Abstract
Huntingtin interacting protein 14 (HIP14, ZDHHC17) is a huntingtin (HTT) interacting protein with palmitoyl transferase activity. In order to interrogate the function of Hip14, we generated mice with disruption in their Hip14 gene. Hip14−/− mice displayed behavioral, biochemical and neuropathological defects that are reminiscent of Huntington disease (HD). Palmitoylation of other HIP14 substrates, but not Htt, was reduced in the Hip14−/− mice. Hip14 is dysfunctional in the presence of mutant htt in the YAC128 mouse model of HD, suggesting that altered palmitoylation mediated by HIP14 may contribute to HD.
INTRODUCTION
Palmitoylation, the reversible thioesterification of palmitic acid onto cysteine residues, is of critical importance in the developing and adult central nervous system, where it plays a role in synaptic plasticity by dynamically influencing protein localization and function (1). Defects in palmitoylation lead to dramatic changes in neuronal excitation and influence neuronal survival (1). Neuronal firing reversibly regulates palmitoylation of post-synaptic density-95 (PSD-95) at the synapse, suggesting a novel mechanism for controlling synaptic strength and plasticity (1).
Palmitoylation is catalyzed by a family of 23 mammalian DHHC-domain containing palmitoyl transferases (PATs) (2) that display distinct as well as overlapping substrate specificities with multiple PATs being able to palmitoylate the same substrates. Huntingtin interacting protein 14 (HIP14, ZDHHC17) is a neuronal PAT that regulates the palmitoylation and trafficking of several synaptic proteins, including huntingtin (HTT), SNAP-25, GAD-65, PSD-95 and synaptotagmin I (3). It is widely expressed but enriched in the brain, where it localizes to the golgi and cytoplasmic vesicles in neurons (4). The closest HIP14 paralog is HIP14L. HIP14 and HIP14L are the only mammalian PATs to have ankyrin repeat domains, a protein interaction domain that is involved in substrate recognition (5). Furthermore, HIP14 and HIP14L show partially overlapping substrate specificities (5).
Absence or dysfunction of PAT activity is associated with several disorders of the nervous system, implicating a crucial role for palmitoylation in neuronal function and health. In Alzheimer's disease, decreased palmitoylation of β-secretase (BACE) alters amyloid precursor protein processing and may result in the formation of harmful protein aggregates (6,7). Several forms of X-linked mental retardation have also been linked to loss of PATs (8–10) and mutations in the ZDHHC8 gene are associated with bipolar disorder and schizophrenia (11,12). In addition, mutations in PPT1, a thioesterase that catalyzes the removal of palmitate, result in neuronal ceroid lipofuscinoses, a group of common childhood neurodegenerative diseases (13). These findings suggest that palmitoylation has a profound impact on neuronal function.
Huntington disease (HD) is a progressive neuropsychiatric disorder caused by an expanded polyglutamine tract in HTT, and is characterized by a motor deficits, psychiatric disturbances and cognitive decline with eventual death 15–20 years after onset of symptoms (14). The hallmark pathology of HD is an early loss of striatal medium spiny neurons (MSNs), followed by loss of deeper layers of the cortex and eventual widespread brain atrophy (14). We developed a HD mouse model, the YAC128 mouse, which contains the entire human HTT gene with 128 CAG repeats, and recapitulates many features of HD, including striatal volume and MSN loss and cognitive and motor dysfunction (15).
HIP14 was first identified in a yeast two-hybrid screen as a HTT-interacting protein that shows significantly reduced interaction with mutant HTT (4). Wild-type (WT) HTT, in addition to being a palmitoylation substrate for HIP14, also influences the PAT function of HIP14 (Huang et al., submitted for publication). Indeed, WT htt potentiates the palmitoylation of HIP14 substrates, and this property is lost in the presence of mutant htt. In addition, palmitoylation of HIP14 substrates is reduced in mice lacking one allele of htt (hdh+/−), indicating a functional role for htt, not merely as a HIP14 substrate, but as a modulator of HIP14 function (Huang et al., submitted for publication).
To gain insights into the function of HIP14 in vivo, we generated and characterized Hip14-deficient (Hip14−/−) mice. We report here that mice lacking the PAT HIP14 share some features of HD, suggesting that disturbed palmitoylation may contribute to the pathogenesis of HD.
RESULTS
Decreased striatal volume and MSN loss in Hip14−/− mice
Hip14−/− mice were generated on the FVB/N strain (Supplementary Material, Fig. S1) to allow comparison to the YAC128 mice that were characterized on this strain (15). Since the mice were generated with a gene-trap insert in intron 5 of the gene, an antibody generated against a peptide corresponding to amino acids 49–60 (RKTHIDDYSTWD) in the N-terminal region (exon 2) of HIP14 was used (4) to determine the absence of Hip14 in the Hip14−/− mice (Supplementary Material, Fig. S1). Brain weights were significantly reduced in Hip14−/− mice (Fig. 1A) at 1, 3 and 12 months of age.
Figure 1.
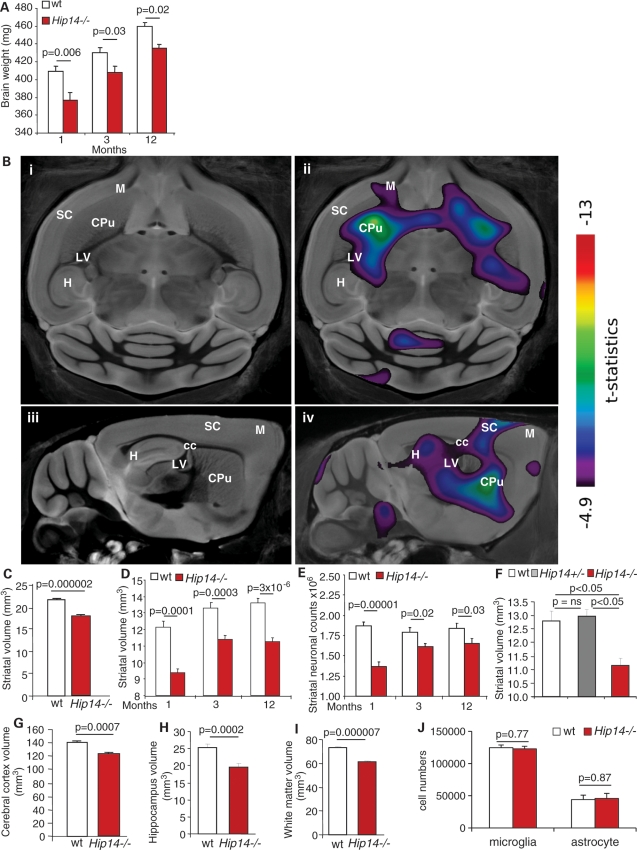
Hip14−/− mice display HD-like neuropathology. (A) The brain weights of Hip14−/− mice were significantly lower than controls (n = 7–11). (B) A predominantly striatal-specific alteration is seen by MRI in Hip14−/− mice (i) shows an axial, and (iii) a sagittal slice from a composite of MRI scans from eight Hip14−/− and eight WT controls at 3 months of age, with (ii) and (iv) showing the same slices with superimposed t-statistics indicating regions of contraction in Hip14−/− mice compared with controls, threshold at a 1% false discovery rate. CPu, caudate/putamen (striatum); cc, corpus callosum; M, motor cortex; LV, lateral ventricle; SC, sensory cortex; H, hippocampus. (C) Quantification of the striatal volume by MRI shows significant reductions in striatal volumes in Hip14−/− mice (n = 8). By immunohistochemistry, significant decreases in striatal volume (D) and (E) neuronal counts are observed in Hip14−/− mice (n = 10–13). (F) No differences in striatal volume are observed in Hip14+/− mice at 12 months (n = 9–11). (G–I) Cortical, hippocampal and white matter volumes are reduced in Hip14−/− brains (n= 8) by MRI. (J) No changes in microglial or astrocyte numbers were observed in the striatum of 3-month-old Hip14−/− mice (n= 7–10).
To identify the brain regions affected by Hip14 deletion, we performed whole-brain magnetic resonance imaging (MRI) analyses of 3-month-old Hip14−/− mice and littermate controls. The most significant and predominant changes occurred in the striatum of the Hip14−/− mice (∼17% decrease, Fig. 1B and C), mimicking the site of hallmark pathology in HD (14).
To determine whether the striatal pathology observed by MRI is due to decreased neuronal cell numbers, we performed immunohistochemistry, followed by stereological analyses of the striatum of Hip14−/− and control mice at 1, 3 and 12 months of age. A significant, but non-progressive ∼17% striatal volume loss (Fig. 1D) was observed, confirming the findings of MRI. In addition, a significant decrease in MSN numbers was observed in Hip14−/− mice (Fig. 1E). Striatal volume was also assessed at 12 months in Hip14+/− mice. No change in striatal volume was observed (Fig. 1F), indicating that 50% Hip14 expression is sufficient for maintaining normal striatal structure.
Quantification of brain region volumes obtained from the MRI analyses also showed decreases in the cerebral cortex, hippocampus and white matter in Hip14−/− mice (Fig. 1G–I). To determine whether only neurons are affected in the Hip14−/− striatum or whether astrocytes and microglial cells are also affected, sections were stained with either glial fibrillary acidic protein (GFAP) or ionized calcium-binding adaptor molecule 1 (IBA1) and cell numbers quantified. No changes in astrocyte or glial numbers were observed in striatum from 3-month-old Hip14−/− mice (Fig. 1J).
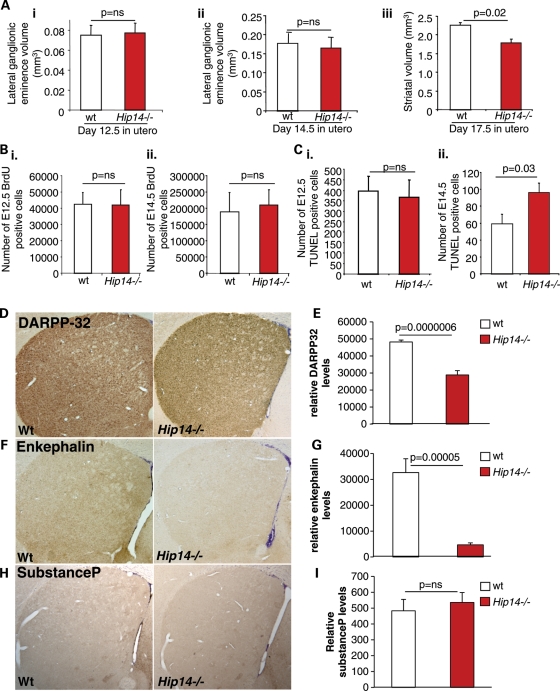
To determine the earliest age of striatal volume loss, we quantified the striatal volumes of embryonic day 12.5 (E12.5), E14.5 and E17.5 Hip14−/− and WT embryos. The striatum arises from the lateral ganglionic eminence (LGE) which is visible at E12.5 in mice. There was no difference in volume of the LGE between Hip14−/− and WT mice at E12.5 and E14.5 (Fig. 2Ai and ii). However, at E17.5, striatal volume was reduced by ∼17% in Hip14−/− mice (Fig. 2Aiii). Thus, the early development of the striatum was normal, but striatal volume failed to develop to the same extent between E14.5 and E17.5 in Hip14−/− mice.
Figure 2.
Increased early striatal cell death and HD-like neurochemical changes in Hip14−/− mice. (Ai) No differences in LGE volumes were observed in E12.5, and (Aii) E14.5 Hip14−/− embryos (n= 5). (Aiii) However, a significant decrease in striatal volume was observed at E17.5 (n= 5). (B) Pregnant females were injected with BrdU, and numbers of BrdU-positive cells were assessed in the LGE of (i) day 12.5 in utero (E12.5) and (ii) E14.5 pups. No differences in number of BrdU-positive cells were observed (n= 6). (C) Adjacent sections were stained for TUNEL to assess apoptosis. (i) No difference was observed in the number of TUNEL-positive striatal neurons at E12.5 (n= 6). However, (ii) a significant increase in the number of TUNEL-positive striatal neurons undergoing apoptosis was observed in E14.5 Hip14−/− embryos (n = 7). (D and E) The majority of MSNs express high levels of DARPP32. DARPP32 levels were significantly decreased in 3-month-old Hip14−/− striatum (n = 7). (F and G) Of the DARPP32-containing neurons, the subset expressing enkephalin is predominantly affected in HD. Similarly, enkephalin levels were significantly decreased in the Hip14−/− striatum. (H and I) No changes in substance P levels were observed in the MSNs of Hip14−/− mice (n = 7).
Increased cell death in the Hip14−/− mouse striatum
Reduced striatal volume in Hip14−/− mice may arise from increased striatal cell death, or from a lack of striatal cell proliferation during development. Therefore, we quantified cell proliferation in Hip14−/− embryos by bromodeoxyuridine (BrdU) staining, and found no difference in the number of proliferating cells at either E12.5 or E14.5 in the LGE (Fig. 2Bi and ii). We next quantified the number of cells undergoing cell death using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) at the same time points. A significant increase in the number of TUNEL-positive cells was observed at E14.5 (Fig. 2Ci and ii). Thus, the absence of Hip14 results in increased striatal cell death.
MSNs expressing DARPP32 and enkephalin are predominantly affected in Hip14−/− mice
The majority of striatal neurons are MSNs, which express dopamine- and cyclic AMP-regulated phosphoprotein (DARPP32) (16). There are two populations of MSNs, expressing D2/met-enkephalin or D1/substance P (16). In HD, levels of both DARPP32 and enkephalin are significantly reduced, even in patients with mild pathology (16). In contrast, substance P is reduced only in advanced disease. To determine whether a similar subset of neurons are affected in Hip14−/− mice, we performed immunohistochemical quantification of these proteins in the Hip14−/− striatum. The Hip14−/− striatum showed significant decreases in both DARPP32 and enkephalin intensity (Fig. 2D–G), while substance P was unchanged (Fig. 2H and I).
Reduced excitatory synapses in Hip14−/− and YAC128 striata
The formation of excitatory (glutamate-mediated) and inhibitory (gamma-aminobutyric acid-mediated) synapses is critical for normal brain physiology. PSD-95, a postsynaptic scaffolding protein that is palmitoylated by HIP14, plays a critical role, especially in excitatory synapses, where it regulates the clustering of 2-amino-3-(5-methyl-3-oxo-1,2- oxazol-4-yl) propanoic acid and N-methyl-D-aspartic acid (NMDA) receptors (17,18). Electron microscopy was used to determine whether the loss of Hip14 altered the numbers of striatal synapses and their structure. A significant decrease in the total number of excitatory but not inhibitory synapses was observed (Fig. 3A and B). To account for the loss of MSNs in Hip14−/− mice, numbers of synapses per neuron were calculated by dividing number of synapses by the average MSN counts obtained from stereology, and were found to be significantly decreased (Fig. 3C). Morphological assessment of synapses revealed no differences in the numbers of reserve pool or docked vesicles, or the area of pre- and postsynaptic compartments (data not shown).
Figure 3.
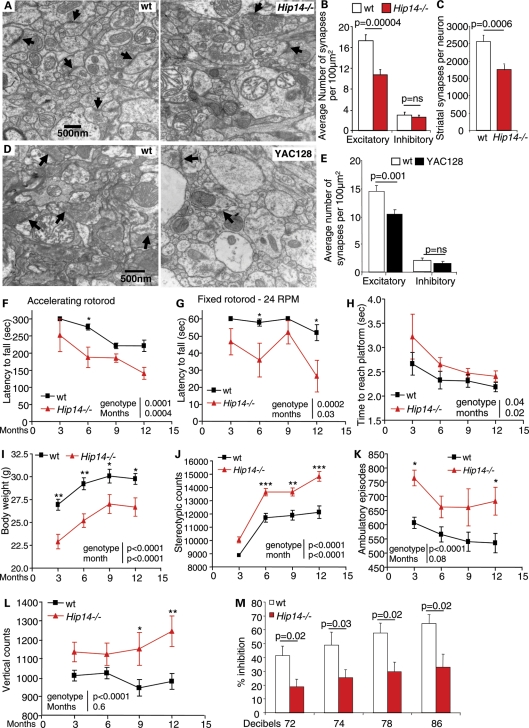
Loss of excitatory synapses and HD-like behavioral changes in Hip14−/− mice. (A) Representative fields of synapses from electron micrographs of striatum show decreases in the numbers of synapses (arrows) in 3-month-old Hip14−/− mice compared with the WT. (B) Quantification revealed a significant reduction in excitatory but not inhibitory synapses (n= 3 mice per genotype; 40 × 100 µm2 fields per genotype). (C) The number of excitatory synapses per neuron is significantly reduced in the Hip14−/− striatum. (D) Electron micrographs showing representative synapses in the striatum from 1-year-old YAC128 and WT mice. As in Hip14−/− mice, a decrease in the total number of synapses is observed. (E) A significant reduction in the number of excitatory synapses is observed in the striatum of YAC128 mice (n = 3 mice per genotype; 40 100 µm2 fields per genotype). All values are mean ± SEM. (F) Hip14−/− mice show a significant deficit on the accelerating and (G) fixed rotorod tests, indicating defects in motor co-ordination at 3, 6, 9 and 12 months of age (n = 7–10). In addition, Hip14−/− mice spend a longer time reaching a platform (H) indicating a decrease in swim speed and further implicating motor co-ordination deficits (n= 7–10). (I) Hip14−/− mice are significantly lower in body weight than controls at all ages tested (n= 15). (J–L) Hip14−/− mice are hyperactive as evidenced by increases in stereotypic, horizontal and vertical movements during open-field testing (n= 15–20). (M) Similar to HD patients and YAC128 mice, Hip14−/− mice show significant decreases in PPI compatible with dysfunction in the cortico-striatal pathway (n= 8).
Since similarities in neuropathological findings were observed between the Hip14−/− mice and HD, the finding of reduced excitatory synapses in the Hip14−/− mice prompted us to assess synapse numbers in 1-year-old, symptomatic YAC128 mice, since this has never been assessed in the YAC128 mice. As in the Hip14−/− mice, both total and excitatory synapse numbers were significantly reduced (Fig. 3D and E), and no changes in the numbers of total and docked synaptic vesicles were observed (data not shown).
Hip14−/− mice show motor co-ordination deficits
Hip14−/− mice displayed lower latency to fall in both accelerating and fixed speed rotorod and showed decreased swim speed (Fig. 3F–H), indicating deficits in motor coordination. Body weights of Hip14−/− mice were significantly reduced (Fig. 3I), similar to that seen in human HD (19). Thus, the disturbed motor function in Hip14−/− mice cannot be attributed to weight gain.
Similar to the YAC128 mice, in dark-phase open-field testing, Hip14−/− mice were hyperactive and showed significantly increased stereotypic (Fig. 3J), horizontal and vertical movements (Fig. 3K and L).
Pre-pulse inhibition (PPI) measures sensorimotor gating which is partly controlled through the cortico-striatal pathway (20). Decreased PPI is observed in HD patients (21) and YAC128 mice (22). Hip14−/− mice also show significant deficits in PPI (Fig. 3M).
Palmitoylation of Hip14 substrates is decreased in Hip14−/− mice
As HIP14 is a PAT, reduced HIP14-mediated palmitoylation is a likely contributor to the phenotypes observed in the Hip14−/− mice. One possibility is that features of HD in Hip14−/− mice could result from reduced palmitoylation of the Hip14 substrate htt (23). We assessed Htt palmitoylation in Hip14−/− brains (Fig. 4A) and found it to be normal. Thus, Hip14−/− phenotypes are not simply due to impaired palmitoylation of Htt.
Figure 4.
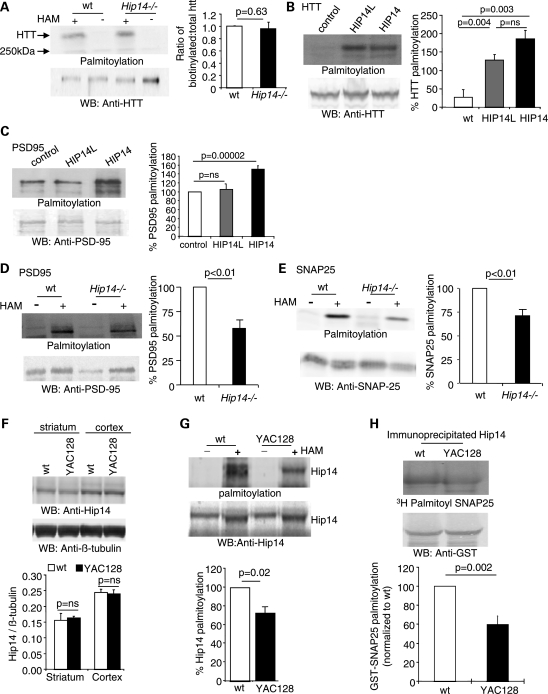
Hip14 is dysfunctional in the presence of mutant HTT. (A) No significant decreases in Htt palmitoylation are observed in Hip14−/− brains compared with the WT (n = 6). (B) HIP14L, a related family member of HIP14, can also act as a PAT for HTT (n= 3). However, (C) HIP14L cannot palmitoylate PSD95 in vitro (n= 5). (D and E) The Hip14 substrates Psd-95 and Snap-25 show significantly reduced palmitoylation in Hip14−/− brains (n= 5). (F) Hip14 protein levels are unaltered in the striatum and cortex from 8-month-old YAC128 mice (n= 3). (G) The palmitoylation of Hip14 is reduced in brains from YAC128 mice indicating a decrease in its ability to act as a PAT (n= 4). (H) Palmitoylation of Snap-25 by Hip14 immunoprecipitated (IP'd) from YAC128 brains is significantly reduced compared with the palmitoylation by Hip14 IP'd from WT mice, indicating Hip14 dysfunction in the presence of mutant HTT (n= 4).
Normal palmitoylation of Htt within the Hip14−/− brain indicates that other PATs are able to compensate for the Hip14 loss. One compensatory PAT may be the HIP14 paralog, HIP14L (ZDHHC13) (4). Indeed, HIP14L robustly palmitoylates HTT in vitro (Fig. 4B), while having no discernible activity towards a second HIP14 substrate, PSD-95 (3) (Fig. 4C). Thus, in Hip14−/− mice, Hip14L may compensate for the loss of Hip14-mediated palmitoylation of Htt, but not of all Hip14 substrates. Clearly, PATs can have overlapping but also discrete patterns of substrates.
We next assessed the palmitoylation status of two known neuronal HIP14 substrates, Psd-95 and Snap-25, in Hip14−/− brains (Fig. 4D and E). Decreased palmitoylation of Snap-25 and Psd-95 was evident in the absence of Hip14, which clearly cannot be compensated for by HIP14L.
Hip14 is dysfunctional in the presence of mutant Htt
Since in the Hip14−/− mice features of HD are replicated in the face of absent Hip14, we wished to assess whether reduced HIP14 function in HD may contribute to some phenotypes of HD. One possibility is that Hip14 expression levels are significantly reduced in the YAC128 striatum. However, no changes in Hip14 protein levels were observed in the YAC128 mice (Fig. 4F). Next, two approaches were used to assess Hip14 palmitoylation activity. Like all PATs, HIP14 itself is palmitoylated (3), and this auto-palmitoylation is a conserved feature that is correlated with PAT activity (3,24). Indeed, the HIP14 homologue in yeast, Akr1p, shows auto-palmitoylation and disruption of the DHHC domain in Akr1p reduces Akr1's auto-palmitoylation and also its PAT activity (24). In YAC128 brains, Hip14 palmitoylation was significantly decreased (Fig. 4G), suggesting that Hip14 PAT activity may be reduced in the presence of mutant HTT.
To further assess whether that Hip14 PAT activity is indeed reduced in the presence of mutant HTT, Hip14 was immunoprecipitated from WT and YAC128 brains, and assayed for its ability to palmitoylate one of its substrates, Snap-25. Hip14 isolated from YAC128 brains showed significantly reduced PAT activity (Fig. 4H), indicating that Hip14 is dysfunctional in the presence of mutant HTT.
DISCUSSION
Characterization of the Hip14−/− mouse reveals behavioral, neurochemical, morphological and neuropathological phenotypes that partially overlap with those seen in HD. However, an obvious difference is the time course with which neuropathology develops. In HD, striatal loss is progressive, with ∼50% volume loss of the caudate and putamen by the late stages of HD (14,25,26). The YAC128 mice display a striatal volume loss of ∼16% at 12 months (15). No striatal volume loss is observed in 1-month-old YAC128 mice, and striatal volume loss is first observed at 3 months (27). In contrast, Hip14−/− mice display a 17% striatal volume loss at E17.5.
Recent MRI analyses on asymptomatic individuals carrying the HD mutation have shown that even at >15 years to predicted disease, significant decreases in whole brain (2.2%), caudate (8.2%), putamen (6.0%) and cerebral white matter volumes (2.4%) are observed (28). These data suggest that the onset of pathology in HD mutation carriers occurs earlier than previously thought, and raise the possibility of a developmental effect of mutant HTT.
In addition to the predominant striatal loss, Hip14−/− mice exhibit a more widespread pathology, with significant volume decreases in the cortex, hippocampus and white matter. Here as well, more recent studies utilizing MRI show that by the mid-stages of HD, significant volume reductions in the cortex (29), hippocampus (up to 18%) (30) and white matter (13%) (25) are observed. Thus, Hip14−/− mice may mimic some features of the later stages of HD.
The neuropathological defects in HD are progressive. However, in Hip14−/− mice, the ∼17% striatal volume loss observed at E17.5 does not progressively worsen. A possible explanation is that in the Hip14−/− mice, Hip14 function is completely lost, resulting in a severe early phenotype with the preferential loss of susceptible neurons. However, in the presence of mutant Htt, the Hip14 protein is rendered partly dysfunctional, resulting in partial but not complete loss of Hip14 function. This may result in the milder course of illness observed in YAC128 mice, resulting from the accumulation over time of neurotoxic insults caused by dysfunctional Hip14. Another possibility for the observed lack of progression of neuropathology is the compensation by alternate PATs as the brain matures.
Our study indicates that the palmitoylation of the HIP14 substrates Psd-95 and Snap-25 are reduced in Hip14−/− mice. If and how reduced palmitoylation of these substrates contribute to the phenotype is unclear. Since there are only 23 putative mammalian PATs, and hundreds of proteins that are palmitoylated, HIP14 likely catalyzes the palmitoylation of many proteins not currently identified as HIP14 substrates. In addition, since the Hip14−/− mice show HD-like phenotypes and Hip14 is dysfunctional in the presence of mutant HTT, the same substrates of HIP14 are likely to show decreased palmitoylation in HD. A comprehensive analysis of the palmitoyl proteome in both the Hip14−/− mice and in HD brains will identify additional HIP14 substrates and may lead to the determination of the substrates with reduced palmitoylation that could contribute to the HD phenotype. In addition, it is possible that the deficits in the Hip14−/− mice may arise from the disruption of a function other than the PAT activity of Hip14. HIP14 has been suggested to act as an Mg++ transporter (31). However, altered Mg++ transport has been associated with some neuronal features in humans, but none of them has any resemblance to HD.
Both HIP14 and HIP14L can act as PATs for HTT (3). Thus, in addition to HIP14, HIP14L may also be dysfunctional in the presence of mutant HTT in HD. Our data suggest that Hip14 dysfunction in the presence of mutant HTT contributes to the phenotypes in the YAC128 mice. However, no striatal volume loss is observed in the Hip14+/− mice, indicating that ∼50% of functional HIP14 is adequate for normal brain development. One possibility for the phenotypes in the YAC128 mice is that >50% of Hip14 is dysfunctional in the presence of mutant HTT in the YAC128 mice. Another, more likely possibility is that in the YAC128 mice and in HD, both Hip14 and Hip14L are dysfunctional, reducing the possibility of compensation by HIP14L. In the Hip14+/− mice, palmitoylation of some important common substrates could still be compensated by the full complement of Hip14L. Generation and characterization of mice with a targeted disruption in their Hip14L gene will shed light on the function of Hip14L in the central nervous system.
Various interventions that serendipitously mirror different aspects of HD have led to fundamental new insights into the pathogenesis of HD and led to new therapeutic approaches. Mice deficient in cortical-derived brain-derived neurotrophic factor (BDNF) display reduced striatal volume and neuronal counts (32). Similarly, mice lacking PGC-1α, a transcriptional co-activator involved in energy metabolism, exhibit striatal atrophy (33). In addition, striatal administration of quinolinic acid, an NMDA receptor agonist, reproduces the neurochemical features of HD, including striatal degeneration and loss of MSNs (34). These findings have identified BDNF deficiency, disrupted bioenergetics and excitotoxicity as important pathways contributing to the pathogenesis of HD and led to new approaches and strategies to treatment (32–34). Our current findings highlight deficits in palmitoylation and HIP14 dysfunction as of potential relevance to HD, and are likely to focus attention on the role of altered HIP14 function in HD.
Taken together, the similarities between the Hip14−/− and YAC128 mice, coupled with the altered function of HIP14 in HD, highlight the potential importance of palmitoylation in the pathogenesis of HD. Furthermore, our data underscore the synapse as a major site of disturbed function in HD.
MATERIALS AND METHODS
Generation of targeted mice
Gene-trapped embryonic stem cells harboring an insertion in intron 5 of the mouse Hip14 gene were purchased from Bay Genomics (RRJ233, California, USA). The targeting of Hip14 was confirmed by sequencing of polymerase chain reaction products generated using gene and targeting vector-specific primers. Chimeras were back-crossed to at least N6 (97%) on the FVB/N strain. All procedures were approved by the University of British Columbia Committee on Animal Care. Hip14 protein levels were assessed as described previously (4).
Neuropathology
All analyses were conducted with the tester blind to genotypes. Neuropathological assessments were performed as described previously (15). For developmental analyses, embryos from pregnant mice were dissected and heads placed in ethanol/acetic acid. Paraffin-embedded sections of 8 µm were generated, stained with cresyl violet and striatal (or LGE) volumes were quantified as above. For astrocyte and glial cell quantification, immunohistochemistry was performed on free-floating sections. The primary antibodies used were a rabbit polyclonal anti-IBA1 (1:1000; Wako Chemicals USA, VA, USA) and mouse monoclonal anti-GFAP (1:500; Sigma, MO, USA). Sections were pre-treated with 10% methanol/1% hydrogen peroxide in phosphate buffered saline (PBS) for 10 min. After a wash in PBS, primary antibody was applied followed by either a goat anti-rabbit or goat anti-mouse horseradish peroxidase (HRP) conjugated secondary antibody (1:500; Jackson ImmunoResearch Labs, PA, USA). Staining was visualized using 3,3′-diaminobenzidine (DAB) in 50 mm Tris-imidazole buffer (pH 7.6). Every eighth coronal section throughout the striatum was collected, mounted on slides, dried overnight and counterstained with 0.5% cresyl violet for 6 min followed by dehydration with a graded series of ethanol solutions (70, 85, 90, 100% for 2 min each). Slides were then treated with Citrisoyl (Fisher, CA, USA) for 2 min, followed by xylene for 10 min and cover slipped with Cytoseal 60 (Richard-Allan Scientific, MI, USA). The striatum was delineated using a Zeiss Imager M2 light microscope (Zeiss, Göttingen, Germany). For counting, stereology software (StereoInvestigator 9.0; MBF Bioscience, VT, USA) was utilized. Microglia and astrocyte number in striatum was determined using the physical fractionator probe with a grid size of 500 × 500 µm and counting frame of 100 × 100 µm. Overall, the coefficient of error was <0.1. The investigator conducted the stereological analysis blind to genotype. All measurements are mean ± SEM. Student’s t-test and analysis of variance (ANOVA) for multiple comparisons were used.
Quantification of DARRP32, enkephalin and substance P staining
Primary antibodies were: mouse anti-DARPP32, #C24-6a (from Dr P. Greengard and Dr. H. Hemming), rabbit anti-enkephalin (Chemicon) and rat anti-substance P (Accurate Chemical). Sections from 1-month-old Hip14−/− and controls were incubated with HRP-conjugated secondary antibodies (Jackson ImmunoResearch) followed by DAB and 0.5% cresyl violet and visualized using a Zeiss Axioplan 2 microscope. Staining was quantified using MetaMorph (Universal Imaging Corporation). Relative levels of staining were calculated as the sum of the integrated optical density divided by the area selected, then multiplied by the sampling interval (×8) and section thickness (25 µm). All values are mean ± SEM. All data were analyzed by GraphPad Prism4 using Student's t-tests.
Magnetic resonance imaging
MRI images of 3-month-old Hip14−/− mice and controls were acquired as previously described (35). A multi-channel 7.0 T MRI scanner (Varian Inc., Palo Alto, CA, USA) with a 6 cm inner bore diameter insert gradient set was used to acquire anatomical images of brains within skulls, and atlas creation was performed as previously described (35). The striatum was segmented on the final atlas resulting from the MR image registration. The transforms from each mouse towards the final atlas were then inverted and the segmented striatum back propagated to each MRI. Volumes were analyzed using two-tailed t-tests.
Behavioral analysis
All behavioral testing was performed with the tester blind to genotypes. Motor coordination was assessed on both fixed and accelerating rotarods as described previously (15). Open-field activity was assessed in an automated open-field activity apparatus (Med Associates) as the number of photobeam breaks in a 60 min period. Activity was measured during the dark cycle. Paired-pulse inhibition was measured as described previously (22). All data were analyzed by GraphPad Prism 4 using ANOVA.
Electron microscopy
Animals were either 3 (WT and Hip14−/−) or 12 months (WT and YAC128) of age. Brains were processed for electron microscopy using standard protocols (36). Synaptic sizes were measured using Image J across six serial sections. Values were averaged to ensure that measurements were not influenced by the two-dimensional position within the synapse. Synapse numbers were quantified in fields of 100 µm2 and classified as either asymmetric (Type I, typically excitatory, thick post-synaptic density and round synaptic vesicles) or symmetric (Type II, typically inhibitory, at least three oval or flattened synaptic vesicles with no thickening of the postsynaptic membrane). To account for the reduction in cells observed in the Hip14−/− striatum, the number of synapses per neuron was calculated by adding the number of synapses counted through a given volume, and then dividing this by number of neurons counted during standard stereomicroscopy. For morphology, >500 synapses were assessed per group. Data were analyzed using Student's t-tests.
Palmitoylation assays
Palmitoylation assays were performed using the acyl-biotin exchange (ABE) methodology (37). The protein of interest was first immunoprecipitated, and ABE chemistry applied. Hydroxylamine cleavage was omitted for one-third of each sample as an assay control. Following biotinylation, the protein was eluted from the antibody and subjected to western analysis with streptavidin conjugated to HRP or Alexa680. Quantifications of western signals were performed using LiCor Odyssey analysis. The signal for palmitoylation (Streptavidin) was divided by the signal for total protein. Whole-brain homogenate from Hip14−/− mice and controls at 3 months of age were used. Data were analyzed using Student's t-tests.
Assessment of HIP14 PAT activity from YAC128 and WT mouse brains
[3H]palmitoyl-CoA and palmitoyl-CoA were synthesized enzymatically from [9,10-3H(N)]palmitic acid (5 mCi/ml; Perkin Elmer Life Sciences) and palmitic acid (Sigma). Synthesized products were purified as previously described (38) and subjected to thin layer chromatography to determine efficiency. Brains were homogenized and normal rabbit IgG or anti-Hip14 antibody (Sigma) was used to immunoprecipitate Hip14. For testing of Hip14 PAT activity, 5 µCi of [3H]palmitoyl-CoA, 0.33 µg/µl of GST-SNAP25 protein, 1 mm ATP, 50 mm 2-(N-morpholino)ethanesulfonic acid, pH 6.4, 0.2 mg/ml bovine liver lipids and 20 µl of the immunoprecipitated Hip14 were incubated for 15 min at 37°C. Samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis. Data were analyzed using Student's t-tests.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
R.R.S. was supported by the Michael Smith Foundation for Health Research (MSFHR). A.J.M. held Canadian Institutes of Health Research (CIHR) and Bluma Tischler Foundation Postdoctoral Research Fellowships. S.S.S. is supported by MSFHR and CIHR. W.N.G. was supported by NIH grants RO1 NS043782, P01 DA019695 and awards from the Peter F. McManus Foundation and the Brain Research Foundation. N.G.D. was supported by NIH R01 GM65525. L.A.R. held CIHR Investigator and MSFHR Senior Scholar awards. M.R.H. is a University Killam professor and holds a Canada Research Chair in human genetics and molecular medicine. This work was funded by grants from CIHR (MOP-84438 to M.R.H. and MOP-12699 to L.A.R.), CIHR emerging team grant GPG-102165, the Huntington Disease Society of America (M.R.H.) and the CHDI Foundation (M.R.H. and L.A.R.).
Supplementary Material
REFERENCES
- 1.El-Husseini AE.-D., Bredt D.S. Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 2002;3:791–802. doi: 10.1038/nrn940. [DOI] [PubMed] [Google Scholar]
- 2.Fukata M., Fukata Y., Adesnik H., Nicoll R.A., Bredt D.S. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Huang K., Yanai A., Kang R., Arstikaitis P., Singaraja R.R., Metzler M., Mullard A., Haigh B., Gauthier-Campbell C., Gutekunst C.A., Hayden M.R., El-Husseini A. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 4.Singaraja R.R., Hadano S., Metzler M., Givan S., Wellington C.L., Warby S., Yanai A., Gutekunst C.A., Leavitt B.R., Yi H., et al. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum. Mol. Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- 5.Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M.R., El-Husseini A. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 2009;23:2605–2615. doi: 10.1096/fj.08-127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjannet S., Elagoz A., Wickham L., Mamarbachi M., Munzer J.S., Basak A., Lazur C., Cromlish J.A., Sisodia S., Checler F., Chretien M., Seidah N.G. Post-translational processing of beta-secretase (beta-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-beta production. J. Biol. Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 7.Sidera C., Parsons R., Austen B. Post-translational processing of beta-secretase in Alzheimer's disease. Proteomics. 2005;5:1533–1543. doi: 10.1002/pmic.200401185. [DOI] [PubMed] [Google Scholar]
- 8.Mansouri M.R., Marklund L., Gustavsson P., Davey E., Carlsson B., Larsson C., White I., Gustavson K.H., Dahl N. Loss of ZDHHC15 expression in a woman with a balanced translocation t(X;15)(q13.3;cen) and severe mental retardation. Eur. J. Hum. Genet. 2005;13:970–977. doi: 10.1038/sj.ejhg.5201445. [DOI] [PubMed] [Google Scholar]
- 9.Ropers H.H. X-linked mental retardation: many genes for a complex disorder. Curr. Opin. Genet. Dev. 2006;16:260–269. doi: 10.1016/j.gde.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Raymond F.L., Tarpey P.S., Edkins S., Tofts C., O'Meara S., Teague J., Butler A., Stevens C., Barthorpe S., Buck G., et al. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus. Am. J. Hum. Genet. 2007;80:982–987. doi: 10.1086/513609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W.Y., Shi Y.Y., Zheng Y.L., Zhao X.Z., Zhang G.J., Chen S.Q., Yang P.D., He L. Case-control study and transmission disequilibrium test provide consistent evidence for association between schizophrenia and genetic variation in the 22q11 gene ZDHHC8. Hum. Mol. Genet. 2004;13:2991–2995. doi: 10.1093/hmg/ddh322. [DOI] [PubMed] [Google Scholar]
- 12.Mukai J., Liu H., Burt R.A., Swor D.E., Lai W.S., Karayiorgou M., Gogos J.A. Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat. Genet. 2004;36:725–731. doi: 10.1038/ng1375. [DOI] [PubMed] [Google Scholar]
- 13.Mitchison H.M., Hofmann S.L., Becerra C.H., Munroe P.B., Lake B.D., Crow Y.J., Stephenson J.B., Williams R.E., Hofman I.L., Taschner P.E., et al. Mutations in the palmitoyl-protein thioesterase gene (PPT; CLN1) causing juvenile neuronal ceroid lipofuscinosis with granular osmiophilic deposits. Hum. Mol. Genet. 1998;7:291–297. doi: 10.1093/hmg/7.2.291. [DOI] [PubMed] [Google Scholar]
- 14.Vonsattel J.P., DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.Z., et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 16.Deng Y.P., Albin R.L., Penney J.B., Young A.B., Anderson K.D., Reiner A. Differential loss of striatal projection systems in Huntington's disease: a quantitative immunohistochemical study. J. Chem. Neuroanat. 2004;27:143–164. doi: 10.1016/j.jchemneu.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 17.El-Husseini A.E., Schnell E., Chetkovich D.M., Nicoll R.A., Bredt D.S. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 18.Noritake J., Fukata Y., Iwanaga T., Hosomi N., Tsutsumi R., Matsuda N., Tani H., Iwanari H., Mochizuki Y., Kodama T., et al. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J. Cell Biol. 2009;186:147–160. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djoussé L., Knowlton B., Cupples L.A., Marder K., Shoulson I., Myers R.H. Weight loss in early stage of Huntington's disease. Neurology. 2002;59:1325–1330. doi: 10.1212/01.wnl.0000031791.10922.cf. [DOI] [PubMed] [Google Scholar]
- 20.Graybiel A.M. The basal ganglia. Curr. Biol. 2000;10:R509–R511. doi: 10.1016/s0960-9822(00)00593-5. [DOI] [PubMed] [Google Scholar]
- 21.Swerdlow N.R., Paulsen J., Braff D.L., Butters N., Geyer M.A., Swenson M.R. Impaired prepulse inhibition of acoustic and tactile startle response in patients with Huntington's disease. J. Neurol. Neurosurg. Psychiatry. 1995;58:192–200. doi: 10.1136/jnnp.58.2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Raamsdonk J.M., Pearson J., Slow E.J., Hossain S.M., Leavitt B.R., Hayden M.R. Cognitive dysfunction precedes neuropathology and motor abnormalities in the YAC128 mouse model of Huntington's disease. J. Neurosci. 2005;25:4169–4180. doi: 10.1523/JNEUROSCI.0590-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanai A., Huang K., Kang R., Singaraja R.R., Arstikaitis P., Gan L., Orban P.C., Mullard A., Cowan C.M., Raymond L.A., et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth A.F., Feng Y., Chen L., Davis N.G. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosas H.D., Koroshetz W.J., Chen Y.I., Skeuse C., Vangel M., Cudkowicz M.E., Caplan K., Marek K., Seidman L.J., Makris N., Jenkins B.G., Goldstein J.M. Evidence for more widespread cerebral pathology in early HD: an MRI-based morphometric analysis. Neurology. 2003;60:1615–1620. doi: 10.1212/01.wnl.0000065888.88988.6e. [DOI] [PubMed] [Google Scholar]
- 26.de la Monte S.M., Vonsattel J.P., Richardson E.P., Jr Morphometric demonstration of atrophic changes in the cerebral cortex, white matter, and neostriatum in Huntington's disease. J. Neuropathol. Exp. Neurol. 1988;47:516–525. doi: 10.1097/00005072-198809000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Carroll J.B., Lerch J.P., Franciosi S., Spreeuw A., Bissada N., Henkelman R.M., Hayden M.R. Natural history of disease in the YAC128 mouse reveals a discrete signature of pathology in Huntington disease. Neurobiol. Dis. 2011;43:257–265. doi: 10.1016/j.nbd.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Paulsen J.S., Nopoulos P.C., Aylward E., Ross C.A., Johnson H., Magnotta V.A., Juhl A., Pierson R.K., Mills J., Langbehn D., Nance M. PREDICT-HD Investigators and Coordinators of the Huntington's Study Group (HSG) Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res. Bull. 2010;82:201–207. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aylward E.H., Nopoulos P.C., Ross C.A., Langbehn D.R., Pierson R.K., Mills J.A., Johnson H.J., Magnotta V.A., Juhl A.R., Paulsen J.S. the PREDICT-HD Investigators and Coordinators of the Huntington Study Group. Longitudinal change in regional brain volumes in prodromal Huntington disease. J. Neurol. Neurosurg. Psychiatry. 2010;82:405–410. doi: 10.1136/jnnp.2010.208264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakar S.N. Correlating changes in QNE scores to quantified changes in hippocampal surface area and volume in Huntington's diease patients. Can. Undergraduate J. Cogn. Sci. 2004:17–22. Fall. [Google Scholar]
- 31.Goytain A., Hines R.M., Quamme G.A. Huntingtin-interacting proteins, HIP14 and HIP14L, mediate dual functions, palmitoyl acyltransferase and Mg2+ transport. J. Biol. Chem. 2008;283:33365–33374. doi: 10.1074/jbc.M801469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baquet Z.C., Gorski J.A., Jones K.R. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J. Neurosci. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St-Pierre J., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jäger S., Handschin C., Zheng K., Lin J., Yang W., et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 34.Beal M.F., Kowall N.W., Ellison D.W., Mazurek M.F., Swartz K.J., Martin J.B. Replication of the neurochemical characteristics of Huntington's disease by quinolinic acid. Nature. 1986;321:168–171. doi: 10.1038/321168a0. [DOI] [PubMed] [Google Scholar]
- 35.Lerch J.P., Carroll J.B., Spring S., Bertram L.N., Schwab C., Hayden M.R., Henkelman R.M. Automated deformation analysis in the YAC128 Huntington disease mouse model. Neuroimage. 2008;39:32–39. doi: 10.1016/j.neuroimage.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 36.Hines R.M., Wu L., Hines D.J., Steenland H., Mansour S., Dahlhaus R., Singaraja R.R., Cao X., Sammler E., Hormuzdi S.G., Zhuo M., El-Husseini A. Synaptic imbalance, stereotypies, and impaired social interactions in mice with altered neuroligin 2 expression. J. Neurosci. 2008;28:6055–6067. doi: 10.1523/JNEUROSCI.0032-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drisdel R.C., Green W.N. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 38.Dunphy J.T., Greentree W.K., Manahan C.L., Linder M.E. G-protein palmitoyltransferase activity is enriched in plasma membranes. J. Biol. Chem. 1996;271:7154–7159. doi: 10.1074/jbc.271.12.7154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.