Abstract
While the distribution of RNA polymerase II (PolII) in a variety of complex genomes is correlated with gene expression, the presence of PolII at a gene does not necessarily indicate active expression. Various patterns of PolII binding have been described genome wide; however, whether or not PolII binds at transcriptionally inactive sites remains uncertain. The two X chromosomes in female cells in mammals present an opportunity to examine each of the two alleles of a given locus in both active and inactive states, depending on which X chromosome is silenced by X chromosome inactivation. Here, we investigated PolII occupancy and expression of the associated genes across the active (Xa) and inactive (Xi) X chromosomes in human female cells to elucidate the relationship of gene expression and PolII binding. We find that, while PolII in the pseudoautosomal region occupies both chromosomes at similar levels, it is significantly biased toward the Xa throughout the rest of the chromosome. The general paucity of PolII on the Xi notwithstanding, detectable (albeit significantly reduced) binding can be observed, especially on the evolutionarily younger short arm of the X. PolII levels at genes that escape inactivation correlate with the levels of their expression; however, additional PolII sites can be found at apparently silenced regions, suggesting the possibility of a subset of genes on the Xi that are poised for expression. Consistent with this hypothesis, we show that a high proportion of genes associated with PolII-accessible sites, while silenced in GM12878, are expressed in other female cell lines.
INTRODUCTION
RNA polymerase II (PolII) is an essential component of the eukaryotic transcriptional machinery. While other RNA polymerases transcribe non-coding genes, PolII is involved in transcription of coding genes as well as non-coding RNA genes (1,2). High-throughput ChIP-seq studies have described the genomic distribution of PolII in a number of model organisms (3–6). PolII localization relative to genes has been observed at transcriptional start sites (TSSs) and is less often distributed along the gene body, beyond the 3′ end, or can be absent all together (7). In addition to its accumulation within actively expressed genes, PolII has also been found at enhancers (8), in intergenic regions (1), and at TSSs of developmentally regulated genes that are periodically turned off and on (3–5).
Promoter regions of highly regulated genes that depend on recruitment of PolII for their activation are often covered with nucleosomes (9) that prevent PolII binding until chromatin remodelers allow the pre-initiation complex (PIC) to assemble (10). In contrast, constitutively transcribed genes maintain their promoter region uncovered (11), thus allowing permanent access to PolII. Upon gaining access to the promoter, a number of steps must occur for PolII to actively engage in transcription. The PIC assembles on the core promoter and induces DNA unwinding, upon which PolII proceeds to a promoter-proximal pause region. At this position, PolII becomes phosphorylated, escapes the promoter-proximal pause region and continues transcription (12). The frequently observed bimodal distribution of TSS-associated PolII signals reflects pausing and possibly also poising at the TSS, as a result of PolII accumulation due to downstream rate-limiting steps (7,13–16). Four general classes of genes have been described based on the presence or absence of the PIC at the promoter region as it relates to transcriptional activity: expressed genes with or without PIC detectable at the promoter, and silenced genes with or without PIC detectable at the promoter (17). PolII can also be observed within the bodies of some genes, reflecting active elongation (18). In addition, PolII peaks can be detected several kilobases downstream from the 3′ end of some genes (1,8), likely reflecting the lack of strong termination signals that allow the enzyme to progress beyond the annotated end of genes (8).
High-throughput ChIP-seq and ChIP-chip technologies have provided valuable information on the spatial and temporal characteristics of various chromatin elements, including PolII, throughout the diploid human genome (18,19). A greater challenge arises when interpreting these data at loci exhibiting allelic imbalance (20), as a number of genes across the human genome are not equally expressed from both alleles due to widespread cis- or trans-determined influences on transcription (21–23) and to effects such as genomic imprinting (24), allelic exclusion (23) and, in females, X chromosome inactivation (25,26). Similarly, associated cis-acting regulatory elements, such as various chromatin marks or components of the transcriptional machinery, can themselves exhibit allele-specific patterns (27,28), and thus, for these genomic loci, the information pertaining to the two alleles must be separated. Examining the allele-specific basis for such signals allows exploration of the nature of cis-regulatory mechanisms on both PolII occupancy and gene expression.
Although the majority of X-linked genes are silenced in mammalian females due to X chromosome inactivation, at least 10% of genes residing on the X chromosome are expressed biallelically in all samples analyzed to date (29–32) and many more exhibit variable expression patterns among different females (29,33,34). It is presently unknown what factors determine the inactivation status of X-linked genes and how important functionally or phenotypically it is for the Xi copy of a given gene to be expressed or silenced. Nonetheless, the evident heterogeneous patterns of gene expression from the Xi provide an opportunity to examine the association among genetic, genomic and/or stochastic signals that may underlie PolII occupancy at different loci.
The facultative heterochromatin of the human Xi consists of at least two distinct types of chromatin marked by a host of components that distinguish them, including various histone modifications or variants, binding proteins and XIST RNA (35,36). While these and other chromatin features have been investigated in the mouse and humans (35–40), none has yet been tightly correlated with the inactivation status of individual genes. Because PolII is required for transcription, its presence or absence would appear to be better suited as a potential indicator for silencing due to X inactivation. In mouse embryonic stem cells, PolII exclusion occurs soon after the induction of differentiation, which triggers expression of the Xist gene (41) and subsequent inactivation of X-linked genes. PolII is also eliminated from the human Barr body (42); however, because the core of the Barr body is composed of repetitive non-coding DNA, while genes (regardless of their expression status) occupy the periphery (42), exclusion of PolII from the Barr body core does not necessarily imply its exclusion from genes.
The process of X inactivation is initiated early in development, and the chromosome to be inactivated is selected at random, such that the resulting female is a mosaic of cells carrying an Xi that is either maternally- (Xim) or paternally derived (Xip) (25,43,44). Because such clonal mosaicism can obscure the detection of Xi-specific features, a number of investigations have turned either to oligoclonal lymphoblast cell lines that deviate from random distribution and are severely skewed toward one Xi chromosome or the other (45,46) or to fibroblast cell lines that exhibit complete non-random inactivation and thus contain pure populations of cells with either Xim or Xip (29,31).
In this paper, we describe the PolII landscape on human female X chromosomes utilizing PolII ChIP-seq data for the GM12878 cell line, a chromosomally normal female lymphoblast line. We selected this cell line because of the opportunity to build on a number of earlier studies that have characterized GM12878 as part of the ENCODE Project and the 1000 Genomes Project (19,47) (Reddy TE, Gertz J, Pauli F, Newberry K, Kucera KS, Wold B, Willard HF, Myers RM, manuscript in preparation). Because of the severe X-inactivation skewing in GM12878 (27), we were able to infer the enrichment of individual alleles on the Xi and Xa and thus analyze these data in an allele-specific manner to compare PolII distribution on the two Xs. Further, for the purpose of elucidating the relationship of PolII-binding and inactivation status of genes, we have focused specifically on PolII sites that could be assigned to particular genes and, where possible, we have determined their inactivation status in homogeneous GM12878 Xip- and Xim-derived cell lines.
RESULTS
The human X chromosome is relatively PolII poor
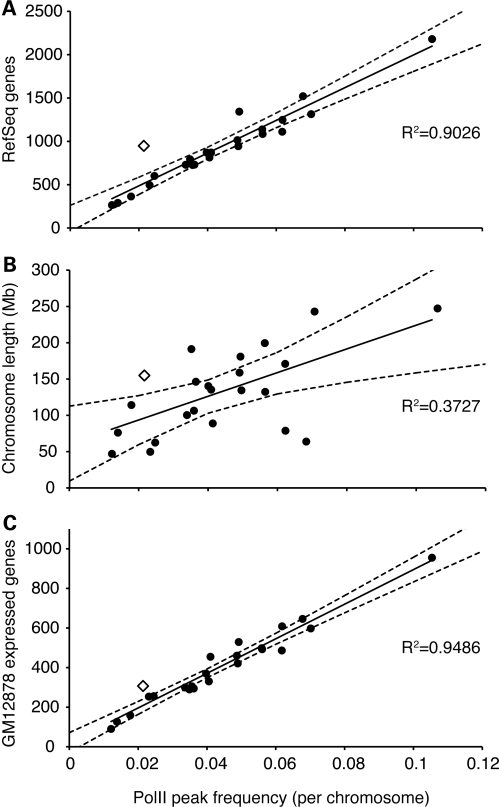
We first sought to measure genomic occupancy of PolII on the X chromosome. To do so, we used a genome-wide ChIP-seq approach and, in two replicate samples, detected an average of 168 sites of PolII enrichment on the X chromosomes in the GM12878 cell line (Table 1). Notably, this reflects a significantly (P<0.001, t-test) lower PolII peak occurrence relative to gene density on the X chromosome compared with autosomes (Fig. 1A). We observed a similar, albeit less extreme, situation for chromosome 11 (P<0.05, t-test) that can likely be attributed to the fact that more than 300 olfactory receptor genes are located on chromosome 11 (48). These genes are not expressed in lymphoblasts, including the Epstein–Barr virus (EBV)-immortalized GM12878 line and make up a substantial proportion of all chromosome 11 genes.
Table 1.
Analysis of PolII-binding sites on the human X chromosome
| PolII peaks called (QuEST)a | PolII-occupied heterozygous SNPsb | |
|---|---|---|
| Total | 168 | 385 |
| TSS ±500 bp | 120 | 37 |
| Intragenic | 138 | 257 |
| Extragenic | 30 | 128 |
| <5 kb from genes | 9 | 60 |
| 5–30 kb from genes | 15 | 17 |
| >30 kb from genes | 6 | 51 |
aNon-allele-specific PolII peaks called by Quest software (64) as a composite of the two X chromosomes.
bPolII occupancy at heterozygous sites; sites with minimum of five reads at one or the other allele were considered.
Figure 1.
PolII peaks per chromosome. Each human chromosome is represented by a circle (autosomes) or an open diamond (X chromosome). Linear regression (solid line) and 97% confidence interval (dashed lines) are shown. Number of PolII peaks is shown relative to (A) RefSeq genes; (B) chromosome length; and (C) expressed genes in GM12878 (expression data from Reddy TE, Gertz J, Pauli F, Newberry K, Kucera KS, Wold B, Willard HF, Myers RM, manuscript in preparation).
The paucity of PolII-binding sites on the X could reflect the number of genes, the number of expressed genes or the frequency of binding as a function of chromosome size. To explore these differences further, we compared the abundance of PolII peaks to chromosome length (Fig. 1B) and to the number of expressed genes for each chromosome in GM12878 (Reddy TE, Gertz J, Pauli F, Newberry K, Kucera KS, Wold B, Willard HF, Myers RM, manuscript in preparation) (Fig. 1C). While PolII binding is loosely correlated with chromosome length (Fig. 1B), it is better correlated with overall gene density (Fig. 1A) and is particularly well correlated with the number of genes expressed in GM12878 (Fig. 1C). Thus, the relative scarcity of PolII peaks on the X chromosome can be at least partially explained by the lower proportion of genes that are expressed from the X chromosome in the GM12878 cell line when compared with autosomes (Fig. 1).
PolII binding is biased toward the active X chromosome
In a previous study (27), we reported 92% skewing of X inactivation in the GM12878 cell line toward the Xip. Because X-inactivation skewing can drift in cell culture over time (45,46), we verified the extent of skewing in GM12878 in RNA samples harvested concurrently with the PolII ChIP-seq samples generated for this study. We detected 95% (SD = 1%) skewing in the same direction as previously described (27), by testing allele-specific expression at XIST (rs1620574) and EBP (rs3048) by SNaPshot assays (29).
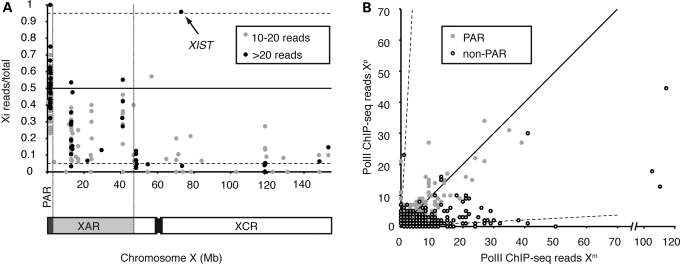
To determine whether PolII binding on the X chromosome is influenced by X-chromosome inactivation, we focused on the subset of individual PolII sites that contain heterozygous single-nucleotide polymorphisms (SNPs) (Supplementary Material, Table S1) and employed an allele-specific alignment approach to distinguish binding on the Xa and Xi chromosomes. We identified 385 heterozygous SNPs (Table 1) that had at least five mapped reads on at least one of the two X chromosomes in the PolII ChIP-seq data set (Supplementary Material, Table S2, Fig. 2). We chose this threshold to limit artifacts due to spurious alignment or non-specific immunoprecipitation of DNA. This allele-specific data set of 385 sites forms the basis for the analysis in this section.
Figure 2.
Allele-specific PolII occupancy on the X chromosome in GM12878. (A) Distribution of heterozygous sites with at least 10 mapped PolII ChIP-seq reads on the X chromosome, displayed as the proportion of sites on the Xi. XAR, X-added region; XCR, X-conserved region; PAR, pseudoautosomal region. (B) PolII occupancy on the Xm versus Xp chromosomes. Dashed axes represent 95% skewing (i.e. monoallelic occupancy) observed in the GM12878 cell line. Solid diagonal represents equal PolII occupancy on Xi and Xa.
PolII binding in the pseudoautosomal region on the distal short arm (PAR1) is especially dense when compared with the non-pseudoautosomal region (Fig. 2A). Furthermore, as expected, we observed no clear bias in occupancy toward Xa or Xi in PAR1, with 47% of the total PolII reads that mapped to the PAR1 aligning to the Xi alleles. A few individual heterozygous PolII-binding sites showed significant bias towards one or the other X (Fig. 2), and these likely represent occasional allele-specific bias similar to that seen throughout the genome (Reddy TE, Gertz J, Pauli F, Newberry K, Kucera KS, Wold B, Willard HF, Myers RM, manuscript in preparation, 27). It remains unknown what significance these PolII sites have and whether they are functionally associated with the neighboring genes.
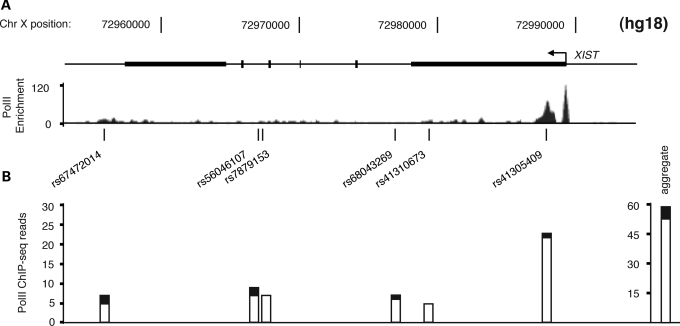
The remaining majority of PolII sites at heterozygous positions on the X were located in the non-pseudoautosomal region of the X chromosome. Two noteworthy observations indicate relative depletion of PolII across the non-pseudoautosomal portion of the Xi. First, considering this region as a whole, we detected about five times more total ChIP-seq reads mapping to the Xa than to Xi in this region (Supplementary Material, Table S2), consistent with reduced PolII binding being associated with X inactivation. Secondly, of the 268 non-pseudoautosomal PolII sites examined, 39% exhibited significant bias (P<0.01, binomial test) in PolII occupancy toward one chromosome or the other (Fig. 2B, Supplementary Material, Table S2). Among these, all but two showed bias toward the Xa allele. Not unexpectedly, the two sites with significant PolII-binding bias toward the Xi were located in the XIST gene (Fig. 3) (49,50). At a PolII site near the 5′ end of XIST, 23 of the 24 sequences (96%) aligned to the paternally derived (Xi) allele, which correlates well with the extent of X-inactivation skewing in the cell line and implies absence of PolII from the XIST allele on the Xa. As PolII is completely depleted from the Xa near the XIST TSS, the relative abundance of PolII at this site is an expression-independent indicator of X-inactivation skewing.
Figure 3.
Xi bias of PolII enrichment at the XIST gene. (A) PolII ChIP-seq enrichment throughout the gene is shown with the highest enrichment near the TSS. Locations and rs numbers of heterozygous SNPs in GM12878 are indicated. (B) Relative PolII occupancy on the two X chromosomes in GM12878, as measured by the number of aligned ChIP-seq reads at each heterozygous SNP, mapping to the Xp (white bars) or Xm (black bars). The aggregated signal from the XIST locus is shown on the far right, indicating significant PolII binding bias toward Xp, consistent with the reported (28) and measured (see Materials and Methods) X-inactivation bias in this cell line.
The observed PolII profile on Xa and Xi (Fig. 2) appears to reflect in part the evolutionary origins of the X chromosome, as the bias is nearly complete in the portion of the X that corresponds to the ancestral conserved region of the X chromosome (XCR), but is decidedly less extreme in the portion of the X that was added more recently during evolution (XAR) (51–53). PolII occupancy also mirrors the reported patterns of X inactivation, as a much greater proportion of genes in XAR escape inactivation than in XCR (29) and thus might be expected to be associated with PolII on both Xa and Xi.
PolII binding on the inactive X chromosome largely mimics inactivation status of genes
Although PolII is generally depleted from the human Xi, many sites deviate from the predicted level of X-inactivation skewing (Fig. 2, Supplementary Material, Table S2). In the non-pseudoautosomal region of the X chromosome, ∼45% of the informative PolII-binding sites showed >10% PolII occupancy on the Xi relative to the Xa, suggesting at least some PolII located on the Xi alleles. Strikingly, at 12% of sites, the level of Xi PolII occupancy exceeded 50% of the PolII levels on Xa. Genes with high PolII occupancy on the Xi were of special interest for considering relative gene expression from Xi and Xa, as they could reflect either biallelic expression or the presence of PolII at silenced genes.
To assess the relationship of PolII binding and gene expression from the Xi, we combined PolII ChIP-seq signal across each annotated gene and its flanking regions (30 kb upstream and 5 kb downstream), considering only those sites that could be unambiguously associated with assayable genes (Supplementary Material, Table S3). While sites upstream from TSSs seem likely to reflect association at regulatory regions for those genes (8) and downstream sites likely result from PolII progression beyond the 3′ end (1,8), we cannot rule out the possibility that, given the currently incomplete state of genome annotation, there is actually no functional association of these sites with the assigned genes.
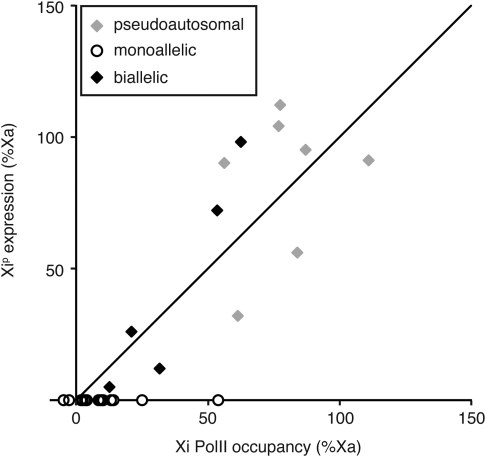
We identified 31 genes with robust and informative PolII binding across the X chromosome (including PAR1), that is, genes containing expressed heterozygous SNPs that could be studied with allele-specific expression assays (Table 2). We assayed allele-specific expression at these 31 genes in a series of apparently clonal cell lines derived from GM12878 that were homogeneous with respect to the Xip or Xim inactive X chromosomes (Supplementary Material, Table S3). All seven pseudoautosomal genes, as expected, exhibited biallelic expression, with mostly similar levels of expression from both alleles corresponding to the PolII occupancy detected at the associated heterozygous-binding sites (Fig. 4A). We noted two pseudoautosomal genes IL3RA and CD99 that were relatively less expressed from the paternally derived copy, regardless of the Xi origin (Supplementary Material, Table S3), which may reflect either a parent-of-origin effect or imbalance due to the particular allelic variants in these genes; further studies in other cell lines would be required to distinguish these possibilities. This situation is not unprecedented on the X chromosome, as such allelic imbalance has been described at the SYBL1 locus in PAR2 (54).
Table 2.
Relationship of expression and PolII occupancy
| Genes | PolII at Xi (no. of genes) | |
|---|---|---|
| Total | 31 | |
| Pseudoautosomal | 7 | 7/7 |
| Non-pseudoautosomal | 24 | |
| Monoallelic Xa | 17 | 8a/17 |
| Monoallelic Xi | 1 | 1/1 |
| Biallelic | 5 | 5/5 |
| Variable | 1 | 0/1 |
aPolII at Xi associated with monoallelically expressed loci is defined as at least three reads mapping to Xi and >15% of Xa PolII occupancy.
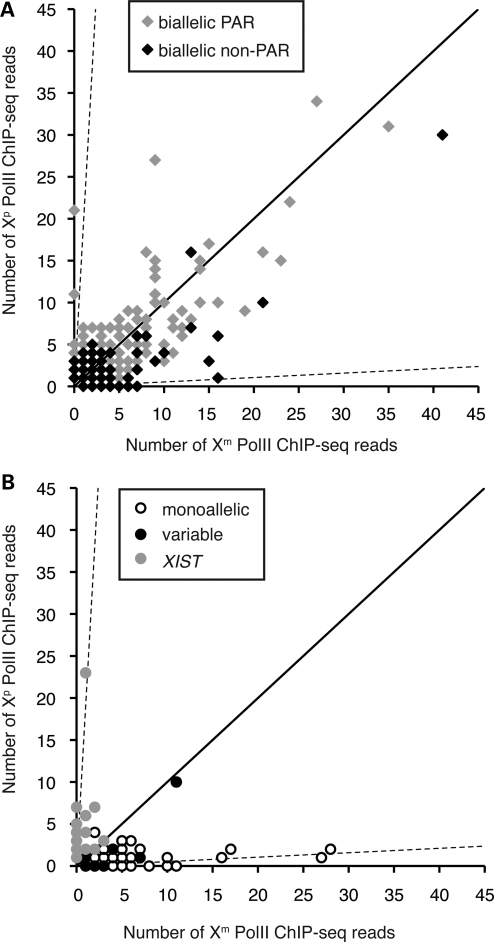
Figure 4.
Expression of genes genomically associated with PolII-occupied heterozygous sites. (A) Allele-specific binding of PolII sites associated with biallelically expressed genes (as designated in the key). (B) Allele-specific binding of PolII sites associated with monoallelically expressed or variable genes (as designated in the key). Dashed axes and diagonal axis are as in Figure 2.
We detected several patterns of expression among the remaining 24 tested genes located on the non-pseudoautosomal portion of the X chromosome (Table 2). As expected, the majority (75%) of the tested genes were expressed monoallelically in both types of Xi isolates (all expressed exclusively from the Xa allele, other than XIST, which was expressed only from the Xi). In addition, five genes (21%) escaped inactivation at varying levels of expression, and one gene (SEPT6) exhibited variable expression between the two types of isolates.
When the gene expression profiles were related back to the individual PolII-occupied heterozygous sites (allele-specific data set of 385 PolII sites) (Fig. 4, Supplementary Material, Table S2), as well as to per-gene PolII occupancy (Fig. 5, Supplementary Material, Table S3), gene silencing on the Xi was found to correlate with PolII depletion at most sites. However, several regions on the Xip showed PolII binding well above the expected ∼5% level corresponding to the X-inactivation skewing ratio. The relationship of PolII binding at a specific site to a particular gene is difficult to establish, especially for sites located in intergenic regions; nevertheless, it appears that PolII can bind at low levels even in regions where silenced genes reside, indicating higher accessibility of the transcriptional machinery to the Xi than previously thought.
Figure 5.
Relationship of PolII levels and gene expression on the inactive X chromosome in GM12878. The relationship of paternal PolII occupancy and Xip gene expression is shown as a percentage of Xa. The diagonal axis represents direct relationship between relative PolII occupancy and expression.
Among the five biallelically expressed genes, Xi expression ranged from 5 to 98% relative to levels detected from the Xa allele (Table 3), showing strong correlation with levels of PolII occupancy at sites associated with those genes (R2=0.9) (Fig. 5). In addition, the levels of expression for these five genes are consistent in the derivative cell lines containing the two different Xi chromosomes (Xip and Xim), as well as in duplicate lines that were cultured separately for several weeks, indicating that the regulation of expression levels of these genes is largely constant and thus presumably reflects stability of the epigenomic environment of the alleles being compared.
Table 3.
Expression and PolII binding on Xi relative to Xa
| Function | Xp PolII (%Xa) | Xip Expression (%Xa) | Y homology | |
|---|---|---|---|---|
| SYN1 | Synaptogenesis and neurotransmitter release | 19% | 5% | — |
| USP9X | Peptidase C19 family, similar to ubiquitin-specific proteases | 36% | 12% | USP9Y |
| TXLNG | Intracellular vesicle trafficking, cell cycle | 28% | 26% | CYorf15 |
| DDX3X | Alteration of RNA secondary structure | 66% | 72% | DDX3Y |
| PRKX | Serine-threonine protein kinase | 75% | 98% | PRKY |
Our analysis of gene expression is limited by the occurrence of transcribed heterozygous SNPs in the cell line under study. Nonetheless, there were a number of PolII sites associated with genes that lacked expressed SNPs, some of which have been studied previously and thus allow for some interpretation. For example, the TIMM17B and RBM3 genes are inactivated in multiple human cell lines (29), which is in agreement with the 95% Xa PolII occupancy in this study, suggesting that both genes are also inactivated in GM12878. In contrast, the UBA1 gene contains three heterozygous PolII sites within its gene body, and, in the aggregate, 33% of reads at these sites map to the Xi allele. This finding is consistent with previous reports that UBA1 escapes inactivation in all samples tested thus far (29,30,55).
PolII occupancy indicates chromatin accessibility and potential for expression
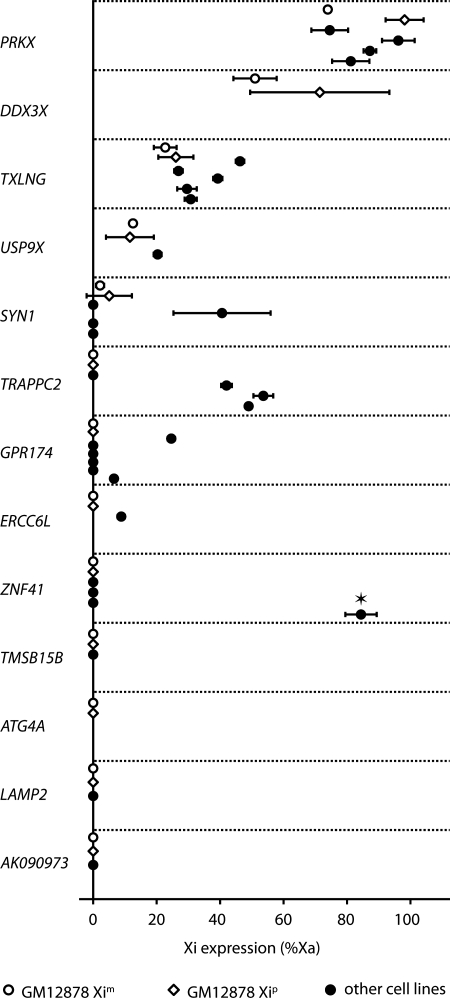
As described above, PolII binding at genes that are not being actively transcribed indicates a level of openness of the local chromatin environment. We hypothesized that such genes have the potential to be expressed in other cell lines either consistently or variably with respect to the population. To test this hypothesis, we generated Xi-homogeneous cell populations from seven additional HapMap CEU cell lines (56,57) and tested expression of 11 genes that were informative for study in these cell lines, 4 genes that were expressed biallelically in GM12878 and 7 that were silenced in GM12878 yet showed significant PolII levels at the Xi allele (Table 2).
In the case of the four genes that are biallelically expressed in GM12878, we confirmed biallelic expression of PRKX, TXLNG and USP9X in all informative female lines tested, indicating that all three genes consistently escape inactivation (Fig. 6). Furthermore, the relative levels of Xi expression of these three genes observed in GM12878 (Table 3) are recapitulated in the additional cell lines, with PRKX exhibiting highest Xi expression (∼85% of Xa levels), TXLNG intermediate (∼35% of Xa) and USP9X lowest expression (∼20% of Xa). In contrast, SYN1 is a gene with variable expression from the Xi. It escapes inactivation in GM12878 at a very low level, but is expressed robustly from Xi in another female cell line and is completely silenced in three other females (Fig. 6).
Figure 6.
Xi gene expression in multiple cell lines. Genes associated with PolII in GM12878 (shown on vertical axis) were tested for expression in GM12878 (open symbols) and additional heterozygous cell lines (closed circles). Error bars represent standard deviation of the mean between biological replicates. Expression of ZNF41 has been tested previously in ten human cell lines (29). It was subject to inactivation in nine informative cell lines (not shown in the figure) and escaped inactivation in one cell line (designated by an asterisk).
Of the seven silenced genes associated with PolII in GM12878 (Table 2), we show that three (ERCC6L, GPR174 and TRAPPC2) are expressed from the Xi in other female cell lines (Fig. 6), and one additional gene (ZNF41) has been reported to escape inactivation previously in at least one female cell line (29). Combined, these data demonstrate that PolII not only binds to the Xi, but also can become engaged and produce a transcript in some individuals.
DISCUSSION
In this study, we have examined the profile of PolII binding on human female X chromosomes. Overall, the frequency of PolII binding on the X chromosome and chromosome 11 is reduced compared with other chromosomes, largely because of the lower proportion of expressed genes in the GM12878 EBV-immortalized lymphoblastoid cell line (Fig. 1). Although the low proportion of genes expressed from the X chromosome relative to autosomes largely explains the reduced frequency of PolII binding, other factors will have to be explored to explain this phenomenon fully. As is evident here, many X-linked genes are not expressed in lymphoblastoid cell lines. These genes might have inactivation profiles relevant to tissue-specific diseases that cannot therefore be addressed in blood-derived cell lines and will have to be explored in other types of cells.
Although overall PolII binding is reduced across the non-pseudoautosomal region of the X chromosome (Fig. 2), the evolutionarily younger XAR spanning most of the Xp arm exhibits higher PolII occupancy than does the Xq arm. This is consistent with the previously documented higher occurrence of genes that escape X inactivation and are expressed from the human Xi in the XAR (29). Here we present another piece of evidence that X inactivation is influenced by the evolutionary origins of the underlying sequences and show that PolII binds more readily at formerly autosomal sequences that were introduced to the X-inactivation system more recently (51–53,58).
Our allele-specific analysis of PolII binding has identified five novel biallelically expressed X-linked genes (Table 3 and Supplementary Material, Table S3), some of which had been hypothesized previously to escape inactivation in studies performed in somatic cell hybrids or on the basis of male/female dosage comparisons (59). For three of these, we were able to verify biallelic expression in multiple individuals (Fig. 6). The proportion of biallelically expressed genes in GM12878 (21%) is consistent with previous estimates of proportion of genes escaping inactivation in the population (29). Also consistent with previous observations is that all five of the biallelically expressed genes are located on Xp, where escape from inactivation occurs more frequently than on Xq (29). Our allele-specific analysis of these five genes expressed from the Xi suggests that, although the phosphorylated form of PolII may be a more precise indicator for expression levels (18), the unphosphorylated PolII queried in our study (see Materials and Methods) is a suitable indicator of inactivation status of genes and relative gene expression levels.
Because CpG methylation and heterochromatin markers have been associated with X-inactivation patterns (35–40), it was of interest to compare our expression and PolII data with other chromatin and genomic features of the X publicly available through the ENCODE database (http://genome.ucsc.edu/ENCODE/). We failed to detect significant association of any particular class of genes with chromatin or genomic features, such as presence of CpG islands or CCCTC-binding factor binding. We did, however, note a slight increase in association of PolII with H3K27me3 in gene bodies, indicating a potential role that this modification could play in marking particular Xi genes for expression in the portions of the Xi that are enriched for this heterochromatin mark. It has been speculated previously that H3K27me3-marked heterochromatin might be less effective at silencing gene expression than is H3K9me3-marked heterochromatin (35,36,60). To facilitate our understanding of X chromosome inactivation, further studies of multiple human females could take advantage of the vast data sets that are being generated by the ENCODE (19) and Human Epigenome (61) Projects. In combination with 1000 Genomes Project (47), future studies will be able to distinguishing features of the two homologous chromosomes in each individual to understand, for example, the role of H3K9me3 and H3K27me3 as well as many other histone modifications and chromatin features potentially implicated in X chromosome inactivation.
We detected increased PolII levels at several sites along the Xi, indicating a potential for expression despite the apparent association with silenced loci in this cell line. In each case, the Xa homologue was actively transcribed, while there was no detectable transcription product from the Xi. PolII occupancy at the Xi allele was particularly striking at the TRAPPC2 gene, where 67% of PolII reads aggregated across the locus mapped to the transcriptionally silenced Xi allele. We hypothesized that TRAPPC2 might be expressed in GM12878 in an unstable and inconsistent manner, and, indeed, testing additional clones derived from GM12878 revealed that TRAPPC2 is in fact occasionally transcribed from the Xi chromosome (Supplementary Material, Fig. S1). In addition, TRAPPC2 clearly escapes inactivation in cell lines from other informative females (Fig. 6); thus it is apparent that the presence of a PolII signal at the TRAPPC2 locus is indeed associated with the potential for expression.
While the finding of sites of PolII occupancy on the Xi near genes that are subject to inactivation may appear surprising, the large number of previously identified genes with variable expression patterns (29,31,33,34) is consistent with the idea that some genes on the Xi are poised for transcription in some individuals and/or cell lineages. Sites with increased PolII binding on the Xi identified in our study were candidate loci for variable expression patterns and indeed, when assayed in additional informative cell lines, we found that a high proportion of genes with Xi PolII association are occasionally expressed from the Xi in the population (Fig. 6).
Most genes that are expressed from the Xi generate RNA output that is reduced relative to the Xa (29), an indication that, although expression is generally suppressed chromosome-wide, the Xi is permissive to the transcriptional machinery. However, it remains unclear whether these low expression levels result from leakiness of the inactive state, with little or no phenotypic consequence in the context of full expression from the Xa copy, or, more provocatively, they reflect, at least in some cases, purposeful turning on the Xi allele to finely tune the overall genetic output. It is likely that there are numerous gene-specific control mechanisms among the ∼1200 genes on the X, superimposed upon the chromosome-wide and/or regional landscapes anticipated by the process of X inactivation. Indeed, the contrast in patterns of PolII distribution and gene expression between the XAR and XCR point to the interplay of evolutionary, genomic and local effects that remain to be fully explored in this context.
MATERIALS AND METHODS
Cell culture and single-cell cloning
GM12878 and other cell lines were grown in a humidified incubator at 37°C and 5% CO2 in RPMI1640 media supplemented with 15% fetal bovine serum and 1% antibiotics according to the ENCODE protocol (27). For single-cell cloning, cells were diluted in 50% conditioned media in a series of dilutions and seeded in 96-well plates. Cells were grown in 50% conditioned media until expansion and fed fresh media thereafter. To prepare conditioned media, cells were grown in fresh media over night and subsequently removed by centrifugation and filtration; the resulting cell-free conditioned media were diluted 1:1 with fresh media to obtain 50% conditioned media. For each presumptive clone, three biological replicates were grown (two for RNA and one for DNA). To confirm homogeneity of each candidate clone with respect to X inactivation, we tested allele-specific expression in monoallelically expressed genes, including XIST (rs1620574) and EBP (rs3048) (29). We further verified the purity of each selected isolate after expansion by the same method. Two confirmed isolates of each type (100% Xim and 100% Xip) were chosen for this study. Additional isolates derived from GM12878 were selected for further TRAPPC2 expression experiments. Isolates from seven additional female cell lines selected from the HapMap CEU population (GM06991, GM10861, GM10831, GM10839, GM12753, GM12802, GM12815) were derived in the same manner. Homogeneity with respect to the Xi was tested as described above in addition to a second XIST SNP (rs1794213) and FHL1 (rs1918), a gene known to be expressed solely from the Xa and not the Xi (29).
ChIP-seq
PolII ChIP was performed on GM12878 cells, and Illumina-based sequencing was carried out as described previously (62,63) with minor modifications. GM12878 cells were fixed, lysed and nuclei were collected. After lysing nuclei, chromatin was sonicated and immunoprecipitated with 8WG16 antibody to PolII (Covance, MMS-126R). Afterward, protein:DNA crosslinks were reversed overnight at 65°C, and DNA was isolated using a Qiagen polymerase chain reaction (PCR) Cleanup column. DNA was prepared for sequencing on an Illumina Genome Analyzer as described previously, modified to eliminate PCR prior to size selection and to use 15 cycles of PCR after size selection. Libraries were then sequenced to a minimum depth of 12 million 36-nucleotide reads per biological replicate. We employed the peak-calling algorithm QuEST (64) to detect sites of enrichment in the genome.
Measuring allele-specific PolII occupancy
To map allele-specific occupancy of PolII sites, we constructed a GM12878 parent-specific version of the human reference genome (hg18), as described previously (Reddy TE, Gertz J, Pauli F, Newberry K, Kucera KS, Wold B, Willard HF, Myers, RM, manuscript in preparation). We aligned ChIP-seq reads to the modified reference genome using the Bowtie aligner (65) and removed any alignments that mismatched at a known heterozygous SNP position.
To detect allelic bias of PolII occupancy at each SNP, we counted the number of reads mapping specifically to a paternal or maternal allele at all heterozygous positions and calculated a P-value according to a binomial model that assumes equal likelihood of each allele.
SNaPshot
A quantitative Q-SNaPshot assay was employed to test the abundance of each allele in the PCR amplicon, using protocols as described previously (29,36). Briefly, DNA was isolated from fresh cells using a Qiagen Gentra Puregene Cell Kit and stored at −20°C. RNA was isolated from fresh cells with a PerfectPure RNA Tissue Kit (5Prime), treated with DNase I (Roche) and stored frozen at −80°C. Quality and concentration of each nucleic acid sample was verified by NanoDrop spectrophotometer and gel electrophoresis. Random primed cDNA was synthesized from 200 to 300 ng of RNA using iScript cDNA synthesis kit (BioRad). In each assay, DNA and cDNA were amplified by a standard 25 μl protocol using Taq DNA Polymerase (Invitrogen). PCR products were then purified (EdgeBio). A third primer was used for the Q-SNaPshot single-nucleotide extension assay with subsequent ABI 3100 sequencer detection. The cDNA readout was normalized to the DNA signal with known 1:1 ratio of the two alleles to correct for biases in fluorescence output.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health/National Human Genome Research Institute (U54HG004576 to R.M.M.); and National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (5T32AR007450 to T.E.R.); K.S.K. was supported by the Duke Institute for Genome Sciences & Policy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Joseph Lucas for statistical advice and members of Willard and Myers labs for helpful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Koch F., Jourquin F., Ferrier P., Andrau J.C. Genome-wide RNA polymerase II: not genes only! Trends Biochem. Sci. 2008;33:265–273. doi: 10.1016/j.tibs.2008.04.006. doi:10.1016/j.tibs.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Brannan C.I., Dees E.C., Ingram R.S., Tilghman S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. doi:10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. doi:10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. doi:10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C., Li X., Hechmer A., Eisen M., Biggin M.D., Venters B.J., Jiang C., Li J., Pugh B.F., Gilmour D.S. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. doi:10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuda N.J., Ardehali M.B., Lis J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. doi:10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B.K., Muller H., Ragoussis J., Wei C.L., Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. doi:10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilchrist D.A., Dos Santos G., Fargo D.C., Xie B., Gao Y., Li L., Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. doi:10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns B.R. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. doi:10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 11.Tirosh I., Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. doi:10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterlin B.M., Price D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. doi:10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. doi:10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 14.Adelman K., Kennedy M.A., Nechaev S., Gilchrist D.A., Muse G.W., Chinenov Y., Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl Acad. Sci. USA. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. doi:10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baugh L.R., Demodena J., Sternberg P.W. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. doi:10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 16.Gilmour D.S. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. doi:10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- 17.Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. doi:10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilchrist D.A., Fargo D.C., Adelman K. Using ChIP-chip and ChIP-seq to study the regulation of gene expression: genome-wide localization studies reveal widespread regulation of transcription elongation. Methods. 2009;48:398–408. doi: 10.1016/j.ymeth.2009.02.024. doi:10.1016/j.ymeth.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. doi:10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zakharova I.S., Shevchenko A.I., Zakian S.M. Monoallelic gene expression in mammals. Chromosoma. 2009;118:279–290. doi: 10.1007/s00412-009-0206-8. doi:10.1007/s00412-009-0206-8. [DOI] [PubMed] [Google Scholar]
- 21.Cheung V.G., Nayak R.R., Wang I.X., Elwyn S., Cousins S.M., Morley M., Spielman R.S. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol. 2010;8:e1000480. doi: 10.1371/journal.pbio.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung V.G., Spielman R.S. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat. Rev. Genet. 2009;10:595–604. doi: 10.1038/nrg2630. doi:10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. doi:10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson L.S., Davies W., Isles A.R. Genomic imprinting effects on brain development and function. Nat. Rev. Neurosci. 2007;8:832–843. doi: 10.1038/nrn2235. doi:10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 25.Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. doi:10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 26.Heard E., Disteche C.M. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. doi:10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 27.McDaniell R., Lee B.K., Song L., Liu Z., Boyle A.P., Erdos M.R., Scott L.J., Morken M.A., Kucera K.S., Battenhouse A., et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. doi:10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasowski M., Grubert F., Heffelfinger C., Hariharan M., Asabere A., Waszak S.M., Habegger L., Rozowsky J., Shi M., Urban A.E., et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. doi:10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. doi:10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 30.Brown C.J., Carrel L., Willard H.F. Expression of genes from the human active and inactive X chromosomes. Am. J. Hum. Genet. 1997;60:1333–1343. doi: 10.1086/515488. doi:10.1086/515488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrel L., Cottle A.A., Goglin K.C., Willard H.F. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl Acad. Sci. USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. doi:10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berletch J.B., Yang F., Disteche C.M. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrel L., Willard H.F. Heterogeneous gene expression from the inactive X chromosome: an X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc. Natl Acad. Sci. USA. 1999;96:7364–7369. doi: 10.1073/pnas.96.13.7364. doi:10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson C.L., Brown C.J. Polymorphic X-chromosome inactivation of the human TIMP1 gene. Am. J. Hum. Genet. 1999;65:699–708. doi: 10.1086/302556. doi:10.1086/302556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chadwick B.P., Willard H.F. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl Acad. Sci. USA. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. doi:10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valley C.M., Pertz L.M., Balakumaran B.S., Willard H.F. Chromosome-wide, allele-specific analysis of the histone code on the human X chromosome. Hum. Mol. Genet. 2006;15:2335–2347. doi: 10.1093/hmg/ddl159. doi:10.1093/hmg/ddl159. [DOI] [PubMed] [Google Scholar]
- 37.Ke X., Collins A. CpG islands in human X-inactivation. Ann. Hum. Genet. 2003;67:242–249. doi: 10.1046/j.1469-1809.2003.00038.x. doi:10.1046/j.1469-1809.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- 38.Marks H., Chow J.C., Denissov S., Francoijs K.J., Brockdorff N., Heard E., Stunnenberg H.G. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. doi:10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brinkman A.B., Roelofsen T., Pennings S.W., Martens J.H., Jenuwein T., Stunnenberg H.G. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7:628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mietton F., Sengupta A.K., Molla A., Picchi G., Barral S., Heliot L., Grange T., Wutz A., Dimitrov S. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol. Cell Biol. 2009;29:150–156. doi: 10.1128/MCB.00997-08. doi:10.1128/MCB.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaumeil J., Le Baccon P., Wutz A., Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. doi:10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clemson C.M., Hall L.L., Byron M., McNeil J., Lawrence J.B. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl Acad. Sci. USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. doi:10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amos-Landgraf J.M., Cottle A., Plenge R.M., Friez M., Schwartz C.E., Longshore J., Willard H.F. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am. J. Hum. Genet. 2006;79:493–499. doi: 10.1086/507565. doi:10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wutz A., Gribnau J. X inactivation Xplained. Curr. Opin. Genet. Dev. 2007;17:387–393. doi: 10.1016/j.gde.2007.08.001. doi:10.1016/j.gde.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Rupert J.L., Brown C.J., Willard H.F. Direct detection of non-random X chromosome inactivation by use of a transcribed polymorphism in the XIST gene. Eur. J. Hum. Genet. 1995;3:333–343. doi: 10.1159/000472322. [DOI] [PubMed] [Google Scholar]
- 46.Plagnol V., Uz E., Wallace C., Stevens H., Clayton D., Ozcelik T., Todd J.A. Extreme clonality in lymphoblastoid cell lines with implications for allele specific expression analyses. PLoS ONE. 2008;3:e2966. doi: 10.1371/journal.pone.0002966. doi:10.1371/journal.pone.0002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durbin R.M., Abecasis G.R., Altshuler D.L., Auton A., Brooks L.D., Gibbs R.A., Hurles M.E., McVean G.A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. doi:10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor T.D., Noguchi H., Totoki Y., Toyoda A., Kuroki Y., Dewar K., Lloyd C., Itoh T., Takeda T., Kim D.W., et al. Human chromosome 11 DNA sequence and analysis including novel gene identification. Nature. 2006;440:497–500. doi: 10.1038/nature04632. doi:10.1038/nature04632. [DOI] [PubMed] [Google Scholar]
- 49.Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. doi:10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 50.Brown C.J., Hendrich B.D., Rupert J.L., Lafreniere R.G., Xing Y., Lawrence J., Willard H.F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. doi:10.1016/0092-8674(92)90520-M. [DOI] [PubMed] [Google Scholar]
- 51.Graves J.A. The origin and function of the mammalian Y chromosome and Y-borne genes—an evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. doi:10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- 52.Kohn M., Kehrer-Sawatzki H., Vogel W., Graves J.A., Hameister H. Wide genome comparisons reveal the origins of the human X chromosome. Trends Genet. 2004;20:598–603. doi: 10.1016/j.tig.2004.09.008. doi:10.1016/j.tig.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 53.Ross M.T., Grafham D.V., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. doi:10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D'Esposito M., Ciccodicola A., Gianfrancesco F., Esposito T., Flagiello L., Mazzarella R., Schlessinger D., D'Urso M. A synaptobrevin-like gene in the Xq28 pseudoautosomal region undergoes X inactivation. Nat. Genet. 1996;13:227–229. doi: 10.1038/ng0696-227. doi:10.1038/ng0696-227. [DOI] [PubMed] [Google Scholar]
- 55.Goto Y., Kimura H. Inactive X chromosome-specific histone H3 modifications and CpG hypomethylation flank a chromatin boundary between an X-inactivated and an escape gene. Nucleic Acids Res. 2009;37:7416–7428. doi: 10.1093/nar/gkp860. doi:10.1093/nar/gkp860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. doi:10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. doi:10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lahn B.T., Page D.C. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. doi:10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 59.Bondy C.A., Cheng C. Monosomy for the X chromosome. Chromosome Res. 2009;17:649–658. doi: 10.1007/s10577-009-9052-z. doi:10.1007/s10577-009-9052-z. [DOI] [PubMed] [Google Scholar]
- 60.Rougeulle C., Chaumeil J., Sarma K., Allis C.D., Reinberg D., Avner P., Heard E. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol. Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. doi:10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.American Association for Cancer Research Human Epigenome Task Force; European Union, Network of Excellence, Scientific Advisory Board. Moving AHEAD with an international human epigenome project. Nature. 2008;454:711–715. doi: 10.1038/454711a. doi:10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. doi:10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 63.Reddy T.E., Pauli F., Sprouse R.O., Neff N.F., Newberry K.M., Garabedian M.J., Myers R.M. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. doi:10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valouev A., Johnson D.S., Sundquist A., Medina C., Anton E., Batzoglou S., Myers R.M., Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. doi:10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.