30e-5p, miR-122a, miR-141, miR-142-5p, miR-146b and miR-203 (±1.4-fold change in our study). These 13 confirmed miRNAs included 6 out of 8 miRNAs that were ±2-fold differentially expressed according to both microarray studies: miR-17, miR-21, miR-31, miR-142-3p, miR-146a/b, miR-200a and miR-203. The remaining 10 miRNAs that were previously reported as ±2-fold differentially expressed in PS were detected by NGS in skin, but showed no evidence for differential expression. We also identified 67 known and 18 novel differentially expressed miRNAs that have not been previously implicated in PS and have validated several of these by stem-loop qRT-PCR. Many of the differentially expressed miRNAs reported in this study were likely to have been missed by previous studies because of their low abundance or because microarray platforms lacked probes that interrogated them.
One tentatively validated miRNA was miR-203, which was reported to be 5.86- and 2.02-fold upregulated in PP versus NN skin, according to the two previous miRNA microarray studies in PS (22,23, respectively). With the DGE analysis described here, miR-203 exhibited a 1.6- and 1.4-fold upregulation in PP skin compared with NN and PN, respectively, which was lower than microarray-based fold changes. We confirmed these modest fold changes with qRT-PCR in our patient cohort (data not shown). The current model for miR-203 function is that its expression in the suprabasal layers of the epidermis limits the proliferative potential of keratinocytes, which establishes a well-defined boundary between proliferating and terminally differentiating keratinocytes (30). Given the hyperproliferative phenotype of psoriatic keratinocytes, the upregulation of the anti-proliferative miR-203 in psoriatic skin is puzzling. However, NGS revealed upregulation of a novel antisense miRNA derived from the miR-203 locus, which we have designated miR-203-AS. Although miR-203-AS was much less abundant than miR-203 in skin, it was the most abundant novel miRNA in the cumulative data set. Furthermore, it was 2.7- and 2.2-fold upregulated in PP and PN skin, respectively, compared with NN. Taken together, our findings more strongly support a role for miR-203-AS in PS, but do not exclude the involvement of miR-203. Functional characterization of miR-203-AS will likely help to reconcile apparent inconsistencies in the biological and pathogenic functions of miR-203.
Significance of low abundance miRNAs
The major advantages of deep sequencing of large cohorts are enhanced sensitivity and dynamic range. The 1442 known, recently described and novel miRNAs we detected in skin produced between 1 and 130 million reads, with the 10 most abundant miRNAs accounting for nearly 70% of all miRNA reads. This finding is consistent with other recently published NGS studies (33,39) and prompts the question of how important the myriad low abundance miRNAs are.
Normal and psoriatic skin biopsies are largely composed of keratinocytes, but contain many other cell types as well, such as fibroblasts and specialized immune cells (40). Some of these are critical for the disease process, but are low in abundance. Consequently, important miRNAs expressed in rare cell types can be drowned out by signals from keratinocyte-derived miRNAs, even if they serve important regulatory functions that influence disease pathogenesis. Indeed, only 2 of the 98 differentially expressed miRNAs represented >1% of miRNA reads in the cumulative data set, indicating that miRNAs of moderate-to-low abundance account for most of the variation in the psoriatic miRNAome. Alternatively, bias introduced during library preparation may have led to an underestimate of miRNA abundance in some cases. It is unlikely that differential expression of these low abundance miRNAs was due to random fluctuations in digital read count because all of these miRNAs exhibited expression changes that were highly reproducible across individuals and statistically significant. Furthermore, differential expression of miRNAs that were represented by as few as 295 reads in the cumulative data set was independently validated with stem-loop qRT-PCR.
Novel miRNA identification
We have reported 284 putative novel miRNA genes, and 22 recently described miRNA genes that were expressed in skin, 3 of which were subjected to extensive experimental validation. Recently described miRNAs were initially characterized as novel, but were independently annotated by other groups while we were analyzing and validating our findings. These annotations were largely due to the recent publication of miRNA profiles from melanoma cell lines and tissues of the female reproductive tract, which each applied similar prediction criteria as the present study (25,32,33). The partial overlap between our study and others provides reassuring validation of our NGS-based in silico method, and suggests that the unprecedented size of our data set was responsible for the identification of such a large number of completely novel miRNAs.
The majority of novel miRNA loci were poorly conserved, which is perhaps not surprising, based on the fact that some miRNA discovery studies have relied on conservation as a prediction criterion. However, in more recent NGS-based studies as well as the present study, conservation was excluded as a prediction criterion in order to obtain a comprehensive profile of all miRNAs that are expressed in human skin, including those that may be newly evolved or evolutionarily transient (25,32,33). Indeed, this approach has led to the discovery of 18 differentially expressed novel miRNAs in psoriatic skin, presenting the intriguing possibility that some newly evolved novel miRNAs may serve human- or primate-specific functions with relevance to PS pathogenesis. According to a recent comparative genomics study, 269 known human miRNAs are primate-specific (41). Consistent with our data, primate-specific miRNAs are generally expressed at low levels in adult tissues, and their computationally predicted targets show functional enrichment for early developmental processes, cell cycle and proliferation (41). Thus, the differential expression of poorly conserved novel miRNAs in psoriatic skin could reflect an innate developmental defect in skin barrier formation.
One small class of novel miRNAs comprised miRNAs derived from the antisense strand at known miRNA loci. To date, only a few known miRNA loci in humans, such as miR-103-1 and miR-103-2, have antisense miRNAs registered in miRBase. However, this phenomenon has been described in detail at the miR-iab-4 locus in Drosophila melanogaster. Both miR-iab-4 and its antisense counterpart miR-iab-8 regulate Hox genes during larval development, but they are functionally distinct based on their discordant expression patterns and unique target profiles (42). Although there were many antisense reads present in our data set, most were consistent with RNA degradation, potential siRNA production or artifacts of palindromic sequence. However, six antisense miRNAs reported in this study showed strongly convincing features of miRNA biogenesis. These sense and antisense miRNAs could functionally interact to create complex regulatory networks or feedback loops.
The largest class of novel miRNAs aligned to introns, which has important implications for biogenesis and function. miRtrons were first described in Drosophila as ∼60 nt introns that resembled pre-miRNAs, and thus do not require Drosha for biogenesis (11). Although such small introns are rare in humans, we did identify 10 putative novel miRtrons ranging from 55–94 nt in length. The remaining intronic miRNAs aligned to longer introns, but were biased towards 3′–5′ intron–exon boundaries. This non-random distribution of intronic novel miRNAs strongly supports spliceosome involvement in the processing of some human miRNAs. The processing of intronic miRNAs may also vary with the expression of alternatively spliced transcripts.
Significance of intronic/intragenic miRNAs
Mounting evidence suggests that intragenic miRNAs, such as intronic miRNAs, functionally interact with their host genes. For example, miR-483, which lies in an intron of the insulin-like growth factor 2 gene (IGF2), has been shown to negatively regulate IGF2 expression via a negative feedback loop (43). Likewise, miR-33, which lies in an intron of the sterol-response-element-binding protein gene (SREBP), regulates cholesterol homeostasis (44). In other cases, a function that has been ascribed to a protein-coding gene may actually be mediated by an intragenic miRNA. For example, the melastatin 1 (MLSN1) gene encodes a protein that was long thought to suppress motility and invasion of melanoma cells, but it has recently been recognized that the intronic miRNA miR-211 is actually responsible for this phenotype (45). Thus, taking into account host gene function of intragenic miRNAs could help to elucidate the pathways and specific targets they regulate.
Notably, five differentially expressed intragenic miRNAs in our data set were encoded within host genes that are differentially expressed in PS (4,5). The upregulated miRNA, miR-147b, is located within the 3′UTR of NMES1 (normal mucosa of esophagus-specific 1), which is upregulated in psoriatic skin. The downregulated miRNAs, miR-10a, miR-100 (and miR-125b, which just narrowly missed our 2-fold cut-off), and novel #342 lie within introns of HOXB3 (homeobox 3), LOC399959 (locus encoding cDNA FLJ34394) and KRT15 (keratin 15), respectively, which are also downregulated in psoriatic skin. LOC399959 encodes an uncharacterized ncRNA, and our findings suggest that this ncRNA likely functions as a polycistronic miRNA precursor. Novel #23 lies within intron 2 of IFI27 (interferon-α inducible protein 27), which is upregulated in psoriatic skin; this miRNA was omitted from our digital DGE because of low abundance, but showed strong evidence for upregulation in psoriatic skin on the basis of normalized digital read counts. Although some intragenic miRNAs have autonomous promoters, the co-regulation of these miRNAs and their host transcripts in PS suggests that these miRNAs are largely dependent on their host gene promoters for transcription. Thus, these differentially expressed intronic miRNAs may be functioning cooperatively with their dysregulated host transcripts to influence PS pathogenesis.
Altered expression of angiomiRs in involved psoriatic skin
miR-21, miR-31 and miR-378 are three of the most abundant and differentially expressed miRNAs in PP skin and are members of a growing class of miRNAs termed ‘angiomiRs’ (46). Pro-angiomiRs promote angiogenesis by targeting negative regulators in angiogenic signaling pathways, whereas anti-angiomiRs inhibit angiogenesis by targeting positive regulators of angiogenesis. The role of these and other differentially expressed angiomiRs in psoriatic skin is of interest, given its proclivity for neovascularization.
Activated keratinocytes mediate angiogenesis through increased synthesis of vascular endothelial growth factor (VEGF), platelet-derived growth factor and other endothelial cell mitogens, and inflammatory skin disease with some features of PS is induced by overexpression of VEGF in murine skin (47–50). miR-378 is proposed to promote VEGF expression by competing with miR-125 (which was modestly downregulated in PP skin) for the same binding site in the VEGF 3′UTR (51). VEGF is upregulated in psoriatic lesions, (52) and has been shown to induce expression of the upregulated miRNAs miR-18a, miR-31 and miR-155 (53).
Interestingly, although a number of pro-angiomiRs were upregulated in PP skin, many miRNAs that would be predicted to inhibit angiogenesis by repressing VEGF were not differentially expressed. This includes miR-15b, miR-16 and miR-20a/b (46). One anti-angiomiR, miR-100, was downregulated in PP skin. miR-100 has been shown to inhibit angiogenesis by repressing the mammalian target of rapamycin (mTOR) in endothelial cells (54). Interestingly, the mTOR-binding partner, regulatory associated protein of mTOR (RAPTOR), is encoded at 17q25, directly under a PS association peak (55), pointing to mTOR upregulation in lesions through loss of RAPTOR activity or decreased levels of miR-100. These findings suggest that therapeutic application of anti-angiomiR mimics might improve symptoms of PS.
miRNA editing
Analysis of single mismatch miRNA reads led to the observation that miRNAs are subject to adenosine and cytidine deamination. There are various examples of functionally important A → I editing of miRNAs catalyzed by adenosine deaminases acting on RNA (9,56,57). For example, targeted A → I editing within the seed region of miR-376 in some human tissues alters the recognition of mRNA targets (57). The role of cytidine deaminases, such as apolipoprotein B mRNA editing enzymes (APOBECs), in miRNA editing is not well understood. However, a recent meta-analysis of small RNA sequences derived from Oryza sativa and Arabidopsis thaliana revealed a similar over-representation of C → T substitutions, suggesting that cytidine deamination may be a predominant mechanism for miRNA editing in eukaryotes (58). Further experiments will be required to determine whether the majority of cytidine deamination events are spontaneous or enzymatically catalyzed. Intriguingly, the cytidine deaminases APOBEC3A and APOBEC3B are expressed in human skin and are upregulated in psoriatic skin (5,59). Although we observed no differences in the global frequency of C → T substitutions in PS, the possibility remains that a small set of miRNAs could be hyper-edited by APOBEC3 enzymes in psoriatic skin or immune cells.
Conclusions
The global patterns of miRNA expression described here have drastically expanded our understanding of miRNAs in normal and psoriatic skin. Furthermore, we have shown that differentially expressed miRNAs are likely to influence many processes that are involved in PS pathogenesis such as angiogenesis (miR-21, miR-31, miR-378), epidermal differentiation (miR-135b, miR-205, miR-203-AS) and inflammation (miR-142-3p). A long-term goal of miRNA research is therapeutic application. Because skin is the most accessible organ in the body, cutaneous diseases such as PS are likely to be on the front line of miRNA therapeutics. The comprehensive profiling of the miRNAome in normal and psoriatic skin as described here represents a critical first step towards this goal.
MATERIALS AND METHODS
Samples
From healthy controls and the uninvolved and involved skin of PS patients, 4–6 mm punch skin biopsies were collected. PS patients enrolled in this study received no systemic, photo or topical therapy in the 4 weeks prior to sample collection. Biopsies were stored in RNAlater (Qiagen) at −80°C prior to RNA extraction.
Small RNA library preparation and sequencing
RNA was extracted with the miRNeasy Mini Kit (Qiagen), with on-column DNase I digestion. RNA was prepared for sequencing on the Illumina GAIIx platform with the Small RNA Sample Prep Kit (Illumina) according to the manufacturer's instructions (protocol v1.5). This protocol required the use of a proprietary 3′ adapter that has a high affinity for Dicer cleavage products. Briefly, 3′ and 5′ adapters were ligated to 1 μg of total RNA. cDNA was synthesized with SuperScript II Reverse Transcriptase (Invitrogen) and subjected to 12 cycles of PCR amplification with high-fidelity Phusion Polymerase (Finnzymes Oy). Each library was loaded on a single Illumina lane at 20 pm and subjected to 36 cycles of sequencing.
Read processing and mapping
Each deep sequencing library was processed independently. Reads with a 3′ adapter substring <6 nt or trimmed sequence length <17 nt were removed from the data set. Trimmed reads were mapped to multiple human sequencing databases with Bowtie: miRNA precursors (miRBase v.16, http://www.mirbase.org/ftp.shtml, last access date: 8-3-11), ncRNAs (fRNAdb, http://www.ncrna.org/frnadb/download, last access date: 8-3-11) and the hg19 build of the human genome (UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgTables?command=start, last access date: 8-3-11; 60–66). Reads that mapped to miRNA precursors were attributed to mature miRNAs if they aligned to the annotated mature sequences with 3 nt up- and downstream extensions.
Novel miRNA prediction
Qualified reads that aligned to the hg19 build of the human genome were subjected to our novel miRNA prediction pipeline. Any reads that mapped to previously described miRNA loci were removed, and loci that shared adjacent reads within a gap of ≤30 nt were merged. For each locus, a series of overlapping DNA sequence segments was extracted for secondary structure analysis with RNAfold (http://www.tbi.univie.ac.at/~ivo/RNA/, last access date: 8-3-11; 67–69). The starting sequence segment extended 220 nt upstream of the locus, and subsequent segments were extracted by a sliding window of 250 nt, with an increment of 100 nt, until the window reached 220 nt downstream of the sequence reads. Segments lacking stems of at least 18 nt and segments lacking reads that mapped to any of their stems were excluded. Candidate miRNAs were prioritized based on (i) the occurrence of sequencing reads on the stem of a predicted hairpin structure (minimum free energy less than −18 kcal/mol); (ii) the presence of miRNA* reads on the opposite stem of the hairpin; (iii) the presence of 3′ overhangs on the highest likelihood miRNA/miRNA* duplex; and (iv) the evidence of spliceosome-mediated precursor processing based on alignment to intron–exon boundaries.
Novel miRNA conservation
Homology searches were performed in eight species: M. mulatta, M. musculus, C. l. familiaris, L. africana, Monodelphis domestica, Gallus gallus, Xenopus tropicalis and Danio rerio. The novel miRNA precursor sequences, defined as the maximal extension of sequences bearing the stem-loop structure, were mapped to each species' genome using blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi/, last access date: 8-3-11) with a minimal word-size of seven. Blastn hits that were reported in any given species shared >90% sequence similarity and had a ratio of alignment length to the precursor length of >0.9. Secondary structure of aligned sequences was determined with RNAfold. Aligned sequences were classified as conserved novel miRNAs if the following criteria were met: (i) the highest likelihood secondary structure was a hairpin with a minimum free energy less than −18 kcal/mol, (ii) the mature miRNA sequence was derived from the hairpin stem, and (iii) the miRNA seed region (nt 2–7) was perfectly conserved.
Digital gene expression
Reads that aligned perfectly to mature known and novel miRNAs with 3 nt extension were subjected to DGE analysis. Reads that mapped to multiple mature miRNAs were attributed to all potential derivative miRNAs. Read counts in each skin category (NN, PN and NN) were normalized to adjust for slight variation in total read count between categories. Let N be the number of qualified reads that aligned to hg19, C the number of categories and M the number of qualified miRNA reads. Thus, the normalized number of reads for each miRNA in a given category is (Ntotal * Mcategory)/(C* Ncategory). A detection filter was applied such that miRNAs that were represented by fewer than 268 raw reads in the cumulative data set were removed. Fold changes were calculated from normalized read counts, and Pearson's χ2 test with Bonferroni correction was applied to determine significance.
qRT-PCR
qRT-PCR of mature miRNAs was performed with TaqMan miRNA assays according to the manufacturer's instructions (Life Technologies). Briefly, 5 ng of total RNA was reverse-transcribed in a 7.5 μl reaction with the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies), and 0.67 μl of cDNA was added to triplicate 10 μl PCR reactions. PCR was performed on a 7900HT thermocycler (Life Technologies). miRNA expression was normalized to the endogenous snoRNA, Z30. Relative expression levels were calculated according to the 2−ΔΔCt method as follows: 100 * 2((Ct Z30) − (Ct miRNA)) (70). Significance was determined with one-way ANOVA and post hoc two tailed t-tests.
miRNA overexpression constructs
miRNA overexpression constructs were generated from the pEP-miR cloning and expression vector (Cell Bio Labs). Briefly, miRNA precursors ±100 nt were amplified from genomic DNA. PCR products were cloned into the BamHI and NheI sites of the vector. Transformants were selected on 1 μg/ml ampicillin and selected transformants were validated by Sanger sequencing.
Transfections
HEK293 cells were cultured in DMEM supplemented with 2 mm l-glutamine, 10 mg/ml penicillin–streptomycin and 10% fetal bovine serum at 37°C and 5% CO2. Transfections were performed in triplicate. Twenty-four hours prior to transfections, 1 × 105 cells were plated in each well of a 24-well plate. Transfections were performed with TransIT-LT1 transfection reagent according to the manufacturer's instructions (Mirus). Briefly, 750 ng of pEP-miR and 3.75 μl of LT-1 were incubated in 46.25 μl of RPMI for 30 min at 22°C before treatment. Cells were collected 48 h post-treatment.
miRNA northern blots
Thirty micrograms of total RNA was mixed with formamide loading dye and incubated at 65°C for 20 min. Samples were loaded on a pre-warmed 12% denaturing polyacrylamide gel (Sequagel), and run at 100 V until bromphenol blue reached the bottom of the gel. RNA was transferred onto a Genescreen Plus membrane (Perkin Elmer) with a Trans-Blot SD semi-dry transfer cell (Bio-Rad) at 250 mA for 15 min. The membrane was baked for 1 h at 80°C, pre-hybridized for 2h in PerfectHyb Plus (Sigma) at hybridization temperature and hybridized overnight with a 32P-labeled DNA probe. miR-203-AS probe sequence was 5′-CCAGTGGTTCTTAACAGTTCAA-3′. The membrane was washed three times in 0.1× SSC, 0.1% SDS at hybridization temperature and exposed for 3 days. For input control, the membrane was stripped with two 20 min applications of boiled 0.1% SDS with gentle agitation at 22°C, and re-hybridized with a 32P-labeled U6 snRNA LNA probe (Exiqon).
Ago2 immunoprecipitations
HEK293 cells were transfected with pEP-miR-null or pEP-miR-novel #117 constructs as described above, except that transfections were scaled up to generate one 10 cm plate per immunoprecipitation. Cells were washed three times in 1× PBS and UV-crosslinked once for 400 mJ/cm2 and again for 200 mJ/cm2, with gentle agitation in between. Cells were pelleted by centrifugation at 4000 r.p.m. for 5 min at 4°C. Cell pellets were washed once in 1× PBS and resuspended in 200 μl of 1× PBS, 0.1% SDS, 0.5% deoxycholate, 0.5% nonidet P-40, supplemented with 1 U/μl RNasin (Promega) and 1× Complete Protease Inhibitor Cocktail (Roche). Lysates were incubated on ice for 10 min, and cleared by centrifugation at 10 000 r.p.m. for 10 min at 4°C. Each cleared lysate was added to 50 μl of protein G-coated Dynabeads (Invitrogen) which had been previously bound to 5 μg of anti-mouse Ago2/eIF2C2 monoclonal antibody (Abcam) or normal rabbit IgG (Cell Signaling Technology), according to the manufacturer's protocol, and incubated for 4 h at 4°C with rotation. Beads were washed three times with 1× PBS, 0.1% SDS, 0.5% deoxycholate, 0.5% nonidet P-40 and three times with 5× PBS, 0.1% SDS, 0.5% deoxycholate, 0.5% nonidet P-40. RNA extraction and qRT-PCR were performed as described above, except that the relative abundance of novel #117 in the Ago2 IP sample was calculated relative to the IgG IP sample, in lieu of an endogenous control.
Western blots
Ten microliters of 2× SDS reducing buffer was added directly to washed beads following immunoprecipitation, and incubated at 95°C for 5 min. Samples were loaded on a pre-warmed 4–20% polyacrylamide gel, run at 200 V for 30 min and wet-transferred for 1 h at 100 V onto a 0.45 μm nitrocellulose membrane. The membrane was blocked in 1× TBST, 5% milk at 22°C for 1 h. A 1:500 dilution of Ago2/eIF2C monoclonal antibody (Abcam) in 1× TBST, 5% milk was applied to the membrane and incubated at 22°C for 3 h. The membrane was washed three times for 10 min in 1× TBST, 5% milk. A 1:5000 dilution of HRP-conjugated secondary antibody in 1× TBST, 5% milk was applied to the membrane and incubated at 22°C for 1 h. The membrane was washed three times for 10 min in 1× TBST, 5% milk. The blot was developed with 1 ml of Supersignal West Femto Chemiluminescent Substrate (Thermo Scientific).
miRNA in situ hybridization
miRNA in situ hybridizations were performed as previously described (71). Briefly, fresh skin biopsies were fixed in 10% formalin for 24–72 h and paraffin-embedded. Six micrometer sections were mounted on glass slides, deparaffinized and treated with 10 μg/ml proteinase K for 20 min at 37°C. Slides were hybridized with 20–60 nm double-DIG-labeled LNA probes (Exiqon) overnight at 57°C (miR-135b, miR-205) or 50°C (miR-142). Slides were washed in 5× SSC, 1× SSC and 0.2× SSC for 10 min at hybridization temperature. Staining was performed with NBT/BCIP (Roche) for 90 min at 32°C followed by nuclear fast red counterstain (Vector Laboratories). An LNA probe with scrambled sequence was used as a negative control (Exiqon).
miRNA editing
Reads that aligned to mature miRNAs with one mismatch were subjected to filters prior to editing analysis. Reads containing a low-quality mismatch [P(sequencing error) > 0.05] based on the single-base Illumina quality score were removed. 3′ terminal N → A or N → T mismatches were also removed. From the remaining pool of one mismatch reads, the relative frequencies of all possible substitutions at positions 1–20 of the miRNA relative to the 5′ end were calculated. Significance was determined with Pearson's χ2 test.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (5RC1AR058681 to A.M.B. and W.Z., 1R01AR050266 to A.M.B.); the National Science Foundation (DBI-0743797 to W.Z.); and the National Human Genome Research Institute (T32HG000045 to C.E.J.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Eli Roberson for insightful discussions and critical comments on this manuscript, Elaine Mardis for the use of the Applied Biosystems 7900HT instrument, and the Genome Technology Access Center at Washington University for GAIIx data generation.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bhalerao J., Bowcock A.M. The genetics of psoriasis: a complex disorder of the skin and immune system. Hum. Mol. Genet. 1998;7:1537–1545. doi: 10.1093/hmg/7.10.1537. doi:10.1093/hmg/7.10.1537. [DOI] [PubMed] [Google Scholar]
- 2.Lowes M.A., Bowcock A.M., Krueger J.G. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. doi:10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 3.Roberson E.D., Bowcock A.M. Psoriasis genetics: breaking the barrier. Trends Genet. 2010;26:415–423. doi: 10.1016/j.tig.2010.06.006. doi:10.1016/j.tig.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou X., Krueger J.G., Kao M.C., Lee E., Du F., Menter A., Wong W.H., Bowcock A.M. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol. Genomics. 2003;13:69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]
- 5.Gudjonsson J.E., Ding J., Johnston A., Tejasvi T., Guzman A.M., Nair R.P., Voorhees J.J., Abecasis G.R., Elder J.T. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J. Invest. Dermatol. 2010;130:1829–1840. doi: 10.1038/jid.2010.36. doi:10.1038/jid.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. doi:10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. doi:10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. doi:10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. doi:10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 11.Ruby J.G., Jan C.H., Bartel D.P. Intronic microRNA precursors that bypass drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. doi:10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berezikov E., Chung W.J., Willis J., Cuppen E., Lai E.C. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. doi:10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. doi:10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 14.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. doi:10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 15.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. doi:10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 16.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. doi:10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 18.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. doi:10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 19.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. doi:10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi R., O'Carroll D., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006;38:356–362. doi: 10.1038/ng1744. doi:10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 21.Yi R., Pasolli H.A., Landthaler M., Hafner M., Ojo T., Sheridan R., Sander C., O'Carroll D., Stoffel M., Tuschl T., et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc. Natl Acad. Sci. US A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. doi:10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonkoly E., Wei T., Janson P.C., Saaf A., Lundeberg L., Tengvall-Linder M., Norstedt G., Alenius H., Homey B., Scheynius A., et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. doi:10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zibert J.R., Lovendorf M.B., Litman T., Olsen J., Kaczkowski B., Skov L. MicroRNAs and potential target interactions in psoriasis. J. Dermatol. Sci. 2010;58:177–185. doi: 10.1016/j.jdermsci.2010.03.004. doi:10.1016/j.jdermsci.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. doi:10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark M.S., Tyagi S., Nancarrow D.J., Boyle G.M., Cook A.L., Whiteman D.C., Parsons P.G., Schmidt C., Sturm R.A., Hayward N.K. Characterization of the melanoma miRNAome by deep sequencing. PLoS One. 2010;5:e9685. doi: 10.1371/journal.pone.0009685. doi:10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marti E., Pantano L., Banez-Coronel M., Llorens F., Minones-Moyano E., Porta S., Sumoy L., Ferrer I., Estivill X. A myriad of miRNA variants in control and huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. doi:10.1093/nar/gkq575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryu S., Joshi N., McDonnell K., Woo J., Choi H., Gao D., McCombie W.R., Mittal V. Discovery of novel human breast cancer microRNAs from deep sequencing data by analysis of pri-microRNA secondary structures. PLoS One. 2011;6:e16403. doi: 10.1371/journal.pone.0016403. doi:10.1371/journal.pone.0016403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. doi:10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lena A.M., Shalom-Feuerstein R., Rivetti di Val Cervo P., Aberdam D., Knight R.A., Melino G., Candi E. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. doi:10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 30.Yi R., Poy M.N., Stoffel M., Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness. Nature. 2008;452:225–229. doi: 10.1038/nature06642. doi:10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. doi:10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 32.Creighton C.J., Benham A.L., Zhu H., Khan M.F., Reid J.G., Nagaraja A.K., Fountain M.D., Dziadek O., Han D., Ma L., et al. Discovery of novel microRNAs in female reproductive tract using next generation sequencing. PLoS One. 2010;5:e9637. doi: 10.1371/journal.pone.0009637. doi:10.1371/journal.pone.0009637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witten D., Tibshirani R., Gu S.G., Fire A., Lui W.O. Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol. 2010;8:58. doi: 10.1186/1741-7007-8-58. doi:10.1186/1741-7007-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregory P.A., Bracken C.P., Bert A.G., Goodall G.J. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. doi:10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 35.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. doi:10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 36.Powell L.M., Wallis S.C., Pease R.J., Edwards Y.H., Knott T.J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. doi:10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 37.Bass B.L. RNA editing and hypermutation by adenosine deamination. Trends Biochem. Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. doi:10.1016/S0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 38.McCormick K.P., Willmann M.R., Meyers B.C. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence. 2011;2:2. doi: 10.1186/1758-907X-2-2. doi:10.1186/1758-907X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fehniger T.A., Wylie T., Germino E., Leong J.W., Magrini V.J., Koul S., Keppel C.R., Schneider S.E., Koboldt D.C., Sullivan R.P., et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20:1590–1604. doi: 10.1101/gr.107995.110. doi:10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haider A.S., Lowes M.A., Suarez-Farinas M., Zaba L.C., Cardinale I., Blumenberg M., Krueger J.G. Cellular genomic maps help dissect pathology in human skin disease. J. Invest. Dermatol. 2008;128:606–615. doi: 10.1038/sj.jid.5701067. doi:10.1038/sj.jid.5701067. [DOI] [PubMed] [Google Scholar]
- 41.Lin S., Cheung W.K., Chen S., Lu G., Wang Z., Xie D., Li K., Lin M.C., Kung H.F. Computational identification and characterization of primate-specific microRNAs in human genome. Comput. Biol. Chem. 2010;34:232–241. doi: 10.1016/j.compbiolchem.2010.08.001. doi:10.1016/j.compbiolchem.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Stark A., Bushati N., Jan C.H., Kheradpour P., Hodges E., Brennecke J., Bartel D.P., Cohen S.M., Kellis M. A single hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. doi:10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma N., Wang X., Qiao Y., Li F., Hui Y., Zou C., Jin J., Lv G., Peng Y., Wang L., et al. Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483-5p as an IGF2 partner. Mol. Cell. Endocrinol. 2011;333:96–101. doi: 10.1016/j.mce.2010.11.027. doi:10.1016/j.mce.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 44.Najafi-Shoushtari S.H., Kristo F., Li Y., Shioda T., Cohen D.E., Gerszten R.E., Naar A.M. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. doi:10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levy C., Khaled M., Iliopoulos D., Janas M.M., Schubert S., Pinner S., Chen P.H., Li S., Fletcher A.L., Yokoyama S., et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell. 2010;40:841–849. doi: 10.1016/j.molcel.2010.11.020. doi:10.1016/j.molcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S., Olson E.N. AngiomiRs—key regulators of angiogenesis. Curr. Opin. Genet. Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. doi:10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansel J.C., Tiesman J.P., Olerud J.E., Krueger J.G., Krane J.F., Tara D.C., Shipley G.D., Gilbertson D., Usui M.L., Hart C.E. Human keratinocytes are a major source of cutaneous platelet-derived growth factor. J. Clin. Invest. 1993;92:671–678. doi: 10.1172/JCI116636. doi:10.1172/JCI116636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown L.F., Harrist T.J., Yeo K.T., Stahle-Backdahl M., Jackman R.W., Berse B., Tognazzi K., Dvorak H.F., Detmar M. Increased expression of vascular permeability factor (vascular endothelial growth factor) in bullous pemphigoid, dermatitis herpetiformis, and erythema multiforme. J. Invest. Dermatol. 1995;104:744–749. doi: 10.1111/1523-1747.ep12606974. doi:10.1111/1523-1747.ep12606974. [DOI] [PubMed] [Google Scholar]
- 49.Viac J., Palacio S., Schmitt D., Claudy A. Expression of vascular endothelial growth factor in normal epidermis, epithelial tumors and cultured keratinocytes. Arch. Dermatol. Res. 1997;289:158–163. doi: 10.1007/s004030050172. doi:10.1007/s004030050172. [DOI] [PubMed] [Google Scholar]
- 50.Xia Y.P., Li B., Hylton D., Detmar M., Yancopoulos G.D., Rudge J.S. Transgenic delivery of VEGF to mouse skin leads to an inflammatory condition resembling human psoriasis. Blood. 2003;102:161–168. doi: 10.1182/blood-2002-12-3793. doi:10.1182/blood-2002-12-3793. [DOI] [PubMed] [Google Scholar]
- 51.Hua Z., Lv Q., Ye W., Wong C.K., Cai G., Gu D., Ji Y., Zhao C., Wang J., Yang B.B., et al. miRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. doi:10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhushan M., McLaughlin B., Weiss J.B., Griffiths C.E. Levels of endothelial cell stimulating angiogenesis factor and vascular endothelial growth factor are elevated in psoriasis. Br. J. Dermatol. 1999;141:1054–1060. doi: 10.1046/j.1365-2133.1999.03205.x. doi:10.1046/j.1365-2133.1999.03205.x. [DOI] [PubMed] [Google Scholar]
- 53.Suarez Y., Fernandez-Hernando C., Yu J., Gerber S.A., Harrison K.D., Pober J.S., Iruela-Arispe M.L., Merkenschlager M., Sessa W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl Acad. Sci. USA. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. doi:10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grundmann S., Hans F.P., Kinniry S., Heinke J., Helbing T., Bluhm F., Sluijter J.P., Hoefer I., Pasterkamp G., Bode C., et al. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation. 2011;123:999–1009. doi: 10.1161/CIRCULATIONAHA.110.000323. doi:10.1161/CIRCULATIONAHA.110.000323. [DOI] [PubMed] [Google Scholar]
- 55.Helms C., Cao L., Krueger J.G., Wijsman E.M., Chamian F., Gordon D., Heffernan M., Daw J.A., Robarge J., Ott J., et al. A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nat. Genet. 2003;35:349–356. doi: 10.1038/ng1268. doi:10.1038/ng1268. [DOI] [PubMed] [Google Scholar]
- 56.Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. doi:10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. doi:10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ebhardt H.A., Tsang H.H., Dai D.C., Liu Y., Bostan B., Fahlman R.P. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. doi:10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vartanian J.P., Guetard D., Henry M., Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. doi:10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 60.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. doi:10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. doi:10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. doi:10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuhn R.M., Karolchik D., Zweig A.S., Wang T., Smith K.E., Rosenbloom K.R., Rhead B., Raney B.J., Pohl A., Pheasant M., et al. The UCSC genome browser database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. doi:10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mituyama T., Yamada K., Hattori E., Okida H., Ono Y., Terai G., Yoshizawa A., Komori T., Asai K. The functional RNA database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 2009;37:D89–D92. doi: 10.1093/nar/gkn805. doi:10.1093/nar/gkn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. doi:10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. doi:10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCaskill J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. doi:10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 69.Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary structure. Monatshefte f. Chemie. 1994;125:167–188. doi:10.1007/BF00818163. [Google Scholar]
- 70.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. doi:10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 71.Nielsen B.S., Jorgensen S., Fog J.U., Sokilde R., Christensen I.J., Hansen U., Brunner N., Baker A., Moller S., Nielsen H.J. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin. Exp. Metastasis. 2011;28:27–38. doi: 10.1007/s10585-010-9355-7. doi:10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.