Abstract
Mutations in NF1 cause neurofibromatosis type I (NF1), a disorder characterized, among other clinical manifestations, by generalized and focal bony lesions. Dystrophic scoliosis and tibial pseudoarthrosis are the most severe skeletal manifestations for which treatment is not satisfactory, emphasizing the dearth of knowledge related to the biology of NF1 in bone cells. Using reporter mice, we report here that the mouse Col2α1-Cre promoter (collagen, type II, alpha 1) is active not only in chondrocytes but also in adult bone marrow osteoprogenitors giving rise to osteoblasts. Based on this finding, we crossed the Col2α1-Cre transgenic and Nf1flox/flox mice to determine whether loss of Nf1 in axial and appendicular osteochondroprogenitors recapitulates the skeletal abnormalities of NF1 patients. By microtomographic and X-rays studies, we show that Nf1Col2−/− mice display progressive scoliosis and kyphosis, tibial bowing and abnormalities in skull and anterior chest wall formation. These defects were accompanied by a low bone mass phenotype, high bone cortical porosity, osteoidosis, increased osteoclastogenesis and decreased osteoblast number, as quantified by histomorphometry and 3D-microtomography. Loss of Nf1 in osteochondroprogenitors also caused severe short stature and intervertebral disc defects. Blockade of the RAS/ERK activation characteristic of Nf1−/− osteoprogenitors by lovastatin during embryonic development could attenuate the increased cortical porosity observed in mutant pups. These data and the skeletal similarities between this mouse model and NF1 patients thus suggest that activation of the RAS/ERK pathway by Nf1 loss-of-function in osteochondroprogenitors is responsible for the vertebral and tibia lesions in NF1 patients, and that this molecular signature may represent a good therapeutic target.
INTRODUCTION
Neurofibromatosis type I (NF1, or Von Recklinghausen disease) is an autosomal dominant genetic disorder with an incidence of approximately 1 in 3000, making it one of the most common genetic disorders (1). Despite complete penetrance, ∼50% of NF1 cases are thought to result from spontaneous mutations in the NF1 gene (2). NF1 encodes neurofibromin, a member of the guanosine triphosphatase-activating proteins known to act as suppressors of the RAS family of proteins. Neurofibromin is a specific suppressor of p21-RAS, and mutations in this gene cause unsuppressed activation of RAS, leading to abnormal cell growth and differentiation and to the clinical features of NF1 (3). These latter include a predisposition to oncogenic transformation leading to neuro-cutaneaous neurofibromas and optic pathway tumors, but also to manifestations not related to cancers, including cognitive defects and skeletal abnormalities.
The etiology of the NF1 skeletal manifestations is unclear and bone cell-autonomous abnormalities as well as nerve-derived or endocrine signals are thought to contribute to these bony defects. Neurofibromin is relatively ubiquitous during development and is later found predominantly within the nervous system and related tissues (4). Mild NF1 skeletal pathologies such as juvenile osteoporosis, shorter than average size, and non-dystrophic scoliosis are thought to result from haploinsufficiency at the NF1 locus (5,6). On the other hand, dystrophic scoliosis and long bone bowing and pseudoarthrosis are focal NF1 defects associated with high morbidity for which treatment or prevention is not satisfactory (7–11). The expression of neurofibromin in adult but also developing skeletal tissues (12–14), the focal, often unilateral and persistent nature of the NF1 dystrophic skeletal lesions, as well as their non-systematic occurrence in this population of patients, led us to hypothesize that complete NF1 loss-of-function in dividing and multipotent bone cells is required for such lesions to occur and persist. Supporting this hypothesis, somatic NF1 loss-of-function has been demonstrated to be at the origin of the development of neurofibroma in NF1 patients (15) and has been detected in a biopsy of a pseudoarthrosis in one NF1 patient (16). In addition, conditional mouse models lacking both alleles of Nf1 specifically in limb osteochondroprogenitors (Nf1Prx model) or mature osteoblasts (Nf1Ob−/− model) displayed bone abnormalities that demonstrated the existence of a cell-autonomous role of Nf1 in the mesenchymal lineage (13,14,17). Whether this loss of NF1 function and the variable occurrence of the NF1 manifestations are caused by somatic loss of heterozygosity, promoter methylation abnormalities or the involvement of modifier genes is currently unclear.
Although these mouse models have been critical to identifying NF1 target kinases and genes (13,14), their use as NF1 pre-clinical models is hampered by the fact that they display bone phenotypes that differ significantly from those of NF1 patients. For instance, neither bone loss nor dystrophic bone lesions were observed in Nf1+/− mice (18,19), indicating that such lesions require NF1 loss-of-function to occur; bowed tibiae, increased osteoid and increased bone resorption, but no axial skeleton abnormalities reminiscent of the vertebral dystrophic lesions observed in NF1 patients, were reported in the Nf1Prx model (14), because the Prx-Cre transgene used in this study is not expressed in the vertebral axis. Lastly, despite increased osteoid and increased bone resorption, the Nf1Ob−/− model displayed a high bone mass phenotype and high bone turnover instead of an expected low bone mass phenotype (13), possibly due to selective Nf1 inactivation in committed mature osteoblasts but not in osteochondroprogenitor cells. Such discrepancies of skeletal defects between these genetic mouse models and NF1 patients thus suggested that the genetic manipulations performed in these mouse models did not occur in the correct cell type, differentiation stage or skeletal element to reproduce the human skeletal defects. Based on these observations, we hypothesized that loss of Nf1 function in a subset of bone marrow mesenchymal stem cells or osteochondroprogenitor cells was at the origin of the NF1 focal dystrophic bone abnormalities.
Type II collagen transcripts (Col2α1) are first detected during embryonic mouse development at 9.5 days post-coitum (d.p.c.) in the sclerotome of the differentiating somites and in the cranial mesenchyme destined to give rise to chondrocytes but also osteoblasts, as well as in the notochord (20). Based on this pattern of expression, we chose the mouse Col2α1-Cre transgenic mice that drives Cre recombinase expression in osteochondroprogenitor cells and their progeny, including chondrocytes and osteoblasts (21–28), to generate mice lacking Nf1 in limb and axial skeleton osteochondroprogenitor cells (herein called Nf1Col2−/− mice), with the expectation that such a mouse model, due to early Nf1 recombination in the mesenchymal lineage, should display a wide spectrum of the NF1 skeletal lesions observed in NF1 patients.
RESULTS
Neurofibromin is expressed in osteochondroprogenitor cells and their progeny
To understand whether loss of Nf1 function in osteochondroprogenitor cells could trigger the aforementioned NF1 skeletal defects, we first examined the expression pattern of Nf1 and its encoded protein neurofibromin in bone primary cells, cell lines and histological sections. Nf1 mRNA expression, assessed by reverse transcription polymerase chain reaction (RT-PCR), was detected in mouse adherent bone marrow stromal osteoprogenitor cells (BMSCs), in primary chondrocytes extracted from ribs as well as in primary undifferentiated and differentiated osteoblasts from calvariae. It was also detected in multiple cell lines, including C3H10T1/2 mesenchymal osteoprogenitors, TMC23 chondrocytes (29) and MC3T3E1 osteoblasts (Fig. 1A). No significant change in Nf1 expression was observed between immature and mature osteoblasts differentiated in vitro (Supplementary Material, Fig. S1). In vivo, neurofibromin immunoreactivity was not detected in resting and proliferative chondrocytes, but was clearly detected in hypertrophic and trabecular osteoblasts from P4 mouse tibiae, as well as in cells of the perichondrium [Fig. 1B and (13)]. This pattern of expression thus supported a cell-autonomous function of Nf1 in the mesenchymal lineage and a potential role of Nf1 in chondrocytes and osteochondroprogenitors.
Figure 1.
Neurofibromin is expressed in osteochondroprogenitor cells and their progeny. (A) Nf1 mRNA expression was detected by RT-PCR in multiple primary cells and cell lines including adherent bone marrow osteochondroprogenitors (BMSCs and C3H10T1/2), chondrocytes (Ch: rib primary chondrocytes and TMC23 cells) and osteoblasts (OB) (calvaria osteoblasts and MC3T3 cells). (B) Neurofibromin expression (brown staining) was detected in growth plate hypertrophic chondrocytes, trabecular osteoblasts and cells of the periosteum and perichondrium (immunohistochemistry).
The Col2α1 collagen promoter recombines Nf1 in osteochondroprogenitor cells
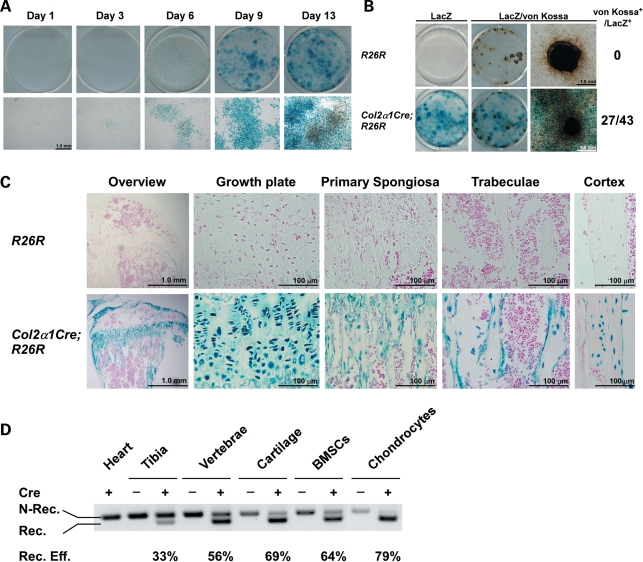
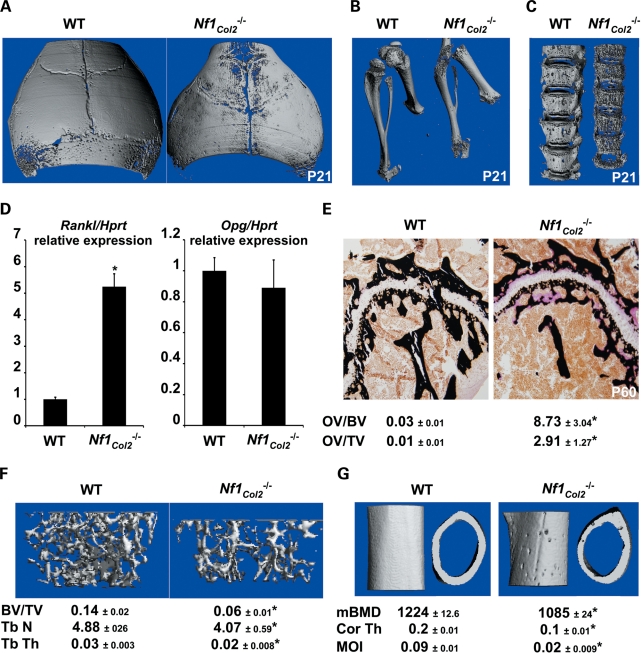
Although the mouse Col2α1-Cre transgenic line has been mainly used with the purpose to delete floxed genes in chondrocytes, several studies have reported that this promoter construct is expressed and active in a subset of osteochondroprogenitor cells that give rise to mature bone marrow osteoblasts (21–28). However, the proportion and biological significance of the recombined osteoblasts generated in this transgenic model remained unclear. To address this question, we generated Rosa26R (R26R) (control) and Col2α1-Cre;R26R reporter mice and grew BMSCs extracted from these mice in osteogenic media. X-gal (5 bromo-4-chloro-3-indoyl-β-d-galactopyranoside) staining was then performed to assess the proportion of LacZ-positive, i.e. recombined cells at various time points. BMSCs from Col2α1-Cre;R26R mice grown in osteogenic media gave rise to adherent LacZ-positive osteoprogenitors that could be observed on the day of bone marrow plating (day 1) and could be found in the majority of the formed nodules at day 3, 6, 9 and 13 in cultures, whereas bone marrow cells from R26R mice were LacZ-negative (Fig. 2A and data not shown). Importantly, the vast majority of cells within each nodule in cultures from double-transgenic mice were LacZ-positive, and 60% of LacZ-positive nodules were calcified following von Kossa staining (Fig. 2B). In vivo, LacZ-positive chondrocytes but also osteoblasts and osteocytes could be detected in adult Col2α1-Cre;R26R mice, but not in control R26R mice (Fig. 2C). The Col2α1-Cre promoter is thus active in a subpopulation of proliferative bone marrow osteoprogenitors in vivo. In addition, it allows recombination in the vertebral axis, as opposed to the use of the Prx-Cre promoter, whose expression is restricted to the limbs. We thus crossed the mouse Col2α1-Cre transgenic mice and the Nf1flox/flox mouse line (30) with the goal to determine the role of Nf1 in osteochondroprogenitor cells and to generate a new NF1 pre-clinical mouse model. This mouse Col2α1-Cre transgenic line was crossed to the Nf1flox/flox mouse line (30) to generate mice heterozygous for the floxed Nf1 allele (genotype: Col2α1-Cre+;Nf1fl/+). These mice had no obvious phenotype and were fertile, and thus were crossed to Nf1flox/flox mice to inactivate both Nf1 alleles by Cre-mediated excision (genotype: Col2α1-Cre+;Nf1flox/flox called Nf1Col2−/− herein). Nf1Col2−/− mice were born at the Mendelian ratio, but only 20–30% survived the weaning period.
Figure 2.
The Col2α1Cre promoter is active in Nf1flox/flox osteochondroprogenitor cells. (A) X-gal staining of BMSCs from Col2α1-Cre;R26R mice at day 1, 3, 6, 9 and 13. (B) X-gal and von Kossa co-staining of BMSC cultures from R26R and Col2α1-Cre;R26R mice following 21 days of osteogenic differentiation. (C) X-gal and nuclear red staining of 2-month-old R26R and Col2α1-Cre;R26R long bones. (D) Non-recombined (N-Rec.) and recombined (Rec.) Nf1 alleles detected by PCR using genomic DNA from bone tissues and primary cells. The ratio of Rec./N.Rec. was calculated following densitometry.
The ability of the Cre recombinase to specifically recombine the floxed Nf1 allele was examined by genomic PCR analysis using tissues and primary bone cells extracted from WT and Nf1Col2−/− mice. The 280 bp amplimer corresponding to the excised floxed Nf1 allele was detectable in Nf1Col2−/− tibiae, vertebrae and cartilage, but not in soft tissues like heart (Fig. 2D). The high ratio of 350 bp (non-recombined)/280 bp (recombined) amplimers in tibiae or vertebrae likely reflects the presence of cells not expressing the Cre recombinase in whole tissues rather than inefficient recombination, since semi-purified chondrocytes and adherent bone marrow osteoprogenitor cells from Nf1Col2−/− mice displayed an efficiency of recombination of 79 and 64%, respectively. Together, these results confirm the existence of Cre-mediated recombination within the Nf1 locus in a subpopulation of bone marrow osteoprogenitor cells, chondrocytes and osteoblasts in Nf1Col2−/− mice.
Nf1 is required for proper endochondral ossification
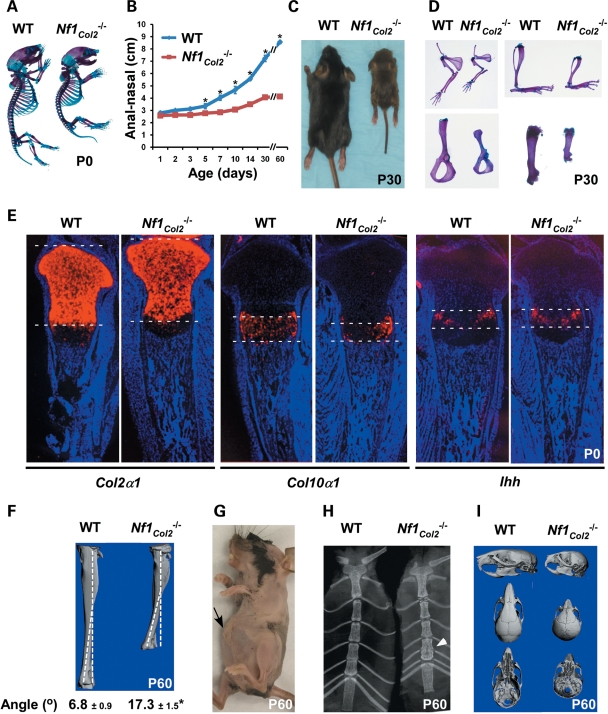
Nf1Col2−/− mutant mice were born with a size only slightly smaller than wild-type (WT) littermates (Fig. 3A) but exhibited impaired growth thereafter, as quantified by reduced anal–nasal length (Fig. 3B) and body weight (data not shown). By one month of age, Nf1Col2−/− mutant mice were half the size of WT littermates (Fig. 3B and C). Shortening of skeletal elements issued from endochondral bone formation, including the forelimbs, hind limbs and vertebrae, was observed in mutant mice (Fig. 3D). A reduction in the size of the growth plate hypertrophic zone was observed in P0 Nf1Col2−/− mutant mice compared with WT littermates, as shown by in situ hybridization for Col10a1 (collagen, type X, alpha 1), whereas Col2α1 and Ihh (Indian Hedgehog) signals were equal between genotypes (Fig. 3E). In addition, all Nf1Col2−/− mice showed evidence of lateral tibia bowing (Fig. 3F), reminiscent of NF1 patients and Nf1Prx mice (14,31), as well as an anterior chest wall anomaly, resulting from the fusion and bending of the most distal sternum bone elements (Fig. 3G and H), which is reminiscent of anterior chest wall anomalies observed in 50% of NF1 patients (32).
Figure 3.
Nf1Col2−/− mice display severe short stature and defects of endochondral bone formation. (A) Nf1Col2−/− mice were born (P0) with a size similar to WT littermates. (B) Increasing growth retardation in Nf1Col2−/− mice over a 2-month-long period. (C) Nf1Col2−/− mice were half the size of WT littermates by 1 month of age. (D) Shortening of humeri, radii, ulnae (top left), scapulae (bottom left), tibiae (top right) and femurs (bottom right) in 1-month-old Nf1Col2−/− mice. (E) Reduced size of the hypertrophic zone in the growth plate of P0 Nf1Col2−/− mice compared with WT littermates, assessed by in situ hybridization for the indicated genes. (F) Anterolateral tibia bowing in Nf1Col2−/− mice. (G and H) Anterior chest wall anomaly in 2-month-old Nf1Col2−/− mice. Arrows point to the protruding and fused anterior bone element. (I) Defect of skull structure in 2-month-old WT and Nf1Col2−/− mice (3D-microtomographic images). *P < 0.01 versus WT mice, n = 5.
Skull formation was affected as well by loss of Nf1 in osteochondroprogenitor cells. Nf1Col2−/− mice were indeed characterized by the presence of a dome-shaped head, a large head-to-body size ratio, a shortened snout and teeth malocclusion, suggestive of defects in the formation of craniofacial bones. Detailed analysis using 3D-microtomographic images suggested that the skull characteristics in mutant mice were caused by abnormalities in bones formed through endochondral formation, whereas the bone elements formed via intramembranous ossification appeared normal. More specifically, the maxilla, zygomatic and squasmosal bones were shorter in 2-month-old mutant mice compared with WT littermates. The frontal bone in mutant mice was shorter and wider and the frontal suture was more patent in mutant mice compared with WT. The length and width of parietal bones were comparable in both mutant and WT, but appeared bent downward compared with WT mice. The premaxilla had comparable height and width in mutant and WT mice. Mutant mice had a prognathic mandible caused by a shorter cranial base and downward bending of parietal bones, explaining the teeth malocclusion observed (Fig. 3I). The difference of skull structure between mice and humans prevents direct comparisons in the structure and organization of skull bones between Nf1Col2−/− mice and NF1 patients, but despite this limitation, the skull defects observed in Nf1Col2−/− mice suggest that proper skull formation is dependent on NF1 function in chondrocytes and/or osteoblasts, and that the skull defects in NF1 patients, including large head circumference, may have a bone cell-autonomous origin.
Nf1 is required for intervertebral disc formation
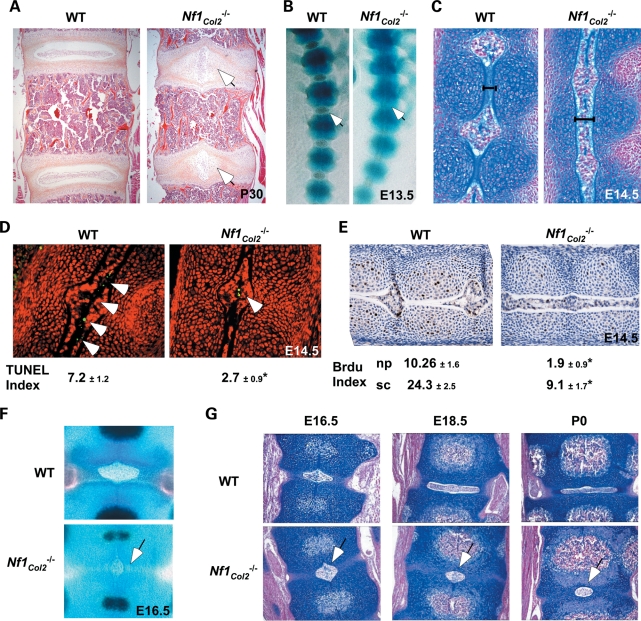
The intervertebral disc (IVD) contains a highly hydrated structure called the nucleus pulposus (NP), which derives from the notochord and contributes to the mechanical function of the spine. Unexpectedly, histological analyses of the axial skeleton revealed the existence of IVD defects in Nf1Col2−/− adult mice, as shown by a 90° rotation of mutant NPs, observed all along the vertebral axis (Fig. 4A). Examination of IVD formation at earlier developmental time points indicated that this abnormal NP structure resulted, at least in part, from a delay in notochord regression. In WT mice, between E13.5 and E16.5, the notochord disappears from the developing vertebral body (VB), possibly extruded by the increasing pressure of the cartilage extracellular matrix, but persists at the center of the IVD, where it forms the NP (33,34). Although Nf1Col2−/− embryos showed a discrete metameric patterning of the spinal column at E14.5, Nf1Col2−/− embryos at that stage showed an unsegmented notochord (Fig. 4B and C). Calculation of apoptotic and proliferative indexes in notochord cells following terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and 5-bromo-2′-deoxyuridine (BrdU) labeling, respectively, indicated that E14.5 Nf1Col2−/− embryos have a significant 3-fold decrease in notochord cells apoptosis compared with WT embryos (Fig. 4D). A significant 5.4- and 2.6-fold decrease in cell proliferation was also observed in the NP and sclerotome area of the Nf1Col2−/− skeletal axis, respectively, when compared with WT embryos (Fig. 4E). At E16.5, the size of the NP was reduced in Nf1Col2−/− embryos compared with WT embryos, but more importantly, this structure as well as the annulus fibrosus surrounding it did not evolve anymore, giving rise to an abnormal IVD structure (Fig. 4F and G). This phenotype was 100% penetrant and in agreement with Col2α1-Cre expression in both notochord and sclerotome (21). The observation that IVD defects have not been reported in NF1 patients suggests that loss of NF1 function in notochordal cells in humans does not occur or is lethal. Alternatively, this defect may have been overlooked in NF1 patients. In any case, this IVD phenotype in Nf1Col2−/− mice reveals a crucial role of NF1 in notochordal cells during IVD development.
Figure 4.
Lack of Nf1 causes IVD formation defects. (A) 90° rotation of the NP in the vertebral axis of 1-month-old Nf1Col2−/− mice (hematoxylin and eosin staining). (B) Discrete patterning of the spinal column in E13.5 Nf1Col2−/− embryos (skeletal preparations). Arrows point to the forming IVD. (C) Unsegmented notochord in E14.5 Nf1Col2−/− embryos (Alcian blue and Fast Red staining). (D) Decreased apoptotic cell number (arrow) in the notochords of E14.5 Nf1Col2−/− embryos (TUNEL assay). (E) Decreased proliferation in the NP and sclerotome area of Nf1Col2−/− embryos (BrdU labeling). np, nucleus pulposus; sc, sclerotome. (F) Reduced size and delayed regression of the nucleus pulposus (arrow) in E16.5 Nf1Col2−/− embryos (skeletal preparations). (G) Abnormal IVD structure (arrow) in Nf1Col2−/− mice from E16.5 to P0 (blue and Fast Red staining). *P < 0.01 versus WT mice, n = 5.
Nf1 is required for proper axial skeleton formation
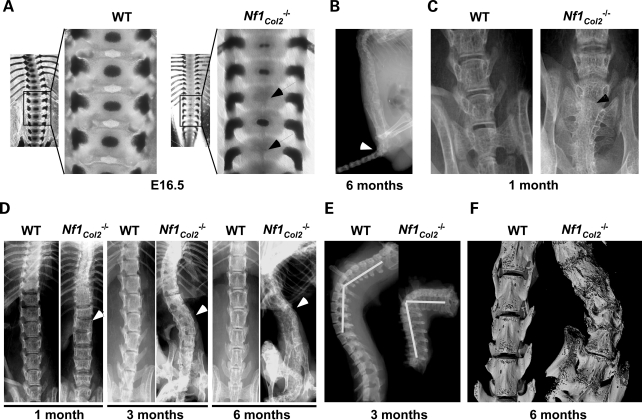
Dystrophic scoliosis is one of the most severe and morbid skeletal manifestation that occurs in NF1 patients and its etiology is unclear. The observation that dystrophic scoliosis is associated with spinal neurofibroma in 70% of the cases (11) led to the belief that adjacent neurofibromas may contribute to this spinal defect, possibly by the release of bone osteolytic factors. Several observations both from the clinic and from the analysis of the Nf1Col2−/− mice suggest, however, that dystrophic scoliosis in NF1 patients might result from a bone cell-autonomous defect. First and obviously, not all cases of dystrophic scoliosis in patients are associated with the presence of neurofibromas, indicating that neurofibromas are not necessary for this defect to occur (35–38); second, gene alterations in osteochondroprogenitor cells can cause developmental vertebral defects, leading to scoliosis/kyphosis in several mouse models, without adjacent neurofibromas (39,40). Following alizarin red/Alcian blue staining, ossification centers were present in every single VB in E16.5 WT embryos but in only a subset of them in Nf1Col2−/− embryos (Fig. 5A), suggestive of a delay of congenital vertebral osteogenesis. By 1 month of age, Nf1Col2−/− displayed a 45° caudal angulation, vertebral fusion at the base of the tail and loss of alignment of vertebral bodies (Fig. 5B–D). Scoliosis and kyphosis was observed by 3 months of age (Fig. 5D and E). Vertebral fusion and severe loss of bone mineral density (BMD) were easily detectable in all mutant animals by X-rays by 6 months of age (Fig. 5D). Importantly, examination of mutant mice did not reveal the presence of adjacent spinal neurofibroma, in agreement with the cell specificity of the genetic manipulation generated in this model. In addition, 3D-microtomographic analyses clearly showed the existence of an abnormal VB shape in Nf1Col2−/− mice, including increased porosity and reduced size (Fig. 5F and see what follows). These observations suggest that the vertebral phenotype observed in Nf1Col2−/− mice, and possibly vertebral dysplasia in NF1 patients, is bone cell-autonomous and has a developmental origin.
Figure 5.
Neurofibromin is required for proper axial skeleton formation. (A) Absence of ossification centers (arrow) in E16.5 Nf1Col2−/− embryonic VB (skeletal preparations). (B) Nf1Col2−/− mice displayed a 45° caudal angulation (arrow) at 6 months of age (X-rays). (C) Vertebral fusion at the base of the tail (arrow) in 1-month-old Nf1Col2−/− mice (X-rays). (D) Misalignments between vertebral bodies by 1 month of age (arrow), scoliosis by 3 months of age (arrow) and vertebral fusion and severe loss of BMD (arrow) by 6 months of age in Nf1Col2−/− mice (X-rays). (E) Kyphosis was observed in Nf1Col2−/− mice by 3 months of age (X-rays). (F) Abnormal VB shape in Nf1Col2−/− mice (3D-microtomography) at 6 months of age.
Nf1 is required for bone remodeling and mechanical properties
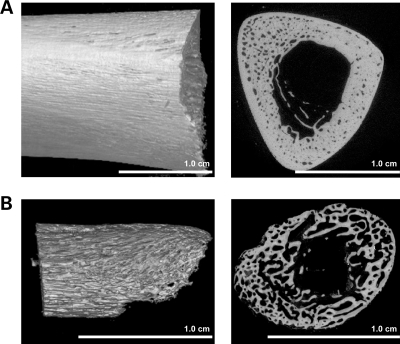
Three-dimensional microtomographic analyses allowed us to gather additional architectural, cortical and trabecular information in the vertebral and appendicular skeletons of mutant mice. One striking observation from these analyses was the existence of a severe cortical porosity in 3 week-old Nf1Col2−/− mice, observed in the skull (Fig. 6A), long bones (Fig. 6B) and most strikingly in vertebrae (Fig. 6C). Vertebral bodies in mutant mice were smaller, displayed irregular ossification and quasi-disappearance of vertebral pedicles. Importantly, our preliminary analyses revealed that this cortical phenotype is also observed in humans with NF1 when looking at surgically discarded samples obtained from NF1 individuals with tibial pseudoarthrosis (Fig. 7). Histological analyses in Nf1Col2−/− mice did not demonstrate any significant increase in blood vessel density within the cortical pores of mutant embryos at E18.5 and P0 (data not shown) but instead the accumulation of non-mineralized collagenous matrix (osteoidosis) (Fig. 6E), as observed in Nf1ob−/− mice (13). Osteoidosis in Nf1Col2−/− mice was of variable intensity and distribution, and not caused by abnormalities in phosphate and calcium homeostasis (Supplementary Material, Fig. S2). A significant decrease in calcified trabecular bone volume per tissue volume (BV/TV), thickness and number, as well as a significant decrease in cortical BMD, thickness and moment of inertia (MOI) was measured in the femurs of 2-month-old Nf1Col2−/− mice compared with littermate controls (Fig. 6F and G). The number and surface of TRAP-positive differentiated multinucleated osteoclasts were significantly increased in adult Nf1Col2−/− femurs compared with WT littermates (Table 1), in agreement with the increase in Rankl (receptor activator of nuclear factor kappa-B ligand) expression observed in Nf1−/− osteoblasts from Nf1ob−/− mice and increased osteoclastogenesis observed in these mice (13) as well as increased Rankl/Opg (Opg, osteoprotegerin) ratio found in bones from Nf1Col2−/− mice (Fig. 6D). In contrast to Nf1ob−/− mice, however, osteoblast number and surface were severely reduced in Nf1Col2−/− mice compared with control littermates (Table 1). The low bone mass observed in Nf1Col2−/− mice is thus caused by increased bone resorption and impaired bone formation and mineralization.
Figure 6.
Lack of Nf1 in osteochondroprogenitors impairs bone structure and remodeling. Cortical porosity in the skull (A), long bones (B) and vertebrae (C) of 3-week-old Nf1Col2−/− mice (3D-microtomography). (D) Rankl and Opg expression in 2-month-old WT and Nf1Col2−/− lumbar vertebrae measured by qPCR, n = 4. (E) Osteoidosis in 2-month-old Nf1Col2−/− femurs. (F) Decreased femoral trabecular BV/TV, thickness and number and (G) decreased cortical mineral BMD (mBMD), thickness (Cor Th) and MOI in 2-month-old Nf1Col2−/− mice (3D-microtomography). *P < 0.01 versus WT mice, n = 7.
Figure 7.
High cortical porosity in an NF1 patient presenting with pseudoarthrosis. Images are of the proximal portion of a surgically discarded tibial sample of a 3-year-old child with NF1 who had a fracture of the tibia with subsequent non-union and pseudoarthrosis. Control sample is from a dissected portion of the tibia near the junction of the middle and distal third segment in a 6-year-old deceased child without NF1 or chronic medical conditions. Fixed bone samples were staged in preparation for scanning and placed in a bench-top microCT scanner (Scanco µCT40, Zurich, Switzerland). The samples were scanned at optimized parameters (including resolution, power and frame averages) to derive the images and metrics for the anatomical region of interest. Three-dimensional volume rendering snapshots (left panel) and 2D cross-section snapshots (right panel) of the tibial samples (A: control individual, B: NF1 individual). Procedures for human sample collection and analyses were approved by the University of Utah Institutional Review Board.
Table 1.
Histomorphometric parameters in 2-month-old WT and Nf1Col2−/− mice
| WT | Nf1Col2−/− | P-value | |
|---|---|---|---|
| Oc.S/BS | 4.89 ± 2.40 | 11.69 ± 3.87 | 0.002 |
| Oc.Nb/BPm | 1.92 ± 0.85 | 5.49 ± 2.01 | 0.001 |
| Ob.S/BS | 8.71 ± 2.13 | 2.37 ± 1.08 | 0.006 |
| Ob.Nb/BPm | 6.53 ± 2.40 | 2.14 ± 0.93 | 0.008 |
Indicated parameters were acquired in vertebral bones from 2-month-old WT and Nf1Col2−/− littermates. Oc.S/BS, osteoclast surface per bone surface; Oc.Nb/BPm, osteoclast number per bone perimeter; Ob.S/BS, osteoblast surface per bone surface; Ob.Nb/BPm, osteoblast number per bone perimeter.
To address whether the low bone mass, accumulation of osteoid and increased cortical porosity in Nf1Col2−/− mice were associated with significant changes in bone mechanical properties, unconfined compression tests and three-point bending tests were performed in 2-month-old WT and Nf1Col2−/− L5 vertebral bodies and femurs, respectively. Nf1Col2−/− vertebrae displayed a significant 74, 68 and 67% reduction in stiffness, yield force and peak force, respectively (Table 2). Similarly, femurs from Nf1Col2−/− mice exhibited a 3-fold decrease in rigidity and maximum moment, indicating strongly compromised bone biomechanical properties in Nf1Col2−/− mice. This deficiency is primarily due to structural and architectural defects in the mutant bones since there was only a marginal, non-significant difference in the estimated bending strength between genotypes (Table 2). Together, these observations indicate that lack of Nf1 in osteochondroprogenitor cells causes severe cellular dysfunctions leading to developmental skeletal abnormalities that deleteriously affect the shape, structure and biomechanical properties of the adult vertebrae and long bones.
Table 2.
Bone mechanical properties in 2-month-old WT and Nf1Col2−/− mice
| WT | Nf1Col2−/− | P-value | |
|---|---|---|---|
| Vertebral stiffness (N/mm) | 179 ± 38 | 45 ± 7 | 0.000001 |
| Vertebral yield force (N) | 27.5 ± 9.9 | 8.8 ± 3.7 | 0.0008 |
| Vertebral peak force (N) | 28.4 ± 9.5 | 9.2 ± 3.8 | 0.0004 |
| Femoral rigidity (kN mm2) | 1.09 ± 0.46 | 0.32 ± 0.18 | 0.001 |
| Femoral peak moment (N mm) | 49.6 ± 16.6 | 13.8 ± 5.7 | 0.0002 |
| Femoral modulus (GPa) | 12.5 ± 4.4 | 12.0 ± 5.3 | 0.83 |
| Femoral bending strength (MPa) | 310 ± 112 | 260 ± 92 | 0.16 |
Indicated parameters were acquired in vertebral or femoral bones from 2-month-old WT and Nf1Col2−/− littermates.
Ras inhibition by lovastatin corrects constitutive extracellular signal-regulated kinase activation in Nf1−/− bone cells and attenuates the cortical porosity in Nf1Col2−/− mice
Constitutive activation of RAS and of the downstream kinase ERK1/2 (ERK, extracellular signal-regulated kinase) in multiple cell types (41–43), including osteoblasts (13,14,18), are thought to underlie most of the NF1-related clinical manifestations. Not surprisingly, we found that ERK1/2 was constitutively activated in Nf1−/− BMSCs (Fig. 8A) and thus hypothesized that Ras or ERK inhibition, if performed at early stages of bone development, could prevent or ameliorate the bone phenotypes observed in Nf1Col2−/− mice. To test this hypothesis, we used lovastatin, which inhibits the mevalonate pathway and subsequently inhibits RAS prenylation, membrane localization and activity (44,45) to attenuate the constitutive activation of RAS and ERK1/2 observed in Nf1−/− osteochondroprogenitors and their progeny. We previously used this approach to successfully ameliorate bone healing in mice lacking Nf1 in mature osteoblasts (17). As an in vitro proof of concept, treatment of BMSCs isolated from Nf1Col2−/− mice with lovastatin (20 µm) for 12 h decreased ERK1/2 activation (Fig. 8A). To address whether this effect of lovastatin could translate in vivo to beneficial bony outcomes, lovastatin slow release microparticles (10 mg/kg) were given intraperitoneally every other day to Col2α1-Cre+;Nf1flox/+ pregnant mothers carrying litters including both WT and Nf1Col2−/− embryos. Treatment was initiated at E10.5, prior to bone development, and pups were analyzed at birth and 3 weeks later. As shown in Figure 8B, lovastatin treatment for 10 days only during early bone development significantly decreased vertebral cortical porosity in Nf1Col2−/− mice compared with control Nf1Col2−/− mice. Because osteoid volume (OV) is barely measurable at birth, we measured the effect of lovastatin by histomorphometry in a group of mice treated as above and sacrificed 3 weeks after birth. In addition, we measured structural parameters by microcomputed tomography (µCT). Our analyses indicated that trabecular BV/TV was not changed between non-treated WT and mutant mice at 3 weeks of age (data not shown); however OV and cortical porosity were increased in mutant mice. The beneficial effect of lovastatin treatment during embryogenesis on cortical porosity and OV was still measurable in mutant bones from 3-week-old mice, although the intensity of the effect was then lower than just after birth (Fig. 8C). These results strongly support the notion that constitutive RAS/ERK activation is the molecular determinant of the cortical porosity and osteoid defects in Nf1Col2−/− mice and suggest that pharmacological approaches targeting this pathway can be considered for the treatment of the NF1 bony abnormalities, including NF1 vertebral dystrophy and pseudoarthrosis.
Figure 8.
Lovastatin treatment attenuates the cortical porosity in Nf1Col2−/− mice. (A) Activated Erk1/2 phosphorylation in Nf1−/− BMSCs was corrected by lovastatin treatment. (B) Lovastatin in utero treatment from E10.5 to P0 significantly decreased cortical porosity in P0 Nf1Col2−/− mice. (C) Increased cortical porosity and OV in P30 Nf1Col2−/− mice, and trend toward a beneficial effect of lovastatin administered in utero in 3-week-old Nf1Col2−/− mice. *P < 0.01 versus WT mice; #P < 0.01 versus Nf1Col2−/− mice, n = 7.
DISCUSSION
The NF1 dystrophic skeletal pathologies, including pseudoarthrosis of the tibia and scoliosis, are associated with high morbidity for which treatment or prevention is not satisfactory (7–11). This lack of satisfactory treatment emphasizes the dearth of knowledge related to the biology of NF1 in bone cells. Through the generation of mice lacking Nf1 in osteochondroprogenitors, we showed in this study that NF1 loss-of-function in a subset of bone marrow mesenchymal osteoprogenitor cells is likely responsible for the vertebral and long-bone osseous defects in NF1 patients, as demonstrated by the similarity between the bony abnormalities of Nf1Col2−/− mice and NF1 patients. This study provides a new NF1 mouse preclinical model to test novel therapeutic approaches. In that regard, the fact that lovastatin administered during early bone development could rescue the bone cortical porosity defect observed in this model constitutes a proof of concept that RAS/ERK signaling is a rationale target for preventive treatment of the NF1 focal bony lesions.
Several observations strongly suggest that Cre-mediated Nf1 deletion in Nf1Col2−/− mice occurred very early during development in mesenchymal osteochondroprogenitors, and not only in chondrocytes, to generate Nf1−/− chondrocytes but also Nf1−/− osteoblasts and osteocytes. Several observations include (i) the formation of LacZ-positive mineralizing osteoblasts using BMSCs from Col2α1-Cre;R26R reporter mice, (ii) the presence of LacZ-positive chondrocytes, osteoblasts and osteocytes in bone sections of these reporter mice, (iii) proof of efficient recombination of the floxed Nf1 locus in bone tissues, semi-purified chondrocytes and adherent bone marrow osteoprogenitors from Nf1Col2−/− mice, and (iv) the presence of typically bony phenotypes in Nf1Col2−/− mice. The existence of shared bone phenotypes among the Nf1Col2−/− and Nf1Prx models further confirms loss of Nf1 function in osteochondroprogenitors and their progeny in Nf1Col2−/− mice. However, despite these observations, we cannot exclude at this point that Nf1−/− chondrocytes indirectly or partially contribute to some of these bony phenotypes.
The comparison between the bone phenotypes of the Nf1Col2−/− mice and other previously described Nf1 mouse models helped to better delineate the role of this gene in the bone mesenchymal lineage and revealed that Nf1 has specific and critical functions at different stages during the differentiation of this lineage. First, Nf1 seems particularly important for the differentiation of osteoprogenitor cells since loss of one (18) or both copies of Nf1 [(14) and our unpublished data] in these cells impairs their differentiation, which might contribute to the low bone mass observed in the Nf1Col2−/− and Nf1Prx models. On the other hand, loss of Nf1 function in mature osteoblasts also impairs their function, i.e. collagen synthesis, matrix mineralization and the regulation of osteoclastogenesis, leading to osteoidosis and increased bone resorption (13). Although Rankl and Atf4 (activating transcription factor 4) are two previously identified target genes of Nf1 in mature osteoblasts and involved in the regulation of osteoclastogenesis and collagen synthesis, respectively (13), the mechanisms whereby lack of Nf1 impairs osteoblast differentiation and matrix mineralization are not yet identified. The absence of cortical fenestrations in Nf1ob−/− mice suggests that this phenotype in Nf1Col2−/− mice is predominantly caused by a defect of Nf1−/− BMSCs during early development.
Tibial dysplasia, typically presenting in a small percentage (3–4%) of NF1 infants (46) as unilateral anterolateral bowing of the tibia, was observed in the Nf1Col2−/− mouse model, though it appeared in mice as lateral instead of anterolateral, and in both tibiae. This difference likely reflects the fact that Nf1Col2−/− mice lack Nf1 function in osteoprogenitor cells in every bone, whereas NF1 patients may lack NF1 function only in a subset of osteoprogenitors restricted most of the time to one tibia. This distinction holds true for the phenotypes in the vertebral axis. What triggers NF1 loss-of-function at this specific skeletal site and why the tibia is the preferential site for bowing and pseudoarthrosis are currently unknown, but load bearing may contribute to it. We did not observe spontaneous fracture in the Nf1Col2−/− mouse model, which is possibly due to the lower body weight compared with WT mice and lower mechanical load quadrupeds experience compared with bipeds, and/or to the fact that mice notoriously show very efficient bone healing capacity compared with humans. Importantly, despite this absence of spontaneous fractures, vertebral and long bones in Nf1Col2−/− mice exhibited weaker biomechanical properties compared with WT littermates. We propose that such altered whole-bone properties may underlie the frequent fracture of bowed tibiae in NF1 children.
Importantly, the Nf1Col2−/− mouse model provides a new model to investigate scoliosis in NF1. This phenotype was not associated with the presence of adjacent plexiform neurofibroma. It became prominent in young mice, as observed in NF1 patients who present with dystrophic scoliosis in pre-adolescent age, and progressed to a more severe phenotype upon aging, including sharp angulation of the vertebral axis and loss of BMD. These observations provide evidence that the vertebral dystrophic lesion in NF1 patients could be a primary bone defect of developmental nature, rather than solely induced by the presence of adjacent plexiform neurofibromas, possibly eroding the vertebral bony structure. The high incidence of neurofibromas associated with dystrophic scoliosis is, however, intriguing and urges further investigation (35–38). The similarities of the skeletal manifestations between the Nf1Col2−/− mouse model and NF1 patients do not prove, but only suggest, that NF1 vertebral dysplasias are caused by NF1 loss-of-function in bone cells, and only a corrective effect of interventions targeting bone cells in patients may prove this point. The observation that RAS inhibition by lovastatin during embryonic development was able to significantly reduce the strong cortical porosity observed in the vertebrae of this mouse model is a first proof of concept that targeting the signaling pathway altered by Nf1 loss-of-function has the potential not only to treat, but also to prevent some of the NF1 skeletal manifestations. Alternative targets and windows of treatment (in particular post-natal treatments) will be necessary to further address this exciting question.
There are a number of observations from this study that raise additional questions. First, whether vertebral fractures or microcracks caused by the impaired bone structure of Nf1Col2−/− mice (including low BMD, high cortical porosity and compromised mechanical properties) contribute to the sharp vertebral axis angulation observed in adult mice is unknown. It is also unclear to what extend the IVD defect of this mouse model contributes to the deterioration of its vertebral axis. The severe abnormalities in shape, cortical structure and mineralization observed in the vertebral bodies of Nf1Col2−/− mice all suggest, however, the existence of an intrinsic bony defect that is independent of the IVD abnormalities. Second, the bone abnormalities observed in Nf1Col2−/− mice occur in the setting of an Nf1+/+ (WT) stroma, as opposed to the NF1+/− stroma characteristic of NF1 patients. Although Nf1+/− mice do not display any bone abnormalities under normal conditions, it is currently unknown whether Nf1+/− bone marrow cells, including cells of the monocytic lineage and other immune cells, could increase the severity of the bony defects observed in Nf1Col2−/− mice. Third, the IVD and severe short stature phenotypes observed in Nf1Col2−/− mice demonstrate that Nf1 is required for proper formation of the IVD and bone growth; however, the observation that IVD defects and severe short stature are not characteristic features of NF1 patients strongly suggests that NF1 loss-of-function in these patients does not occur in notochordal cells or chondrocytes during development, or that this event is embryonic lethal. Fourth, the different skull morphology between rodents and humans may explain the apparently dissimilar skull malformations observed between NF1 patients and Nf1Col2−/− mice. The latter may nevertheless represent a mouse equivalent to the sphenoid wing dysplasia and macrocephalia observed in ∼10% of NF1 patients. In any case, these observations raise the hypothesis that the skull bony defects observed in NF1 patients may be independent of the presence of a neighboring plexiform neurofibroma which is sometimes observed in NF1 patients with sphenoid wing dysplasia.
From these results, we hypothesize that the relative low incidence and variable severity of the dystrophic vertebral defects in NF1 patients may be explained by multiple and independent factors, possibly related to the variable occurrence of NF1 loss-of-function in different populations of vertebral bone osteoprogenitor cells. The mechanisms underlying NF1 loss-of-function are still unclear and may involve ‘second hit’ mutations, somatic mosaicism, modifier genes or epigenetic events. The progression of these dystrophic lesions, on the other hand, could be caused by an impaired mechanical response of these abnormal bones during growth and aging, and/or defects in bone remodeling or healing, as supported by the severe loss of BMD in this model, and healing defects in Nf1+/−, Nf1ob−/− and Nf1Prx mice (17,47,48).
In summary, this study characterizes the critical role of Nf1 during endochondral bone formation and strongly suggests that the most severe bone abnormalities in NF1 patients are caused by a bone cell-autonomous loss of NF1 function during early development that triggers tissue abnormalities progressively causing abnormal bone shape, architecture, mechanical properties and remodeling. Many of the similarities of bone phenotypes between the Nf1Col2−/− mouse model and NF1 patients make it a useful pre-clinical model to test rational strategies aimed at preventing or reducing the skeletal burden associated with NF1. The beneficial effect of lovastatin administrated during embryonic development on the Nf1Col2−/− cortical bone phenotype supports the notion that pharmacological interventions targeting the specific molecular defects of NF1−/− cells could be a promising avenue to prevent skeletal dysplasiae in NF1 patients.
MATERIAL AND METHODS
Animals
WT and Nf1Col2−/− mice were generated by crossing Col2α1-Cre;Nf1flox/+ mice and Nf1flox/flox mice. Col2α1-Cre;R26R mice were generated by crossing Col2α1-Cre mice to ROSA26 reporter mice (49). Embryos, pups and adult mice were harvested at indicated stages, fixed with 10% formalin or 95% alcohol and then processed as indicated in what follows. Administration of lovastatin microparticles (10 mg/kg) (17) was performed by intraperitoneal injection every other day to pregnant females from 10.5 d.p.c. to E20.5. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the Vanderbilt University Medical Center.
Primary Cell Cultures
BMSCs were extracted from 2-month-old mice tibia by cutting off the epiphyses and spinning down the diaphyses at 2000g for 10 min. The cells were then counted, plated and grown for 10 days before the addition of ascorbic acid and β-glycerophosphate (osteogenic differentiation medium) or with STEMPRO® Chondrogenesis Differentiation Kit medium (Invitrogen Life Science, USA). Primary chondrocytes were extracted from 4-day-old pup rib cages. The cartilaginous part of the rib was dissected and the soft tissue removed, then digested by collagenase D (3 mg/ml, Roche, USA) and 0.25% of 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA) (Gibco, USA) in DMEM for 3 h. Extracted cells were cultured in DMEM with 10% FBS and passaged no more than twice.
RT-PCR, genomic PCR and qPCR
Total RNA was extracted using TRIzol (Invitrogen), and cDNAs were synthesized following DNase I treatment using the high-capacity cDNA reverse-transcription kit (Applied Biosystems, USA). Nf1 mRNA was detected by RT-PCR using forward primer 5′-GTATTGAATTGAAGCACCTTTGTTTGG-3′ and reverse primer 5′-CTGCCCAAGGCTCCCCCAG-3′, 35 cycles at the melting temperature of 60°C.
Detection of recombination of the floxed alleles was performed by PCR from 100 ng of genomic DNA from indicated tissues or cells using primers P1, P2 and P4, as described by Zhu et al. (30), resulting in a 280 bp Cre-mediated recombination band and a 350 bp non-recombined band. Genomic DNA was isolated by phenol–chloroform extraction and ethanol precipitation.
Total RNA was extracted from calvarial osteoblasts, bone marrow osteoblasts and 2-month-old mice lumbar vertebrae, and qPCR reactions were performed using TaqMan gene expression assays on an ABI 7300 Real-Time PCR instrument (Applied Biosystems). The primers for Rankl (Mm00441908_m1), Opg (Mm00441906_m1), Nf1 (Mm00812424_m1) and the normalizer Hprt (Mm00446968_m1) were obtained from Applied Biosystems.
In situ hybridization
Paraformaldehyde-fixed samples were decalcified in 0.5 m EDTA (pH 8.0) overnight, dehydrated in graded series of ethanol, cleared and embedded in paraffin. Five-micrometer sagittal sections were cut and used for in situ hybridization analysis. Probes for Col2α1, Ihh and Col10α1 were prepared as described previously (50,51). Antisense cDNAs were used for riboprobe synthesis with RNA polymerases (Invitrogen) and [35S]-uridine triphosphate (Perkin Elmer, USA).
Western blot analyses
Lysates from BMSCs were prepared in RIPA buffer in the presence of protease and phosphatase inhibitors (Roche). Proteins were separated by reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane using standard protocols. Western blot analyses were performed using anti-total and anti-phospho Erk1/2 antibodies (Cell Signaling, USA), and reaction was detected by chemoluminescence. All the reagents, if not stated, were from Sigma (USA).
Skeletal preparations and histology
Whole-mount skeletal preparations were prepared by removing the skin and internal organs of the mice before immersion in 95% ethanol overnight (embryos or young pups) or for 1 week (adult). Specimens were stained with 0.015% Alcian Blue 8GX (Sigma) in 80% ethanol/20% acetic acid, and 0.005% Alizarin Red S (Sigma) in 1% KOH after digestion by 2% KOH overnight. Specimens were cleared with 1% KOH/20% glycerol solution and were stored in a 1:1 mix of glycerol and 95% ethanol.
For histology, formalin-fixed samples were decalcified in 20% EDTA (pH 7.2) for one week, dehydrated in graded series of ethanol, cleared and embedded in paraffin. Five-micrometer sagittal sections were cut and stained with hematoxylin and eosin or Alcian blue and fast red. Immunocytochemistry was performed according to standard protocols using an antibody against Neurofibromin (sc-67, Santa Cruz, USA) or a non-immune IgG antibody. Apoptotic cells were detected by TUNEL assay using the In Situ Cell Death Detection Kit (Roche) following the manufacturer's instructions. In vivo proliferation assays were performed by intraperitoneally injecting pregnant mice with BrdU (0.1 mg/g) 2 h prior to sacrifice. Embryos were then harvested and processed for embedding and sectioning. BrdU was detected with a BrdU staining kit (Zymed Laboratories, USA) following the manufacturer's instructions. Undecalcified bone samples were fixed in formalin, processed and embedded in polymethyl methacrylate following standard procedures (52). Seven-micrometer-thick sagital sections were obtained and stained with toluidine blue, TRAP staining or von Kossa/van Gieson following standard protocols.
LacZ staining was performed using standard protocols. Briefly, the cells were rinsed with DPBS and fixed with 0.25% glutaraldehyde for 5 min on ice, and then stained with X-gal staining solution (2 mm MgCl2, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide and 1 mg/ml X-Gal).
Whole-mount LacZ staining was performed on tibiae fixed in fresh 4% paraformaldehye/PBS (pH 7.0–7.5) for 1h on ice. Bones were split sagittally before staining with X-gal staining solution overnight at 37°C water bath to improve penetration. The samples were further fixed and processed for paraffin sections as stated earlier.
Histomorphometric measurements were performed using the Bioquant Analysis System (Nashville, TN, USA).
Mouse X-Ray and µCT analysis
Radiographs were obtained by using a digital cabinet X-ray system (LX-60, Faxitron X-Ray, USA). µCT analyses were performed using a Scanco µCT 40 system (Scanco Medical, Bassersdorf, Switzerland). Tomographic images were acquired at 55 kVp and 145 mA with an isotropic voxel size of 12 µm and at an integration time of 250 ms with 500 projections collected per 180° rotation. During the scan, the bones were immersed in 70% ethanol or phosphate buffered saline in case the bones were subsequently subjected to biomechanical testing.
Serum calcium and phosphate
Blood samples were collected from WT and Nf1Col2−/− mice at sacrifice. Serum calcium and phosphate levels were determined using the Calcium L3K® reagent and Phosphorus-SL Assay Kit (Diagnostic Chemicals Ltd, USA) following the manufacturer's instructions.
Mouse biomechanical testing
Each femur was placed on the lower support points of a three-point bending fixture with the anterior side down (i.e. bending about the medial–lateral plane). With the span between the lower supports set to 6 or 7 mm for KO and WT, respectively, the hydrated bones were loaded to failure at a rate of 3.0 mm/min using a material-testing system (Dynamight 8841, Instron, Canton, OH, USA). The force–displacement data were converted to a moment (force × span/4) versus normalized deflection (12 × displacement/span2), and the structural properties were rigidity or the slope of the linear portion of the curve (kN mm2) and peak moment or the maximum moment endured by the bone (N mm). To account for the structural contribution to rigidity and strength, we estimated the material properties, modulus and bending strength, using the flexural equations from beam theory as well as the μCT-derived measurement of MOI and the distance between neutral axis of bending and the outermost point in the anterior–posterior direction. After removing the end plates and trimming the transverse processes with a scalpel, a flat, cylindrical platen (20 mm diameter) attached to the material-testing machine compressed the VB to failure at a rate of 3 mm/min. From the resulting force–displacement curve, we determined the biomechanical properties as follows: stiffness was the slope of the linear portion of the curve (N/mm), yield force (N) occurred when the slope of the aforementioned curved deviated by 15% and peak force (N) was the maximum force endured by the VB.
Statistical analysis
For statistical comparison, the two-tailed, paired Student's t-test or one-way analysis of variance was used, with P-values of <0.05 considered significant.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by the Department of Defense (W81XWH-09-01-0207), NIAMS/National Institutes of Health (1R01AR055966-01A1), and the Children's Tumor Foundation (YIA 2007-01-008 to W.W.). D.A.S. is supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award, the Shriners Hospitals for Children Research Foundation, the Thrasher Research Fund, the University of Utah Clinical Genetics Research Program, and the National Institute of Neurological Disorders and Stroke (K23NS052500).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Intermountain Donor Services, the NF Orthopedic Core Facility (NOCF) and their associated members including Dr John Carey, Dr David Viskochil and Dr Jacques D'Astous for help in tissue collection. We thank Dr Scott Miller and Dr Jill Shea for help in microCT imaging and processing of human samples. We thank Dr Perrien for help with the mouse microCT studies, and Dr C. Mooney for help with the skull analyses.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Friedman J.M. Epidemiology of neurofibromatosis type 1. Am. J. Med. Genet. 1999;89:1–6. doi:10.1002/(SICI)1096-8628(19990326)89:1<1::AID-AJMG3>3.0.CO;2-8. [PubMed] [Google Scholar]
- 2.Upadhyaya M., Roberts S.H., Maynard J., Sorour E., Thompson P.W., Vaughan M., Wilkie A.O., Hughes H.E. A cytogenetic deletion, del(17)(q11.22q21.1), in a patient with sporadic neurofibromatosis type 1 (NF1) associated with dysmorphism and developmental delay. J. Med. Genet. 1996;33:148–152. doi: 10.1136/jmg.33.2.148. doi:10.1136/jmg.33.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu G.F., O'Connell P., Viskochil D., Cawthon R., Robertson M., Culver M., Dunn D., Stevens J., Gesteland R., White R., et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. doi:10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 4.Daston M.M., Scrable H., Nordlund M., Sturbaum A.K., Nissen L.M., Ratner N. The protein product of the neurofibromatosis type 1 gene is expressed at highest abundance in neurons, Schwann cells, and oligodendrocytes. Neuron. 1992;8:415–428. doi: 10.1016/0896-6273(92)90270-n. doi:10.1016/0896-6273(92)90270-N. [DOI] [PubMed] [Google Scholar]
- 5.Brunetti-Pierri N., Doty S., Hicks J., Phan K., Mendoza-Londono R., Blazo M., Tran A., Carter S., Lewis R., Plon S., et al. Generalized metabolic bone disease in neurofibromatosis type 1. Mol. Genet. Metab. 2008;94:105–111. doi: 10.1016/j.ymgme.2007.12.004. doi:10.1016/j.ymgme.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seitz S., Schnabel C., Busse B., Schmidt H.U., Beil F.T., Friedrich R.E., Schi4nke T., Mautner V.F., Amling M. High bone turnover and accumulation of osteoid in patients with neurofibromatosis 1. Osteoporos. Int. 2010;21:119–127. doi: 10.1007/s00198-009-0933-y. doi:10.1007/s00198-009-0933-y. [DOI] [PubMed] [Google Scholar]
- 7.Crawford A.H., Schorry E.K. Neurofibromatosis in children: the role of the orthopaedist. J. Am. Acad. Orthop. Surg. 1999;7:217–230. doi: 10.5435/00124635-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Viskochil D., Buchberg A.M., Xu G., Cawthon R.M., Stevens J., Wolff R.K., Culver M., Carey J.C., Copeland N.G., Jenkins N.A. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. doi:10.1016/0092-8674(90)90252-A. [DOI] [PubMed] [Google Scholar]
- 9.Cawthon R.M., Weiss R., Xu G.F., Viskochil D., Culver M., Stevens J., Robertson M., Dunn D., Gesteland R., O'Connell P. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. doi:10.1016/0092-8674(90)90253-B. [DOI] [PubMed] [Google Scholar]
- 10.Marchuk D.A., Saulino A.M., Tavakkol R., Swaroop M., Wallace M.R., Andersen L.B., Mitchell A.L., Gutmann D.H., Boguski M., Collins F.S. cDNA cloning of the type 1 neurofibromatosis gene: complete sequence of the NF1 gene product. Genomics. 1991;11:931–940. doi: 10.1016/0888-7543(91)90017-9. doi:10.1016/0888-7543(91)90017-9. [DOI] [PubMed] [Google Scholar]
- 11.Elefteriou F., Kolanczyk M., Schindeler A., Viskochil D.H., Hock J.M., Schorry E.K., Crawford A.H., Friedman J.M., Little D., Peltonen J., et al. Skeletal abnormalities in neurofibromatosis type 1: approaches to therapeutic options. Am. J. Med. Genet. A. 2009;149A:2327–2338. doi: 10.1002/ajmg.a.33045. doi:10.1002/ajmg.a.33045. [DOI] [PubMed] [Google Scholar]
- 12.Kuorilehto T., Nissinen M., Koivunen J., Benson M.D., Peltonen J. NF1 tumor suppressor protein and mRNA in skeletal tissues of developing and adult normal mouse and NF1-deficient embryos. J. Bone Miner. Res. 2004;19:983–989. doi: 10.1359/JBMR.040130. doi:10.1359/JBMR.040130. [DOI] [PubMed] [Google Scholar]
- 13.Elefteriou F., Benson M.D., Sowa H., Starbuck M., Liu X., Ron D., Parada L.F., Karsenty G. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab. 2006;4:441–451. doi: 10.1016/j.cmet.2006.10.010. doi:10.1016/j.cmet.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolanczyk M., Kossler N., Kuhnisch J., Lavitas L., Stricker S., Wilkening U., Manjubala I., Fratzl P., Sporle R., Herrmann B.G., et al. Multiple roles for neurofibromin in skeletal development and growth. Hum. Mol. Genet. 2007;16:874–886. doi: 10.1093/hmg/ddm032. doi:10.1093/hmg/ddm032. [DOI] [PubMed] [Google Scholar]
- 15.Cichowski K., Shih T.S., Schmitt E., Santiago S., Reilly K., McLaughlin M.E., Bronson R.T., Jacks T. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. doi:10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 16.Stevenson D.A., Zhou H., Ashrafi S., Messiaen L.M., Carey J.C., D'Astous J.L., Santora S.D., Viskochil D.H. Double inactivation of NF1 in tibial pseudoarthrosis. Am. J. Hum. Genet. 2006;79:143–148. doi: 10.1086/504441. doi:10.1086/504441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W., Nyman J.S., Moss H.E., Gutierrez G., Mundy G.R., Yang X., Elefteriou F. Local low-dose lovastatin delivery improves the bone-healing defect caused by Nf1 loss of function in osteoblasts. J. Bone Miner. Res. 2010;25:1658–1667. doi: 10.1002/jbmr.42. doi:10.1002/jbmr.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X., Chen S., Potter O.L., Murthy S.M., Li J., Pulcini J.M., Ohashi N., Winata T., Everett E.T., Ingram D., et al. Neurofibromin and its inactivation of Ras are prerequisites for osteoblast functioning. Bone. 2005;36:793–802. doi: 10.1016/j.bone.2005.01.022. doi:10.1016/j.bone.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Yang F.C., Chen S., Robling A.G., Yu X., Nebesio T.D., Yan J., Morgan T., Li X., Yuan J., Hock J., et al. Hyperactivation of p21ras and PI3K cooperate to alter murine and human neurofibromatosis type 1-haploinsufficient osteoclast functions. J. Clin. Invest. 2006;116:2880–2891. doi: 10.1172/JCI29092. doi:10.1172/JCI29092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheah K.S., Lau E.T., Au P.K., Tam P.P. Expression of the mouse alpha 1(II) collagen gene is not restricted to cartilage during development. Development. 1991;111:945–953. doi: 10.1242/dev.111.4.945. [DOI] [PubMed] [Google Scholar]
- 21.Ovchinnikov D.A., Deng J.M., Ogunrinu G., Behringer R.R. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. doi:10.1002/(SICI)1526-968X(200002)26:2<145::AID-GENE14>3.0.CO;2-C. [PubMed] [Google Scholar]
- 22.Terpstra L., Prud'homme J., Arabian A., Takeda S., Karsenty G., Dedhar S., St-Arnaud R. Reduced chondrocyte proliferation and chondrodysplasia in mice lacking the integrin-linked kinase in chondrocytes. J. Cell Biol. 2003;162:139–148. doi: 10.1083/jcb.200302066. doi:10.1083/jcb.200302066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karreth F., Hoebertz A., Scheuch H., Eferl R., Wagner E.F. The AP1 transcription factor Fra2 is required for efficient cartilage development. Development. 2004;131:5717–5725. doi: 10.1242/dev.01414. doi:10.1242/dev.01414. [DOI] [PubMed] [Google Scholar]
- 24.Jacob A.L., Smith C., Partanen J., Ornitz D.M. Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev. Biol. 2006;296:315–328. doi: 10.1016/j.ydbio.2006.05.031. doi:10.1016/j.ydbio.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ford-Hutchinson A.F., Ali Z., Lines S.E., Hallgrimsson B., Boyd S.K., Jirik F.R. Inactivation of Pten in osteo-chondroprogenitor cells leads to epiphyseal growth plate abnormalities and skeletal overgrowth. J. Bone Miner. Res. 2007;22:1245–1259. doi: 10.1359/jbmr.070420. doi:10.1359/jbmr.070420. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh S.C., Chen N.T., Lo S.H. Conditional loss of PTEN leads to skeletal abnormalities and lipoma formation. Mol. Carcinog. 2009;48:545–552. doi: 10.1002/mc.20491. doi:10.1002/mc.20491. [DOI] [PubMed] [Google Scholar]
- 27.Downey C.M., Horton C.R., Carlson B.A., Parsons T.E., Hatfield D.L., Hallgrimsson B., Jirik F.R. Osteo-chondroprogenitor-specific deletion of the selenocysteine tRNA gene, Trsp, leads to chondronecrosis and abnormal skeletal development: a putative model for Kashin-Beck disease. PLoS Genet. 2009;5:e1000616. doi: 10.1371/journal.pgen.1000616. doi:10.1371/journal.pgen.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maes C., Kobayashi T., Selig M.K., Torrekens S., Roth S.I., Mackem S., Carmeliet G., Kronenberg H.M. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. doi:10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C., Ji X., Harris M.A., Mundy G.R., Harris S.E. A clonal chondrocytic cell line derived from BMP-2/T antigen-expressing transgenic mouse. In Vitro Cell. Dev. Biol. Anim. 1998;34:359–363. doi: 10.1007/s11626-998-0015-z. doi:10.1007/s11626-998-0015-z. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y., Romero M.I., Ghosh P., Ye Z., Charnay P., Rushing E.J., Marth J.D., Parada L.F. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. doi:10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevenson D.A., Birch P.H., Friedman J.M., Viskochil D.H., Balestrazzi P., Boni S., Buske A., Korf B.R., Niimura M., Pivnick E.K., et al. Descriptive analysis of tibial pseudarthrosis in patients with neurofibromatosis 1. Am. J. Med. Genet. 1999;84:413–419. doi: 10.1002/(sici)1096-8628(19990611)84:5<413::aid-ajmg5>3.0.co;2-1. doi:10.1002/(SICI)1096-8628(19990611)84:5<413::AID-AJMG5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 32.Riccardi V.M. Neurofibromatosis: Phenotype, Natural History, and Pathogenesis. Baltimore and London: The Johns Hopkins University Press; 1999. pp. 250–273. [Google Scholar]
- 33.Theiler K. Vertebral malformations. Adv. Anat. Embryol. Cell Biol. 1988;112:1–99. doi: 10.1007/978-3-642-73775-6. [DOI] [PubMed] [Google Scholar]
- 34.Aszodi A., Chan D., Hunziker E., Bateman J.F., Fassler R. Collagen II is essential for the removal of the notochord and the formation of intervertebral discs. J. Cell Biol. 1998;143:1399–1412. doi: 10.1083/jcb.143.5.1399. doi:10.1083/jcb.143.5.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casselman E.S., Mandell G.A. Vertebral scalloping in neurofibromatosis. Radiology. 1979;131:89–94. doi: 10.1148/131.1.89. [DOI] [PubMed] [Google Scholar]
- 36.Winter R.B., Moe J.H., Bradford D.S., Lonstein J.E., Pedras C.V., Weber A.H. Spine deformity in neurofibromatosis. A review of one hundred and two patients. J. Bone Joint Surg. Am. 1979;61:677–694. [PubMed] [Google Scholar]
- 37.Crawford A.H. Pitfalls of spinal deformities associated with neurofibromatosis in children. Clin. Orthop. Relat. Res. 1989;245:29–42. [PubMed] [Google Scholar]
- 38.Alwan S., Tredwell S.J., Friedman J.M. Is osseous dysplasia a primary feature of neurofibromatosis 1 (NF1)? Clin. Genet. 2005;67:378–390. doi: 10.1111/j.1399-0004.2005.00410.x. doi:10.1111/j.1399-0004.2005.00410.x. [DOI] [PubMed] [Google Scholar]
- 39.Behrens A., Haigh J., Mechta-Grigoriou F., Nagy A., Yaniv M., Wagner E.F. Impaired intervertebral disc formation in the absence of Jun. Development. 2003;130:103–109. doi: 10.1242/dev.00186. doi:10.1242/dev.00186. [DOI] [PubMed] [Google Scholar]
- 40.Murakami S., Balmes G., McKinney S., Zhang Z., Givol D., de Crombrugghe B. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 2004;18:290–305. doi: 10.1101/gad.1179104. doi:10.1101/gad.1179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brannan C.I., Perkins A.S., Vogel K.S., Ratner N., Nordlund M.L., Reid S.W., Buchberg A.M., Jenkins N.A., Parada L.F., Copeland N.G. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. doi:10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 42.Klesse L.J., Parada L.F. p21 ras and phosphatidylinositol-3 kinase are required for survival of wild-type and NF1 mutant sensory neurons. J. Neurosci. 1998;18:10420–10428. doi: 10.1523/JNEUROSCI.18-24-10420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parrinello S., Noon L.A., Harrisingh M.C., Digby P.W., Rosenberg L.H., Cremona C.A., Echave P., Flanagan A.M., Parada L.F., Lloyd A.C. NF1 loss disrupts Schwann cell-axonal interactions: a novel role for semaphorin 4F. Genes Dev. 2008;22:3335–3348. doi: 10.1101/gad.490608. doi:10.1101/gad.490608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghittoni R., Patrussi L., Pirozzi K., Pellegrini M., Lazzerini P.E., Capecchi P.L., Pasini F.L., Baldari C.T. Simvastatin inhibits T-cell activation by selectively impairing the function of Ras superfamily GTPases. FASEB J. 2005;19:605–607. doi: 10.1096/fj.04-2702fje. [DOI] [PubMed] [Google Scholar]
- 45.Cerezo-Guisado M.I., Garcia-Roman N., Garcia-Marin L.J., Alvarez-Barrientos A., Bragado M.J., Lorenzo M.J. Lovastatin inhibits the extracellular-signal-regulated kinase pathway in immortalized rat brain neuroblasts. Biochem. J. 2007;401:175–183. doi: 10.1042/BJ20060731. doi:10.1042/BJ20060731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman J.M., Birch P.H. Type 1 neurofibromatosis: a descriptive analysis of the disorder in 1,728 patients. Am. J. Med. Genet. 1997;70:138–143. doi: 10.1002/(sici)1096-8628(19970516)70:2<138::aid-ajmg7>3.0.co;2-u. doi:10.1002/(SICI)1096-8628(19970516)70:2<138::AID-AJMG7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Schindeler A., Little D.G. Recent insights into bone development, homeostasis, and repair in type 1 neurofibromatosis (NFI) Bone. 2008;42:616–622. doi: 10.1016/j.bone.2007.11.006. doi:10.1016/j.bone.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Kolanczyk M., Kuehnisch J., Kossler N., Osswald M., Stumpp S., Thurisch B., Kornak U., Mundlos S. Modelling neurofibromatosis type 1 tibial dysplasia and its treatment with lovastatin. BMC Med. 2008;6:21. doi: 10.1186/1741-7015-6-21. doi:10.1186/1741-7015-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. doi:10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 50.Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. doi:10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 51.Takeda S., Bonnamy J.P., Owen M.J., Ducy P., Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467–481. doi: 10.1101/gad.845101. doi:10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erben R.G. Embedding of bone samples in methylmethacrylate: an improved method suitable for bone histomorphometry, histochemistry, and immunohistochemistry. J. Histochem. Cytochem. 1997;45:307–313. doi: 10.1177/002215549704500215. doi:10.1177/002215549704500215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.