Abstract
Background and Aims
Brachypodium distachyon is a temperate grass with a small stature, rapid life cycle and completely sequenced genome that has great promise as a model system to study grass-specific traits for crop improvement. Under iron (Fe)-deficient conditions, grasses synthesize and secrete Fe(III)-chelating agents called phytosiderophores (PS). In Zea mays, Yellow Stripe1 (ZmYS1) is the transporter responsible for the uptake of Fe(III)–PS complexes from the soil. Some members of the family of related proteins called Yellow Stripe-Like (YSL) have roles in internal Fe translocation of plants, while the function of other members remains uninvestigated. The aim of this study is to establish brachypodium as a model system to study Fe homeostasis in grasses, identify YSL proteins in brachypodium and maize, and analyse their expression profiles in brachypodium in response to Fe deficiency.
Methods
The YSL family of proteins in brachypodium and maize were identified based on sequence similarity to ZmYS1. Expression patterns of the brachypodium YSL genes (BdYSL genes) were determined by quantitative RT–PCR under Fe-deficient and Fe-sufficient conditions. The types of PS secreted, and secretion pattern of PS in brachypodium were analysed by high-performance liquid chromatography.
Key Results
Eighteen YSL family members in maize and 19 members in brachypodium were identified. Phylogenetic analysis revealed that some YSLs group into a grass-specific clade. The Fe status of the plant can regulate expression of brachypodium YSL genes in both shoots and roots. 3-Hydroxy-2′-deoxymugineic acid (HDMA) is the dominant type of PS secreted by brachypodium, and its secretion is diurnally regulated.
Conclusions
PS secretion by brachypodium parallels that of related crop species such as barley and wheat. A single grass species-specific YSL clade is present, and expression of the BdYSL members of this clade could not be detected in shoots or roots, suggesting grass-specific functions in reproductive tissues. Finally, the Fe-responsive expression profiles of several YSLs suggest roles in Fe homeostasis.
Keywords: Brachypodium distachyon, Zea mays, iron homeostasis, phytosiderophore, nicotianamine, Yellow Stripe-Like, YSL, YS1
INTRODUCTION
Grasses acquire iron (Fe) from the soil using a chelation strategy known as Strategy II. Under Fe-deficient conditions, grasses synthesize and secrete Fe(III)-chelating agents called phytosiderophores (PS), also known as mugineic acids (MAs). PS are derivatives of the non-proteinogenic amino acid nicotianamine (NA), which also functions as a transition metal chelator in plants. After chelation by PS, insoluble Fe(III) in the rhizosphere becomes soluble as Fe(III)–PS complexes. Then, through the action of Yellow Stripe1 (YS1) transporters at the plasma membrane, grasses move these Fe(III)–PS complexes into the root cells (Romheld and Marchner, 1986; von Wiren et al., 1994; Curie et al., 2001; Schaaf et al., 2004; Murata et al., 2006). The Zea mays ys1 mutant, which lacks the ability to take up Fe(III)–PS complexes, shows severe Fe deficiency chlorosis (yellowing between the veins), and is ultimately lethal, indicating that uptake of Fe(III)–PS is an essential process for maize (Bell et al., 1962; von Wiren et al., 1994; Curie et al., 2001). In contrast, plants with T-DNA insertions in Oryza sativa Yellow Stripe-Like15 (OsYSL15), which encodes an Fe(III)–PS transporter in rice that appears to be orthologous to ZmYS1, exhibit Fe deficiency chlorosis, but are viable (Inoue et al., 2009; Lee et al., 2009). This is consistent with rice's ability to acquire Fe both as Fe(III)–PS and as Fe(II) via IRT-type transporters (Cheng et al., 2007; Walker and Connolly, 2008; Ishimaru et al., 2006).
The Yellow Stripe-Like (YSL) proteins show strong sequence similarity to ZmYS1, and several are known to be transporters of metal–NA complexes in both monocotyledonous and dicotyledonous plants (DiDonato et al., 2004; Koike et al., 2004; Gendre et al., 2007). There are eight YSL family members in arabidopsis, while rice has 18 YSL genes (Gross et al., 2003; Koike et al., 2004). The pattern of expression of the AtYSL genes indicates that transport of metal–NA occurs in diverse cell types in roots and shoots, stems, flowers, fruits and seeds, suggesting that metals are regularly imported into cells as NA complexes (DiDonato et al., 2004; Waters et al., 2006). Expression of several of the AtYSL genes changes in response to Fe deficiency. Interestingly, the most common change is a decrease in expression during nutrient stress. This pattern is quite different from the pattern exhibited by genes involved in uptake of Fe from the soil, such as ZmYS1 and OsYSL15, which are upregulated by Fe deficiency, presumably as an attempt to increase nutrient uptake during deficiency (Curie et al., 2001; Inoue et al., 2009). Downregulation may reflect an attempt to stop Fe sequestration in older tissues and allow Fe to be moved into other, less nutrient-replete plant parts (DiDonato et al., 2004; Waters et al., 2006). Expression levels of AtYSL1 and AtYSL3 increase in senescing leaves, suggesting a role in Fe remobilization from old leaves to reproductive parts (DiDonato et al., 2004; Waters et al., 2006; Chu et al., 2010). Likewise, the activity of some rice YSL promoters in floral parts and developing seeds suggests functions in translocation of Fe into the seeds (Bughio et al., 2002; Ishimaru et al., 2006, 2007; Aoyama et al., 2009; Lee et al., 2009).
A complete genome sequence, short generation time, fecundity, small stature and a diverse array of molecular genetic tools developed by a large research community have made Arabidopsis thaliana a key species for the discovery of molecular mechanisms of plant growth and development (Arabidopsis Genome Initiative, 2000). However, as a dicotyledonous plant, arabidopsis is only distantly related to the grass family (Poaceae) and lacks many biological features of monocotyledonous grass crops. In particular, the distinct characteristics of dicots related to agricultural traits can be difficult to apply to the domesticated cereal crops. Rice has been developed as a model grass species and is a major resource to study grass crop genomics (International Rice Genome Sequencing Project, 2005). Although the rice genome sequence is available, and molecular genetic resources are rapidly developing, rice has disadvantages as a model organism for functional genomics owing to its large stature and long life cycle. Although the mechanisms of Fe uptake and translocation in rice are undeniably important because of rice's status as a major grain crop, rice is imperfect as a model for grass-specific processes related to Fe nutrition and uptake studies. Although rice synthesizes (Kobayashi et al., 2005), secretes (Nozoye et al., 2011) and uses (Walker and Connolly, 2008) PS, rice is normally grown under submerged conditions where Fe(II) is more abundant than Fe(III), and has thus evolved an Fe(II) transport mechanism that is similar to the Strategy I Fe uptake mechanism of non-grasses (Bughio et al., 2002; Ishimaru et al., 2006). Therefore, there is a need for a grass species that has the characteristics of a model organism, and at the same time represents the Fe homeostasis characteristics of important grain crops such as wheat, barley and corn.
Brachypodium distachyon (hereafter referred to as brachypodium) has many physical and genetic attributes that make this species a very attractive model system to study grass-specific biological processes. It is evolutionarily closely related to wheat and barley, and can efficiently be transformed (Kellogg, 2001; Vogel and Hill, 2008). Maintenance requirements for brachypodium are undemanding and growth is simple under controlled environments. The rapid life cycle of brachypodium is similar to the short generation time of arabidopsis. It has a small nuclear genome and the complete genome sequence is available (International Brachypodium Initiative, 2010). Therefore, brachypodium fills a gap in grass crop research.
Our aim is to establish brachypodium as a model plant to study the comparative genomics of Fe distribution and translocation in grasses. For this purpose, members of the YSL family of transporter proteins in brachypodium (BdYSLs) were identified. A total of 19 predicted brachypodium proteins that showed strong, full-length similarity to the maize YS1 protein were identified. These sequences were used in a phylogenetic analysis that shows four distinct YSL clades. Three of the clades are found in both grasses and in the non-grasses arabidopsis, Glycine max and Medicago truncatula, while one of the clades is specific to grass species (brachypodium, rice and maize). Expression of brachypodium YSL genes in the roots and shoots of Fe-sufficient and Fe-deficient plants was analysed. We also characterized PS secretion rhythm and the types of PS released from brachypodium roots under Fe-deficient conditions.
MATERIALS AND METHODS
Plant material
Seeds of brachypodium inbred line Bd21-3 were sterilized in 15 % bleach plus 0·1 % Triton X-100 for 30 min. After soaking in the bleach solution the seeds were rinsed with sterile distilled water (Huo et al., 2006). Sterilized seeds were cold treated at 4 °C for 7 d in order to synchronize germination. The seeds then were germinated on moistened clay balls (Hydroton) in mesh pots. When seedlings emerged, the pots were placed in a semi-aeroponic culture set-up in which modified Hoagland's medium [1 mm KH2PO4; 3·75 mm KOAc; 5 mm Ca(NO3)2; 1·25 mm KNO3; 2 mm MgSO4; 3·75 mm NH4OAc; 46 µm H3BO3; 9·1 µm MnCl2; 0·77 µm ZnSO4; 0·32 µm CuSO4; 0·83 µm H2Mo4; 100 µm FeSO4; 100 µm EDTA] was continuously sprayed onto the growth substrate. Nutrient solutions were changed every 2 d. For the first 2 d, plants were grown in half-strength nutrient solutions in order to avoid root burn in the young seedlings; after 2 d, full-strength nutrient solutions were used. Plants that were used for the Fe deficiency experiment received Fe for the first week of growth to allow strong root development. Then the plants were grown in nutrient solution with no added Fe for 10 d. Plants were grown at 20 °C with 20 h of light at 200 µE, and 4 h of darkness in controlled growth chambers. After 17 d of growth, the shoots and roots were harvested separately and used for downstream expression analysis.
Identification of BdYSLs
Brachypodium YSL family members were identified by comparing six frame translations of the 8× complete, assembled brachypodium genome sequence with the maize YS1 protein sequence using the TBLASTN algorithm (Altschul et al., 1990, 1997). For each of the predicted proteins identified, N- and C- terminal amino acid sequence similarity analysis was further examined to make certain that we obtained full-length protein sequences for all the brachypodium YSL family members. Gene models were carefully examined by analysing multiple sequence alignments of brachypodium YSL protein sequences with maize, rice and arabidopsis YSLs. The accuracy of gene models was also confirmed by searching full-length genomic DNA sequences against brachypodium expressed sequence tag (EST) sequences in GenBank. Full-length cDNAs corresponding to two candidate ZmYS1 orthologues, BdYS1A (GenBank accession no. HM443950) and BdYS1B (GenBank accession no. HM443951), and to BdYSL9 (GenBank accession no. HM443952) were cloned. Full-length cDNA fragments of each gene were amplified from brachypodium root cDNA using primers: BdYS1A-forward 5'CTGCGCGGAGACAAGACAGGAAG3', BdYS1A-reverse 5'AAGGTCGAGCACCAAGGAACCTG3', BdYS1B-forward 5'CGCGGAGACTGAGGAGAGCAAG3', BdYS1B-reverse 5'TTTCAGTTGAACATCCATGAAATTGA3', BdYSL9-forward 5'TAAGGAGGCGGAGGGAGCAAAC3' and BdYSL9-reverse 5'TGCACAACTTCAGACATGGTCCAC3'. After cloning into the pCR8/GW/TOPO TA vector (Invitrogen, Carlsbad, CA, USA) full-length cDNAs of each gene were fully sequenced.
Yeast functional complementation
Full-length cDNAs for BdYS1A and BdYS1B in the pCR8/GW/TOPO TA vector were used as entry clones in Gateway LR recombination reactions (Invitrogen). Gateway reading frame cassette A was introduced into the yeast expression vector pYES6/CT using the NotI site, and used as the destination vector in LR reactions. Iron transport-deficient Saccharomyces cerevisiae strain DEY1453 (MATa/MATa ade2/ADE2 can1/can1 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 fet3-2::HIS3/fet3-2::HIS3 fet4-1::LEU2/fet4-1::LEU2) was transformed with pGEV-Trp (Gao and Pinkham, 2000) together with pYES6/CT or pYES6/CT expressing BdYS1A, BdYS1B or OsYSL15 as a positive control (Lee et al., 2009).
For complementation assays, SD-Trp medium was made with a yeast nitrogen base formulated without added Fe, and buffered with 25 mm MES at pH 6·0. A 17 µL aliquot of 7·4 mm FeCl3 and 25 µL of 10 mm 2'-deoxymugineic acid (DMA) were placed in the centre of each empty plate and incubated at room temperature for 10 min. A 25 mL aliquot of molten SD-Trp with 10 nm β-oestradiol was then added to the plates before solidification.
Suspensions were prepared from 3-day-old yeast colonies that were removed from the plates and suspended in sterile H2O. The OD550 of the resulting suspension was measured, and the suspension was brought to 0·1 OD550. Serial dilutions (1:10, 1:100 and 1:1000) of the suspension were prepared, and 8 µL of each dilution was spotted on the plates. Plates were then grown at 27 °C for 5 d.
Identification of maize YSL genes
Using the ZmYS1 complete protein sequence as a query, TBLASTN (Altschul et al., 1990, 1997) was used to identify maize YSL genes represented on bacterial artificial chromosome (BAC) clones in the Genbank High Throughput Genomic Sequences (HTGS) data set. The N- and C-termini of each matching sequence were carefully examined to determine that all identified genes were full length. Four of the originally identified putative maize genes contained mutations encoding frameshifts and/or stop codons. These were deemed to be pseudogenes (Table 3), and were not included in downstream analyses. Putative full-length cDNAs were identified from the maize EST collections, and were obtained from the Arizona Genomics Institute (http://www2.genome.arizona.edu/genomes/maize). Complete sequencing of maize cDNAs corresponding to ZmYSL1 (GenBank accession no. BT063894), ZmYSL2, ZmYSL3 (GenBank accession no. BT086561), ZmYSL6 (GenBank accession no. BT034471), ZmYSL11 (GenBank accession no. HM444829), ZmYSL11A (GenBank accession no. BT084965), ZmYSL12 (GenBank accession no. HM444830), ZmYSL14A (GenBank accession no. HM444831) and ZmYSL17 (GenBank accession no. HM444832) was performed to determine the sequence of these ZmYSL proteins. Neither of the ZmYSL2 cDNA clones sequenced (ZM_BFb0209B17 and ZM_BFb0331015) was found to be full length. Gene models were developed for the other maize YSL genes (ZmYSL5, ZmYSL10, ZmYSL13, ZmYSL14 and ZmYSL18) by comparing the genomic sequences either with EST sequences corresponding to each gene (when available) or with YSL proteins from other species.
Table 3.
YSL family members identified in maize
| Gene name | BAC clone | EST | AGI cDNA Clone | ZmYS1 similarity |
|---|---|---|---|---|
| ZmYSL1 | ZMMBBc236J17 | Yes | ZM_BFc0116O09 | Positives = 542/679 (79 %) |
| ZmYSL2 | ZMMBBb0160K05 | Yes | NFL | |
| ZmYSL3 | ZMMBBc0188N03 | Yes | ZM_BFc0183J03 | Positives = 580/683 (84 %) |
| ZmYSL5 | ZMMBBc0304K02 | No | NFL | |
| ZmYSL6 | ZMMBBb0104J01 | Yes | ZM_BFc0173F20 | Positives = 395/545 (72 %) |
| ZmYSL10 | ZMMBBc0253M12 | No | NFL | |
| ZmYSL11 | ZMMBBc0253M12 | Yes | ZM_BFb0369O22 | Positives = 472/687 (68 %) |
| ZmYSL11A | ZMMBBc0387I20 | Yes | ZM_BFb0216K02 | Positives = 394/599 (65 %) |
| ZmYSL12 | ZMMBBc0253M12 | Yes | ZM_Bfb0208H07 | Positives = 479/680 (70 %) |
| ZmYSL13 | ZMMBBb0399B15 | Yes | NFL | |
| ZmYSL14 | ZMMBBc0381P05 | Yes | NFL | |
| ZmYSL14A | ZMMBBc0559K13 | Yes | ZM_BFc0143N23 | Positives = 474/695 (68 %) |
| ZmYSL17 | ZMMBBc0116H02 | Yes | ZM_BFb0095P17 | Positives = 393/687 (57 %) |
| ZmYSL18 | ZMMBBc0169J03 | Yes | NFL | |
| Pseudogene A | ZMMBBc0104G21 | Unclear | NFL | |
| Pseudogene B | ZMMBBc0116C01 | Yes | NFL | |
| Pseudogene C | ZMMBBc0130M13 | No | NFL | |
| Pseudogene D | ZMMBBc0217D07 | Yes | NFL |
Maize YSL cDNA clones sequenced in this study are listed, along with amino acid sequence similarity to maize YS1.
NFL, no full-length cDNA clone available.
Phylogenetic tree
Multiple alignment of protein sequences was performed using ClustalW (Thompson et al., 1994). Alignments of the amino acid sequences were checked and refined by eye. The unrooted tree was generated by the Neighbor–Joining method using MEGA5 (Tamura et al., 2011), and 1000 bootstrap replicates were performed. Also incluced in the analysis were the YSL proteins from Physcomitrella patens (Pp111567 and Pp122023), Selaginella moellendorffii (Sm179245 and Sm269976), M. truncatula (Mt1g008570, Mt1g008580, Mt1g008620, Mt1g008640, Mt3g123340, Mt5g098780, Mt6g089710 and Mt7g018260) and G. max (Gm04g41020, Gm06g13820, Gm09g29410, Gm10g31610, Gm11g31870, Gm13g10410, Gm16g05850, Gm16g33840 and Gm17g26520).
PCR
Total RNA was extracted from frozen tissues using the Qiagen RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). RNA samples were then treated with RNase-free DNase I (Ambion, Austin, TX, USA) to remove contaminating genomic DNA. The integrity of RNA samples was examined by reverse transcription–PCR (RT–PCR) before carrying out quantitative real-time PCRs (qRT-PCRs) as described previously (Hong et al., 2008). First-strand cDNA was synthesized from 600 ng of total RNA using SuperScriptIII reverse transcriptase (Invitrogen) and oligo(dT) primers. Reactions without reverse transcriptase were also used for each sample in order to detect any genomic DNA contamination. Equal amounts of each reverse transcription reaction were used as templates for PCR amplifications.
BdUBC18 and BdGAPDH were used as reference genes to normalize qRT-PCR gene expression data. For qRT-PCR analyses of BdYSL genes, the QuantPime primer design tool (Arvidsson et al., 2008) was used whenever possible. Primer sets used for amplification of BdYS1A and BdYS1B were designed using Primer3 software (Rozen and Skaletsky, 2000) because these genes are incorrectly annotated in the brachypodium reference genome sequence as a single gene, making the primers specified in QuantPrime incorrect. Primers used in this study are listed in Table 1. Primer efficiencies of each gene and housekeeping genes were determined empirically by amplifying serial dilutions of the appropriate cDNA template with each primer set.
Table 1.
Primer sequences used in quantitative RT-PCR analysis.
| Sequence (5'–3') | |
|---|---|
| BdYS1A | |
| Forward | CTCATCTGTGTCATGAGTGTGG |
| Reverse | GAACCTGGAATACAGCAAACAC |
| BdYS1B | |
| Forward | AGGCATTTTCGTAGCGATTG |
| Reverse | AATGACAGAAAGGACGAGCAC |
| BdYSL1 | |
| Forward | TCATGGAGATGCAATGGCAAAGC |
| Reverse | AGCTCCAGAGGAAGCTGATTGC |
| BdYSL2 | |
| Forward | TCGTAAGCATGTCAAGAGAGCTG |
| Reverse | TCGCGTTGCATATCATCAATGGC |
| BdYSL3 | |
| Forward | TGCTCCCTCTCAGAAAGGTACTGG |
| Reverse | TATGAGAACGGCAGTCGCAGTC |
| BdYSL4 | |
| Forward | ATCGGTGCATTCTTCGGCGTTG |
| Reverse | TCCTGTCGATCTTCTGCCACAC |
| BdYSL5 | |
| Forward | ATCAACAGCTTCCACACTCCTCAG |
| Reverse | CAGCGTCTTCACCTGTCTCTTG |
| BdYSL6A | |
| Forward | CCCTTGTCCAGGTTCAGGATGATG |
| Reverse | CCAGGGAGGGATACTGTCTTTCAC |
| BdYSL6B | |
| Forward | ACTGCGCAAGGTGATGGTAATTG |
| Reverse | TCAACATAGCTGTGGCAGTTCCG |
| BdYSL9 | |
| Forward | CAGGGAGATGAAGTGGCAAAGATG |
| Reverse | AGCTCCAGAAGAAGCTGATTGC |
| BdYSL10 | |
| Forward | TCAACAGCTTCCATACACCTCAAG |
| Reverse | GGATCATCATCGACGTTTGCTTCG |
| BdYSL11 | |
| Forward | TGGACTGCAGGCTTACAAGGTG |
| Reverse | CGTCAGAGACAGGAAGTGCCTTTG |
| BdYSL12 | |
| Forward | ATCGCCTTCAGCGGTGGATTTG |
| Reverse | TGTTTGATTGGCGATGGTGTCG |
| BdYSL13 | |
| Forward | TCTGGTTCTCTGCCGATCTTGC |
| Reverse | ATGGCTATGAACACCCGGTAGC |
| BdYSL14 | |
| Forward | GCCAAGCTTGCAAAGAAGCAAGTG |
| Reverse | TGGGAAGTTCTTGAAACCGCAGTC |
| BdYSL15 | |
| Forward | CGTTGCGGAAGCTTATGATACTGG |
| Reverse | TCCAGCGATAGCTGAACCAGTTG |
| BdYSL16 | |
| Forward | TAGTGTGCCGTTGAACCAGGTG |
| Reverse | TGAGCTTGAGCTGTCCCAGTAG |
| BdYSL17 | |
| Forward | CACGGCGCATCTCATCAATAGC |
| Reverse | TCGACACCTGCTGCTTTGCTTG |
| BdYSL18 | |
| Forward | ATCGCCATGGTTTGGCTGAGAAGC |
| Reverse | AGGTCCGTCAGATCGGCAATGTAG |
| BdUBC18 | |
| Forward | TCACCCGCAATGACTGTAAG |
| Reverse | ACCACCATCTGGTCTCCTTC |
| BdGAPDH (qRT-PCR) | |
| Forward | GCTCCCATGTTTGTTGTCG |
| Reverse | GACCCTCAACAATGCCAAAG |
| BdGAPDH (RT-PCR) | |
| Forward | ATGGGCAAGATTAAGATCGGAATCAACGG |
| Reverse | AGTGGTGCAGCTAGCATTTGAGACAAT |
Brilliant II SYBR Green QPCR Master Mix (Stratagene, La Jolla, CA, USA) was used to carry out qRT-PCRs. The two-step thermal cycling profile used was 15 min at 95 °C, followed by 40 cycles of 10 s at 95 °C, 30 s at 60 °C. Following amplification, melt curves were performed to verify that a single product was amplified. All qRT-PCRs were carried out using at least three technical replicates.
The final threshold cycle (Ct) values were the mean of at least three replicates. The comparative ΔCt method was used to evaluate the relative quantities of each amplified product by comparing the Ct values of the samples of interest with a reference (calibrator) sample. The Ct values of both the reference samples and the samples of interest were normalized to BdUBC18 and BdGAPDH, and primer efficiencies were taken into account using the geNorm algorithm (Vandesompele et al., 2002). A negative control with water instead of cDNA was included for each qRT-PCR set.
Collection and analysis of root exudates and identification of phytosiderophores
Plants were germinated in the dark at 22 °C on a mesh in contact with deionized water. After germination, seedlings were cultured in 1/5 Hoagland solution in a growth chamber (22 °C day/18 °C night, 14 h/10 h). The nutrient solution was renewed every 3 d. After 16 d of culture, the seedlings were transferred to 1/5 Hoagland with or without Fe and used for the following experiments.
The root exudates were collected from 45-day-old seedlings, which had been subjected to Fe deficiency for 2 weeks. On the day before exudate collection, roots were washed with deionized water twice, and then placed in a pot containing 1·2 L of deionized water. Collection of the root exudates was started at 1730 h in a growth chamber (20 °C day/15 °C night). At 2 h intervals during the day, the exudate solution was removed and roots were transferred to fresh deionized water for a total period of 24 h. No antimicrobial reagent was used during collection of root exudates since this could affect the biosynthesis of PS. Instead, the roots were kept very clean by changing the solution frequently. The root exudates collected were immediately passed through a cation exchange column (16 mm × 14 cm) filled with Amberlite IR 120B (H+ form, Organo Co., Tokyo, Japan) and eluted with 2 m NH4OH (Ma et al., 2003). The eluates were concentrated by using a rotary evaporator at 40 °C. After the residues were dissolved in 1 mL of distilled water, the type and amounts of PS were determined by high-performance liquid chromatography (HPLC) using a cation exchange column (Shim-Pack, Amino-Li; Shimadzu Co., Kyoto, Japan) according to Ueno et al. (2007). Phytosiderophores [epi-3-hydroxy-2'-deoxymugineic acid (epiHDMA), HDMA, epi-hydroxymugineic acid (epiHMA), HMA, MA and DMA] used for standards were obtained from our previous studies (Ma et al., 2003; Ueno et al., 2007). Their purity and structures have been confirmed by nuclear magnetic resonance (NMR). The concentration was calculated based on the peak area of HDMA on the HPLC traces. Purified PS were used for comparison of retention times.
RESULTS
Identification of brachypodium and maize YSL family members
The amino acid sequence of ZmYS1 shows strong similarity to 19 predicted brachypodium proteins (Table 2). Brachypodium YSLs showed 57–88 % sequence similarity to ZmYS1 (Table 2). In the maize genome, 18 potential YSL family members were identified. Nine of these had apparently full-length cDNA clones available, and these were sequenced (Table 3). Sequencing revealed that four YSLs in the reference maize line, B73, contain frameshift mutations leading to early stop codons, which would presumably prevent translation of a functional protein. Therefore, these were not included in further analyses, and are listed as pseudogenes (Table 3). Maize YSLs showed 54–87 % sequence similarity to YS1 (Table 3).
Table 2.
List of putative brachypodium YSL (BdYSL) genes and BdYSL amino acid sequence similarities toZmYS1
| Gene name | JGIv1·0 name | ZmYS1 similarity |
|---|---|---|
| BdYS1A | Bradi3g50270 | Positives = 589/669 (88 %) |
| BdYS1B | Bradi3g50270 | Positives = 389/455 (85 %) |
| BdYSL1 | Bradi5g17220 | Positives = 538/673 (79 %) |
| BdYSL2 | Bradi3g50260 | Positives = 576/683 (84 %) |
| BdYSL3 | Bradi5g17230 | Positives = 559/667 (83 %) |
| BdYSL4 | Bradi5g08280 | Positives = 436/646 (67 %) |
| BdYSL5 | Bradi5g25990 | Positives = 432/652 (66 %) |
| BdYSL6A | Bradi5g08260 | Positives = 461/643 (71 %) |
| BdYSL6B | Bradi5g08250 | Positives = 459/639 (71 %) |
| BdYSL9 | Bradi5g17210 | Positives = 521/680 (76 %) |
| BdYSL10 | Bradi2g53950 | Positives = 412/659 (62 %) |
| BdYSL11 | Bradi5g16190 | Positives = 462/649 (71 %) |
| BdYSL12 | Bradi5g16170 | Positives = 475/676 (70 %) |
| BdYSL13 | Bradi5g16160 | Positives = 486/702 (69 %) |
| BdYSL14 | Bradi3g49520 | Positives = 461/648 (71 %) |
| BdYSL15 | Bradi2g08270 | Positives = 402/665 (60 %) |
| BdYSL16 | Bradi2g31720 | Positives = 381/668 (57 %) |
| BdYSL17 | Bradi3g01520 | Positives = 438/658 (66 %) |
| BdYSL18 | Bradi3g49490 | Positives = 355/538 (65 %) |
The nomenclature of YSL proteins in the literature is imperfect. The eight arabidopsis YSL proteins were first identified according to their similarity to maize YS1 protein and named AtYSL1–AtYSL8 (Curie et al., 2001). Originally, 18 rice YSL genes were identified based on sequence similarity to maize YS1 and designated OsYSL1–OsYSL18 (Gross et al., 2003) but in a subsequent study of the OsYSL family in rice they were renamed (again as OsYSL1–OsYSL18) (Koike et al., 2004). These are the names that have been commonly used in the literature. However, arabidopsis and rice YSL family members are not given names in accordance with their phylogenetic relatedness, or their functional similarities. In this study we aimed to overcome this disorder in the literature by designating names for brachypodium and maize YSL members as closely as possible according to their phylogenetic relationship to the arabidopsis and rice YSLs. We have named the two genes most closely related to ZmYS1, HvYS1 and OsYSL15 as BdYS1A and BdYS1B. The remainder of the family members are called YSLs and are given numbers that best match the numbers of the the closest related genes in arabidopsis or rice. In cases where two very closely related genes are found in a species, we designated the genes ‘A’ and ‘B’, e.g. BdYSL6A and BdYSL6B, which are present as a tandem repeat on brachypodium chromosome 5.
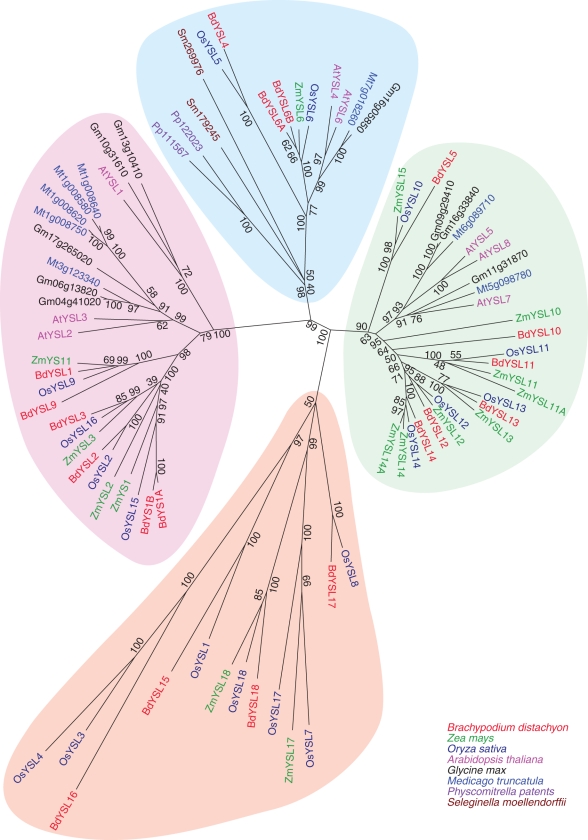
The complete genome sequences of O. sativa, Z. mays and B. distachyon are available. We included the YSLs of these three species as well as those from the model dicot A. thaliana, M. truncatula and G. max in a phylogenetic analysis. The moss P. patens (Pp111567 and Pp122023) and the lycopodium S. moellendorffii (Sm179245 and Sm269976) each have two YSL genes, which were also included in the analysis. Four clades were observed, one of which is found only in the grass species (Fig. 1, orange clade). Most of the BdYSLs identified are closely related to a single rice YSL (OsYSL) and a single maize YSL (ZmYSL), indicating the close relationship between individual members of the family in these grasses. Grass YSLs and dicot YSLs fall into discrete sub-clades within two of the major clades containing both grass and non-grass members (Fig. 1, pink and green clades). One of the largest clades (Fig. 1, pink) contains the YSLs that are most closely related to the founding member of the family, ZmYS1. The proteins from P. patens and S. moellendorffii fall into a single clade, which is thus apparently the most basal clade of the YSL family (Fig. 1, blue clade). The single grass-specific clade is phylogenetically distant from the other three clades (Fig. 1, orange clade).
Fig. 1.
Phylogenetic analysis of the family of YSL proteins from brachypodium (BdYSL), maize (ZmYSL), rice (OsYSLs), arabidopsis (AtYSL), Medicago truncatula and soy. Maize YS1 (ZmYS1), the two Physcomitrella patens and the two Selaginella moellendorffii YSLs are also included in the analysis. Values indicate the number of times (as a percentage of 1000 replicates) that each branch topology was found during bootstrap analysis.
Brachypodium YS1 orthologues
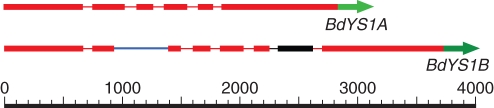
The in silico analysis of the YSL family of brachypodium revealed that there are two genes in brachypodium that are most likely to be YS1 orthologues. We designated these proteins as BdYS1A and BdYS1B. Both of these genes show strong sequence similarity and are closely related to ZmYS1, HvYS1 (Murata et al., 2006) and OsYSL15 (Fig. 1). Moreover, the map positions of these genes (on chromosome 3 of brachypodium) are syntenic with OsYSL15 of rice (on choromosome 2) and with ZmYS1 of maize (on chromosome 5). BdYS1A and BdYS1B share strong DNA-level similarity (88 %) that includes both coding sequences, most introns (the exception is a single intron in BdYS1B that is not present in BdYS1A) and the 5′ untranslated regions (UTRs; Fig. 2). The 3′ UTRs of the two genes are distinct.
Fig. 2.
Structures of BdYS1A and BdYS1B – the orthologues of ZmYS1. The extra intron (blue) and duplicated segment (black) of BdYS1B are shown. Red areas show strong DNA similarity. The 3′ UTRs of the genes are unique (green).
One of the two putative brachypodium YS1 genes (BdYS1B) may be non-functional. Two features lead us to this conclusion. First, this gene has an unusual, approx. 400 bp long duplication of sequence that encompasses all of the final intron and the 5′ portion of the final exon of the gene (Fig. 2). Using RT–PCR, cDNA clones of BdYS1B were obtained that have the duplicated exon sequence, which adds approx. 100 amino acids to the protein. It is not known whether such a large insertion of sequence would cause the encoded protein to become non-functional. A second unusual feature of BdYS1B is the retained intron that is found in the majority of cDNAs we have cloned. The presence of the intron would cause a premature stop in the protein, which would almost certainly lead to a non-functional product. However, we were able to detect and clone the fully spliced version of BdYS1B mRNA, so the intron is not always retained.
BdYS1A and BdYS1B are regulated by iron
The Fe status of the plant influences the expression of the YS1 genes in maize, barley and rice (Curie et al., 2001; Koike et al., 2004; Murata et al., 2006; Inoue et al., 2009; Lee et al., 2009; Ueno et al., 2009). ZmYS1 mRNA and protein levels were strongly elevated in both leaves and roots of Fe-starved plants (Roberts et al., 2004; Ueno et al., 2009), and OsYSL15 appears likewise to be upregulated in both leaves and roots during Fe deficiency (Lee et al., 2009). The barley YS1 orthologue HvYS1 was expressed exclusively in the roots, and expression increased in Fe-deficient roots (Murata et al., 2006; Ueno et al., 2009).
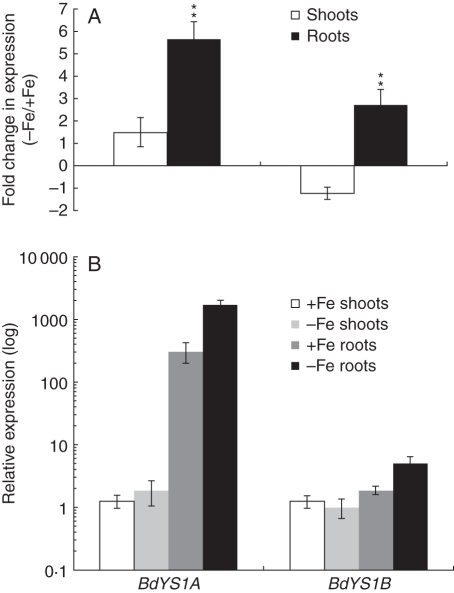
To determine whether Fe regulates the expression of BdYS1A and BdYS1B, we performed qRT-PCR analysis. Since there is strong sequence similarity between the two genes, we designed primer pairs corresponding to the 3′ UTR of each gene to ensure gene-specific amplification. After a period of 10 d of Fe starvation, plants were harvested and transcript levels of both genes were measured in shoots and roots of Fe-sufficient and Fe-deficient plants. Levels of both genes were extremely low in shoots, and there was no notable increase in the transcript levels in the shoots under Fe-deficient conditions for either BdYS1 gene (Fig. 3A). Quantitative analysis of mRNA levels of both BdYS1 genes in roots revealed that the transcript level of BdYS1A is significantly higher than that of BdYS1B (165-fold higher in Fe-sufficient roots, P < 0·01; 340-fold higher in Fe-deficient roots, P < 0·01; Fig. 3B). Expression of both BdYS1A and BdYS1B was upregulated in the roots under Fe-deficient conditions, as expected for a YS1 gene (Fig. 3A). Expression of BdYS1A was prominently induced in the roots of Fe-deficient plants compared with Fe-sufficient plants (5·7-fold, P < 0·01). On the other hand, BdYS1B expression was only moderately elevated (2·7-fold; P < 0·01; Fig. 3A).
Fig. 3.
Real-time PCR quantification of BdYS1A and BdYS1B expression levels during Fe deficiency. One-week-old plants were transferred for 10 d to Fe-deficient medium or, as a control, to Fe-containing medium, before harvesting. BdYS1 values were normalized using BdGAPDH and BdUBC18 as internal standards. Two representations of the data are shown. (A) Fold change in mRNA level during iron deficiency. Note the linear scale. (B) Relative expression levels. The condition with the lowest expression level (BdYS1B shoots grown without Fe) was set to 1, and all other values are set relative to this. Note the log10 scale. Data are means ± s.d. (n = 5). * Minimum 2-fold difference and P < 0·05; **minimum 2-fold difference and P < 0·01.
Transport activities of BdYS1A and BdYS1B
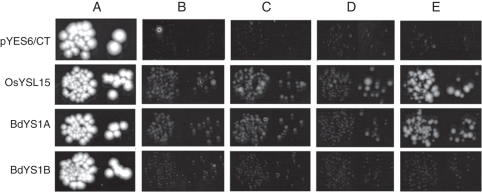
The Fe transport abilities of BdYS1A and BdYS1B were tested using the Fe uptake-defective yeast strain fet3fet4. The mutant strain was transformed with BdYS1A- or BdY S1B-expressing plasmids, or with the OsYSL15-expressing plasmid as a positive control (Lee et al., 2009). The empty vector pYES6/CT was used as a negative control. In order to control the level of expression in yeast, a β-oestradiol-inducible system was used (Gao and Pinkham, 2000). Viability of the strains was shown by growing them on medium with 50 µm iron citrate permissive conditions (Fig. 4A). To test whether BdYS1A and BdYS1B are able to transport Fe3+, the strains were grown on medium with FeCl3 (no PS and no YS1 gene expression; Fig. 4B), FeCl3 plus β-oestradiol (no PS and the YS1 gene is expressed; Fig. 4C), FeCl3 plus DMA (PS present, but no YS1 gene expression; Fig. 4D) and FeCl3 plus DMA and β-oestradiol (PS present and the YS1 gene is expressed; Fig. 4E). The results indicate that BdYS1A complemented yeast growth only when FeCl3 was supplied along with DMA and β-oestradiol in the medium (Fig. 4E). When β-oestradiol was withheld from the medium the yeast strain was unable to grow, showing its dependence on BdYS1A expression (Fig. 4D). BdYS1B did not complement yeast growth in any of the test conditions, indicating that BdYS1B is not capable of transporting Fe–PS complexes, and is, as suspected, a defective gene (pseudogene).
Fig. 4.
Functional complementation of fet3fet4 yeast. DEY1453-derived yeast strains transformed with pGEV-TRP and constructs containing OsYSL15, BdYS1A, BdYS1B or empty pYES6/CT were grown on synthetic defined media containing variable conditions for Fe(III), chelator (DMA) and β-oestradiol (BE). Pairs of spots correspond to 100-fold and 1000-fold dilutions of the original cultures. (A) Iron citrate (50 µm) (B) FeCl3 (5 µm). (C) FeCl3 (5 µm) with 10 nm β-oestradiol. (D) FeCl3 (5 µm) with 10 µm DMA. (E) FeCl3 (5 µm) with 10 µm DMA and 10 nm β-oestradiol (E).
Expression patterns of BdYSLs in response to iron
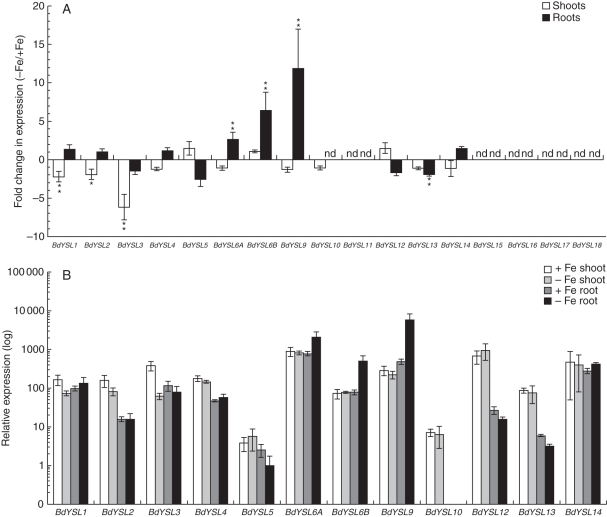
Among the 19 YSL genes, BdYS1A, BdYS1B, BdYSL1, BdYSL2, BdYSL3 and BdYSL9 are phylogenetically grouped together in a clade that contains both grass and non-grass members, and includes the founding member ZmYS1 (Fig. 1, pink clade). BdYSL6A, BdYSL6B and BdYSL4 genes belong to a distinct clade that is also conserved in both non-grasses and grasses (Fig. 1, blue clade). This clade appears to be the basal clade for the YSL family, since the YSLs from P. patens and S. moellendorffii are also members of this clade. BdYSL5, BdYSL10, BdYSL11, BdYSL12, BdYSL13 and BdYSL14 are grouped together in a third large clade containing both grass and non-grass members (Fig. 1, green clade). Finally, BdYSL15, BdYSL16, BdYSL17 and BdYSL18 are members of the grass-specific clade (Fig. 1, orange clade). Expression of the BdYSL genes was evaluated by qRT-PCR analysis in the roots and shoots of Fe-sufficient and Fe-deficient plants (Fig. 5).
Fig. 5.
Real-time PCR quantification of BdYSL mRNA levels in Fe-deficient shoots and roots. One-week-old plants were transferred for 10 d to Fe-deficient medium or, as a control, to Fe-containing medium, before harvesting. BdYS1 values were normalized using BdGAPDH and BdUBC18 as internal standards. Two representations of the data are shown. (A) Fold change in mRNA level during iron deficiency. Note the linear scale. (B) Relative expression levels. The condition with the lowest expression level (BdYSL5 Fe-deficient roots) was set to 1, and all other values are set relative to this. Note the log10 scale. Data are means ± s.d. (n = 3). * Minimum 2-fold difference and P < 0·05; **minimum 2-fold differenceand P < 0·01.
In shoots, expression of BdYSLs was either downregulated by Fe deficiency or unchanged. BdYSL3 showed the strongest change in shoots in response to deficiency, with a 6·2-fold (P < 0·01) reduction in mRNA level. Smaller reductions in the transcript levels of BdYSL1 (2·2-fold, P < 0·01) and BdYSL2 (1·9-fold, P < 0·05) were also observed in Fe-deficient shoots. This pattern of downregulation is similar to that of arabidopis YSL1, YSL2 and YSL3 genes, which belong to the same clade. This pattern may be indicative of a role in unloading of Fe from vascular regions (DiDonato et al., 2004). BdYSL11, BdYSL15, BdYSL16, BdYSL17 and BdYSL18 transcripts were not detected in shoots. Other BdYSL genes did not display notable changes in expression in Fe-deficient shoots (Fig. 5A).
In the roots, Fe-regulated expression was observed for BdYSL6A, BdYSL6B, BdYSL9 and BdYSL13 (Fig. 5A). BdYSL6A, BdYSL6B and BdYSL9 mRNA levels were elevated by 2·6- (P < 0·01), 6·5- (P < 0·01) and 11·9-fold (P < 0·01), respectively, in Fe-starved roots (Fig. 5A). The BdYSL13 mRNA level was reduced 1·9-fold (P < 0·01) in response to Fe deficiency. No expression was detected for BdYSL10, BdYSL11, BdYSL15, BdYSL16, BdYSL17 and BdYSL18 in roots.
The relative level of expression for each YSL was examined in order to determine which BdYSL genes are most abundantly expressed in each organ (Fig. 5B). BdYSL6A and BdYSL9 mRNA levels are highest among the members of the BdYSL family (excluding BdYS1A and BdYS1B). BdYSL9 had the highest transcript level of any BdYSL under Fe-deficient conditions in roots (Fig. 5B), indicating a potential role in Fe acquisition. However, we note that BdYS1A is expressed at higher levels than BdYSL9 (4·8-fold higher in Fe-sufficient roots, P < 0·01; 2·3-fold higher in Fe-deficient roots, P < 0·01). The BdYSL6A expression level was high under all conditions tested, with the highest expression in Fe-deficient roots. BdYSL5 and BdYSL10 mRNA levels were the lowest among the BdYSL genes (Fig. 5B). Expression of various BdYSL genes differs by up to 5000-fold.
Phytosiderophores in brachypodium
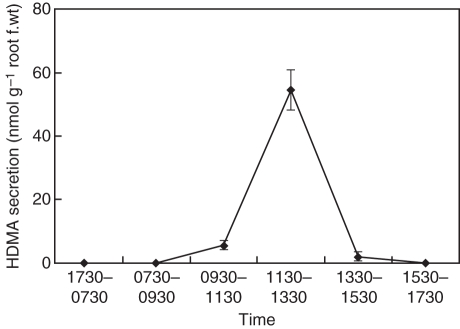
In Strategy II plants, there is a positive correlation between the amount of PS released and the tolerance of plants to Fe deficiency (Marschner, 1995). Rice, sorghum and maize secrete only small amounts of DMA, while barley secretes large amounts of PS including MA, HMA and epiHMA (Mori et al., 1987; Kawai et al., 1988; Ma and Nomoto, 1993). For several grasses, it has been demonstrated that secretion of PS follows a distinct diurnal rhythm (Tagaki et al., 1984; Romheld and Marchner, 1986; Ueno et al., 2009). To characterize brachypodium as a model species to study Fe homeostasis in grasses, we investigated the secretion rhythm and the types of PS secreted by brachypodium. In Fe-deficient brachypodium, PS secretion from roots follows a diurnal rhythm (Fig. 6). With daybreak occurring at 0730 h, secretion reached a peak during 1130–1330 h.
Fig. 6.
Diurnal changes of phytosiderophore secretion in Fe-deficient brachypodium. The collection of root exudates was conducted every 2 h from 0730 to 1730 h at 20 °C. Lights on was at 0730 h. The amount of HDMA was determined by HPLC. Error bars represent ± s.d. (n = 3).
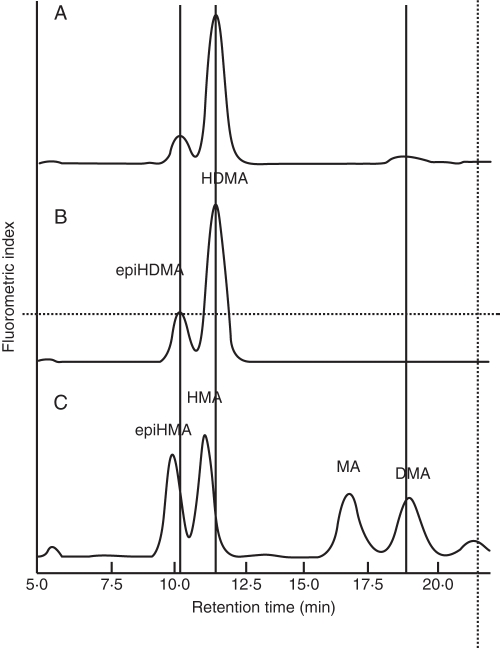
The types of PS secreted by brachypodium plants were also identified using HPLC analysis. Comparison of the retention times with those of identified PS showed that brachypodium secreted HDMA, epiHDMA and DMA, with HDMA being the major Fe(III)-chelating compound secreted (Fig. 7).
Fig. 7.
High-performance liquid chromatography (HPLC) spectra of the root exudates from Fe-deficient brachypodium. (A) Spectra of brachypodium root exudates. (B, C) Standards of 3-epihydroxy-2′-deoxymugineic acid (epiHDMA), 3-epihydroxymugineic acid (epiHMA), 3-hydroxymugineic acid (HMA), 2′-deoxymugineic acid (DMA), mugineic acid (MA) and 3-hydroxy-2′-deoxymugineic acid (HDMA).
DISCUSSION
The emerging model species brachypodium is an extremely promising model that can be used to enhance research into grass-specific traits. Since the Fe uptake strategy (Strategy II) and Fe translocation in grasses differ from those of non-grasses, brachypodium represents a good model system for elucidation of grass-specific aspects of Fe homeostasis. In this study we aimed to identify YSL family members in brachypodium and maize that show strong sequence similarity to the maize YS1 protein, and are believed to be involved in maintaining proper homeostasis of Fe and other transition metals in both grasses and non-grasses. Our analysis has revealed that there are 19 members of this family in brachypodium (Table 2) and 18 members in maize (Table 3).
The phylogenetic analysis presented here includes the complete YSL families of three grass species: brachypodium, rice and maize, thus expanding a previous phylogenetic analysis using rice as the sole representitive of a grass (Curie et al., 2009). According to phylogenetic analysis, land plant YSLs can be grouped into four major clades. Only one of these clades is grass specific, and includes YSLs from brachypodium, maize and rice (Fig. 1, orange clade). This clade is also phylogenetically distant from the other three clades, which suggests that these YSLs might perform exclusive functions in grasses. Information about members of this clade is currently very limited, as only one, OsYSL18, has been studied. Interestingly, OsYSL18 appears to transport Fe(III)–PS, but not complexes of other metals or metals with NA (Aoyama et al., 2009). In a recent microarray study in which genome-wide expression data throughout the life cycle of rice plants were generated, OsYSL genes of this grass-specific clade (Fig. 1, orange clade) were shown to be expressed only in reproductive tissues, mainly in stamens (Wang et al., 2010). These results correspond to our findings that none of these BdYSL genes could be detected in shoots or roots, and suggest unique roles for these YSLs in reproductive tissues. In addition to their potential role(s) in reproductive structures, the YSLs in this clade are used only under very specific conditions such as heavy metal stress, or other biotic or abiotic stresses. The other three major clades include YSLs from both grasses and non-grasses. The largest group includes YSLs that cluster with the founding member of the YSL family, ZmYS1 (Fig. 1, pink clade). AtYSLs in this group (AtYSL1, AtYSL2 and AtYSL3) transport Fe–NA complexes (DiDonato et al., 2004; Chu et al., 2010). Interestingly, the expression patterns of BdYSL1, BdYSL2 and BdYSL3 were well correlated with those of arabidopsis YSL1, YSL2 and YSL3 (Fig. 5A). These genes are downregulated in response to Fe deficiency (DiDonato et al., 2004; Waters et al., 2006), particularly in shoots. In arabidopsis, this downregulation is hypothesized to indicate a role in xylem unloading, which is downregulated in mature leaves to allow Fe to be carried more efficiently into younger tissues. Future studies regarding the cell type-specific expression and mutant phenotypes of BdYSL1, BdYSL2 and BdYSL3 may elucidate whether these genes have similar roles in Fe homeostasis in brachypodium.
Previous studies have identified maize YS1 orthologues in barley (Murata et al., 2006) and rice (Inoue et al., 2009; Lee et al., 2009). ZmYS1 and HvYS1 are localized at the root epidermal cells (Murata et al., 2006; Ueno et al., 2009) and are responsible for transport of Fe(III)–PS complexes. Expression analysis showed that HvYS1 is mainly expressed in the roots, and expression levels were significantly elevated in response to Fe deficiency (Murata et al., 2006). In rice, OsYSL15 was identified as the Fe–PS transporter that is responsible for Fe acquisition. It has been demonstrated that OsYSL15 expression was strongly upregulated in Fe-deficient roots, but not in leaves (Inoue et al., 2009). In contrast, Lee et al. (2009) showed that in a different rice cultivar, expression of OsYSL15 is upregulated in both shoots and roots. Genetic variation between cultivars may explain the differences between the two studies. We identified two different genes, BdYS1A and BdYS1B, as orthologues of ZmYS1. Both genes are upregulated under Fe-deficient conditions only in roots. However, transcript levels of BdYS1B are much lower than those of BdYS1A (Fig. 3B), and BdYS1B is not capable of complementing the growth defect of Fe uptake-deficient yeast (Fig. 4). The presence of a repeated exon sequence, a retained intron, low expression levels and weak induction of BdYS1B in response to Fe deficiency show that BdYS1B is most probably non-functional. BdYS1A is the Fe(III)–PS transporter that is responsible for iron uptake in brachypodium.
Many of the brachypodium YSL genes identified are closely related to a single maize and single rice gene, suggesting functional conservation of each YSL family member. Surprisingly, the patterns of expression observed for several BdYSL genes did not correlate well with the patterns observed for the corresponding rice YSL genes reported in the literature. OsYSL2 and BdYSL2 have opposite patterns of regulation during Fe deficiency: OsYSL2 is upregulated (Koike et al., 2004), whereas BdYSL2 is downregulated by Fe deficiency. Apparent orthologues OsYSL16 and BdYSL3 (Fig. 1) again show marked differences: BdYSL3 showed a 6-fold downregulation in Fe-deficient shoots, while OsYSL16 expression did not change in shoots or roots (Lee et al., 2009) or was even increased in roots (Inoue et al., 2009). Both BdYSL1 and BdYSL9 are phylogenetically grouped with OsYSL9 (Fig. 1). OsYSL9 transcript levels increase under Fe deficiency, particularly in shoots (Lee et al., 2009), while BdYSL1 transcript levels decreased in Fe-deficient shoots. BdYSL9 expression did increase in response to Fe deficiency, but the increase was observed in roots, not in shoots (Fig. 4). Iron deficiency slightly reduced expression of OsYSL6 in rice roots (Koike et al., 2004; Inoue et al., 2009), but both BdYSL6A and BdYSL6B expression levels were higher in Fe-deficient roots (Fig. 5A). The differences in expression profiles of some of the related rice and brachypodium YSL genes may be due to rice's ability to use either Fe(II)–PS (taken up through OsIRT1 and OsIRT2; Bughio et al., 2002; Ishimaru et al., 2006) or Fe(III)–PS (taken up through OsYSL15; Inoue et al., 2009; Lee et al., 2009). Therefore, the form of Fe translocated throughout the rice plant may vary, necessitating specific and distinct patterns of YSL expression in each species.
The brachypodium and rice members of the second largest clade (Fig. 1, green clade) generally seem to exhibit very similar expression profiles. BdYSL12 and BdYSL14 and OsYSL12 and OsYSL14 transcripts were detected in shoots and roots, and Fe status did not affect their expression (Fig. 5A; Koike et al., 2004; Inoue et al., 2009). Transcript levels of BdYSL5, BdYSL10 and BdYSL11 and orthologous rice members OsYSL10 and OsYSL11 were either very low or not detected (Inoue et al., 2009). BdYSL13 and OsYSL13 expression in both roots and shoots was reduced in response Fe deficiency (Fig. 5A) (Koike et al., 2004). The similar expression profiles of grass-specific YSL genes in this clade suggest that these YSLs could be functioning in a similar fashion among grass species.
Our aim in this study was to introduce and establish brachypodium as a new model system to study Fe deficiency responses in grasses. Therefore, we analysed the pattern of PS secretion, and the types of PS secreted by brachypodium plants under Fe starvation. Like wheat and barley, brachypodium secretes PS with a distinct diurnal rhythm (Tagaki et al., 1984; Romheld and Marchner, 1986; Ueno et al., 2009). Furthermore, our HPLC analysis revealed that brachypodium plants secrete three types of PS, i.e. epiHDMA, DMA and HDMA, the latter of which is the major Fe(III)-chelating compound secreted by brachypodium (Fig. 7). These types are similar to those secreted by Lolium perenne (Ueno et al., 2007). It is supposed that epiHDMA and HDMA are synthesized from DMA by hydroxylation at the C-3 position by the activity of IDS2 [the protein encoded by Iron Deficiency Specific Clone 2 (Nakanishi et al., 2000)]. The protein encoded by Iron Deficiency Specific Clone 3 (IDS3) hydroxylates the C-2' position of DMA, converting DMA to MA (Nakanishi et al., 2000; Kobayashi et al., 2001). Our results demonstrated that MA is not one of the types of PS secreted by brachypodium. Accordingly, a TBLASTN search using the amino acid sequence of barley IDS3 against the brachypodium genome indicated that brachypodium lacks IDS3 orthologues.
Brachypodium is a new model plant that will serve as a model for functional genomics studies in grasses. Here we demonstrate that brachypodium represents a facile model to investigate Fe homeostasis in grasses. Our phylogenetic analysis described the members of the BdYSL family and their phylogenetic relatedness to YSL families of rice and maize. Furthermore, the low expression of all grass-specific BdYSLs in vegetative tissues suggests that these YSLs could have highly specialized roles, perhaps in reproduction, that do not occur in non-grass species. Regulation of the BdYSL genes by Fe status suggested roles for these transporters in Fe metabolism.
ACKNOWLEDGEMENTS
This work was supported by grants from the USDA (AFRI grant no. 2009-02268 to E.L.W.), and the National Science Foundation (grant no. IOS0847687 to E.L.W.), by the Program of Promotion of Basic Research Activities for Innovative Biosciences (BRAIN to J.F.M), a Sunbor grant and the Ohara Foundation for Agricultural Science. We gratefully thank Teddi Bloniarz for her expert assistance with growth chambers and in the greenhouse.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Reseasrch. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Kobayashi T, Takahashi M, et al. OsYSL18 is a rice iron(III)–deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Molecular Biology. 2009;70:681–692. doi: 10.1007/s11103-009-9500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Arabidopsis Genome Initiative: analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–813. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riano-Pachon DM, Mueller-Roeber B. QuantPrime – a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008;9:465. doi: 10.1186/1471-2105-9-465. doi:10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell WD, Bogorad L, McIlrath WJ. Yellow-stripe phenotype in maize. I. Effects of ys1 locus on uptake and utilization of iron. Botanical Gazette. 1962;124:1–8. [Google Scholar]
- Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S. Cloning an iron-regulated metal transporter from rice. Journal of Experimental Botany. 2002;53:1677–1682. doi: 10.1093/jxb/erf004. [DOI] [PubMed] [Google Scholar]
- Cheng L, Wang F, Shou H, et al. Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiology. 2007;145:1647–57. doi: 10.1104/pp.107.107912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H-H, Chiecko J, Punshon T, et al. Successful reproduction requires the function of Arabidopsis YELLOW STRIPE-LIKE1 and YELLOW STRIPE-LIKE3 metal–nicotianamine transporters in both vegetative and reproductive structures. Plant Physiology. 2010;154:197–210. doi: 10.1104/pp.110.159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- Curie C, Cassin G, Couch D, et al. Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Annals of Botany. 2009;103:1–11. doi: 10.1093/aob/mcn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato R, Jr, Roberts L, Sanderson T, Eisley R, Walker E. Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine–metal complexes. The Plant Journal. 2004;39:403–414. doi: 10.1111/j.1365-313X.2004.02128.x. [DOI] [PubMed] [Google Scholar]
- Gao CY, Pinkham JL. Tightly regulated, beta-estradiol dose-dependent expression system for yeast. Biotechniques. 2000;29:1226–1231. doi: 10.2144/00296st02. [DOI] [PubMed] [Google Scholar]
- Gendre D, Czernic P, Conejero G, et al. TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine–Ni/Fe transporter. The Plant Journal. 2007;49:1–15. doi: 10.1111/j.1365-313X.2006.02937.x. [DOI] [PubMed] [Google Scholar]
- Gross J, Stein RJ, Fett-Neto AG, Fett JP. Iron homeostasis related genes in rice. Genetics and Molecular Biology. 2003;26:477–497. [Google Scholar]
- Hong S-Y, Seo PJ, Yang M-S, Xiang F, Park C-M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biology. 2008;8:112. doi: 10.1186/1471-2229-8-112. doi:10.1186/1471-2229-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo N, Gu YQ, Lazo GR, et al. Construction and characterization of two BAC libraries from Brachypodium distachyon, a new model for grass genomics. Genome. 2006;49:1099–1108. doi: 10.1139/g06-087. [DOI] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, et al. Rice OsYSL15 is an iron-regulated iron(III)–deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. Journal of Biological Chemistry. 2009;284:3470–3479. doi: 10.1074/jbc.M806042200. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, et al. Rice plants take up iron as an Fe3 + -phytosiderophore and as Fe2 + The Plant Journal. 2006;45:335–46. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Kim S, Tsukamoto T, et al. Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Proceedings of the National Academy of Sciences, USA. 2007;104:7373–7378. doi: 10.1073/pnas.0610555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S, Takagi S, Sato Y. Mugineic acid-family phytosiderophores in root-secretions of barley, com and sorghum varieties. Journal of Plant Nutrition. 1988;11:633–642. [Google Scholar]
- Kellogg EA. Evolutionary history of the grasses. Plant Physiology. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nakanishi H, Takahashi M, Kawasaki S, Nishizawa NK, Mori S. In vivo evidence that Ids3 from Hordeum vulgare encodes a dioxygenase that converts 2′-deoxymugineic acid to mugineic acid in transgenic rice. Planta. 2001;212:864–871. doi: 10.1007/s004250000453. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Suzuki M, Inoue H, et al. Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. Journal of Experimental Botany. 2005;56:1305–1316. doi: 10.1093/jxb/eri131. [DOI] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, et al. OsYSL2 is a rice metal–nicotianamine transporter that is regulated by iron and expressed in the phloem. The Plant Journal. 2004;39:415–424. doi: 10.1111/j.1365-313X.2004.02146.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Chiecko JC, Kim SA, et al. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiology. 2009;150:786–800. doi: 10.1104/pp.109.135418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Nomoto K. Two related biosynthetic pathways for mugineic acids in gramineous plants. Plant Physiology. 1993;102:373–378. doi: 10.1104/pp.102.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Ueno H, Ueno D, Rombola AD, Iwashita T. Characterization of phytosiderophore secretion under Fe deficiency stress in Festuca rubra. Plant and Soil. 2003;256:131–137. [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. New York: Academic Press; 1995. [Google Scholar]
- Mori S, Nishizawa N, Kawai S, Sato Y, Takagi S. Dynamic state of mugineic acid and analogous phytosiderophores in Fe-deficient barley. Journal of Plant Nutrition. 1987;10:1003–1011. [Google Scholar]
- Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T. A specific transporter for iron(III)–phytosiderophore in barley roots. The Plant Journal. 2006;46:563–572. doi: 10.1111/j.1365-313X.2006.02714.x. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Yamaguchi H, Sasakuma T, Nishizawa NK, Mori S. Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgare are involved in the biosynthesis of mugineic acid family phytosiderophores. Plant Molecular Biology. 2000;44:199–207. doi: 10.1023/a:1006491521586. [DOI] [PubMed] [Google Scholar]
- Nozoye T, Nagasaka S, Kobayashi T, et al. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. Journal of Biological Chemistry. 2011;286:5446–5454. doi: 10.1074/jbc.M110.180026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LA, Pierson AJ, Panaviene Z, Walker EL. Yellow stripe1. Expanded roles for the maize iron–phytosiderophore transporter. Plant Physiology. 2004;135:112–120. doi: 10.1104/pp.103.037572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romheld V, Marchner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiology. 1986;80:175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, Wirén NV. ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. Journal of Biological Chemistry. 2004;279:9091–9096. doi: 10.1074/jbc.M311799200. [DOI] [PubMed] [Google Scholar]
- Tagaki S, Nomoto K, Takemoto T. Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. Journal of Plant Nutrition. 1984;7:469–477. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011 doi: 10.1093/molbev/msr121. in press. doi:10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno D, Rombola A, Iwashita T, Nomoto K, Ma JF. Identification of two novel phytosiderophores secreted from perennial grasses. New Phytologist. 2007;174:304–310. doi: 10.1111/j.1469-8137.2007.02056.x. [DOI] [PubMed] [Google Scholar]
- Ueno D, Yamaji N, Ma JF. Further characterization of ferric–phytosiderophore transporters ZmYS1 and HvYS1 in maize and barley. Journal of Experimental Botany. 2009;60:3513–3520. doi: 10.1093/jxb/erp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034. doi:10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Hill T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Reports. 2008;27:471–478. doi: 10.1007/s00299-007-0472-y. [DOI] [PubMed] [Google Scholar]
- Walker EL, Connolly EL. Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Current Opinion in Plant Biology. 2008;11:530–535. doi: 10.1016/j.pbi.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Wang L, Xie W, Chen Y, et al. A dynamic gene expression atlas covering the entire life cycle of rice. The Plant Journal. 2010;61:752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- Waters BM, Chu H-H, Didonato RJ, et al. Mutations in Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiology. 2006;141:1446–1458. doi: 10.1104/pp.106.082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wiren N, Mori S, Marschner H, Romheld V. Iron inefficiency in maize mutant ys1 (Zea mays L. cv Yellow-Stripe) is caused by a defect in uptake of iron phytosiderophores. Plant Physiology. 1994;106:71–77. doi: 10.1104/pp.106.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]