Abstract
Most primary CNS lymphomas (PCNSL) are diffuse large B-cell lymphomas (DLBCL). However, clinical behavior and prognosis differ considerably from those for nodal DLBCL (nDLBCL), and their pathogenesis is still not fully understood. Micro-RNAs (miRNAs) have been associated with cancer development and progression. We investigated a large miRNA panel for differential expression in PCNSL and nDLBCL, to determine new mechanisms potentially involved in PCNSL pathogenesis. Using paraffin-embedded biopsy specimens from 21 HIV-negative patients with newly diagnosed PCNSL (n = 11) and nDLBCL (n= 10), we measured the expression of 365 miRNA species by quantitative real-time PCR using low-density PCR arrays. We found that 18 miRNAs were differentially expressed: median expression levels of 13 miRNAs were 2.1–13.1 times higher in PCNSL, and median expression levels of 5 miRNAs were 2.6–3.3 times higher in nDLBCL. MiRNAs upregulated in PCNSL were associated with the Myc pathway (miR-17-5p, miR-20a, miR-9), with blocking of terminal B-cell differentiation (miR-9, miR-30b/c), or with upregulation by inflammatory cytokines (miR-155). Putative tumor-suppressor miRNAs (miR-199a, miR-214, miR-193b, miR-145) were downregulated in PCNSL. There was no overlap of miRNAs dysregulated in PCNSL with those differentially expressed between immunohistologically defined germinal center B cell–like (GCB) and non-GCB types or, apart from miR-9, with miRNAs known to be overexpressed in human brain. We conclude that PCNSL exhibits a distinct pattern of miRNA expression compared with nDLBCL. This argues for the involvement of different molecular mechanisms in the pathogenesis of these two lymphoma types.
Keywords: micro-RNA, Myc, primary CNS lymphoma
Primary CNS lymphoma (PCNSL) is a rare subtype of extranodal non-Hodgkin's lymphoma that is confined to the CNS in the absence of systemic disease. Most cases are classified as diffuse large B-cell lymphoma (DLBCL).1 PCNSL is characterized by a poor prognosis compared with systemic lymphoma with similar histology.2 Mechanisms leading to the development of a DLBCL in the CNS, which is normally devoid of a lymphatic system, are still not fully understood.
Recently, the widespread dysregulation of the expression of micro-RNAs (miRNAs) in cancer cells has become evident.3 MiRNAs are small (18 to 24 nucleotides) noncoding RNAs that regulate gene expression by binding to partially complementary target sites in mRNA 3′ untranslated regions, which results in degradation of the target mRNAs or translational repression of the encoded proteins.4 Expression of specific miRNAs has been shown to promote tumorigenesis, whereas other miRNAs exert tumor-suppressor properties.5 MiRNA expression signatures are increasingly used for the classification of cancer and have been associated with prognosis in some cases.6,7
Human B-cell lymphomas including DLBCL overexpress members of the miR-17-92 cluster and also miR-155, both of which are thought to be involved in B-cell lymphomagenesis.8–16 Furthermore, B-cell lymphomas and DLBCL subtypes exhibit different miRNA signatures, reflecting their differentiation stages.17,18
In this work, we investigated the expression of a wide array of miRNA species in PCNSL to search for a differential expression compared with nodal DLBCL (nDLBCL), in an effort to determine pathways potentially involved in PCNSL pathogenesis.
Materials and Methods
Immunohistochemistry
Formalin-fixed and paraffin-embedded (FFPE) biopsy samples from HIV-negative and not otherwise immunosuppressed patients with newly diagnosed PCNSL participating in a controlled therapy trial19 and from patients with nDLBCL were obtained with informed consent and in accordance with the Declaration of Helsinki. The study was approved by the local ethics committee. Histological diagnosis of DLBCL was confirmed by central pathological review in all cases.19 Tissue sections were further stained for cluster of designation (CD) 10, B-cell lymphoma 6 (BCL6) protein, and multiple myeloma oncogene 1 (MUM1) (cutoff ≥30% positive cells) using standard methods to establish the cell-of-origin (COO) subtype according to the Hans classifier.20 Immunohistochemical analysis for Myc protein was performed, and the number of positive cells was estimated in 25% steps.
RNA Extraction
For RNA analysis, we selected only samples with a tumor cell content of at least 80%. Under RNase-free conditions, 4 to 6 serial sections (20 µm) were cut from FFPE tissue blocks. Sections were deparaffinized twice with xylene (3 min, 50°C), dehydrated with 100% ethanol, and air dried. RNA was extracted using the RecoverAll Kit (Ambion) following the manufacturer's instructions (proteinase K digestion for 3 h at 50°C) and eluted in 100 µL of nuclease-free water (95°C). Remaining DNA was digested using the RNeasy Mini Kit (Qiagen). RNA was finally eluted in 40 µL of RNase-free water and stored at −20°C. A NanoDrop 1000 Spectrophotometer (NanoDrop Technologies) was used to determine RNA concentration.
MiRNA Expression Analysis
For miRNA analysis, 100 ng of total RNA was reverse transcribed in each case using 8 predefined primer pools containing up to 48 multiplex reverse transcription primers each. MiRNA levels were measured by quantitative real-time PCR (qPCR) using commercially available TaqMan low-density arrays (TLDAs) together with the 7900HT (high throughput) Fast Real-Time PCR System according to the manufacturer's recommendations (TaqMan Human MicroRNA Array v1.0; Applied Biosystems). Each TLDA contained specific primer-probe combinations for 365 miRNA species. Each qPCR was analyzed in duplicate. Threshold cycle (Ct) values were determined using Sequence Detection System software (v2.3, Applied Biosystems). Only miRNAs with a Ct value of ≤37 in both technical replicates were considered as being present in the sample. To compare the expression of miRNAs not present in a group of samples and putatively expressed in other samples, all Ct values >37 were set to Ct = 38 according to the method of Kubista et al.21 The TLDAs contained 3 internal controls: small nucleolar RNAs relative nucleotide unit (RNU) 44, RNU48, and RNU6B. Because Ct values for RNU48 differed significantly in PCNSL and nDLBCL (P = .005, Mann–Whitney U-test), RNU48 was excluded as an internal control. Data were then independently normalized to RNU44 and to RNU6B, and expression levels were calculated using the 2−▵Ct method. Only miRNAs that were differentially expressed after normalization to both internal controls are reported here.
Statistical Analysis
The Mann–Whitney U-test was used to compare the expression of each miRNA between PCNSL and nDLBCL and between germinal center B cell–like (GCB) and non-GCB subtypes. Age distribution in PCNSL and nDLBCL was compared using the t-test, and sex as well as COO subtype were compared using the chi-square test. For bivariate correlations, Pearson's R was calculated. All tests were two-sided, with P < .05 indicating a significant difference. Statistical analyses were performed using Predictive Analytics SoftWare, version 18. To perform hierarchical clustering of cases and differentially expressed miRNAs, we used Partek Genomics Suite 6.5 beta (Pearson correlation, complete linkage).
Results
Patient Characteristics
MiRNA expression was analyzed in 21 patients: 11 with PCNSL and 10 with nDLBCL. Patient characteristics were balanced between the PCNSL and nDLBCL groups. The median patient age was 65 years (range 53–78) in the PCNSL group and 69 years (63–78) in the nDLBCL group (t-test, P = .39). The male:female ratio was 4:7 in PCNSL and 5:5 in nDLBCL (chi-square test, P = .53), and the non-GCB:GCB subtype ratio was 4:7 in PCNSL and 5:5 in nDLBCL (P = .53). Five patients with nDLBCL had stage III or IV disease, and 5 patients had stage I or II.
MiRNA Expression Levels in PCNSL and DLBCL
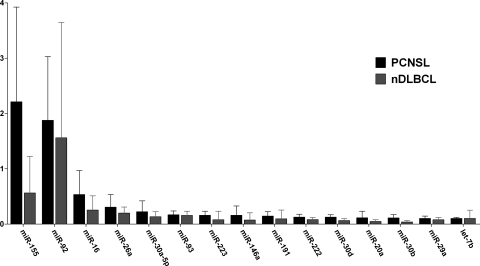
Of 365 miRNA species detectable by TLDA, 29% and 33% were present in more than half of the samples of nDLBCL and PCNSL, respectively. The most abundant miRNAs in both lymphoma types were miR-155, miR-92, miR-16, miR-26a, miR-30a-5p, and miR-93 (Fig. 1). We found 18 miRNAs that were differentially expressed between PCNSL and nDLBCL. The expression of 13 miRNAs was significantly higher in PCNSL, whereas 5 miRNAs showed a reduced expression. The expression ratios of specific miRNAs in PCNSL compared with nDLBCL ranged from 13.1-fold overexpression (miR-9) to 3.3-fold reduced expression (miR-145) (Table 1).
Fig. 1.
Expression levels (relative to RNU44) of the 15 most abundant miRNAs in PCNSL and their respective expressions in nDLBCL. Bars depict median expression and interquartile range.
Table 1.
Micro-RNAs with differential expression in PCNSL and nDLBCL and according to their cell of origin (immunohistochemically determined)
| Micro-RNA | PCNSL/DLBCL expression ratioa | P |
|---|---|---|
| miR-9 | 13.11 | .006 |
| miR-20b | 5.97 | .017 |
| miR-155 | 3.93 | .009 |
| miR-340 | 3.72 | .002 |
| miR-17-5p | 3.60 | .029 |
| miR-148a | 3.39 | .014 |
| miR-30b | 2.86 | .002 |
| miR-27b | 2.65 | .020 |
| miR-26b | 2.50 | .035 |
| miR-146b | 2.44 | .011 |
| miR-20a | 2.40 | .035 |
| miR-30c | 2.30 | .029 |
| let-7g | 2.10 | .007 |
| miR-199a | 0.38 | .011 |
| miR-214 | 0.36 | .006 |
| miR-432 | 0.36 | .041 |
| miR-193b | 0.32 | .005 |
| miR-145 | 0.30 | .024 |
| Micro-RNA | GCB/non-GCB expression ratioa | P |
| miR-296 | 5.68 | .003 |
| miR-361 | 4.38 | .013 |
| miR-301 | 3.33 | .016 |
| miR-642 | 2.84 | .039 |
| miR-29c | 1.49 | .009 |
aRatio of median expression levels sorted in descending order.
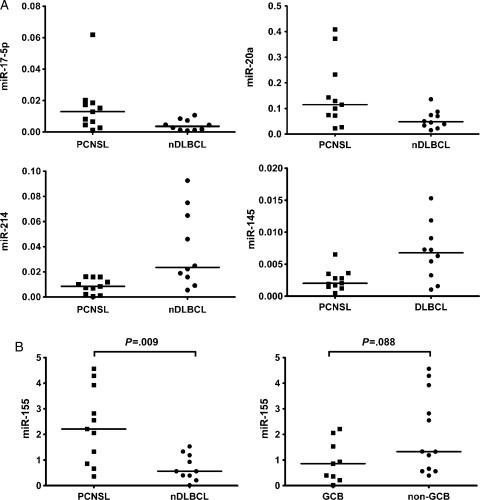
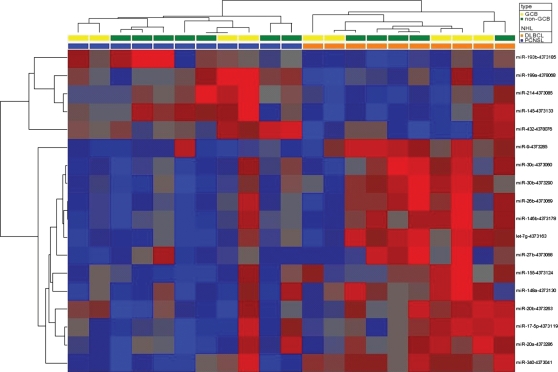
In PCNSL we found higher expression of miRNAs belonging to the miR-17-92-cluster (miR-17-5p and miR-20a) and of miR-155, whereas putative tumor-suppressor miRNAs (miR-214, miR-199a, miR-193b, and miR-145) were expressed at lower levels than in nDLBCL (Fig. 2A). Supervised hierarchical clustering employing the differentially expressed miRNAs clearly separated PCNSL and nDLBCL (Fig. 3). An unsupervised analysis did not result in a reliable distinction between PCNSL and DLBCL.
Fig. 2.
(A) Differential expression of specific miRNAs in PCNSL and nDLBCL: examples of significant overexpression (miR-17-5p and miR-20a) and downregulation (miR-214 and miR-145) in PCNSL (expression levels relative to RNU48; the line indicates the median, all P < .05). (B) MiR-155 is overexpressed in PCNSL compared with nDLBCL, whereas no significantly different expression for miR-155 is seen between GCB- and non–GCB-type lymphomas (expression levels relative to RNU44; the line indicates the median).
Fig. 3.
Supervised cluster analysis of miRNA levels in PCNSL and nDLBCL. Heatmap of differentially expressed miRNAs. Data normalized to RNU44 (dCt) were hierarchically clustered (Pearson correlation, complete linkage). Red indicates a decrease relative to all data in this set; blue indicates an increase relative to all data in this set.
When we compared miRNA expression across all samples according to their immunohistologically defined GCB and non-GCB types, we found 5 miRNAs differentially expressed. None of these miRNAs was among those with differential expression in PCNSL and nDLBCL (Table 1). There was a trend toward higher miR-155 expression in only non-GCB cases (P = .088, Fig. 2B).
Expression of Brain-Enriched MiRNAs
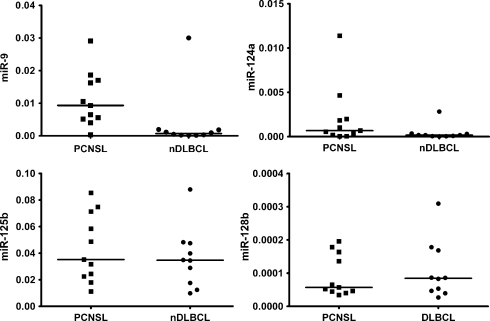
The expression of several miRNAs has been reported to be greatly enriched in the human CNS but not in most other organs. They play an important role in neuronal development and function and therefore are called brain-specific or brain-enriched miRNAs.22,23,57 These include miR-9, miR-124, miR-128, miR-125, and possibly further miRNA species.22,23 Of these, only miR-9 exhibited a significantly higher expression in PCNSL than in nDLBCL, whereas the higher expression of miR-124 in PCNSL was of borderline significance (P = .05). MiR-128 and miR-125 (Fig. 4) and an additional 19 putatively brain-enriched miRNAs showed similar expression levels in PCNSL and nDLBCL (data not shown). MiR-9 expression in PCNSL did not correlate with tumor cell content (R = −0.34, P = .3).
Fig. 4.
Expression of brain-enriched miRNAs in PCNSL and nDLBCL. MiR-9 was significantly higher expressed in PCNSL (P = .006), and miR-124a was nonsignificantly higher in PCNSL (P = .05); expressions of miR-125b and miR-128b were similar in both entities (P = .48 and 0.73).
Expression of Clustered MiRNAs
Although formalin fixation is known to have detrimental effects on mRNA quality, it does not affect miRNA analyses, due to the smallness of the molecules.24 To demonstrate the validity of our miRNA analyses, we tested some of the miRNAs known to be clustered and thus co-regulated for the correlation of their expression in the available lymphoma samples. Members of the miR-17-92 cluster (miR-17-5p, miR-18a, miR-19a, miR-19b, mir-20a, and miR-92)10,12 all showed a significant (P < .05) correlation of their expression, with R-values ranging from 0.473 to 0.953 (median 0.671), irrespective of their differential expression in PCNSL and nDLBCL. The expression of the clustered miRNAs miR-199a and miR-214 was also highly correlated (Online Supplementary Fig. S1).25
Myc Protein Expression
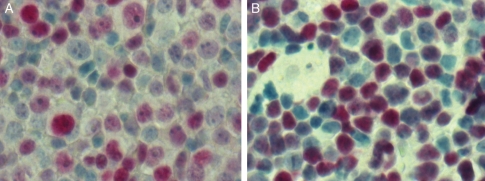
Several miRNAs (miR-9, miR-17-5p, miR-20a, miR-20b, and miR145) that were differentially expressed in PCNSL are able to contribute to an increased expression of MYC, which is a common oncogenic event in cancer pathogenesis. We therefore looked at Myc protein expression using immunohistochemistry.12,26,27 Tissue sections of 8 nDLBCL and 8 PCNSL were stained for Myc. All cases were Myc positive, exhibiting a heterogeneous staining pattern. In both nDLBCL and PCNSL, the proportion of positive cells was at least 50% (Fig. 5).
Fig. 5.
Immunohistochemistry for Myc protein. Representative sections of a nodal DLBCL (A) and a PCNSL (B) (40×) demonstrate Myc positivity in both entities.
Discussion
Whether PCNSL and nDLBCL differ in their molecular features and pathogenesis is still uncertain. Gene expression analyses were able to distinguish PCNSL and systemic DLBCL. However, the magnitude of the effect of the CNS background is a matter of ongoing debate.28–30 The importance of miRNA dysregulation in cancer etiology, progression, and metastasis has become evident in recent years.5 Studying miRNAs that regulate gene expression at the posttranscriptional level may reveal features specific for PCNSL in contrast to nDLBCL and thus improve our understanding of PCNSL pathogenesis. However, data on miRNA expression in PCNSL are very scarce. MiRNAs have recently been utilized as cerebrospinal fluid biomarkers for the detection of PCNSL.31 To date, only one study has reported on the differential expression of 15 miRNAs (miR-15a, miR-15b, miR-16, miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, miR-21, miR-92, miR-127, miR-155, miR-181a, and miR-221); this study, which examined 9 PCNSL, 19 nDLBCL, 11 extranodal DLBCL, and 11 primary testicular DLBCL, found miR-17-5p to be overexpressed in PCNSL compared with nDLBCL.32
In our study, we examined the expression of a large panel of 365 miRNAs in PCNSL using a qPCR-based array technique. The RNA investigated had been extracted from FFPE samples. The feasibility of this approach had been demonstrated before and is also shown by the strongly correlated expression of clustered miRNAs in our study.18,24,33
Approximately one-third of the analyzed miRNAs were detectable in PCNSL. MiR-155 and miR-92, a member of the miR-17-92 cluster, exhibited the highest expression levels in both nDLBCL and PCNSL, a finding in line with previous reports describing high expression and a possible role in lymphomagenesis for these miRNAs in systemic DLBCL. In mice, an augmented B-cell proliferation and transformation by miR-155 and an accelerated B-cell lymphoma development driven by miR-17-92 have been demonstrated.10,13–15 When we compared miRNA expression in nDLBCL and PCNSL, we found that 18 miRNAs were differentially expressed, 13 were overexpressed, and 5 were downregulated in PCNSL. In addition to a known miR-17-5p overexpression in PCNSL, we found miR-20a, which is also a member of the miR-17-92 polycistron on chromosome 13, and miR-20b, a member of the miR-106a-92 cluster on chromosome X homologous to miR-17-92, to be significantly overexpressed in PCNSL. MiR-17-92 is upregulated by the proto-oncogene MYC and thus accelerates MYC-induced lymphoma development.12 Correspondingly, miR-145, which directly represses Myc expression (thus mediating the activity of the tumor suppressor p53) was downregulated in our PCNSL samples.27
Furthermore, we found miR-9 and members of the miR-30 family (miR-30b and miR-30c) to be upregulated in PCNSL. Both are overexpressed in germinal center B cells, in which they downregulate the transcription factor positive regulatory domain (PRDM)-1 (Blimp-1), an essential regulator of plasma cell differentiation.17,34 Blimp-1 is able to repress MYC and thereby facilitates the exit of B cells from the cell cycle and their terminal differentiation into plasma cells.35 Downregulation of Blimp-1 by mutation or epigenetical silencing of the PRDM-1 gene has already been recognized as an oncogenic mechanism in DLBCL and also in one report on PCNSL.36–38 Thus, overexpression of miR-9 and miR-30 may contribute to lymphomagenesis by blocking B-cell terminal differentiation through PRDM-1 silencing. Recently, miR-9 activation by MYC has been demonstrated in breast cancer.26 In PCNSL, the data concerning the role of MYC are few and conflicting.39–42 When we looked for Myc expression in PCNSL by immunohistochemistry, we found a strong expression. We did not perform a direct comparison of the immunohistologically determined MYC expression in PCNSL and nDLBCL. However, recent gene expression analyses demonstrated a higher expression of MYC mRNA in PCNSL than in nDLBCL.29 Consequently, an activation of the MYC pathway in PCNSL seems likely. Nonetheless, further targets of the dysregulated miRNAs are known and might be relevant in PCNSL. Members of the miR-17-92 cluster downregulate a number of proapoptotic and tumor-suppressive factors, such as CDKN1A/p21, phosphatase and tensin homolog (PTEN), E2F1, and Bim.8,9,11 For instance, downregulation of Bim leads to overexpression of BCL2, which is a prominent feature in PCNSL.41,43 Other miRNAs that were downregulated in PCNSL, namely miR-199a, miR-214, and miR-193b, have been demonstrated to act as tumor suppressors via downregulation of certain oncogenes in different solid cancers.44–47
Another striking finding of our study was the significant overexpression of miR-155 in PCNSL. This is in contrast to findings of the previously published miRNA study on PCNSL and is possibly explained by a different sample preparation, varying RNA quality, and low sample size in both our study and theirs.32 MiR-155 possesses an oncogenic potential, as demonstrated in a lymphoma model in mice.13 Physiologically it is expressed in activated B and T cells and monocytes and controls the germinal center reaction by regulating the production of certain cytokines such as tumor necrosis factor (TNF).48,49 Normal CNS lacks the formation of germinal centers, but under certain circumstances, specifically in inflammatory CNS diseases, ectopic germinal centers have been observed within the meninges.50,51 An association of inflammatory processes and lymphoma development in PCNSL pathogenesis has been hypothesized. MiR-155 has been reported to be upregulated by immune stimuli such as TNF-α and on the other hand can render nDLBCL resistant to growth-inhibitory cytokines such as transforming growth factor–β.52–55 However, conclusive data regarding the possible role of inflammation in PCNSL pathogenesis are still lacking, and further studies are necessary.
An overexpression of miR-155, as well as the differential expression of a number of further miRNAs in immunohistologically defined non-GCB type DLBCL, has been reported by some authors.14,53,56 According to gene expression data, PCNSL are heterogeneous, in particular regarding their COO type. Notwithstanding the limitations of the Hans classifier, which furthermore has not been tested in PCNSL, we used this immunohistological classification to avoid a potential bias introduced by a predominance of one COO type in the PCNSL or nDLBCL samples examined. Still, this and other molecular signatures might have superimposed the specific differences between PCNSL and nDLBCL detectable by statistical approaches.
We cannot exclude the possibility that our miRNA expression data for PCNSL may in part be derived from the CNS background. As noted, miR-9, which was most strongly overexpressed in PCNSL, belongs to the group of brain-specific or brain-enriched miRNAs.22,23,57 However, miR-9 is also strongly involved in B-cell maturation, and together with miR-124 was the only brain-enriched miRNA highly expressed in PCNSL in our study, whereas other members of this group exhibited similar expression levels in nDLBCL and PCNSL.
In summary, in this study we were able to detect a differential miRNA expression pattern in PCNSL. Dysregulation of specific miRNAs leading to, for instance, an activation of the MYC pathway or to a blockade of germinal center exit in B cells by Blimp-1 downregulation might play a role in PCNSL pathogenesis. Further studies are warranted to better describe the targets of the identified miRNAs.
Supplementary Material
Conflict of interest statement. None declared.
Funding
No external funding for this study.
Supplementary Material
Acknowledgments
None.
References
- 1.Miller DC, Hochberg FH, Harris NL, et al. Pathology with clinical correlations of primary central nervous system non-Hodgkin's lymphoma. The Massachusetts General Hospital experience 1958–1989. Cancer. 1994;74:1383–1397. doi: 10.1002/1097-0142(19940815)74:4<1383::aid-cncr2820740432>3.0.co;2-1. doi:10.1002/1097-0142(19940815)74:4<1383::AID-CNCR2820740432>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 2.DeAngelis LM, Seiferheld W, Schold SC, Fisher B, Schultz CJ. Combination chemotherapy and radiotherapy for primary central nervous system lymphoma: Radiation Therapy Oncology Group Study 93-10. J Clin Oncol. 2002;20:4643–4648. doi: 10.1200/JCO.2002.11.013. doi:10.1200/JCO.2002.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. doi:10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. doi:10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. doi:10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. doi:10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. doi:10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Inomata M, Tagawa H, Guo YM, et al. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. doi:10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 9.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. doi:10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 10.Ota A, Tagawa H, Karnan S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. doi:10.1158/0008-5472.CAN-03-3773. [DOI] [PubMed] [Google Scholar]
- 11.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. doi:10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. doi:10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eµ-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. doi:10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. doi:10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kluiver J, Poppema S, de Jong D, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. doi:10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 16.Lawrie CH, Chi J, Taylor S, et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. J Cell Mol Med. 2009;13:1248–1260. doi: 10.1111/j.1582-4934.2008.00628.x. doi:10.1111/j.1582-4934.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Jima DD, Jacobs C, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. doi:10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malumbres R, Sarosiek KA, Cubedo E, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. doi:10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiel E, Korfel A, Martus P, et al. High-dose methotrexate with or without whole brain radiotherapy for primary CNS lymphoma (G-PCNSL-SG-1): a phase 3, randomised, non-inferiority trial. Lancet Oncol. 2010;11:1036–1047. doi: 10.1016/S1470-2045(10)70229-1. doi:10.1016/S1470-2045(10)70229-1. [DOI] [PubMed] [Google Scholar]
- 20.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. doi:10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 21.Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. doi:10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. doi:10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- 23.Sempere LF, Freemantle S, Pitha-Rowe I, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. doi:10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson PT, Baldwin DA, Kloosterman WP, et al. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. doi:10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YB, Bantounas I, Lee DY, et al. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37:123–128. doi: 10.1093/nar/gkn920. doi:10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. doi:10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montesinos-Rongen M, Brunn A, Bentink S, et al. Gene expression profiling suggests primary central nervous system lymphomas to be derived from a late germinal center B cell. Leukemia. 2008;22:400–405. doi: 10.1038/sj.leu.2405019. doi:10.1038/sj.leu.2405019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubenstein JL, Fridlyand J, Shen A, et al. Gene expression and angiotropism in primary CNS lymphoma. Blood. 2006;107:3716–3723. doi: 10.1182/blood-2005-03-0897. doi:10.1182/blood-2005-03-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tun HW, Personett D, Baskerville KA, et al. Pathway analysis of primary central nervous system lymphoma. Blood. 2008;111:3200–3210. doi: 10.1182/blood-2007-10-119099. doi:10.1182/blood-2007-10-119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140–3146. doi: 10.1182/blood-2010-09-308684. doi:10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 32.Robertus JL, Harms G, Blokzijl T, et al. Specific expression of miR-17-5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod Pathol. 2009;22:547–555. doi: 10.1038/modpathol.2009.10. doi:10.1038/modpathol.2009.10. [DOI] [PubMed] [Google Scholar]
- 33.Roehle A, Hoefig KP, Repsilber D, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. doi:10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 34.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. doi:10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 35.Lin KI, Lin Y, Calame K. Repression of c-myc is necessary but not sufficient for terminal differentiation of B lymphocytes in vitro. Mol Cell Biol. 2000;20:8684–8695. doi: 10.1128/mcb.20.23.8684-8695.2000. doi:10.1128/MCB.20.23.8684-8695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Courts C, Montesinos-Rongen M, Brunn A, et al. Recurrent inactivation of the PRDM1 gene in primary central nervous system lymphoma. J Neuropathol Exp Neurol. 2008;67:720–727. doi: 10.1097/NEN.0b013e31817dd02d. doi:10.1097/NEN.0b013e31817dd02d. [DOI] [PubMed] [Google Scholar]
- 37.Pasqualucci L, Compagno M, Houldsworth J, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. doi:10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tam W, Gomez M, Chadburn A, et al. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. doi:10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 39.Cady FM, O'Neill BP, Law ME, et al. Del(6)(q22) and BCL6 rearrangements in primary CNS lymphoma are indicators of an aggressive clinical course. J Clin Oncol. 2008;26:4814–4819. doi: 10.1200/JCO.2008.16.1455. doi:10.1200/JCO.2008.16.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang CC, Kampalath B, Schultz C, et al. Expression of p53, c-Myc, or Bcl-6 suggests a poor prognosis in primary central nervous system diffuse large B-cell lymphoma among immunocompetent individuals. Arch Pathol Lab Med. 2003;127:208–212. doi: 10.5858/2003-127-208-EOPMOB. [DOI] [PubMed] [Google Scholar]
- 41.Cobbers JM, Wolter M, Reifenberger J, et al. Frequent inactivation of CDKN2A and rare mutation of TP53 in PCNSL. Brain Pathol. 1998;8:263–276. doi: 10.1111/j.1750-3639.1998.tb00152.x. doi:10.1111/j.1750-3639.1998.tb00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nozaki M, Tada M, Mizugaki Y, et al. Expression of oncogenic molecules in primary central nervous system lymphomas in immunocompetent patients. Acta Neuropathol. 1998;95:505–510. doi: 10.1007/s004010050831. doi:10.1007/s004010050831. [DOI] [PubMed] [Google Scholar]
- 43.Braaten KM, Betensky RA, de Leval L, et al. BCL-6 expression predicts improved survival in patients with primary central nervous system lymphoma. Clin Cancer Res. 2003;9:1063–1069. [PubMed] [Google Scholar]
- 44.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. doi:10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 45.Kim S, Lee UJ, Kim MN, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–18166. doi: 10.1074/jbc.M800186200. doi:10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 46.Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–3948. doi: 10.1038/onc.2009.245. doi:10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 47.Rauhala HE, Jalava SE, Isotalo J, et al. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int J Cancer. 2010;127:1363–1372. doi: 10.1002/ijc.25162. doi:10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
- 48.Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. doi:10.1016/S0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 49.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. doi:10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 50.Narayan K, Dail D, Li L, et al. The nervous system as ectopic germinal center: CXCL13 and IgG in lyme neuroborreliosis. Ann Neurol. 2005;57:813–823. doi: 10.1002/ana.20486. doi:10.1002/ana.20486. [DOI] [PubMed] [Google Scholar]
- 51.Serafini B, Rosicarelli B, Magliozzi R, Stigliano E, Aloisi F. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 2004;14:164–174. doi: 10.1111/j.1750-3639.2004.tb00049.x. doi:10.1111/j.1750-3639.2004.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pedersen IM, Otero D, Kao E, et al. Onco-miR-155 targets SHIP1 to promote TNFα-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1:288–295. doi: 10.1002/emmm.200900028. doi:10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rai D, Karanti S, Jung I, Dahia PL, Aguiar RC. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet Cytogenet. 2008;181:8–15. doi: 10.1016/j.cancergencyto.2007.10.008. doi:10.1016/j.cancergencyto.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischer L, Korfel A, Pfeiffer S, et al. CXCL13 and CXCL12 in central nervous system lymphoma patients. Clin Cancer Res. 2009;15:5968–5973. doi: 10.1158/1078-0432.CCR-09-0108. doi:10.1158/1078-0432.CCR-09-0108. [DOI] [PubMed] [Google Scholar]
- 55.Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 56.Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. doi:10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 57.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. doi:10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.