Abstract
Background and Aims
The genus Nicotiana includes diploid and tetraploid species, with complementary ecological, agronomic and commercial characteristics. The species are of economic value for tobacco, as ornamentals, and for secondary plant-product biosynthesis. They show substantial differences in disease resistance because of their range of secondary products. In the last decade, sexual hybridization and transgenic technologies have tended to eclipse protoplast fusion for gene transfer. Somatic hybridization was exploited in the present investigation to generate a new hybrid combination involving two sexually incompatible tetraploid species. The somatic hybrid plants were characterized using molecular, molecular cytogenetic and phenotypic approaches.
Methods
Mesophyll protoplasts of the wild fungus-resistant species N. debneyi (2n = 4x = 48) were electrofused with those of the ornamental interspecific sexual hybrid N. × sanderae (2n = 2x = 18). From 1570 protoplast-derived cell colonies selected manually in five experiments, 580 tissues were sub-cultured to shoot regeneration medium. Regenerated plants were transferred to the glasshouse and screened for their morphology, chromosomal composition and disease resistance.
Key Results
Eighty-nine regenerated plants flowered; five were confirmed as somatic hybrids by their intermediate morphology compared with parental plants, cytological constitution and DNA-marker analysis. Somatic hybrid plants had chromosome complements of 60 or 62. Chromosomes were identified to parental genomes by genomic in situ hybridization and included all 18 chromosomes from N. × sanderae, and 42 or 44 chromosomes from N. debneyi. Four or six chromosomes of one ancestral genome of N. debneyi were eliminated during culture of electrofusion-treated protoplasts and plant regeneration. Both chloroplasts and mitochondria of the somatic hybrid plants were probably derived from N. debneyi. All somatic hybrid plants were fertile. In contrast to parental plants of N. × sanderae, the seed progeny of somatic hybrid plants were resistant to infection by Peronospora tabacina, a trait introgressed from the wild parent, N. debneyi.
Conclusions
Sexual incompatibility between N. × sanderae and N. debneyi was circumvented by somatic hybridization involving protoplast fusion. Asymmetrical nuclear hybridity was seen in the hybrids with loss of chromosomes, although importantly, somatic hybrids were fertile and stable. Expression of fungal resistance makes these somatic hybrids extremely valuable germplasm in future breeding programmes in ornamental tobacco.
Keywords: Chloroplast DNA (cpDNA), protoplasts, electrofusion, fungal resistance, genomic in situ hybridization (GISH), mitochondrial DNA (mtDNA), Nicotiana debneyi, N. × sanderae, Peronospora tabacina, random amplified polymorphic DNA (RAPD), somatic hybridization
INTRODUCTION
As in other agricultural species, trait introgression from ‘wild’ species of the genus Nicotiana has been used to improve the cropped species, and characters from at least 13 different species have been transferred into tobacco (Lewis, 2011). As well as smoking tobacco from N. tabacum, several species in the genus are popular ornamental plants. The high concentrations of nicotine, a poison of acetocholine receptors that is toxic to all species with neuromuscular junctions (Baldwin, 2001), provide excellent resistance to herbivorous pests. However, many Nicotiana species are susceptible to fungal and virus diseases as seedlings or adult plants. Economic and environmental considerations require reductions in the use of agrochemicals, increasing interest in exploiting genetic resistance in both field and horticultural crops.
The genus Nicotiana is complex with some 75 species currently recognized (Chase et al., 2003; Kapp et al., 2004). Of these, 35 are allotetraploids (Clarkson et al., 2010). Kovarik et al. (2008) and Lim et al. (2007) investigated the repetitive DNA (rDNA) evolution and chromosome constitution of various hybrid species using genomic in situ hybridization (GISH). These results showed that not only are rDNA gene arrays silenced and lost in the hybrids, but that there is homogenization of rDNA between ancestral genomes, which can be identified by the increasing similarity of the GISH signal with the ancestral genomes used as a probe as age of the polyploids increases. Anssour et al. (2009) examined both allo- and auto-tetraploid Nicotiana species, and showed that there were substantial changes both in morphology and in the genome which are found in the hybrid species compared with their ancestral diploids. Some of these changes have adaptive significance.
Nicotiana × sanderae (section: Alatae), a sexual hybrid between N. alata and N. forgetiana [both 2n = 2x = 18; with the internal transcriber spacer (ITS) of the nuclear rDNA matching the maternal parent, N. alata (Chase et al., 2003)], is cultivated for its attractive flowers that range from white, through green to pink and red, lines of uniform colour being selected by breeders for commercial production. Pigmentation in N. sanderae is derived from N. forgetiana. Nicotiana × sanderae and its parental species are not resistant to blue mould (blue mold), caused by Peronospora tabacina (Clayton, 1945), a fungal disease that devastates ornamental tobaccos in bedding schemes and under glass. This disease is now prevalent in many parts of the world; plants readily succumb to infection and are lost from floral displays. However, Nicotiana germplasm includes species with agronomically important traits, such as tolerance to abiotic stress, including salt and drought (Komari et al., 2000) and resistance to P. tabacina. Thus, N. debneyi Domin (section Suaveolentes), a wild Australian species indigenous to Rockingham Bay in Queensland, was identified as a potential source of resistance to P. tabacina (Wark, 1970; Milla et al., 2005) for transfer to recipient species. Nicotiana debneyi is an allotetraploid (2n = 4x = 48, XXYY genome designation) that has two genomes distinct from those of tobacco (2n = 4x = 48, SSTT) (Goodspeed, 1954; Bai et al., 1995). The Germplasm Resources Information Network (GRIN) database (USDA, 2010) recognizes the species as a gene source for disease resistance and a tertiary genetic relative of tobacco. Resistance to black root rot (Chalara elegans), a common fungal disease of Canadian tobacco, has been transferred from N. debneyi to tobacco by sexual hybridization and backcrossing (Bai et al., 1995, 1996); resistance from this species is likely to be present in some Canadian flue-cured tobacco lines and US burley tobacco lines (R. Lewis, North Carolina State University, Raleigh, NC, USA, pers. comm.). Despite contrary evidence from pedigrees, marker genotyping indicates that blue mould resistance in various commercial tobacco lines has originated from N. debneyi (Milla et al., 2005).
Many horticultural crops do not have the resistances to biotic and abiotic stresses that are present in their wild relatives, the pool of uncultivated germplasm. Sexual hybridization is often impossible because of pre- or post-fertilization barriers (Tezuka et al., 2010). The genetic basis of the resistance and the genes involved are unknown, preventing any transgenic approach. Sexual incompatibility limits the genetic combinations achievable by conventional hybridization. Somatic hybridization, involving protoplast fusion, can be exploited to circumvent sexual incompatibility at both the pre- and post-zygotic stages of development, providing an approach to exploit naturally occurring biodiversity. Additionally, the extensive nuclear and cytoplasmic combinations resulting from protoplast fusion, with potential incorporation of plastids and mitochondria from both parents, generate novel germplasm for incorporation into breeding programmes (Horsman et al., 2001; Davey et al., 2005; Pati et al., 2005). Somatic hybrids are rare that are symmetric at the nuclear level with the complete nuclear complements of both parents, since culture of fusion-treated protoplasts and shoot regeneration from heterokaryon-derived tissues is often accompanied by the loss of chromosomes of one or both fusion partners to produce asymmetric nuclear hybrids (Liu et al., 2005).
Since the pioneering publication describing the first somatic hybrid plants between N. tabacum and N. glauca, which was also the first proof-of-principle of somatic hybridization (Carlson et al., 1972), several somatic hybrid and cybrid plants have been reported in the genus Nicotiana, most of which are cited by the authors listed here in more recent publications (IIcheva et al., 2000). Fitter et al. (2005) fused protoplasts of N. tabacum with those of N. suaveolens to generate three male sterile cybrid plants with recombinant mitochondrial DNA, the DNA sequences correlating with floral morphology. The flowers resembled those of N. tabacum. Chloroplasts were either of the N. tabacum or the N. suaveolans type. Sun et al. (2005) also fused mesophyll protoplasts of N. tabacum with those of N. repanda, generating cybrids with the nuclei of N. tabacum, but mitochondria from N. repanda. These experiments, like those of Fitter et al. (2005), demonstrated the possibility of introgressing cytoplasmic male sterility, carried by mitochondrial DNA, from a wild species into the cultivated crop. The same research group also generated a novel fertile symmetric somatic hybrid between N. tabacum and N. glauca with, unusually, a chromosome number equal to the summation of the complements of the parents. Morphologically, the flowers of the somatic hybrid were similar to those of N. tabacum, but petals were comparable to those of N. glauca (Sun et al., 2007). In other investigations, Sytnik et al. (2005) exploited somatic hybridization to transfer transformed chloroplasts from a transplastomic plant of N. tabacum into Lycium barbarum, demonstrating that organelles can be transferred to remote species by protoplast fusion.
Although N. debneyi can be crossed with N. tabacum, the species is sexually incompatible with N. × sanderae making it impossible to transfer disease resistance into the ornamental N. × sanderae by conventional breeding. As in several other Nicotiana species, plant regeneration from protoplasts has been achieved in N. debneyi (Scowcroft and Larkin, 1980; Kumashiro and Kubo, 1986). The present investigation aimed to generate somatic hybrid plants by protoplast fusion between N. × sanderae and N. debneyi, circumventing sexual incompatibility, and characterizing somatic hybrid plants at the morphological and molecular levels. This study also aimed to examine the fertility of the somatic hybrids, to confirm their chromosomal compositions, which included four genomes from the ancestors present in the hybrid originating from the two tetraploids, N. × sanderae and N. debneyi, and to evaluate seed progeny for their resistance to P. tabacina.
MATERIALS AND METHODS
Glasshouse-grown plants and sexual hybridization
Seeds of Nicotiana × sanderae Hort. ex W.Watson ‘Avalon Red Improved’ (2n = 2x = 18) and N. debneyi (2n = 4x = 48) were supplied by Floranova, Foxley, Dereham, UK, sown on Levington M3 soil-less compost (Scotts Ltd, Ipswich, UK) and maintained to flowering under glasshouse conditions. Natural daylight in the glasshouse was supplemented with 16 h of fluorescent illumination (195 µmol m−2 s−1; TLD/58 W 35 V ‘Daylight’ fluorescent tubes; Phillips, Croydon, UK) with day and night temperatures of 25 ± 1 °C.
Three plants of each parent for sexual hybridization were grown in isolation. Buds of recipient plants were opened before anthesis and emasculated. Pollen from dehisced anthers was applied manually to receptive stigmas; reciprocal crosses were performed. Two plants of each parent were also self-pollinated. All pollinated flowers were labelled.
Axenic shoot cultures
Seeds were immersed in 10 % (v/v) ‘Domestos’ bleach solution (John Diversey Lever, Northampton, UK) for 5–8 min, followed by three washes in autoclaved reverse osmosis water. Plants were established in vitro by germinating seeds on Murashige and Skoog (MS)-based culture medium (Murashige and Skoog, 1962) with 30 g L−1 sucrose, but lacking growth regulators, and semi-solidified with 8·0 g L−1 agar (Type IV; Sigma-Aldrich). Culture medium was contained in 9-cm-diameter Petri dishes (Bibby Sterilin, Stone, UK) under a 16-h photoperiod (90 µmol m−2 s−1; ‘Daylight’ fluorescent illumination) at 25 ± 2 °C. Seedlings were transferred to clip-lid axenic plastic vessels (Ashwood, London, UK; one seedling per vessel) each vessel containing 200 mL of the same MS-based medium without growth regulators, to establish axenic shoot cultures. The latter were subcultured every 28 d by excising proliferating shoot apices (each 2 cm in height) and inserting the apices into new medium of the same composition (five shoots per vessel).
Isolation of leaf protoplasts
Protoplasts were isolated from expanded leaves excised three to six nodes from the apex of axenic shoots. Excised leaves were cut transversely into strips (each 0·5–1·0 mm wide) after removing the main veins. Leaf tissues were plasmolysed by immersion in CPW salts solution (Frearson et al., 1973) containing 9 % (w/v) mannitol (CPW9M) for 1 h in 9-cm Petri dishes (1 g f.wt. tissues/10 mL solution). The plasmolysing solution was replaced with an enzyme mixture consisting of 0·5 % (w/v) Cellulase R-10 and 0·5 % Macerozyme R-10 for N. debneyi, and 1 % (w/v) Cellulase R-10 and 0·5 % Macerozyme R-10 for N. × sanderae (enzymes from Duchefa Biochimie B.V., BH-Haarlem, The Netherlands) and dissolved in CPW9M solution, pH 5·8. Incubation was in the dark at 25 ± 2 °C on a horizontal rotary shaker (40 rpm) for 12–16 h. Mesophyll protoplasts were filtered through a nylon mesh (64 µm pore size) and washed twice by gentle centrifugation (700 rpm for 5 min in 16-mL screw-capped tubes; MSE Centaur bench centrifuge) in CPW9M solution overlaying CPW solution containing 25 % (w/v) sucrose. Protoplasts were removed using a Pasteur pipette from the interface of the two solutions and were resuspended in EF9M electrofusion solution [9 % (w/v) mannitol, 0·5 mm CaCl2] at 1·0 × 105 protoplasts mL−1. Protoplast viability was determined by staining with fluorescein diacetate (Widholm, 1972).
Electrofusion of protoplasts
The electrofusion apparatus was described by Jones et al. (1994). Isolated protoplasts of the two parental species were mixed in equal numbers. Aliquots (1 mL) of the protoplast mixture were dispensed into the inner nine wells of a 5 × 5 square grid dish (Bibby Sterilin Ltd), and the protoplasts allowed to settle for 5–10 min. A multi-electrode block of seven brass plates, each separated by 2·7-mm Perspex spacers, was inserted sequentially into each well. Protoplasts were aligned into pearl chains in an AC field (70 V cm−1, 1·0 MHz, 4 s) which was increased to 400 V cm−1 (0·5 s). Protoplasts were fused by 2 DC pulses (each 890 V cm−1) of 1·5-ms and 500-μs duration, separated by a period of 1 s. Parental protoplasts were self-fused. Other parental protoplast preparations were not electrofused, but mixed or cultured separately as controls.
Protoplast culture and plant regeneration
Protoplasts were cultured in liquid MS-based medium containing 9 % (w/v) mannitol, 2·0 mg L−1 α-naphthaleneacetic acid (NAA) and 0·5 mg L−1 6-benzylaminopurine (benzyladenine; BAP) at a density of 1·0 × 105 protoplasts mL−1 (8 mL/9-cm Petri dish). After 21 d, the medium was replaced with the same MS-based medium, but with mannitol reduced to 4·5 % (w/v). Individual protoplast-derived cell colonies were selected randomly and transferred manually when 2–4 mm in size to 5·5-cm-diameter Petri dishes, the latter each containing 4 mL of MS-based medium with 1·0 mg L−1 BAP, 2·0 mg L−1 indole acetic acid (IAA), 4·5 % (w/v) mannitol and 0·8 % (w/v) agar (Type IV; Sigma-Aldrich), pH 5·8 (ten tissues/dish). Green, nodular tissues were sub-cultured when approx. 1 cm2 in size to the same medium, but lacking mannitol (5 tissues/9 cm Petri dish; 8 mL medium). Regenerated shoots were excised and transferred to semi-solid MS-based medium lacking growth regulators for root induction.
Regenerated, rooted plants (R0 generation) were potted in Levington M3 soil-less compost when 6–8 cm in height and maintained under high humidity for 10–14 d by covering the plants and pots with plastic bags. The bags were opened gradually before removal and exposing the plants to growth room conditions (25 ± 1 °C; ‘Daylight’ fluorescent illumination; 90 µmol m−2 s−1; 16-h photoperiod). Regenerated plants were grown to flowering; any plants that resembled N. debneyi were discarded.
Five putative somatic hybrid plants were retained, selfed manually with flowers from the same plant, and the flowers bagged after selfing. Seeds were collected from these five putative somatic hybrid plants, ten seeds were selected at random from each seed batch and germinated under glasshouse conditions to give R1 generation plants. R2 and R3 generation plants were established in the same way. Morphological characteristics of five R1, R2 and R3 generation protoplast-derived plants were recorded 75 d after seed sowing (Table 1). The same characteristics of five seed-derived plants of both N. × sanderae and N. debneyi were also recorded, but 90 d after seed sowing. Statistical analysis employed Minitab 15® software.
Table 1.
Characteristics of N. × sanderae, N. debneyi and their somatic hybrid plants
| Parental plants |
Somatic hybrids |
||||||
|---|---|---|---|---|---|---|---|
| Characters | N. × sanderae | N. debneyi | SH28 | SH31 | SH354 | SH411 | SH447 |
| Plant height (cm) | |||||||
| R1 | 58·2 ± 2·0 | 73·3 ± 1·7 | 63·0 ± 0·2 | 65·1 ± 0·3 | 68·8 ± 1·0 | 72·0 ± 0·3 | 70·3 ± 1·0 |
| R2 | 58·0 ± 6·1 | 108·3 ± 4·4 | 111·7 ± 4·4 | 125·0 ± 2·9 | 100·0 ± 5·8 | 96·7 ± 3·3 | 80·0 ± 4·0 |
| R3 | 61·7 ± 4·4 | 108·6 ± 4·7 | 110·0 ± 5·8 | 128·3 ± 1·7 | 108·7 ± 0·7 | 98·9 ± 2·8 | 81·7 ± 4·4 |
| Mean leaf index | |||||||
| R1 | 1·9 ± 0·1 | 2·0 ± 0·1 | 1·3 ± 0·1 | 1·9 ± 0·1 | 1·4 ± 0·1 | 1·3 ± 0·2 | 1·3 ± 0·1 |
| R2 | 1·6 ± 0·0 | 1·9 ± 0·1 | 1·3 ± 0·1 | 1·4 ± 0·0 | 1·7 ± 0·1 | 1·5 ± 0·0 | 1·6 ± 0·0 |
| R3 | 1·6 ± 0·0 | 1·8 ± 0·1 | 1·3 ± 0·0 | 1·4 ± 0·0 | 1·8 ± 0·1 | 1·5 ± 0·0 | 1·6 ± 0·0 |
| Internode length (cm) | |||||||
| R1 | 5·8 ± 1·0 | 9·4 ± 1·5 | 6·6 ± 1·1 | 7·4 ± 1·6 | 6·2 ± 1·1 | 9·0 ± 1·7 | 7·5 ± 1·5 |
| R2 | 6·7 ± 0·3 | 9·0 ± 0·6 | 11·0 ± 1·2 | 11·3 ± 0·7 | 9·0 ± 0·0 | 10·0 ± 0·6 | 10·3 ± 0·9 |
| R3 | 5·7 ± 0·3 | 8·7 ± 0·7 | 11·0 ± 0·6 | 11·3 ± 0·7 | 9·0 ± 0·6 | 9·3 ± 0·7 | 11·0 ± 0·0 |
| Flower length (cm) | |||||||
| R1 | 4·5 ± 0·0 | 1·5 ± 0·0 | 3·0 ± 0·1 | 3·0 ± 0·2 | 3·0 ± 0·2 | 3·0 ± 0·2 | 3·5 ± 0·0 |
| R2 | 4·5 ± 0·0 | 1·8 ± 0·0 | 3·5 ± 0·0 | 3·3 ± 0·0 | 3·3 ± 0·0 | 3·0 ± 0·0 | 3·5 ± 0·0 |
| R3 | 4·5 ± 0·0 | 1·8 ± 0·0 | 3·5 ± 0·0 | 3·4 ± 0·0 | 3·3 ± 0·0 | 3·0 ± 0·0 | 3·5 ± 0·0 |
| Flower width (cm) | |||||||
| R1 | 5·0 ± 0·0 | 1·2 ± 0·1 | 2·4 ± 0·0 | 2·5 ± 0·1 | 3·0 ± 0·0 | 3·0 ± 0·0 | 3·0 ± 0·1 |
| R2 | 5·0 ± 0·0 | 1·1 ± 0·1 | 2·6 ± 0·0 | 2·7 ± 0·1 | 2·5 ± 0·0 | 3·0 ± 0·0 | 3·0 ± 0·0 |
| R3 | 5·0 ± 0·0 | 1·2 ± 0·0 | 2·7 ± 0·0 | 2·5 ± 0·0 | 2·5 ± 0·0 | 3·0 ± 0·0 | 3·0 ± 0·0 |
| Pollen viability (%) | |||||||
| R1 | 78·2 ± 2·0 | 80·0 ± 0·0 | 82·0 ± 0·1 | 78·8 ± 1·5 | 86·9 ± 3·0 | 67·6 ± 1·8 | 80·7 ± 1·0 |
| R2 | 75·0 ± 2·9 | 82·3 ± 1·5 | 86·7 ± 1·7 | 77·3 ± 3·7 | 85·7 ± 2·6 | 62·7 ± 4·8 | 78·7 ± 2·0 |
| R3 | 73·0 ± 2·1 | 82·3 ± 1·9 | 84·3 ± 2·9 | 75·3 ± 2·6 | 82·3 ± 1·5 | 55·0 ± 2·9 | 77·3 ± 3·7 |
| Mean no. of seeds/pod | |||||||
| R1 | 502·4 ± 1·8 | 263·3 ± 1·3 | 82·3 ± 0·1 | 79·6 ± 5·6 | 83·7 ± 2·3 | 42·3 ± 0·3 | 92·6 ± 3·1 |
| R2 | 510·0 ± 5·6 | 220·0 ± 11·6 | 78·3 ± 1·7 | 75·7 ± 3·5 | 85·0 ± 2·9 | 45·0 ± 2·9 | 103·3 ± 3·3 |
| Seed germination (%) | |||||||
| R1 | 90·1 ± 0·1 | 99·4 ± 2·4 | 66·2 ± 1·8 | 40·4 ± 2·5 | 95·4 ± 2·0 | 82·0 ± 1·5 | 60·7 ± 2·2 |
| R2 | 81·7 ± 4·4 | 93·3 ± 3·3 | 56·7 ± 3·3 | 40·0 ± 5·6 | 86·7 ± 3·3 | 83·3 ± 3·3 | 60·0 ± 0·0 |
| R3 | 80·0 ± 5·6 | 90·0 ± 0·0 | 53·3 ± 3·3 | 43·3 ± 3·3 | 76·6 ± 3·2 | 76·6 ± 3·3 | 63·3 ± 2·3 |
| Flower colour R1, R2 and R3 | Dark red (61 A/B)* | White/cream (155 D)* | Red with faint white colour in centre (all somatic hybrid lines) (64 A/B)* | ||||
Values are mean ± s.e. of the mean. n = 5 throughout.
Mean leaf index = Ratio of leaf length to width.
* Flower colour assessed using the Royal Horticultural Society of London Colour Charts, 5th edn.
Cytology and genomic in situ hybridization
Root tips, each 0·5 cm in length, were excised from R0 generation protoplast-derived plants and from parental plants maintained in the growth room, pre-treated in 2 mm 8-hydroxyquinoline solution for 2–4 h in the dark, and fixed in 3 : 1 (v/v) ethanol : glacial acetic acid for 24 h. Preparations were hydrolysed in 1 m HCl (60 °C, 10 min), and stained with aqueous 1 % (w/v) aceto-orcein. Cells were examined by light microscopy for metaphase chromosomes. Ten preparations were examined per plant; chromosome numbers were recorded.
Root tips for in situ hybridization were excised from glasshouse-grown plants (R0 generation protoplast-derived), pre-treated with water-saturated α-bromonaphthalene for 24 h at 4 °C, fixed in freshly prepared 3 : 1 (v/v) ethanol : glacial acetic acid (24 h), and stored at 4 °C until use. Root tips were digested by incubating in a mixture of 2 % (w/v) Cellulase Onozuka RS and 2 % (w/v) Pectolyase Y23 (Duchefa) in citrate buffer at 37 °C for 30–45 min, transferred to glass slides and the meristematic tissue macerated in 45 % or 60 % (v/v) acetic acid (Schwarzacher et al., 1989). Slides were screened for cells with visible chromosomes prior to GISH.
Total genomic DNA of parental Nicotiana species, used as probes, was labelled with biotin-11-dUTP (GIBCO-BRL) and/or digoxigenin-11-dUTP (Boehringer Mannheim) using a nick translation kit (Bioprime CGH; Invitrogen). The hybridization mixture (30–40 µL per slide) contained 50 % (v/v) deionized formamide, 10 % (w/v) dextran sulphate, 0·125 % (w/v) sodium dodecyl sulphate, 1·0 µg mL−1 salmon sperm DNA and 2× SSC (Schwarzacher and Heslop-Harrison, 2000). The denaturation of chromosomes, hybridization conditions, stringency washing, amount of probe DNA, blocking DNA and hybridization mixture, were as described (Schwarzacher and Heslop-Harrison, 2000), with the most stringent wash being carried out in 0·2× SSC at 42 °C. Digoxigenin-labelled probe hybridization sites were detected using fluorescein isothiocyanate (FITC; Sigma-Aldrich) conjugated to sheep anti-digoxigenin antibody (Boehringer Mannheim) simultaneously to biotin-labelled sites with Cy3-streptavidin (Sigma-Aldrich). Chromosomes were observed using a Zeiss epifluorescence microscope, and images overlayed and processed, using only functions affecting the whole image equally (except drawing identification lines, cropping and filling edges outside crops) with Photoshop CS3.
Nuclear DNA, cpDNA and mtDNA analyses
Total DNA was isolated from leaves using a GenElute plant genomic DNA miniprep kit (Sigma-Aldrich) following the manufacturer's instructions. InterRetroelement Amplified Polymorphism (IRAP) and Retroelement-Microsatellite Amplified Polymorphism (REMAP) analyses were carried out following Saeidi et al. (2008) and Kalendar et al. (1999) using the primer sequences Nikita (5′-CGCATTTGTTCAAGCCTAAACC-3′) with the microsatellite sequence REMAP-GA, (GA)9C and Sukkula (5′-GATAGGGTCGCATCTTGGGCGTGAC-3′) with LTR6149 (5′-CTCGCTCGCCCACTACATCAACCGCGTTTATT-3′). Briefly, 50-ng genomic DNA samples were amplified in a 20-μL PCR mixture containing 1× PCR buffer (Kapa Biosystems), 2 mm MgCl2, 5 pmol of each primer, 200 mm dNTP mix and 1 U Kapa Taq polymerase. The annealing temperatures were optimized using gradients and the PCR programme consisted of 95 °C (2 min); 30 cycles of 95 °C (60 s), annealing at the optimized temperature for 60 s, ramp of 0·5 °C s−1 to 72 °C, and 72 °C for 2 min, adding 3 s per cycle, with a final extension at 72 °C for 10 min. PCR products were analysed by electrophoresis on 2 % (w/v) agarose gels and detected by ethidium bromide staining.
For RAPD analysis, PCR was carried out using the primer B07 (5′-GGTGACGCA-3′; Operon Technologies Inc.) in a programmed thermal cycler (Techne PHC-3) using 40 cycles of 94 °C (3 min), 94 °C (1 min), 35 °C (1 min), 72 °C (1 min), 72 °C (7 min), followed by 4 °C. PCR products were separated by 1·5 % (w/v) agarose gel electrophoresis and detected by ethidium bromide staining.
For cpDNA analysis, the trnL-ndhD fragment of the chloroplast genome was amplified by PCR (Sytnik et al., 2005) using the primers 5′-GTAGACACGCTGCTCTTAGG-3′ (specific to the trnL gene) and 5′ -CGCCAGATGTTCTATGGATAC- 3′ (specific to the ndhD gene; MWG-Biotech AG) to amplify a product of 1536 bp. The PCR programme was 35 cycles of 94 °C (3 min), 94 °C (20 s), 60 °C (20 s), 72 °C (1·5 min), 72 °C (5 min) and 4 °C. The amplified fragment was digested with AluI restriction endonuclease and the products analysed as above.
For mtDNA analysis, the forward primer 5′-CACGGGTCGCCCTCGTTCCG-3′ (rps14) and the reverse primer 5′-GTGTGGAGGATATAGGTTGT-3′ (cob; MWG – Biotech AG), were used to amplify mitochondrial DNA sequences (Demesure et al., 1995). PCR involved 40 cycles of 95 °C (5 min), 94 °C (30 s), 53 °C (30 s), 72 °C (2 min), 72 °C (7 min) and 4 °C. Reaction products were analysed as above.
Evaluation of disease resistance
Seeds harvested from the R1 generation of the five somatic hybrid plants SH28, SH31, SH354, SH411 and SH447 were sown on Levington M3 soil-less compost and individual R2 generation seedlings transferred 14 d after seed sowing to 13-cm-diameter plastic pots of the same compost. Plants were given a liquid feed (N : P : K, 3 : 2 : 3, v/v/v; Vitax) at every watering. Twelve plants of each somatic hybrid were evaluated for their fungal resistance against the same number of plants of N. debneyi, N. × sanderae ‘Avalon Red Improved’ and two other lines of N. × sanderae, namely ‘Avalon Formula Mixed’ and ‘Perfume Formula Mixed’ (Floranova). Plants were randomized in the glasshouse (natural daylight; maximum day temperature 25–30 °C; minimum night temperature 10–15 °C) and sprayed to run off on all their surfaces when 49 and 63 d old, with an aqueous suspension of spores prepared from leaves harvested from field-grown plants of N. × sanderae exhibiting extensive infestation by P. tabacina. Disease symptoms were recorded on glasshouse-grown plants when the latter were at the early flowering stage, 12 d after the second spraying with the spore suspension.
RESULTS
Sexual hybridization
Of 20 flowers on two plants of N. × sanderae and the same number on N. debneyi that were selfed, all except one flower of N. × sanderae produced pods. Each pod had 510·0 ± 5·6 and 220·0 ± 11·6 (mean ± s.d.) seeds for N. × sanderae and N. debneyi, respectively. Pollination of 20 flowers of N. × sanderae with pollen from N. debneyi and the same number of flowers in the reciprocal cross, failed to produce pods, confirming the unpublished but known sexual incompatibility of the parental species (L. Garland and N. Belfield-Smith, Floranova, http://floranova.co.uk, pers. com.).
Protoplast isolation, fusion and selection of putative somatic hybrid plants
Protoplast yields (mean ± standard deviation; n = 5) of 2·0 ± 0·5 × 105 and 4·0 ± 1·0 × 106 g−1 f. wt., were obtained from leaves of cultured shoots of N. × sanderae and N. debneyi, respectively. Protoplasts in all preparations, including those subjected to electrofusion treatment, had a viability of approx. 80 % and protoplasts commenced division within 7 d of culture. Protoplasts developed into cell colonies, each approx. 1 mm diameter, within 35 d in liquid MS-based medium containing 9 % (w/v) mannitol, 2·0 mg L−1 NAA and 0·5 mg L−1 BAP. Protoplasts of N. × sanderae that were electrofused alone produced tissues but, importantly, the latter failed to regenerate shoots, providing a means of eliminating the N. × sanderae parental material. In contrast, tissues derived from protoplasts of N. debneyi electrofused alone and those not exposed to electrofusion, regenerated shoots following transfer to MS-based medium of the same composition, but with mannitol reduced to 4·5 % (w/v) and, subsequently, to MS-based medium with 1·0 mg L−1 BAP, 2·0 mg L−1 IAA and lacking mannitol.
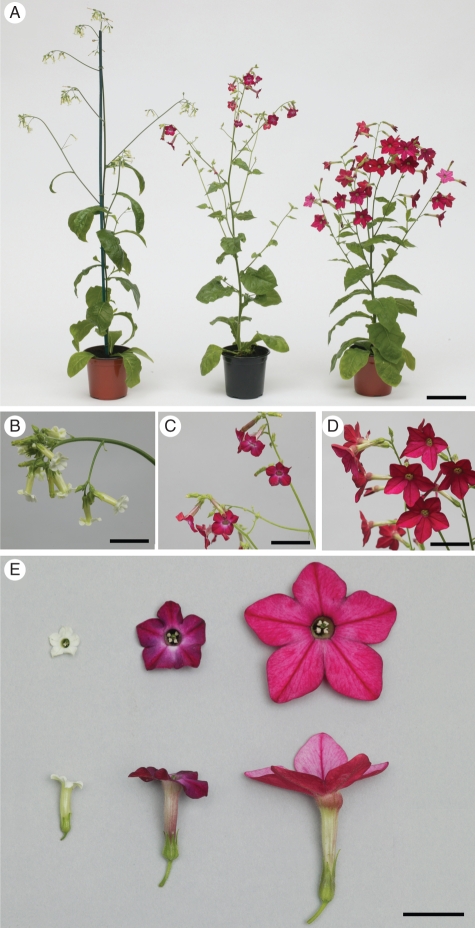
Tissues from the electrofusion of protoplasts of N. debneyi with those of N. × sanderae also regenerated shoots. Of 1570 protoplast-derived cell colonies selected manually using forceps and transferred to MS-based medium with 1·0 mg L−1 BAP, 2·0 mg L−1 IAA, 4·5 % (w/v) mannitol and 0·8 % (w/v) agar, 580 developed into tissues. Eighty-nine tissues regenerated shoots when cultured on the same medium without mannitol. All the plants regenerated from the electrofusion of protoplasts of N. debneyi electrofused with those of N. × sanderae were screened for their phenotypic characteristics at flowering. Five plants that exhibited mainly intermediate characteristics compared with seed-derived parental plants, were retained, but the remaining 84 plants resembled N. debneyi and were discarded. The five somatic hybrids (designated SH28, SH31, SH354, SH411 and SH447; R0 generation) were taller than plants of N. × sanderae, but with a spreading habit comparable to that of N. debneyi. Their dominant flower colour, 64 A/B, was similar to that of N. × sanderae (61 A/B; Fig. 1). Importantly, plants were self-fertile. As in N. × sanderae, trichomes were present on both surfaces of the leaves of putative somatic hybrid plants. In contrast, trichomes were present only on the midrib at the base of the laminae of the leaves of N. debneyi.
Fig. 1.
A seed-derived plant of N. debneyi (left), a typical, putative somatic hybrid (SH 354, centre), and a seed-derived plant of N. × sanderae (right) showing their morphology (A), inflorescences (B–D) and flowers in face and side views (E). Scale bars: (A) = 13 cm; (B) = 3 cm; (C) = 4 cm; (D) = 6 cm; (E) = 2·5 cm.
Characterization of R1, R2 and R3 generation plants
The parameters evaluated for the subsequent R1, R2 and R3 seed generations, and for seed-derived parental plants, are summarized in Table 1. The mean leaf index and pollen viability of parental and somatic hybrid plants were similar in three seed generations. In contrast, flower length and flower width of all somatic hybrid plants were intermediate in size compared with those of parental plants; these parameters were consistent over the three generations assessed. Two somatic hybrid plants, SH411 and SH447, were closer to the N. debneyi parent in the R1, R2 and R3 generations in terms of plant height and internode length. In contrast, in somatic hybrid plants SH28, SH31 and SH354, such characters were closer to N. × sanderae in the R1 generation, but this stability was not maintained in the R2 and R3 generations when these parameters were closer to those of the N. debneyi parent. The flower colour of the somatic hybrids was stable in the R1, R2 and R3 generations and was consistent with that of the R0 generation plants. The number of seeds per pod was less for somatic hybrids compared with parental plants, with the lowest seed germination in SH31.
Cytology and molecular analysis of regenerated putative somatic hybrid plants
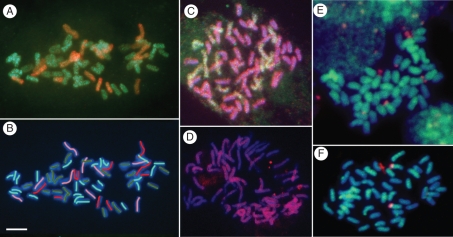
The chromosome number of each of the five somatic hybrids was 60 (SH31) or 62 (SH28, SH354, SH411 and SH447), fewer than the 66 expected from addition of 18 from N. × sanderae and 48 from N. debneyi (Fig. 2A). GISH analysis using labelled total genomic DNA as a probe to chromosomes of the R0 generation somatic hybrids (Fig. 2A–D), enabled the parental chromosomes to be distinguished in root tip cells at mitotic metaphase by probing simultaneously with total genomic DNA from N. × sanderae (labelled with biotin) and N. debneyi (labelled with digoxigenin). All metaphases showed 18 chromosomes of N. × sanderae origin. Somatic hybrids SH28, SH411 and SH447 had 44 chromosomes from N. debneyi. In situ hybridization also differentiated chromosome sets from the two parents, themselves both of sexual hybrid origin; the differentiation was particularly clear in SH447 (Fig. 2A, B). Among the 44 chromosomes of N. debneyi origin, there were 24 with more label from one ancestor and 20 with less label from the other ancestor, indicating that all four chromosomes that were missing in the somatic hybrid plant SH447 came from the same ancestral genome. The chromosome complements of the original somatic hybrid plants were retained in their R1, R2 and R3 seed progeny.
Fig. 2.
In situ hybridization showing the genomic origins of chromosomes in root-tip meristems of N. × sanderae (+) N. debneyi somatic hybrids (A–D) and N. debneyi (E, F). (A) Chromosomes of hybrid SH447 (2n = 2x + 2x = 62) labelled with genomic DNA of N. × sanderae in red and N. debneyi in green. (B) The metaphase from A stained with DAPI and overlayed with a drawing to indicate the chromosomes labelled strongly or weakly with DNA from N. × sanderae (nine bright red and nine pink chromosomes) and N. debneyi (24 cyan and 20 olive green chromosomes). (C) A metaphase from SH31 hybridized with digoxigenin-stained N. debneyi DNA (green) and biotin-labelled N. × sanderae DNA (red). (D) A metaphase from SH411 with 44 chromosomes more strongly labelled red with N. debneyi DNA and 18 counterstained blue with DAPI. In all three metaphases, the full chromosome complement (24) is present from N. × sanderae, but four more weakly labelled chromosomes of N. debneyi (20 not 24) are missing. (E, F) Two metaphases from N. debneyi (2n = 4x = 48) labelled green with genomic N. debneyi DNA with half the chromosomes (24) strongly labelled and half more weakly labelled showing blue DAPI counterstain. Six sites of 45S rDNA are seen red in (E); two major and one minor 5S rDNA are shown red in (F). Scale bar in (B) = 12 µm.
In metaphases of N. debneyi hybridized with its own DNA, half of the chromosomes were labelled more strongly than the remaining chromosomes, and some of these had terminal sites which were brighter, thus confirming the amphipolyploid hybrid origin of N. debneyi (Fig. 2E, F), with substantial differentiation of the two genomes, although there remains speculation about their origin (Khan and Narayan, 2007; Clarkson et al., 2010).
Molecular analysis of DNA
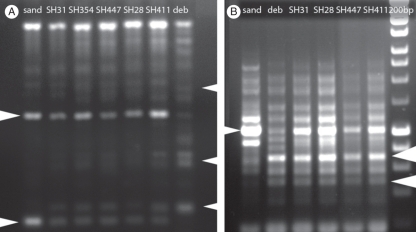
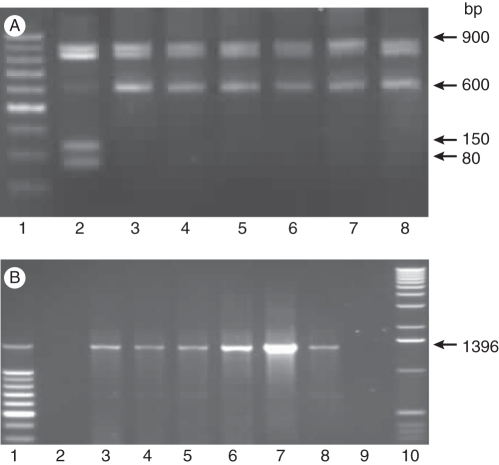
The REMAP and IRAP analysis with genomic DNA extracted from the parental species and the five somatic hybrids (R0 generation) showed that the hybrids had a pattern generally representing the sum of both parents (Fig. 3A, B). Although four or six of the chromosomes of N. debneyi origin were missing in the somatic hybrids, all but one band seen in N. debneyi were also detected in the hybrids (Fig. 3B); primer competition may have reduced amplification of this and another band which is less clear in SH447 and SH411 which showed weaker amplification. Two other primer pairs were also used. REMAP-GA with the primer Sukkula showed shared bands and one specific to N. × sanderae that was present in all hybrids; primer Nikita with LTR6149 showed little amplification with N. × sanderae DNA, but four bands in N. debneyi and the hybrids (not shown).
Fig. 3.
REMAP and IRAP analysis of somatic hybrids and their parents showing hybrids with an additive combination of parental bands: (A) with primers Nikita and REMAP-GA; (B) with primers Sukkula and LTR6149. Patterns from parents and somatic hybrid plants with bands originating uniquely from N. debneyi or N. × sanderae are indicated by arrowheads (left and right, respectively). Abbreviations: deb, N. debneyi; sand, N. × sanderae; SH, somatic hybrid plants; 200 bp, ladder with 200-bp steps from bottom.
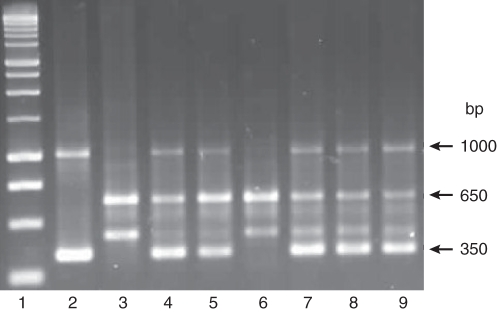
Primer OPB07 used for RAPD analysis generated distinct bands for the parents (Fig. 4, lanes 2, 3 and 6) with somatic hybrids having a summation of the parental bands (lanes 4, 5 and 7–9). The bands obtained for the cpDNA of N. × sanderae (Fig. 5A, lane 2) differed from those of N. debneyi (lane 8) and the bands of the somatic hybrids (lanes 3–7). The cpDNA banding pattern of somatic hybrid plants resembled the pattern of N. debneyi, indicating that plastids in the somatic hybrids were derived from the wild N. debneyi parent. mtDNA analysis revealed a band of 1396 bp present in the somatic hybrid plants (Fig. 5B, lanes 3–7) and in N. debneyi (lane 8). This band was absent from N. × sanderae (lane 2), suggesting that the mitochondrial genomes of the five somatic hybrid plants were probably derived from N. debneyi.
Fig. 4.
PCR products generated by primer OPB07. Lane 1, 1-kb ladder; lane 2, N. × sanderae DNA; lanes 3 and 6, N. debneyi DNA; lanes 4, 5 and 7–9, DNA from somatic hybrid plants SH28, SH31, SH354, SH411 and SH447, respectively, showing characteristic bands from both parents.
Fig. 5.
Analysis of cytoplasmic genomes in Nicotiana parental plants and somatic hybrids by PCR amplification of specific sequences. (A) cpDNA bands specific for trnL-ndhD gene fragments after digestion of the latter with Alu1. Lane 1, 100-bp DNA ladder; lane 2, cpDNA of N. × sanderae. The banding in lanes 3–7 from somatic hybrid plants SH28, SH31, SH354, SH411 and SH447, respectively, is the same as the banding pattern of N. debneyi (lane 8), confirming that chloroplasts are from the wild parent. (B) Amplification using mtDNA-specific primers rps14 and cob. Lane 1, 10- to 100-bp DNA ladder; lane 2, N. × sanderae mtDNA; lanes 3–7, banding of mtDNA from somatic hybrid plants resembling the banding pattern of N. debneyi (lane 8); lane 9, water control.
Disease resistance
All plants of N. × sanderae ‘Avalon Red Improved’, ‘Avalon Formula Mixed’ and ‘Avalon Perfume Formula Mixed’ (n = 12 for each cultivar) showed characteristic and extensive infection when evaluated 12 d after the second spraying with a spore suspension of P. tabacina. Infection was accompanied by yellowing and twisting of infected leaves (Fig. 6A, B), with thick mats of sporulating mycelium on the lower surfaces of infected leaves (Fig. 6C) and subsequent death of the plants. In contrast, leaves of all plants of N. debneyi (Fig. 6D, E; n = 12) and 12 R2 generation seedlings of each of the somatic hybrids N. × sanderae (+) N. debneyi (60 plants in total) were all completely resistant to P. tabacina and did not exhibit any signs of fungal infection (Fig. 6F, G).
Fig. 6.
A leaf from a seed-derived plant of N. × sanderae infected with Peronospora tabacina showing curling and chlorosis on the upper (A) and lower surfaces (B), with sporulating fungal hyphae on the lower leaf surface (C). In contrast, upper and lower surfaces of leaves from a seed-derived plant of N. debneyi (D, E) and a seed-derived plant (R2 generation) of somatic hybrid SH354 (F, G) are free of fungal infection. Scale bars: (A, B) = 6 cm; (C) = 50 µm; (D, E) = 5 cm; (F, G) = 4·5 cm.
DISCUSSION
Wild germplasm frequently has novel characteristics that are valuable for breeding, including resistances to biotic or abiotic stress and quality characters. Interspecific sexual hybridization has been used extensively and, increasingly, transgenic approaches have been exploited for gene transfer. However, there are formidable regulatory obstacles relating to transformation technology and the products generated, adding to the laboratory costs to this latter strategy. In the genus Nicotiana, despite the presence of many hybrid species, it was impossible to obtain seed from sexual crosses between N. × sanderae (Nicotiana section Alatae), an ornamental tobacco affected by blue mould, and N. debneyi (section Suaveolentes), a wild species which has been used in breeding fungal resistance into tobacco by sexual hybridization. Tezuka et al. (2010) examined sexual crosses within the section Suaveolentes and found both prezygotic barriers to fertilization and post-zygotic failures during seed development, some of which could be overcome by ovule culture. Hybrid lethality, where seedlings die soon after germination, was also frequent in the progeny of the successful sexual crosses of Tezuka et al. (2010). In contrast, the somatic hybrids between N. × sanderae and N. debneyi flowered (Fig. 1) and produced seed, enabling assessments of subsequent generations. It would be interesting to determine whether hybrid lethality occurs in seedlings from somatic hybrids of those species combinations in the successful sexual crosses reported by Tezuka et al. (2010), provided such somatic hybrids can be generated by the fusion of diploid parental cells.
The shoot regeneration potential of cultured tissues in the genus Nicotiana was evaluated by Li et al. (2003), who showed that organogenesis was poor in N. × sanderae. The present investigation confirmed that although cells derived from electrofusion-treated protoplasts of N. × sanderae commenced division during the first 7 d of culture, they failed to regenerate shoots. This inability of protoplast-derived tissues of N. × sanderae to regenerate was exploited as a half selection procedure to eliminate the N. × sanderae parent from electrofusion-treated mixtures of parental protoplasts. In contrast, and in agreement with the results of Scowcroft and Larkin (1980), tissues that developed from electrofusion-treated protoplasts of N. debneyi regenerated shoots under the same conditions. Thus, any hybrid cells that developed from heterokaryons following electrofusion of protoplasts of N. × sanderae with those of N. debneyi would also be expected to develop through heterosis and to regenerate shoots under the same conditions; the conspicuous morphology of the somatic hybrids compared with that of N. debneyi allowed the somatic hybrids to be identified.
Somatic hybrids and their seed progeny retained a flower colour similar to that of the N. × sanderae parent (Fig. 1), making them attractive ornamentals in their own right. Other morphological characteristics of the five somatic hybrid plants and their seed-derived progeny were mainly intermediate compared with those of the parental plants. Plastids and mitochondria in the somatic hybrids were probably from the wild N. debneyi parent (Fig. 5). Loss of cytoplasmic genomes from one partner is not uncommon in somatic hybrids, with organelles from the other fusion partner becoming dominant (Davey et al., 2005). For example, in Solanum bulbocastanum (+) S. tuberosum plastids and mitochondria were from the cultivated parent (S. tuberosum) in most of the hybrid plants (Lovene et al., 2007).
DNA and GISH analysis of metaphase chromosomes revealed that the somatic hybrids of N. × sanderae (+) N. debneyi included nuclear genomes from both parents (Figs 2, 3 and 4), although four to six chromosomes from N. debneyi were absent from these hybrids (Fig. 2). Loss of parental chromosomes in somatic hybrids is not uncommon (Liu et al., 2005). In fact, most somatic hybrid plants generated between N. tabacum and wild Nicotiana species have been asymmetric at the nuclear level with only a limited number being symmetric hybrids (Sun et al., 2007). Yang et al. (2007) also reported a range of chromosome numbers from 54 to 74 in somatic hybrids between Gossypium hirsutum and G. klozschianum. Despite a loss of chromosomes, the somatic hybrid plants of N. sanderae (+) N. debneyi were morphologically stable as demonstrated by the retention of characteristics that were scored for the original individual somatic hybrid plants (R0 generation) and their seed-derived progeny over the R1, R2 and R3 generations. The five somatic hybrids that were generated were self-fertile, an extremely important attribute, enabling characterization of seed progeny. Whilst characteristics of the somatic hybrids were generally intermediate between those of the parental plants, some of the seed-derived progeny exhibited characteristics that were more similar to those of one parent, as in the case of plant height and internode length. Somatic hybrid plants and their seed progeny also exhibited novel floral morphology and a consistent flower colour, the latter being slightly more intense than that of N. × sanderae as judged using the universally accepted colour charts of The Royal Horticultural Society of London. Segregation of characteristics was not observed in the R1 to R3 seed generations, with 60–80 % seed germination, except in SH31 in which seed germination was approx. 40 %.
Seed-derived progeny were resistant to P. tabacina, this resistance being introgressed from the wild species, N. debneyi. These experiments emphasize the relevance of somatic hybridization through protoplast fusion to transfer complex genetic traits from wild species (Fig. 6) into sexually incompatible recipients without the requirement for identification of the genes involved and recombinant DNA technology. Indeed, N. debneyi may be a useful protoplast fusion partner in attempts to introgress fungal resistance into other sexually incompatible Nicotiana species of value as ornamentals, or for the production of secondary plant products, including pharmaceuticals, where complex disease resistance traits could be combined with simple monogenic traits carried by transgenes. This approach may be exploited more in the future, especially as P. tabacina appears to be more widespread and, for example, has become endemic in the UK in the last 20 years. The fertility and stability of somatic hybrid plants is crucial for the incorporation of this germplasm into breeding programmes. Future experiments will also evaluate transfer of fungal resistance from somatic hybrids of N. × sanderae (+) N. debneyi by sexual backcrossing to lines of N. × sanderae and, possibly, to other ornamental species of Nicotiana. Whilst the semi-dwarf stature of N. × sanderae may have been lost in the somatic hybrid plants generated in these experiments, in their seed progeny and in any future back-cross progeny, additional genetic manipulation of these somatic hybrids and back-cross progeny by the introduction and expression of genes controlling gibberellin metabolism and, hence, stature (Dijkstra et al., 2008) could enable restoration of the dwarf habit characteristic of N. sanderae.
ACKNOWLEDGEMENTS
The authors thank L. Garland and N. Belfield-Smith (Floranova) for seeds of N. × sanderae and N. debneyi, for financial assistance to D.P., and for experiments relating to assessments of fungal resistance. Dr K. Pyke (Plant and Crop Sciences Division, University of Nottingham) assisted with the formatting of Figs 1 and 4–6.
LITERATURE CITED
- Anssour S, Krügel T, Sharbel TF, Saluz HP, Bonaventure G, Baldwin IT. Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of Nicotiana attenuata and Nicotiana obtusifolia. Annals of Botany. 2009;103:1207–1217. doi: 10.1093/aob/mcp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai D, Reeleder R, Brandle JE. Identification of two RAPD markers tightly linked with the Nicotiana debneyi gene for resistance to black root rot of tobacco. Theoretical and Applied Genetics. 1995;91:1184–1189. doi: 10.1007/BF00220927. [DOI] [PubMed] [Google Scholar]
- Bai D, Reeleder R, Bramble JE. Production and characterization of tobacco addition lines carrying Nicotiana debneyi chromosomes with a gene for resistance to black root rot. Crop Science. 1996;36:852–857. [Google Scholar]
- Baldwin T. An ecologically motivated analysis of plant–herbivore interactions in native tobacco. Plant Physiology. 2001;127:1449–1458. [PMC free article] [PubMed] [Google Scholar]
- Carlson PS, Smith HH, Dearing RD. Parasexual interspecific plant hybridization. Proceedings of the National Academy of Sciences of the USA. 1972;69:2292–2294. doi: 10.1073/pnas.69.8.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Knapp S, Cox AV, et al. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae) Annals of Botany. 2003;92:107–127. doi: 10.1093/aob/mcg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson JJ, Kelly LJ, Leitch AR, Knapp S, Chase MW. Nuclear glutamine synthetase evolution in Nicotiana: phylogenetics and the origins of allotetraploid and homoploid (diploid) hybrids. Molecular Phylogenetics and Evolution. 2010;55:99–112. doi: 10.1016/j.ympev.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Clayton EE. Resistance of tobacco to blue mould (Peronospora tabacina) Journal of Agricultural Research. 1945;70:79–87. [Google Scholar]
- Davey MR, Anthony P, Power JB, Lowe KC. Plant protoplasts: status and biotechnological perspectives. Biotechnology Advances. 2005;23:131–171. doi: 10.1016/j.biotechadv.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Demesure B, Sodzi N, Petit RJ. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- Dijkstra C, Adams E, Bhattacharya A, et al. Over-expression of a gibberellin 2-oxidase gene from Phaseolus coccineus L. enhances gibberellin inactivation and induces dwarfism in Solanum species. Plant Cell Reports. 2008;27:463–470. doi: 10.1007/s00299-007-0471-z. [DOI] [PubMed] [Google Scholar]
- Fitter JT, Thomas MR, Niu C, Rose RJ. Investigation of Nicotiana tabacum (+) N. suaveolens cybrids with carpelloid stamens. Journal of Plant Physiology. 2005;162:225–235. doi: 10.1016/j.jplph.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Frearson EM, Power JB, Cocking EC. The isolation, culture and regeneration of Petunia leaf protoplasts. Developmental Biology. 1973;33:130–137. doi: 10.1016/0012-1606(73)90169-3. [DOI] [PubMed] [Google Scholar]
- Goodspeed TH. The genus Nicotiana: origins, relationships, and evolution of its species in the light of their distribution, morphology, and cytogenetics. Waltham, MA, USA: Chronica Botanica; 1954. [Google Scholar]
- Horsman K, Gavrilenko T, Bergervoet JEM, Huigen D, Joe ATW, Jacobsen E. Alteration of the genomic composition of Solanum nigrum (+) potato backcross derivatives by somatic hybridization: selection of fusion of hybrids by DNA measurements and GISH. Plant Breeding. 2001;120:201–207. [Google Scholar]
- IIcheva V, San LH, Dimitrov B, Zagorska N. Morphological and cytological characteristics of somatic hybrids of Nicotiana tabacum L. (+) N. megalosiphon Heurk. et Mull. In Vitro Cellular and Developmental Biology – Plant. 2000;36:69–73. [Google Scholar]
- Jones B, Lynch PT, Handley GJ, et al. Equipment for the large-scale electromanipulation of plant protoplasts. BioTechniques. 1994;16:312–321. [PubMed] [Google Scholar]
- Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A. IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theoretical and Applied Genetics. 1999;98:704–711. [Google Scholar]
- Kapp S, Chase MW, Clarkson JJ. Nomenclatural changes and a new classification in Nicotiana (Solanaceae) Taxon. 2004;53:73–82. [Google Scholar]
- Khan MQ, Narayan RKJ. Phylogenetic diversity and relationships among species of genus Nicotiana using RAPDs analysis. African Journal of Biotechnology. 2007;6:148–162. [Google Scholar]
- Komori T, Myers PN, Yamada S, Kubo T, Imaseki H. Comparative study of Nicotiana with respect to water deficit tolerance during the early growth. Euphytica. 2000;116:121–130. [Google Scholar]
- Kovarik A, Dadejova M, Lim YK, et al. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Annals of Botany. 2008;101:815–823. doi: 10.1093/aob/mcn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumashiro T, Kubo T. Cytoplasm transfer of Nicotiana debneyi to Nicotiana tabacum by protoplast fusion. Japanese Journal of Breeding. 1986;36:39–48. [Google Scholar]
- Lewis R. In: Nicotiana. In. Kole C, editor. 2011. Wild crop relatives: genomic and breeding resources, plantation and ornamental crops. Berlin: Springer, in press. [Google Scholar]
- Li B, Huang W, Bass T. Shoot production per responsive leaf explant increases exponentially with explant organogenic potential in Nicotiana species. Plant Cell Reports. 2003;22:231–238. doi: 10.1007/s00299-003-0679-5. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, et al. The sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytologist. 2007;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- Liu JH, Xu XY, Deng XX. Intergeneric somatic hybridization and its application to crop genetic improvement. Plant Cell Tissue and Organ Culture. 2005;82:19–44. [Google Scholar]
- Lovene M, Savasrese S, Cardi T, et al. Nuclear and cytoplasmic genome composition of Solanum bulbocastanum (+) S. tuberosum somatic hybrids. Genome. 2007;50:443–450. doi: 10.1139/g07-024. [DOI] [PubMed] [Google Scholar]
- Milla SR, Levin JS, Lewis RS, Rufty RC. RAPD and SCAR markers linked to an introgressed gene conditioning resistance to Peronospora tabacina D.B. Adam. in tobacco. Crop Science. 2005;45:2346–2354. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Pati PK, Sharma M, Ahuja PS. Extra thin alginate film: an efficient technique for protoplast culture. Protoplasma. 2005;226:217–221. doi: 10.1007/s00709-005-0096-4. [DOI] [PubMed] [Google Scholar]
- Saeidi H, Rahiminejad MR, Heslop-Harrison JS. Retroelement insertional polymorphisms, diversity and phylogeography within diploid, D-Genome Aegilops tauschii (Triticeae, Poaceae) sub-taxa in Iran. Annals of Botany. 2008;101:855–861. doi: 10.1093/aob/mcn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS. Oxford: BIOS Scientific Publishers; 2000. Practical in situ hybridization. [Google Scholar]
- Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. In situ localization of parental genomes in a wide hybrid. Annals of Botany. 1989;64:315–324. [Google Scholar]
- Scowcroft WR, Larkin PJ. Isolation, culture and plant regeneration from protoplasts of Nicotiana debneyi. Australian Journal of Plant Physiology. 1980;7:635–644. [Google Scholar]
- Sun Y-H, Xue Q-Z, Ding C-M, et al. Somatic cybridization between Nicotiana tabacum and N. repanda based on a single inactivation procedure of nuclear donor parental protoplasts. Plant Science. 2005;168:303–308. [Google Scholar]
- Sun Y, Xue Q, Zhang X, et al. Morphological, cytological and molecular characterization of a novel symmetric somatic hybrid between N. tabacum and N. glauca. Plant Biosystems. 2007;141:129–133. [Google Scholar]
- Sytnik E, Komarnytsky I, Gleba Y, Kuchuk N. Transfer of transformed chloroplasts from Nicotiana tabacum to the Lycium barbarum plants. Cell Biology International. 2005;29:71–75. doi: 10.1016/j.cellbi.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Tezuka T, Kuboyama T, Matsuda T, Marubashi W. Seven of eight species in Nicotiana section Suaveolentes have common factors leading to hybrid lethality in crosses with Nicotiana tabacum. Annals of Botany. 2010;106:267–276. doi: 10.1093/aob/mcq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA. ARS, National Genetic Resources Program. 2010 Germplasm Resources Information Network – (GRIN) [Online Database]. National Germplasm Resources Laboratory, Beltsville, Maryland. URL: http://www.ars-grin.gov/cgi-bin/npgs/html/taxon.pl?25266. (accessed 28 July 2010) [Google Scholar]
- Wark DC. Development of flue-cured tobacco cultivars resistant to a common strain of blue mold. Tobacco Science. 1970;14:147–150. [Google Scholar]
- Widholm J. The use of FDA and phenosafranine for determining viability of cultured plant cells. Stain Technology. 1972;47:186–194. doi: 10.3109/10520297209116483. [DOI] [PubMed] [Google Scholar]
- Yang X-Y, Zhang X-L, Jin S-X, Fu L-L, Wang L-G. Production and characterization of asymmetric hybrids between upland cotton Coker 201 (Gossypium hirsutum) and wild cotton (G. klozschianum) Plant Cell, Tissue and Organ Culture. 2007;89:225–235. [Google Scholar]