Abstract
Background
High input costs and environmental pressures to reduce nitrogen use in agriculture have increased the competitive advantage of legume crops. The symbiotic relationship that legumes form with nitrogen-fixing soil bacteria in root nodules is central to this advantage.
Scope
Understanding how legume plants maintain control of nodulation to balance the nitrogen gains with their energy needs and developmental costs will assist in increasing their productivity and relative advantage. For this reason, the regulation of nodulation has been extensively studied since the first mutants exhibiting increased nodulation were isolated almost three decades ago.
Conclusions
Nodulation is regulated primarily via a systemic mechanism known as the autoregulation of nodulation (AON), which is controlled by a CLAVATA1-like receptor kinase. Multiple components sharing homology with the CLAVATA signalling pathway that maintains control of the shoot apical meristem in arabidopsis have now been identified in AON. This includes the recent identification of several CLE peptides capable of activating nodule inhibition responses, a low molecular weight shoot signal and a role for CLAVATA2 in AON. Efforts are now being focused on directly identifying the interactions of these components and to identify the form that long-distance transport molecules take.
Keywords: Legume nodulation, AON, signalling, hormone, plant peptide, receptor kinase, symbiosis
INTRODUCTION
Nitrogen is a component of many biological molecules, making its availability critical to sustained plant growth and reproduction. Atmospheric nitrogen gas is plentiful but is unavailable to most organisms. Many legumes overcome this limitation by initiating a symbiotic relationship with soil bacteria, collectively referred to as rhizobia. These rhizobia are capable of biological nitrogen fixation where atmospheric nitrogen (N2) is fixed by the nitrogenase enzyme complex of the endocytotic bacteria when they reside inside legume organs called nodules. Nodules provide the rhizobia with an energy source from photoassimilates (as malate; Udvardi et al., 1988) while maintaining the low oxygen environment required for efficient nitrogen fixation. The process of nodule formation and subsequent nitrogen fixation is balanced with the plant's own energy requirements in a process termed autoregulation of nodulation (AON; see Caetano-Anollés and Gresshoff, 1991; Ferguson et al., 2010). Nodulation is also regulated in response to nitrogen availability in the soil (Carroll et al., 1985; Ferguson and Mathesius, 2003).
This review highlights significant progress that has recently been made in the identification of new genes and factors controlling nodulation. Further understanding of the mechanisms that allow plants to balance nitrogen resources with their energy demands may enable nitrogen use optimization in important legume crops, including the most widely grown soybean. Significant agricultural, economic and environmental benefits stand to be gained by further reducing nitrogen fertilizer inputs while maintaining or improving legume yields.
AUTOREGULATION OF NODULATION
Nodulation occurs in a distinct pattern where nodules form in the region having susceptible root hairs at the time of inoculation [zone of nodulation (ZON); Bhuvaneswari et al., 1981; Calvert et al., 1984]. AON causes a nodulation phenotype where the majority of nodules form near the crown of the root system. It is unclear which stage of nodule development AON inhibits, although approach grafting in Pisum sativum indicated that the onset of AON is triggered before extensive cell divisions are observed (Li et al., 2009). This supports anatomical observations in soybean where a significant reduction of nodule development stages occurs along the root (Mathews et al., 1989) and split-root experiments where inoculation is delayed by 3–4 d (Kosslak and Bohlool, 1984; Olsson et al., 1989). It has been proposed that AON reduces the speed of cortical cell divisions to restrict nodulation at the early stages when these cell divisions are occurring (Mathews et al., 1989).
The precise nature and co-ordination of these regulatory cues remain to be elucidated; however, the maintenance of autoregulation in spontaneous nodulation mutants indicates that a bacterial-derived signal is not the elicitor of AON and that it is not entirely dependent on nitrogen fixation in the nodule (Caetano-Anollés et al., 1990; Tirichine et al., 2006, 2007). The precise onset of AON is also not known, although experimental evidence suggests that AON is effective as early as 4 d after inoculation (Kosslak and Bohlool, 1984; Olsson et al., 1989; Suzuki et al., 2008).
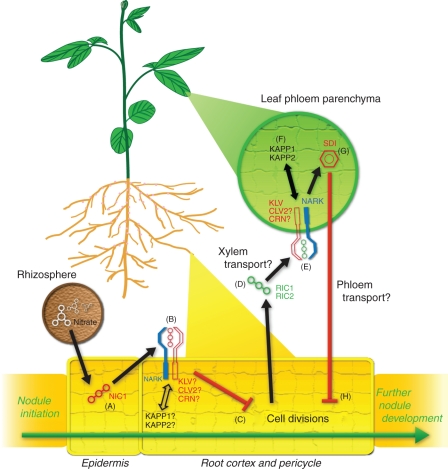
Studies using grafting and split-root techniques have shown that AON is induced systemically by a graft-transmissible signal and is controlled by the shoot (Delves et al., 1986, 1992). These experiments led to a model where a root-derived nodulation signal is the cue (Q) for the onset of AON. Subsequently, a shoot-derived inhibitor (SDI) is transported back down to the roots where it regulates further nodulation events (Fig. 1D–H).
Fig. 1.
A working model of root and shoot mechanisms in autoregulation of nodulation (AON). Legumes regulate nodulation in response to pre-existing infections and soil nitrogen levels. Nitrate induces the production of a nitrate-induced CLE peptide (NIC1; A) that acts locally in the root via the AON receptor kinase, NARK (B; or its orthologues in other species), to inhibit nodule progression (C). NARK may act in concert with other components to perceive NIC1. Rhizobia-induced CLE peptides (RICs) are induced at several stages of nodule development and may be transported via the xylem (D) to the shoot. In the shoot, NARK and possibly also CLV2, KLV and CRN are required for the perception of these putative ligands (E). Two kinase-associated protein phosphatases (KAPP1/2) are phosphorylated by NARK and in turn dephosphorylate the NARK kinase (F). An equilibrium of phosphorylation between these components may be required preceding the production of the shoot-derived inhibitor (SDI; G). SDI is transported via the phloem to the roots where it inhibits further nodule progression and cell divisions (H). A compound similar to SDI may also be involved in the nitrate pathway that acts locally to inhibit the progression of nodule formation (C).
In addition to the systemic AON mechanism where plants regulate nodulation in response to existing infection events, legumes also regulate nodulation in response to environmental nitrogen availability. This may represent a means of preferentially obtaining nitrogen from sources that are energetically favourable relative to the energy costs of nodulation. Nodulation tolerant to high soil nitrate levels has been used as a screening method to identify nodule regulation mutants (e.g. nod3, Jacobsen and Feenstra, 1984; nts, Carroll et al., 1985). The nts mutants of soybean exhibit a supernodulation phenotype in both high and low nitrate conditions, indicating that the AON and nitrate regulation pathways share genetic components (Day et al., 1986). Evidence exists for both local and systemic regulatory mechanisms functioning in response to nitrate, and it is likely that multiple mechanisms are acting in concert (Hinson, 1975; Cho and Harper, 1991; Okamoto et al., 2009; Jeudy et al., 2010; Reid et al., 2011).
ROOT-DEPENDENT COMPONENTS OF AON
Autoregulation of nodulation can be divided into root- and shoot-dependent components (Fig. 1). The former components are likely to include factors involved in the response to initial cell divisions that lead to the induction of Q. A second class of root-dependent components are those which act downstream of shoot signalling to perceive the SDI signal and/or that inhibit further nodule progression.
Several mutants have been isolated that exhibit an increased nodulation phenotype, known as hyper- or supernodulation (see Table 1). Those functioning in the root include rdh1 (Ishikawa et al., 2008), rdn1 (Schnabel et al., 2011), nod3 (Jacobsen and Feenstra, 1984), too much love (Magori et al., 2009), plenty (Yoshida et al., 2010), efd-1 (Vernié et al., 2008), astray (Nishimura et al., 2002b) and sickle (Penmetsa et al., 2008). Each of these mutants forms an overabundance of nodules, though some differences are noted in their pattern and extent of nodulation. In addition, not all of these factors function directly in the AON pathway.
Table 1.
Genes/mutants and products involved in legume regulation of nodule number
| Gene/mutant | Gene product | Site of production | Site of action | Comments | References |
|---|---|---|---|---|---|
| GmNARK; GsNARK; LjHAR1; MtSUNN; PsSYM29 | LRR-RK | Shoot/root | Shoot/root | Acts in shoot (AON) and root (NO3− inhibition) (LRR-RK) | Sagan and Duc (1996); Krusell et al. (2002); Men et al. (2002); Nishimura et al. (2002a); Searle et al. (2003); Schnabel et al. (2005) |
| GmNIC1 | CLE | Root | Root | NO3− induced (CLE pre-propeptide) | Reid et al. (2011) |
| GmRIC1/2; LjCLE-RS1/2; MtCLE12/13 | CLE | Root | Probably the shoot | Rhizobia-induced (CLE prepropeptide) | Okamoto et al. (2009); Mortier et al. (2010); Reid et al. (2011); Lim et al. (2011) |
| LjASTRAY | bZIP TF | Root | Also acts in photomorphogenesis (transcription factor) | Nishimura et al. (2002b) | |
| LjCLV2; PsSYM28 | CLV2 | Shoot/root | Shoot/root? | May interact with other AON LRR RKs (truncated LRR-receptor protein) | Sagan and Duc (1996); Krusell et al. (2011) |
| LjETR1 | ETR1 | Shoot/root | Shoot/root | Ethylene receptor (two-component receptor) | Gresshoff et al. (2009); Lohar et al. (2009) |
| LjKLV | LRR-RK | Shoot/root? | Shoot/root? | May interact with other AON LRR RKs | Oka-Kira et al. (2005) |
| LjPLENTY | Unknown | Root | Root | Hypernodulation phenotype | Yoshida et al. (2010) |
| LjRDH1 | Unknown | Root | Root | Ishikawa et al. (2008) | |
| LjTML | Unknown | Root | Root | Magori et al. (2009) | |
| MtEFD | AP2-EREBP TF | Root | Root | Positively regulates CK levels (transcription factor) | Vernié et al. (2008) |
| MtLSS | Unknown | Shoot/root? | Shoot/root? | Possible epigenetic factor of MtSUNN | Schnabel et al. (2010) |
| MtSKL | EIN2 | Root | Root | \ethylene response factor | Penmetsa and Cook (1997); Penmetsa et al. (2008) |
| PsNOD1 and 2 | Unknown | Gelin and Blixt (1964) | |||
| PsNOD3; MtRDN1 | RDN1 | Root | Root | Affects CLE synthesis and/or transport | Jacobsen and Feenstra (1984); Engvild (1987); Novák et al. (1997); Li et al. (2009); Schnabel et al. (2011) |
| PsNOD4 and 5 | Unknown | Shoot | Sidorova and Shumnyi (1998, 2003) | ||
| PsNOD6 | Unknown | Shoot | Sidorova and Shumnyi (1998) |
Approach grafting indicated that PsNOD3, the homologue of MtRDN1, may function prior to the shoot responses, possibly in the production or transmission of the root-derived signal (Li et al., 2009). In contrast, TOO MUCH LOVE inhibits nodulation locally and may act downstream of the shoot components, possibly as a receptor for the SDI signal or in a related function (Magori et al., 2009).
CLE PEPTIDES IN AON
The first genes to be identified in AON were those encoding a group of orthologous leucine-rich repeat (LRR) receptor kinases similar to CLAVATA1 (CLV1; Clark et al., 1997) in arabidopsis (LjHAR1, Krusell et al., 2002; Nishimura et al., 2002a; PsSYM29, Krusell et al., 2002; GmNARK, Searle et al., 2003; Men et al., 2002; and MtSUNN, Schnabel et al., 2005). Mutations in these genes reduce the plant's ability to regulate nodule numbers and, in all cases tested, nodulation is tolerant to otherwise inhibitory high nitrate conditions.
The similarity of these AON receptor kinases to CLV1 prompted searches for ligands related to the CLV3 peptide, the ligand of CLV1 (Fletcher et al., 1999; Kondo et al., 2006, 2008; Oelkers et al., 2008; Ogawa et al., 2008). CLE peptides responding to inoculation were identified which can systemically reduce nodule numbers in Lotus japonicus (Okamoto et al., 2009). Related peptides with AON receptor-dependent activity have since been identified in Medicago truncatula (Mortier et al., 2010; Saur et al., 2011) and Glycine max (Reid et al., 2011; Mortier et al., 2011; Lim et al., 2011). The expression of these CLE peptide-encoding genes suggests they are induced in response to inoculation and the onset of nodule development. However, the peptides themselves have yet to be directly detected in planta and they have not been confirmed to move long distances as is expected if they are perceived in the shoot. Some of these CLE peptide-encoding genes (LjCLE-RS2 and GmNIC1) were also found to be responsive to nitrate, highlighting the mechanistic and functional similarities between AON and the nitrate regulation of nodulation. GmNIC1 differed from the other CLE peptides so far identified as it functions locally via NARK to regulate nodulation (Fig. 1A–C) and does not appear to be induced by the rhizobia.
There appears to be some functional divergence between the inoculation-responsive CLE peptides as the timing of their induction was found to be variable. The expression of LjCLE-RS1/2, MtCLE13 and GmRIC1 is induced early in response to inoculation, whereas GmRIC2, MtCLE12 and MtCLE13 were persistent later when mature nodules were present. Li and associates (2009) showed that AON activation requires signalling at several nodule developmental stages, indicating that multiple signals may be required for activation and maintenance.
The secondary structure of CLE pre-propeptides can be characterized by the presence of a 5′ signal peptide which is likely to be required for the cellular export and localization properties of the peptide (Meng et al., 2010). CLE peptides also possess a 12–13 amino acid motif deemed to represent the final active peptide which is cleaved from close to the 3′ end of the initial protein (Oelkers et al., 2008). Outside of the signal peptide and the CLE motif there appears to be little sequence conservation within CLE proteins. However, the CLE peptides capable of regulating nodulation share some common features outside of these more general CLE characteristics (Okamoto et al., 2009; Mortier et al., 2010; Reid et al., 2011). Within the signal peptide region, a well-conserved motif was identified, although apart from the predicted export role the exact function of this remains obscure. Several of these CLE peptides also possess a 5–7 amino acid conserved extension beyond the 3′ CLE motif. CLE peptides require processing to generate the final product and several protease cleavage events may be required (Ni and Clark, 2006; Ni et al., 2011). The extracellular fluid of legumes has been demonstrated to possess factors with proteolytic activity capable of producing biologically active CLE peptides (Djordjevic et al., 2011). The activity of CLE peptides has also been shown to be dependent on post-translational modifications, including hydroxylation of proline residues and glycosylation of key residues (Kondo et al., 2006; Ohyama et al., 2009). In arabidopsis, CLV3 and CLE2 (which share a similar CLE domain sequence to the nodulation CLE peptides) were identified with three β-1,2-linked arabinose moieties bound to Hyp7 of the 13 amino acid CLE peptide (Ohyama et al., 2009).
Due to the systemic signalling requirement in AON, it is presumed that long-distance transport of the Q signal is required, probably via the xylem (Fig. 1D). Efforts have therefore been undertaken to characterize the protein and metabolite components of the xylem sap of legume plants. The soybean xylem sap proteome identified several protein components, although none of these differed between plants with or without nodules (Djordjevic et al., 2007). There were, however, changes observed in the xylem sap proteome of inoculated soybeans at the seedling stage, though a distinct role for these changes in nodulation was not identified (Subramanian et al., 2009).
SHOOT-DEPENDENT COMPONENTS IN AON
The secondary structure of NARK includes an N-terminal signal peptide and an extracellular LRR domain, which is the proposed binding site for Q. Transmembrane and intracellular kinase domains are also key features of NARK and are essential for membrane localization, protein–protein interactions and downstream phosphorylation and signalling events. Supernodulation phenotypes result from mutations in NARK in either the LRR or kinase domains, indicating that both are required for AON signalling and/or stabilizing the signalling complex (Searle et al., 2003). Modelling of the NARK LRR domain indicates that it may form a boomerang shape that acts to perceive the AON ligand (Reid et al., 2011). Two known missense mutants in soybean, nod4 and nod3-7, display severe supernodulation phenotypes resembling those of deletion (Men et al., 2002) and nonsense (Carroll et al., 1985; Searle et al., 2003) mutants despite having only single amino acid substitutions within the proposed ligand-binding site (Reid et al., 2011).
Mutants that affect the expression or localization of the AON receptor might also be predicted to cause supernodulation phenotypes. Shoot-controlled nodule regulation is lost in the lss (like-SUNN supernodulator) mutant in M. truncatula (Schnabel et al., 2010). The LSS locus maps in a region close to the SUNN gene; however, sequencing of SUNN and the surrounding regions indicates that there is no mutation within the 20 kbp SUNN region. SUNN expression is greatly reduced in lss and epigenetic factors may be responsible for loss of SUNN activity.
As mentioned above, NARK and its orthologues share a high degree of similarity with CLV1 in arabidopsis (75 % amino acid similarity; Searle et al., 2003), which is required for maintenance of the shoot apical meristem (SAM). Several protein interactions which may be relevant to the activity of NARK in AON have been reported with CLV1, including CLAVATA2 (CLV2; Jeong et al., 1999) and CORYNE (CRN; Muller et al., 2008). CLV2 is a receptor-like protein that lacks an intracellular kinase domain, whereas CRN is a kinase-like protein that lacks an extracellular LRR domain. CRN appears to lack effective kinase activity and may be required as a structural component in a CLV1 complex or for facilitating the inclusion of other components including CLV2 in a receptor complex (Nimchuck et al., 2011).
Additional shoot-controlled supernodulation mutants have recently been genetically characterized and represent further components associated with the CLV signalling pathway (Fig. 1B, E). The shoot-dependent supernodulation mutant sym28 in pea and LjCLV2 in L. japonicus are the orthologues of AtCLV2 (Krusell et al., 2011). Experiments to determine if CLV2 forms a heterodimer complex with HAR1 in a similar manner to the CLV2–CLV1 complex in the SAM were unable to establish an interaction. KLAVIER (KLV) is a receptor-like kinase similar to RPK2/TOAD2 in arabidopsis which is required for CLV3-dependent meristem regulation (Kinoshita et al., 2010; Miyazawa et al., 2010). KLV was shown to form homo- or heterodimer complexes with itself and HAR1, respectively, suggesting that a receptor complex may be required for the perception of Q (Miyazawa et al., 2010). This work serves to highlight the extent to which nodule regulation activity utilizes the machinery of SAM regulation. Further investigation of CLV signalling components which may function in AON will be of interest, including whether a CRN-like protein plays a role in AON.
TRANSMISSION OF AON SIGNALLING IN THE LEAF
Knowledge of signal transduction mechanisms acting downstream of CLV in arabidopsis has assisted in the identification of homologous elements in AON. Two kinase-associated protein phosphatases (KAPP1/2) were identified in soybean that are phosphorylated by NARK in vitro and subsequently dephosphorylate the NARK kinase (Miyahara et al., 2008). This may indicate that a sufficient equilibrium of phosphorylation states between NARK and KAPP1/2 must exist before a downstream AON response is generated (Fig. 1F).
In arabidopsis, the primary function of CLV signalling activity in the SAM is the restriction of WUSCHEL (WUS) production. This acts to maintain an appropriate balance between differentiated and undifferentiated cells via a constant feedback between WUS and CLV3 (Schoof et al., 2000). WUS-related homeobox (WOX) genes have been identified in other CLE peptide/LRR receptor systems including the regulation of vascular differentiation (Hirakawa et al., 2010; Ji et al., 2010) and in the root apical meristem (Kamiya et al., 2003; Sarkar et al., 2007). Likewise, a WOX component may be involved in AON signalling, though this remains to be determined.
To identify components of AON acting downstream of NARK in the leaf, transcriptional profiling using Affymetrix GeneChips or subtractive hybridization techniques has been undertaken in soybean (Seo et al., 2007; Kinkema and Gresshoff, 2008). Both of these studies identified components of the jasmonic acid (JA) biosynthesis or response pathways being regulated following rhizobia inoculation. Foliar application of methyl jasmonate in L. japonicus inhibited nodulation in both wild-type and har1 plants, further indicating that JA may play a role in nodule regulation (Nakagawa and Kawaguchi, 2006).
The identification of additional nodule regulation mutants through candidate gene selection in TILLING populations or through screening of traditional mutant populations may be useful for identifying further downstream components of AON.
SDI AND EFFORTS TO IDENTIFY IT
Shoot-derived inhibitor is produced in the shoot following the perception of Q (Fig. 1G). Phloem transport of the SDI signal from the shoot to the root would appear to be the most probable mechanism based on the timing and direction of its response. It is then predicted to be perceived in the root, where it acts to prevent cell divisions required for nodule development (Fig. 1H). A bioassay approach has been exploited to characterize the nature of SDI partially through petiole feeding of plant extracts into the phloem of intact plants (Lin et al., 2010, 2011). Using this technique, leaf extracts from wild-type plants inhibited nodulation in hypernodulating nts mutants that are unable to produce SDI. Various pre-treatments of the leaf extract showed that SDI is probably a small, heat-stable molecule that is not a protein or RNA. The inhibitory capacity of the leaf extracts was also dependent on NARK and on nod factor signalling (Lin et al., 2010, 2011). The petiole feeding bioassay technique has also been exploited to show that nitrogen fixation in nodules may be systemically regulated through phloem transport of the amino acid asparagine (Sulieman et al., 2010).
CONCLUSIONS AND FUTURE PROSPECTS
Plants maintain appropriate growth and development via constant feedback to environmental and internal conditions. The regulation of nodulation in legumes is one such system, where systemic signalling ensures a balance between nodule formation and energy requirements. Knowing the identity of both the root- and shoot-derived mobile signals in AON would be of immense value to the field. The identification of CLE peptides capable of nodule regulation and a role for CLV2 and KLV in AON have emphasized the similarities that exist between AON and other CLE peptide ligand–receptor systems, particularly that of the CLAVATA signalling pathway in the SAM. Ongoing research on AON will draw on these similarities and will in turn contribute to the better understanding of other environmental and developmental regulation responses occurring in the plant. The decreasing cost and increased availability of high-throughput sequencing technology continue to drive discoveries in plant genetics. Moreover, the recent sequencing of the soybean, L. japonicus and M. truncatula genomes means that three largely complete legume genomes are now publicly available (Young et al., 2005; Sato et al., 2008; Schmutz et al., 2010). These resources will considerably support future advances in understanding the molecular mechanisms underlying nodule regulation.
ACKNOWLEDGEMENTS
This work was supported by the University of Queensland and the Australian Research Council through a Centre of Excellence grant. Additional support was provided to D.E.R. by an Australian Government DIISR Australian Postgraduate Award. We thank Meng-Han Lin, Alina Tollenaere, Dongxue Li and Alvin van Niekerk for continued support.
LITERATURE CITED
- Bhuvaneswari TV, Bhagwat AA, Bauer WD. Transient susceptibility of root cells in four common legumes to nodulation by Rhizobia. Plant Physiology. 1981;68:1144–1149. doi: 10.1104/pp.68.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles G, Gresshoff P. Plant genetic control of nodulation. Annual Review of Microbiology. 1991;45:345–382. doi: 10.1146/annurev.mi.45.100191.002021. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G, Joshi PA, Gresshoff PM. Spontaneous nodules induce feedback suppression of nodulation in alfalfa. Planta. 1990;183:77–82. doi: 10.1007/BF00197570. [DOI] [PubMed] [Google Scholar]
- Calvert H, Pence M, Pierce M, Malik N, Bauer W. Anatomical analysis of the development and distribution of Rhizobium infections in soybean roots. Canadian Journal of Botany. 1984;62:2375–2384. [Google Scholar]
- Carroll BJ, McNeil DL, Gresshoff PM. Isolation and properties of soybean [Glycine max (L) MERR] mutants that nodulate in the presence of high nitrate concentrations. Proceedings of the National Academy of Sciences, USA. 1985;82:4162–4166. doi: 10.1073/pnas.82.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M-J, Harper JE. Effect of localized nitrate application on isoflavonoid concentration and nodulation in split-root systems of wild-type and nodulation-mutant soybean plants. Plant Physiology. 1991;95:1106–1112. doi: 10.1104/pp.95.4.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Day DA, Lambers H, Bateman J, Carroll BJ, Gresshoff PM. Growth comparisons of a supernodulating soybean (Glycine max) mutant and its wild-type parent. Physiologia Plantarum. 1986;68:375–382. [Google Scholar]
- Delves AC, Mathews A, Day DA, Carter AS, Carroll BJ, Gresshoff PM. Regulation of the soybean–Rhizobium nodule symbiosis by shoot and root factors. Plant Physiology. 1986;82:588–590. doi: 10.1104/pp.82.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delves A, Higgins A, Gresshoff P. Shoot apex removal does not alter autoregulation of nodulation in soybean. Plant, Cell and Environment. 1992;15:249–254. [Google Scholar]
- Djordjevic MA, Oakes M, Li DX, Hwang CH, Hocart CH, Gresshoff PM. The Glycine max xylem sap and apoplast proteome. Journal of Proteome Research. 2007;6:3771–3779. doi: 10.1021/pr0606833. [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Oakes M, Wong CE, et al. Border sequences of Medicago truncatula CLE36 are specifically cleaved by endoproteases common to the extracellular fluids of Medicago and soybean. Journal of Experimental Botany. 2011 doi: 10.1093/jxb/err185. in press. doi:10.1093/jxb/err185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engvild KC. Nodulation and nitrogen fixation mutants of pea, Pisum sativum. Theoretical and Applied Genetics. 1987;74:711–713. doi: 10.1007/BF00247546. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Mathesius U. Signaling interactions during nodule development. Journal of Plant Growth Regulation. 2003;22:47–72. [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, et al. Molecular analysis of legume nodule development and autoregulation. Journal of Integrative Plant Biology. 2010;52:61–76. doi: 10.1111/j.1744-7909.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Gelin O, Blixt S. Root nodulation in peas. Agricultural and Horticultural Genetics. 1964;22:149–159. [Google Scholar]
- Gresshoff PM, Lohar D, Chan PK, et al. Genetic analysis of ethylene regulation of legume nodulation. Plant Signaling and Behaviour. 2009;4:818–823. doi: 10.4161/psb.4.9.9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson K. Nodulation responses from nitrogen applied to soybean half-root systems. Agronomy Journal. 1975;67:799–804. [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H. TDIF peptide signalling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. The Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Yokota K, Li YY, et al. Isolation of a novel root-determined hypernodulation mutant rdh1 of Lotus japonicus. Soil Science and Plant Nutrition. 2008;54:259–263. [Google Scholar]
- Jacobsen E, Feenstra WJ. A new pea mutant with efficient nodulation in the presence of nitrate. Plant Science Letters. 1984;33:337–344. [Google Scholar]
- Jeong S, Trotochaud A, Clark S. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. The Plant Cell. 1999;11:1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy C, Ruffel S, Freixes S, et al. Adaptation of Medicago truncatula to nitrogen limitation is modulated via local and systemic nodule developmental responses. New Phytologist. 2010;185:817–828. doi: 10.1111/j.1469-8137.2009.03103.x. [DOI] [PubMed] [Google Scholar]
- Ji J, Strable J, Shimizu R, Koenig D, Sinha N, Scanlon MJ. WOX4 promotes procambial development. Plant Physiology. 2010;152:1346–1356. doi: 10.1104/pp.109.149641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M. Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. The Plant Journal. 2003;35:429–441. doi: 10.1046/j.1365-313x.2003.01816.x. [DOI] [PubMed] [Google Scholar]
- Kinkema M, Gresshoff PM. Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Molecular Plant-Microbe Interactions. 2008;21:1337–1348. doi: 10.1094/MPMI-21-10-1337. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- Kondo T, Nakamura T, Yokomine K, Sakagami Y. Dual assay for MCLV3 activity reveals structure–activity relationship of CLE peptides. Biochemical and Biophysical Research Communications. 2008;377:312–316. doi: 10.1016/j.bbrc.2008.09.139. [DOI] [PubMed] [Google Scholar]
- Kosslak RM, Bohlool BB. Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side. Plant Physiology. 1984;75:125–130. doi: 10.1104/pp.75.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420:422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- Krusell L, Sato N, Fukuhara I, et al. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. The Plant Journal. 2011;65:861–871. doi: 10.1111/j.1365-313X.2010.04474.x. [DOI] [PubMed] [Google Scholar]
- Li D, Kinkema M, Gresshoff PM. Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. Journal of Plant Physiology. 2009;166:955–967. doi: 10.1016/j.jplph.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Lim CW, Lee YW, Hwang CH. Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant and Cell Physiology. 2011 doi: 10.1093/pcp/pcr091. in press. doi:10.1093/pcp/pcr091. [DOI] [PubMed] [Google Scholar]
- Lin Y-H, Ferguson BJ, Kereszt A, Gresshoff PM. Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent low molecular mass fraction. New Phytologist. 2010;185:1074–1086. doi: 10.1111/j.1469-8137.2009.03163.x. [DOI] [PubMed] [Google Scholar]
- Lin Y-H, Lin M-H, Gresshoff PM, Ferguson BJ. An efficient petiole-feeding bioassay for introducing aqueous solutions into dicotyledonous plants. Nature Protocols. 2011;6:36–45. doi: 10.1038/nprot.2010.171. [DOI] [PubMed] [Google Scholar]
- Lohar D, Stiller J, Kam J, Stacey G, Gresshoff PM. Ethylene insensitivity conferred by a mutated Arabidopsis ethylene receptor gene alters nodulation in transgenic Lotus japonicus. Annals of Botany. 2009;104:277–285. doi: 10.1093/aob/mcp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magori S, Oka-Kira E, Shibata S, et al. TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Molecular Plant-Microbe Interactions. 2009;22:259–268. doi: 10.1094/MPMI-22-3-0259. [DOI] [PubMed] [Google Scholar]
- Mathews A, Carroll BJ, Gresshoff PM. Development of Bradyrhizobium infections in supernodulating and non-nodulating mutants of soybean (Glycine max [L.] Merrill) Protoplasma. 1989;150:40–47. [Google Scholar]
- Men AE, Laniya TS, Searle IR, et al. Fast neutron mutagenesis of soybean (Glycine soja L.) produces a supernodulating mutant containing a large deletion in linkage group H. Genome Letters. 2002;1:147–155. [Google Scholar]
- Meng L, Ruth KC, Fletcher JC, Feldman L. The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Molecular Plant. 2010;3:760–772. doi: 10.1093/mp/ssq021. [DOI] [PubMed] [Google Scholar]
- Miyahara A, Hirani TA, Oakes M, et al. Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro. Journal of Biological Chemistry. 2008;283:25381–91. doi: 10.1074/jbc.M800400200. [DOI] [PubMed] [Google Scholar]
- Miyazawa H, Oka-Kira E, et al. The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development. 2010;137:4317–4325. doi: 10.1242/dev.058891. [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, et al. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology. 2010;153:222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, Fenta B, Martens C, Rombauts S, Holsters M, Kunert K, Goormachtig S. Search for nodulation-related CLE genes in the genome of Glycine max. Journal of Experimental Botany. 2011;62:2571–2583. doi: 10.1093/jxb/erq426. [DOI] [PubMed] [Google Scholar]
- Muller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. The Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kawaguchi M. Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant and Cell Physiology. 2006;47:176–180. doi: 10.1093/pcp/pci222. [DOI] [PubMed] [Google Scholar]
- Ni J, Clark SE. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiology. 2006;140:726–733. doi: 10.1104/pp.105.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Guo Y, Jin H, Hartsell J, Clark S. Characterization of a CLE processing activity. Plant Molecular Biology. 2011;75:67–75. doi: 10.1007/s11103-010-9708-2. [DOI] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Meyerowitz EM. An evolutionarily conserved pseudokinase mediates stem cell production in plants. The Plant Cell. 2011;23:851–854. doi: 10.1105/tpc.110.075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002a;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Fujita H, Kawaguchi M. A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proceedings of the National Academy of Sciences, USA. 2002b;99:15206–15210. doi: 10.1073/pnas.222302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak K, Skrdleta V, Kropacova M, Lisa L, Nemcova M. Interaction of two genes controlling symbiotic nodule number in pea (Pisum sativum L.) Symbiosis. 1997;23:43–62. [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology. 2008;8:1. doi: 10.1186/1471-2229-8-1. doi:10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chemical Biology. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- Oka-Kira E, Tateno K, Miura K, et al. klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. The Plant Journal. 2005;44:505–515. doi: 10.1111/j.1365-313X.2005.02543.x. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, et al. Nod factor, nitrate-induced CLE genes that drive systemic regulation of nodulation. Plant and Cell Physiology. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- Olsson J, Nakao P, Bohlool B, Gresshoff PM. Lack of systemic suppression of nodulation in split root systems of supernodulating soybean (Glycine max [L.] Merr.) mutants. Plant Physiology. 1989;90:1347–1352. doi: 10.1104/pp.90.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–30. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Uribe P, Anderson J, et al. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. The Plant Journal. 2008;55:580–595. doi: 10.1111/j.1365-313X.2008.03531.x. [DOI] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Molecular Plant-Microbe Interactions. 2011;24:606–618. doi: 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]
- Sagan M, Duc G. Sym28 and Sym29, two new genes involved in regulation of nodulation in pea (Pisum sativum L.) Symbiosis. 1996;20:229–245. [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, et al. Genome structure of the legume. Lotus japonicus. DNA Research. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur IML, Oakes M, Djordjevic MA, Imin N. Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytologist. 2011;190:865–874. doi: 10.1111/j.1469-8137.2011.03738.x. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. Genome sequence of the paleopolyploid soybean (Glycine max (L.) Merr.) Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet E-P, Carvalho-Niebel Fd, Duc G, Frugoli J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Molecular Biology. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- Schnabel E, Mukherjee A, Smith L, Kassaw T, Long S, Frugoli JA. The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiology. 2010;154:1390–1402. doi: 10.1104/pp.110.164889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E, Kassaw T, Smith L, Marsh J, Oldroyd GE, Long SR, Frugoli J. ROOT DETERMINED NODULATION 1 regulates nodule number in M. truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiology. 2011 doi: 10.1104/pp.111.178756. in press. doi: 10.1104/pp.111.178756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, et al. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Seo HS, Li J, Lee S-Y, et al. The hypernodulating nts mutation induces jasmonate synthetic pathway in soybean leaves. Molecules and Cells. 2007;24:185–193. [PubMed] [Google Scholar]
- Sidorova K, Shumnyi V. Analysis of pea (Pisum sativum L.) supernodulating mutants. Russian Journal of Genetics. 1998;34:1233–1235. [Google Scholar]
- Sidorova KK, Shumnyi VK. A collection of symbiotic mutants in pea (Pisum sativum L.): creation and genetic study. Russian Journal of Genetics. 2003;39:406–413. [PubMed] [Google Scholar]
- Subramanian S, Cho U-H, Keyes C, Yu O. Distinct changes in soybean xylem sap proteome in response to pathogenic and symbiotic microbe interactions. BMC Plant Biology. 2009;9:119. doi: 10.1186/1471-2229-9-119. doi:10.1186/1471-2229-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulieman S, Fischinger S, Gresshoff PM, Schulze J. Asparagine as a major factor in the N-feedback regulation of N2 fixation in Medicago truncatula. Physiologia Plantarum. 2010;140:21–31. doi: 10.1111/j.1399-3054.2010.01380.x. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Hara H, Kinoue T, et al. Split-root study of autoregulation of nodulation in the model legume Lotus japonicus. Journal of Plant Research. 2008;121:245–249. doi: 10.1007/s10265-007-0145-5. [DOI] [PubMed] [Google Scholar]
- Tirichine Ll, Imaizumi-Anraku H, Yoshida S, et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Price GD, Gresshoff PM, Day DA. A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Letters. 1988;231:36–40. [Google Scholar]
- Vernié T, Moreau S, de Billy F, et al. EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. The Plant Cell. 2008;20:2696–2713. doi: 10.1105/tpc.108.059857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C, Funayama-Noguchi S, Kawaguchi M. plenty, a novel hypernodulation mutant in Lotus japonicus. Plant and Cell Physiology. 2010;51:1425–1435. doi: 10.1093/pcp/pcq115. [DOI] [PubMed] [Google Scholar]
- Young ND, Cannon SB, Sato S, et al. Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiology. 2005;137:1174–1181. doi: 10.1104/pp.104.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]