Abstract
Several studies on molecular profiling of oligodendrogliomas (OGs) in adults have shown a distinctive genetic pattern characterized by combined deletions of chromosome arms 1p and 19q, O6-methylguanine-methyltransferase (MGMT) methylation, and isocitrate dehydrogenase 1 (IDH1) mutation, which have potential diagnostic, prognostic, and even therapeutic relevance. OGs in pediatric and young adult patients are rare and have been poorly characterized on a molecular and biological basis, and it remains uncertain whether markers with prognostic significance in adults also have predictive value in these patients. Fourteen cases of OGs in young patients (age, ≤25 years) who received a diagnosis over 7 years were selected (7 pediatric patients age ≤18 years and 7 young adults aged 19–25 years). The cases were evaluated for 1p/19q status, MGMT promoter methylation, p53 mutation, and IDH1 mutation. None of the pediatric cases showed 1p/19q deletion. In young adults, combined 1p/19q loss was observed in 57% and isolated 1p loss in 14% of cases. The majority of cases in both subgroups (71% in each) harbored MGMT gene promoter methylation. TP53 and IDH1 mutations were not seen in any of the cases in both the groups. To our knowledge, this is the first study to show that molecular profile of OGs in pediatric and young adult patients is distinct. Further large-scale studies are required to identify additional clinically relevant genetic alterations in this group of patients.
Keywords: adolescent, IDH1, MGMT, oligodendrogliomas, 1p/19q, pediatric, p53, TP53, young adults
Over the past decade, a plethora of cytogenetic and molecular genetic alterations in gliomas have been intensely studied and exploited to facilitate glioma classification, prognostication, and therapeutic monitoring. Oligodendroglial tumors have attracted great interest in both basic and clinical neuro-oncology, because they have a better overall prognosis and longer survival times than the diffuse astrocytic tumors.
Combined loss of 1p and 19q, seen in 40%–80% of oligodendrogliomas (OGs), has been established as a molecular signature of oligodendroglial tumors.1–3 Assessment of allelic status of chromosomes 1p and 19q plays a crucial role in the diagnosis, treatment, and prognostic assessment of adult patients with OGs.4–6 Thus, unambiguous classification of oligodendroglial tumors has become vital for improved clinical management in the field of neuro-oncology.
O6-methylguanine-methyltransferase (MGMT) promoter methylation has been established as an independent predictor for therapeutic response to chemotherapy in high-grade astrocytic tumors treated with temozolomide.7,8 A few recent studies have reported MGMT promoter methylation in majority of adult OGs and also documented its significant positive correlation with 1p/19q loss.9–11 These authors have also suggested that MGMT promoter methylation in oligodendroglial tumors could possibly be associated with chemosensitivity and prolonged survival.9,12
In 2008, in a genome-wide sequencing study by Parsons et al, mutations in a gene-encoding isocitrate dehydrogenase (IDH) were identified in 12% of glioblastomas (GBMs).13 Subsequent studies have shown these mutations in a significant proportion of diffuse gliomas (astrocytic, oligodendroglial, and oligoastrocytic) and secondary GBMs.14,15 The frequency in astrocytic and oligodendroglial tumors is almost similar.16–18 Various studies have shown a strong association of isocitrate dehydrogenase 1 (IDH1) mutation with p53 mutations in astrocytomas and 1p/19q deletion in OGs.17–19 Recent clinical trials have recognized IDH1 mutation as an independent diagnostic and prognostic marker in both astrocytic and oligodendroglial tumors and have shown a strong association of IDH1 mutation with better overall survival (OS) and progression-free survival (PFS).13,20–22
Another molecular alteration that is seen in a majority of astrocytic tumors but extremely rare in OGs is TP53 mutation.3,23,24 Various studies have shown an inverse correlation between 1p/19q deletion and TP53 gene mutation.25,26
Thus, the molecular profile of adult OGs has been extensively studied, and various studies have given an insight into the molecular pathogenesis of these tumors.
On the other hand, OGs in younger age group are extremely rare and comprise only 1%–3% of pediatric central nervous system neoplasms.27,28 Their molecular profile has not been well characterized.
There are only a few studies available in the English literature that have suggested that molecular alterations in pediatric OGs are distinctly different from their adult counterparts.29,30 There is no study to date on MGMT promoter methylation and IDH1 mutation status in pediatric cases. Therefore, this study was undertaken with the aim of comprehensive analysis of frequency of various molecular alterations in OGs in pediatric and young adult patients. On careful search of the neuropathology record of the past 7 years, we found 14 cases of OGs in patients aged ≤25 years. We analyzed these tumors for frequency of 1p/19q loss, MGMT promoter methylation status, TP53, and IDH1 mutations.
Materials and Methods
Clinical Data
One hundred eighty cases of OGs diagnosed from January 2003 through December 2009 were identified from a detailed review of the neuropathology records of the All India Institute of Medical Sciences (New Delhi, India). Age and sex of all patients were noted. Approximately 8% of these (14 cases) were reported in young patients (age, ≤25 years). The patients were divided into 2 subgroups: pediatric (age, ≤18 years) and young adults (age, 19–25 years).
Histopathological Examination
The original hematoxylin and eosin (H&E) slides were re-evaluated independently by 3 neuropathologists (C.S., M.C.S., and V.S.). The diagnosis was reconfirmed in accordance with the recent World Health Organization classification.31 Age- and sex-matched normal cortical brain tissue samples from patients with epilepsy surgery were chosen as controls.
Fluorescence In Situ Hybridization Analysis of 1p and 19q
Dual-probe fluorescence in situ hybridization (FISH) assay was performed on paraffin-embedded sections, with locus-specific probes for 1p36 and 19q13 paired, respectively, with the reference probes for 1q25 and 19p13 (Vysis). Deparaffinization of the sections was performed with 3 10-minute immersions in xylene, followed by 2 3-minute immersions in 100% ethanol. After rinsing in water, target retrieval was achieved by immersing the slides in citrate buffer (pH, 6.0) and boiling in a microwave for 20 min. Slides were exposed to 0.04% pepsin (P-7000; Sigma- Aldrich) digestion for 30 min at 37°C, fixed, and dehydrated. Probe mixture (10 µL per slide) was applied on each section. Simultaneous probe/specimen denaturation at 73°C for 5 min, with subsequent overnight incubation at 37°C, was performed in a Thermobrite hybridization chamber (Vysis). The sections were washed the next day in 2× saline sodium citrate (SSC) buffer (2 min at 73°C), followed by 0.5× SSC (2 min at room temperature); counterstained with 4,6-diamidino-2-phenylindole (Vysis); and visualized under a fluorescent microscope. Signals were scored in at least 200 nonoverlapping, intact nuclei. Sections form nonneoplastic cortical tissue obtained from epilepsy surgery specimens were used as a control for each probe pair. At least 50% nuclei had to show a ratio of 1:2 for test versus reference probes to be scored as deletion. The cutoff value was estimated by calculating the mean plus 3 standard deviations (SDs) of deletions shown by nonneoplastic brain.
Methylation-Specific Polymerase Chain Reaction for MGMT Promoter
Genomic DNA was isolated from formalin-fixed paraffin-embedded (FFPE) sections after conformation of histology. One section of each paraffin block was stained with H&E, and areas with maximum amount of tumor tissue without necrosis were marked out. Subsequently, 4 20-µm sections were serially cut from the same block and taken on separate sterile slides. Tumor area matched from H&E sections was dissected with a disposable scalpel to ensure that all the sections contained >80% tumor tissue. All these sections were then pooled in a sterile eppendorf vial. DNA was extracted using genomic DNA extraction kit (Real Genomics).
DNA methylation pattern of the MGMT gene promoter was determined using methylation-specific polymerase chain reaction (PCR). This procedure involves chemical modification of unmethylated, cytosine to uracil, followed by a nested, 2-stage PCR. Genomic DNA (∼500 ng) isolated from paraffin blocks of each sample was modified by sodium bisufite treatment (EZ Gold DNA methylation kit; Zymo Research). Enzymatically methylated DNA was used as positive methylation control, whereas normal lymphocytic DNA served as unmethylation control. Methylation-specific primers were used for PCR. First-stage primer recognizes the bisulfite-modified template flanking the MGMT gene but does not discriminate between methylated and unmethylated alleles. The PCR amplification procedure for stage 1 was as follows: an initial denaturation step of 5 min at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at 52°C, and 30 s at 72°C, and a final elongation step of 7 min at 72°C in DNA Engine Peltier Thermal Cycler (Bio Rad) with use of recombinant Taq DNA polymerase (Fermentas Life Sciences). A 25-µL volume was used in all PCRs. The stage-1 PCR product was diluted 20-fold, and 2 µL of this dilution was subjected to a stage-2 PCR. Methylation and unmethylation primers were used separately for each test. The PCR protocol for stage 2 was as follows: an initial denaturation step of 5 min at 94°C, followed by 35 cycles of 15 s at 94°C, 15 s at 62°C, and 15 s at 72°C, and final elongation step of 7 min at 72°C. Methylation-specific PCR was performed to amplify 83 base pair fragment of the methylated MGMT gene promoter and 91 base pair fragment of unmethylated product. Amplified products were separated on 4% agarose gel, ethidium bromide stained, and visualized under UV illumination.
Mutation Analysis of TP53 and IDH1 by Sequencing
For Frozen Samples
In 6 cases, fresh frozen tissue was available in the tumor bank. For DNA isolation, the frozen tumor tissue was cryosectioned at 5 µm and stained H&E. These sections of each collected specimen were reviewed to verify adequacy of the tumor area (ie, minimal contamination with nonneoplastic elements) and to assess the extent of tumoral necrosis and cellularity. The subsequent 15 serial sections of 40 µm were taken separately and stored in liquid nitrogen–chilled vials. The sections were used for DNA isolation by Genelute Mammalian DNA Miniprep Kit (Sigma Aldrich) according to the manufacturer's instructions. In them, TP53 and IDH1 mutational analysis was performed.
All TP53 coding region from exon 5–8 were investigated by direct sequencing protocol, as described by IARC p53 database.32 The following primer sequences were used for exon 5–6: forward 5′ TGTTCACTTGTGCCCTGACT 3′ and reverse 5′ TTAACCCCTCCTCCCAGAGA 3′; exon 7: forward 5′ CTTGCCACAGGTCTCCCCAA 3′ and reverse 5′ AGGGGTCAGCGGCAAGCAGA 3′; and exon 8: forward 5′ TTGGGAGTAGATGGAGCCT 3′ and reverse 5′ AGGCATAACTGCACCCTTGG 3′.
Mutations in exon 4 of IDH1 were determined using direct sequencing protocol. Primer sequences used were forward 5′ AATGAGCTCTATATGCCATCACTG 3′ and reverse 5′ TTCATACCTTGCTTAATGGGTGT 3′. PCR amplification was performed in a total of 10 µL of reaction mixture containing 50 ng of tumor DNA, 1 µL of 10× PCR buffer, 0.8 µL of 10 mM dNTPs, 0.25 µL of each forward and reverse primers, and 0.2 µL of AmpliTaq Gold PCR Master Mix (Applied Biosystems). The reaction mixture was subjected to an initial denaturation of 95°C for 5 min, followed by 35 cycles of amplification consisting of denaturation at 95°C for 1 min, annealing at 57°C for 45 s, and extension 72°C for 2 min. Bidirectional sequencing was performed using ABI 3730 sequencer (Applied Biosystems).
For FFPE Samples
In 8 cases in which only formalin fixed paraffin embedded (FFPE) blocks were available, DNA was isolated as described earlier for MGMT gene promoter methylation assay. TP53 and IDH1 sequencing on FFPE DNA was performed using comparatively smaller product length.
TP53 sequencing was performed as described by IARC p53 database.32 The primers used were exon-5: forward 5′ TTCAACTCTGTCTCCTTCCT 3′ and reverse 5′ CAGCCCTGTCGTCTCTCCAG 3′; exon-6: forward 5′ GCCTCTGATTCCTCACTGAT 3′, and reverse 5′ TTAACCCCTCCTCCCAGAGA 3′; exon-7: forward 5′ AGGCGCACTGGCCTCATCTT 3′ and reverse 5′ TGTGCAGGGTGGCAAGTGGC 3′; and exon-8: forward 5′ TTCCTTACTGCCTCTTGCTT 3′ and reverse 5′ AGGCATAACTGCACCCTTGG 3′.
For IDH1 mutational analysis, a fragment of 129 base pairs corresponding to exon 4 of IDH1 was amplified using the following primer sequence: forward 5′ CGGTCTTCAGAGAAGCCATT 3′ and reverse 5′ GCAAAATCACATTATTGCCAAC 3′. The composition of the 10× PCR buffer used was 100 mM Tris-HCl (pH, 8.3), 15 mM MgCl2, and 500 mM KCl. PCR was performed in 50-μL volume containing 50 ng of genomic DNA, 1× PCR buffer, 250 μM dNTPs (NEB), 100 nM of each primer (Sigma), and 1.5 U of Taq DNA polymerase (NEB). Reactions were performed in a thermal cycler (MyCycler; Biorad) as follows: 95°C for 10 min, 37 cycles (95°C for 30 s, 56°C for 30 s, and 72°C for 30 s), and 72°C for 10 min. Sequencing was performed using the forward primer used for PCR on ABI 3730 sequencer (Applied Biosystems).
Results
A summary of the clinical features and results of each of the 14 cases (7 pediatric and 7 young adults) is shown in Table 1. The age of the patients ranged from 3 to 25 years, with a mean of 16.4 years. There were 9 male and 5 female patients. On histopathological analysis, 7 of the 14 cases were diagnosed as OG, World Health Organization grade II, whereas the remaining 7 were classified as anaplastic OG, World Health Organization grade III. All the cases showed classical histological features, similar to those of adults. These included cells with round to oval nuclei, perinuclear halos, fine branching capillary network, and presence of microgemistocytes. Anaplastic OGs showed prominent mitotic activity and focal endothelial cell proliferation. Necrosis was seen in 2 cases.
Table 1.
Details of the oligodendrogliomas (OGs) in pediatric and young adult patients
| Case no. | Diagnosis | Age/Sex | 1p deletion (FISH) | 19q deletion (FISH) | MGMT methylation status (PCR) | TP53 mutation | IDH1 mutation |
|---|---|---|---|---|---|---|---|
| 1 | OG II | 4/M | − | − | Methylated | − | − |
| 2 | OG II | 15/F | − | − | Methylated | − | − |
| 3 | OG III | 18/F | − | − | Methylated | − | − |
| 4 | OG III | 20/F | − | − | Unmethylated | − | − |
| 5 | OG II | 20/M | + | + | Methylated | − | − |
| 6 | OG II | 19/M | + | + | Methylated | − | − |
| 7 | OG III | 9/M | − | − | Methylated | − | − |
| 8 | OG III | 11/M | − | − | Unmethylated | − | − |
| 9 | OG III | 18/M | − | − | Methylated | − | − |
| 10 | OG II | 3/M | − | − | Unmethylated | − | − |
| 11 | OG III | 25/F | + | + | Methylated | − | − |
| 12 | OG III | 23/M | + | − | Methylated | − | − |
| 13 | OG II | 25/M | + | + | Methylated | − | − |
| 14 | OG II | 20/F | − | − | Unmethylated | − | − |
Abbreviations: FISH, fluorescence in situ hybridization; IDH1, isocitrate dihydrogenase 1; PCR, polymerase chain reaction.
1p/19q Status
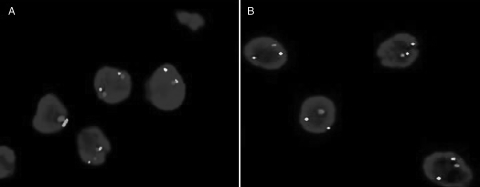
None of the pediatric cases showed 1p and/or 19q deletions. Of interest, combined 1p/19q loss (4 [57%] of 7 cases) was observed in majority of young adults. Isolated 1p deletion was noted in 1 case (14%) (Fig 1B).
Fig. 1.
Hetrozygosity status of 1p chromosome, as determined by FISH). (A) A case showing 2 red (test: 1p36) and 2 green (control: 1q 25) signals, implying no 1p deletion. (B) A case showing 1 red (test) and 2 green (control) signals, implying 1p deletion.
IDH1 Mutation
None of the cases in either of the subgroups showed this alteration.
TP53 Mutation
TP53 mutation was also not seen in any of the 14 cases analyzed.
MGMT Promoter Methylation
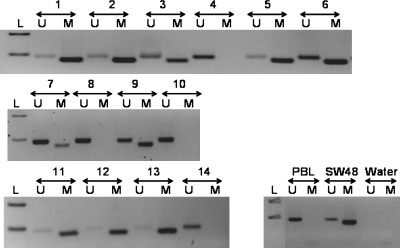
The majority of cases in both subgroups (5 [71%] of 7 in each) showed methylation of MGMT promoter (Fig 2).
Fig. 2.
Methylation status of the MGMT promoter, as determined by methylation-specific PCR assay. DNA from normal peripheral blood lymphocytes (PBL) was used as a control for the unmethylated MGMT promoter (U), enzymatically methylated lymphocytic DNA (SW48) served as a positive control for the methylated MGMT promoter (M), and water was used as a negative control for the PCR. A 100-bp marker ladder (L) was loaded to estimate molecular size. M: PCR product amplified by methylation-specific primers; U: PCR product amplified by unmethylated-specific primers; L: ladder; SW48: methylated control DNA; PBL: unmethylated control DNA. Samples 1, 2, 3, 5, 6, 7, 9, 11, 12, and 13 were methylated with presence of band in methylated “M” lane. Band in “U” lane is because of lymphocytes and normal tissue present with tumor tissue. Cases 4, 8, 10, and 14 were unmethylated with no band in “M” lane. In control samples, PBL had a band only in “U” lane, whereas SW48 had a band only in “M” lane.
Discussion
The pathological evaluation and subtyping of gliomas is sometimes very challenging. Although morphologically classical OGs are easy to diagnose, there are a small fraction of grade II and III diffuse gliomas that show ambiguous histology.33,34 In addition, dysembryoplastic neuroepithelial tumor, extraventricular neurocytoma, clear cell ependymoma, and pilocytic astrocytomas with focal oligodendroglial differentiation can sometimes pose diagnostic dilemma.33
In this regard, assessment of 1p/19q has been shown to represent a potentially useful marker in establishing diagnosis in small biopsy specimens or diagnostically challenging cases.35 Assessment for 1p/19q status has objectivized the diagnosis of oligodendroglial tumors in adults.34 Furthermore, association of this molecular signature with better prognosis and chemosensitivity in adult OGs has generated a lot of interest in use of this marker in routine neuropathology practice.33,34
There are only a few isolated studies on 1p/19q status in oligodendroglial tumors in young patients (Table 2). The largest series is by Raghavan et al on 23 pediatric cases. The authors showed combined 1p/19q deletion in 12% and isolated 1p deletion in 7.7% of cases.29 In another series of 13 cases, only 1 case showed isolated 1p loss.30 In our study, none of the cases in the pediatric age group showed 1p/19q loss. However, a significant number of young adults (4 [57%] of 7) showed combined 1p and 19q deletion. In a previous study, we observed a frequency of 63% in adult patients with OG.3
Table 2.
Comparison of results of 1p/19q chromosomal status in oligodendrogliomas (OGs) involving pediatric and young adult patients with the available literature.
| Reference, year [No.] | No. of cases | Age range (years) | Age <10 years | Isolated 1p deletion | Isolated 19q deletion | Combined 1p/19q deletion | Method used |
|---|---|---|---|---|---|---|---|
| Raghavan et al, 2003 [29]** | 26 | 2–18 | 15 | 2/26 (7.7%) | 0/25 | 3/25 (12%) | FISH |
| Myal et al, 2003 [36]** | 3 | 2–17 | 1 | None | None | None | PCR |
| Kreiger et al, 2005 [30]** | 13 | 0–19 | 4 | 1/13 (7.7%) | None | None | FISH |
| Hergersberg et al, 2006 [37] | 4* | <25 | − | − | − | 1/4 (25%)* | PCR |
| Hyder et al, 2007 [38]** | 7 | 0.3–16.8 | 5 | 1/7 (14.3%) | 1/7 (14.3%) | None | PCR |
| 0ur series | 14 | 3–25 | 3 | 1/14 (7.1%)*** | None | 4/14 (28.6%)*** | FISH |
| TOTAL | 67 | − | 28 | 5/67 (7.5%) | 1/66 (1.5%) | 8/66 (12.1%) | − |
Abbreviations: FISH, fluorescence in situ hybridization; PCR, polymerase chain reaction.
*Derived from figure in the paper. **No case with 1p 19q deletion in first decade. ***All the cases with 1p &/or 19q loss were young adults.
Raghavan et al raised a possibility that a high-definition technique, such as loss of heterozygosity analysis, should be performed in pediatric tumors to rule out small interstitial losses that may not be apparent by FISH assay.29 However, 3 studies in which PCR assay was used found that the frequency of this alteration is equally rare even with this technique (Table 2).36–38
After compiling data from all the series, a total of 67 pediatric and young patients with oligodendroglial tumors were assessed for 1p/19q status. Combined 1p/19q loss was observed in 12.1%, isolated 1p loss in 7.5%, and isolated 19q loss in 1.5%. There were 28 cases in the first decade of life, and none of these had this alteration (Table 2). Although the number of cases analyzed is very small, these observations suggest that this molecular alteration does not occur in the very young patients (first decade) and is infrequent until the age of 18.
MGMT hypermethylation is another molecular alteration that has been established as an independent predictor of response to chemotherapy with alkylating agents in malignant astrocytomas.7,8,39 Few recent studies have documented MGMT promoter methylation in 60%–90% of adult OGs.9–11,40 We have also observed a frequency of 84% in adult OGs.41
A significant positive correlation of this alteration with 1p and 19q loss has also been reported.9–11 Some authors have suggested that MGMT promoter methylation is associated with frequent sensitivity of oligodendroglial tumors to alkylating agents and better prognosis.9,11,40 In the Neuro-Oncology Working Group-04 trial, on anaplastic gliomas, hypermethylation of MGMT promoter was associated with prolonged PFS in both radiotherapy and chemotherapy arms.42
In the present series, the majority of pediatric and young adult patients (71% each) showed MGMT promoter methylation. There is no previous study on methylation status of pediatric OGs. In a study by Möllemann et al on assessment of hypermethylation of MGMT gene in oligodendroglial tumors, there were 3 pediatric cases, 2 of which showed promoter methylation.9 Although the frequency of this alteration in pediatric cases seems to be similar to that reported in adults,9,12 rarity of 1p/19q loss observed in pediatric population suggests that events involving 1p/19q loss and MGMT promoter methylation are independent.
In a genome-wide sequencing analysis by Parsons et al in 2008, somatic mutations in the gene encoding IDH were identified in 12% of GBMs at position 395 (amino acid 132) of the IDH1 transcript, and most were a G-A change with amino acid substitution of arginine to histidine.13 This revolutionary discovery led to an increase of studies on various aspects of IDH1 mutation in gliomas of all types and grades. These studies have documented a very high frequency of IDH1 mutations in grade II and grade III gliomas (astrocytic, oligodendroglial, and oligoastrocytic) and secondary GBMs.16–18,43
A positive correlation of IDH mutation has been noted with p53 mutation in astrocytic tumors and 1p/19q loss in oligo/oligoastrocytic tumors.17–19 The presence of mutations was shown in most of the studies to be associated with younger age of the patients and longer survival.13,20 Van den Bent et al determined the effect of IDH1 mutations on PFS and OS and its correlation with other clinical and molecular features in the prospective randomized European Organisation for Research and Treatment of Cancer study 26951 on adjuvant procarbazine, lomustine, and vincristine (PCV) therapy in anaplastic OGs.22 The presence of IDH1 mutation was found to carry a very strong prognostic significance for PFS and OS, but no evidence was found of a predictive significance for outcome to PCV chemotherapy. A few studies have also shown mutations of enzyme isoform 2 (IDH2) in a small fraction of gliomas.17,22,43
In the present study, none of the pediatric or young adult cases showed IDH1 mutation. In a recent study on GBMs, we did not find IDH1 mutation in any of the 15 pediatric cases. Only 1 (age, 28 years) of 4 patients in the age group of 19–30 years showed this alteration.44 Even Yan et al in 2009 (15 patients with GBM aged <21 years) and Antonelli et al in 2010 (27 pediatric high-grade gliomas) did not find IDH1 mutation in any of the cases.17,45 Thus, it appears that IDH1 mutation is conspicuously absent in pediatric gliomas.
In contrast, in a recent study of 100 gliomas, we observed IDH1 mutation in 18 (66.7%) of 27 adult cases with oligodendroglial and mixed oligoastrocytic phenotype.46
Two recent studies have suggested that IDH1 mutations are very early events in gliomagenesis, which occur before p53 mutations or 1p/19q loss, and may affect a common stem cell population that can cause both oligodendrocytes and astrocytes.17,47 On the contrary, presence of 1p/19q deletion along with complete absence of IDH1 mutation in young adults observed in the present study implies that the sequence of molecular events in this age group may not be similar to that in their adult counterparts.
TP53 gene mutation and p53 protein expression is more common in astrocytic tumors than in oligodendroglial tumors.3,23,24 Various studies have shown an inverse correlation between 1p/19q deletion and TP53 gene mutation.25,26
In the present series, none of the pediatric or young adult cases showed p53 mutation, suggesting that this alteration is extremely rare even in these age groups.
To conclude, molecular pathogenesis of OGs arising in pediatric and young adult patients appears to be noticeably distinct from that in adults. The 1p/19q deletion, the molecular signature of adult OGs, is extremely rare in pediatric patients and virtually absent in the first decade. However, the majority of young adults show this alteration. The IDH1 mutation, which occurs at very high frequency in adult OGs, seems to be virtually absent in patients <25 years of age. High frequency of MGMT promoter methylation and absence of TP53 mutation are the only alterations shared by OGs of all age groups. Thus, the results of studies on prognostication and clinical management in adult OGs cannot be translated to pediatric cases. Multicentric studies with genome-wide expression profiling and therapeutic response monitoring should be performed to delineate the unique molecular characteristics of OGs arising in young patients.
Funding
This work was supported by the Indian Council of Medical Research, Neuro Sciences Centre, and the Department of Pathology.
Acknowledgments
The authors thank the Indian Council of Medical Research, Neuro Sciences Centre, and the Department of Pathology for funding; Sandor Proteomics, Hyderabad, and M. Kiran Kumar, for help in IDH1 mutational analysis; all consultants from the Department of Neurosurgery, All India Institute of Medical Sciences (AIIMS); and all technical staff from the neuropathology laboratory, AIIMS.
Conflict of interest statement. None declared.
References
- 1.Smith JS, Alderete B, Minn Y, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18:4144–4152. doi: 10.1038/sj.onc.1202759. doi:10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 2.Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am. J. Pathol. 1994;145:1175–1190. [PMC free article] [PubMed] [Google Scholar]
- 3.Shukla B, Agarwal S, Suri V, et al. Assessment of 1p/19q status by fluorescence in situ hybridization assay: A comparative study in oligodendroglial, mixed oligoastrocytic and astrocytic tumors. Neurol. India. 2009;57:559–566. doi: 10.4103/0028-3886.57795. doi:10.4103/0028-3886.57795. [DOI] [PubMed] [Google Scholar]
- 4.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl. Cancer. Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. doi:10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 5.Ino Y, Betensky RA, Zlatescu MC, et al. Molecular subtypes of anaplastic oligodendroglioma: implications for patient management at diagnosis. Clin. Cancer Res. 2001;7:839–845. [PubMed] [Google Scholar]
- 6.Smith JS, Perry A, Borell TJ, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J. Clin. Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 7.Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. doi:10.1158/1078-0432.CCR-03-0384. [DOI] [PubMed] [Google Scholar]
- 8.Kamiryo T, Tada K, Shiraishi S, Shinojima N, Kochi M, Ushio Y. Correlation between promoter hypermethylation of the O6-methylguanine-deoxyribonucleic acid methyltransferase gene and prognosis in patients with high-grade astrocytic tumors treated with surgery, radiotherapy, and 1-(4-amino-2-methyl-5-pyrimidinyl)methyl-3-(2-chloroethyl)-3-nitrosourea-based chemotherapy. Neurosurgery. 2004;54:349–357. doi: 10.1227/01.neu.0000103422.51382.99. doi:10.1227/01.NEU.0000103422.51382.99. [DOI] [PubMed] [Google Scholar]
- 9.Möllemann M, Wolter M, Felsberg J, Collins VP, Reifenberger G. Frequent promoter hypermethylation and low expression of the MGMT gene in oligodendroglial tumors. Int. J. Cancer. 2005;113:379–385. doi: 10.1002/ijc.20575. doi:10.1002/ijc.20575. [DOI] [PubMed] [Google Scholar]
- 10.Dong SM, Pang JC, Poon WS, et al. Concurrent hypermethylation of multiple genes is associated with grade of oligodendroglial tumors. J Neuropathol. Exp. Neurol. 2001;60:808–816. doi: 10.1093/jnen/60.8.808. [DOI] [PubMed] [Google Scholar]
- 11.Brandes AA, Tosoni A, Cavallo G, et al. ICNO. Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: a prospective GICNO study. J. Clin. Oncol. 2006;24:4746–4753. doi: 10.1200/JCO.2006.06.3891. doi:10.1200/JCO.2006.06.3891. [DOI] [PubMed] [Google Scholar]
- 12.Ishii D, Natsume A, Wakabayashi T, et al. Efficacy of temozolomide is correlated with 1p loss and methylation of the deoxyribonucleic acid repair gene MGMT in malignant gliomas. Neurol. Med. Chir. (Tokyo) 2007;47:341–349. doi: 10.2176/nmc.47.341. discussion 350 doi:10.2176/nmc.47.341. [DOI] [PubMed] [Google Scholar]
- 13.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. doi:10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age:a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. doi:10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 15.Kloosterhof NK, Bralten LB, Dubbink HJ, French PJ, van den Bent MJ. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol. 2010;12(1):83–91. doi: 10.1016/S1470-2045(10)70053-X. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Sonoda Y, Kumabe T, Nakamura T, et al. Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci. 2009;100:1996–1998. doi: 10.1111/j.1349-7006.2009.01270.x. doi:10.1111/j.1349-7006.2009.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. doi:10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichimura K, Pearson DM, Kocialkowski S, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. doi:10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gravendeel LA, Kloosterhof NK, Bralten LB, et al. Segregation of non-p.R132H mutations in IDH1 in distinct molecular subtypes of glioma. Hum. Mutat. 2010;31:E1186–199. doi: 10.1002/humu.21201. doi:10.1002/humu.21201. [DOI] [PubMed] [Google Scholar]
- 20.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin. Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. doi:10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 21.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J. Clin. Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. doi:10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 22.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin. Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. doi:10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 23.von Deimling A, Eibl RH, Ohgaki H, et al. p53 mutations are associated with 17p allelic loss in grade II and grade III astrocytoma. Cancer Res. 1992;52:2987–2990. [PubMed] [Google Scholar]
- 24.Hagel C, Lacking G, Laas R, et al. Demonstration of p53 protein and TP53 gene mutation in oligodendrogliomas. Eur. J. Cancer. 1996;32:2242–2248. doi: 10.1016/s0959-8049(96)00259-6. doi:10.1016/S0959-8049(96)00259-6. [DOI] [PubMed] [Google Scholar]
- 25.Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. J. Neuropathol Exp. Neurol. 2003;62:111–126. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- 26.Jeon YK, Park K, Park CK, Paek SH, Jung HW, Park S. Chromosome 1p and 19q status and p53 and p16 expression patterns as prognostic indicators of oligodendroglial tumors: A clinicopathological study using fluorescence in situ hybridization. Neuropathol. 2007;27:10–20. doi: 10.1111/j.1440-1789.2006.00735.x. doi:10.1111/j.1440-1789.2006.00735.x. [DOI] [PubMed] [Google Scholar]
- 27.Dohrmann GJ, Farwell JR, Flannery JT. Oligodendrogliomas in children. Surg. Neurol. 1978;10:21–25. [PubMed] [Google Scholar]
- 28.Razack N, Baumgartner J, Bruner J. Pediatric oligodendrogliomas. Pediatr. Neurosurg. 1998;28:121–129. doi: 10.1159/000028635. doi:10.1159/000028635. [DOI] [PubMed] [Google Scholar]
- 29.Raghavan R, Balani J, Perry A, et al. Pediatric oligodendrogliomas: a study of molecular alterations on 1p and 19q using fluorescence in situ hybridization. J. Neuropathol. Exp. Neurol. 2003;62:530–537. doi: 10.1093/jnen/62.5.530. [DOI] [PubMed] [Google Scholar]
- 30.Kreiger PA, Okada Y, Simon S, Rorke LB, Louis DN, Golden JA. Losses of chromosomes 1p and 19q are rare in pediatric oligodendrogliomas. Acta Neuropathol. 2005;109:387–392. doi: 10.1007/s00401-004-0976-2. doi:10.1007/s00401-004-0976-2. [DOI] [PubMed] [Google Scholar]
- 31.Kleihues P, Louis DN, Wiestler OD, Burger PC, Scheithauer BW. WHO grading of tumours of the central nervous system. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO Classification of Tumours of the Central Nervous System. 4th ed. Lyon: IARC; 2007. pp. 10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. doi:10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 33.Aldape K, Burger PC, Perry A. Clinicopathologic aspects of 1p/19q loss and the diagnosis of oligodendroglioma. Arch. Pathol. Lab. Med. 2007;131:242–251. doi: 10.5858/2007-131-242-CAOQLA. Review. [DOI] [PubMed] [Google Scholar]
- 34.Korshunov A, Sycheva R, Golanov A. Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells: the utility of fluorescence in situ hybridization. Clin. Cancer Res. 2004;10:7820–7826. doi: 10.1158/1078-0432.CCR-04-0710. doi:10.1158/1078-0432.CCR-04-0710. [DOI] [PubMed] [Google Scholar]
- 35.Fuller CE, Schmidt RE, Roth KA, et al. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/astrocytic features. J. Neuropathol Exp. Neurol. 2003;62:1118–1128. doi: 10.1093/jnen/62.11.1118. [DOI] [PubMed] [Google Scholar]
- 36.Myal Y, Del Bigio MR, Rhodes RH. Age-related differences in 1p and 19q deletions in oligodendrogliomas. BMC Clin. Pathol. 2003;3:6. doi: 10.1186/1472-6890-3-6. doi:10.1186/1472-6890-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hergersberg M, Mariani L, Vassella E, et al. Age at diagnosis and loss of heterozygosity on chromosome 1p and 19q in oligodendroglial tumors. J. Neurooncol. 2006;80:215–217. doi: 10.1007/s11060-006-9177-2. doi:10.1007/s11060-006-9177-2. [DOI] [PubMed] [Google Scholar]
- 38.Hyder DJ, Sung L, Pollack IF, et al. Anaplastic mixed gliomas and anaplastic oligodendroglioma in children: results from the CCG 945 experience. J. Neurooncol. 2007;83:1–8. doi: 10.1007/s11060-006-9299-6. doi:10.1007/s11060-006-9299-6. [DOI] [PubMed] [Google Scholar]
- 39.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. doi:10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 40.van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J. Clin. Oncol. 2009;27:5881–5886. doi: 10.1200/JCO.2009.24.1034. doi:10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jha P, Suri V, Jain A, et al. O6-methylguanine DNA methyltransferase gene promoter methylation status in gliomas and its correlation with other molecular alterations: first Indian report with review of challenges for use in customized treatment. Neurosurgery. 2010;67(6):1681–1691. doi: 10.1227/NEU.0b013e3181f743f5. Review doi:10.1227/NEU.0b013e3181f743f5. [DOI] [PubMed] [Google Scholar]
- 42.Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J. Clin. Oncol. 2009;27:5874–5880. doi: 10.1200/JCO.2009.23.6497. doi:10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 43.Felsberg J, Wolter M, Seul H, et al. Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol. 2010;119:501–507. doi: 10.1007/s00401-010-0647-4. doi:10.1007/s00401-010-0647-4. [DOI] [PubMed] [Google Scholar]
- 44.Jha P, Suri V, Singh G, et al. Characterization of molecular genetic alterations in GBMs highlights a distinctive molecular profile in young adults. Diagn. Mol. Pathol. doi: 10.1097/PDM.0b013e31821c30bc. In press. [DOI] [PubMed] [Google Scholar]
- 45.Antonelli M, Buttarelli FR, Arcella A, et al. Prognostic significance of histological grading, p53 status, YKL-40 expression, and IDH1 mutations in pediatric high-grade gliomas. J Neurooncol. 2010;99:209–215. doi: 10.1007/s11060-010-0129-5. doi:10.1007/s11060-010-0129-5. [DOI] [PubMed] [Google Scholar]
- 46.Jha P, Suri V, Sharma V, et al. IDH1 mutations in gliomas: first series from a tertiary care centre in India with comprehensive review of literature. Exp. Mol. pathol. 2011;91:385–393. doi: 10.1016/j.yexmp.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. doi:10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]