Abstract
Background and Aims
Most cooking banana and several desert bananas are interspecific triploid hybrids between Musa acuminata (A genome) and Musa balbisiana (B genome). In addition, M. balbisiana has agronomical characteristics such as resistance to biotic and abiotic stresses that could be useful to improve monospecific acuminata cultivars. To develop efficient breeding strategies for improving Musa cultivars, it is therefore important to understand the possibility of chromosome exchange between these two species.
Methods
A protocol was developed to prepare chromosome at meiosis metaphase I suitable for genomic in situ hybridization. A series of technical challenges were encountered, the main ones being the hardness of the cell wall and the density of the microsporocyte's cytoplasm, which hampers accessibility of the probes to the chromosomes. Key parameters in solving these problems were addition of macerozyme in the enzyme mix, the duration of digestion and temperature during the spreading phase.
Results and Conclusions
This method was applied to analyse chromosome pairing in metaphase from triploid interspecific cultivars, and it was clearly demonstrated that interspecific recombinations between M. acuminata and M. balbisiana chromosomes do occur and may be frequent in triploid hybrids. These results provide new insight into Musa cultivar evolution and have important implications for breeding.
Keywords: Musa, banana, genomic in situ hybridization, meiosis, homoeologous chromosomes pairing, interspecific hybrid, polyploidy
INTRODUCTION
Cultivated bananas (bananas and plantains) are seedless parthenocarpic clones, selected by early farmers in South-East Asia and maintained by vegetative propagation (Simmonds, 1962). They represent the fourth most important crop in developing countries (Lassois et al., 2009). They are important as an export crop, providing a vital source of income for many countries and also playing a major role in local food security (Chalmin, 2009). Cultivars are derived from natural intra- and interspecific hybridization between wild fertile diploid Musa accessions. Most cultivars are triploid, some are diploid and a few are tetraploid. The main species involved are Musa acuminata Colla (A genome, 2n = 2x = 22) and Musa balbisiana Colla (B genome, 2n = 2x = 22) (Cheesman, 1947; Simmonds and Shepherd, 1955). Cultivars were classified based on morphological characters and chromosome number into genome groups AA, AB, AAA, AAB or ABB (Cheesman, 1947; Simmonds and Shepherd, 1955). More recently, molecular markers confirmed and refined this classification (Ude et al., 2002; Carreel et al., 2002; Creste et al., 2004; Perrier et al., 2009; Hřibová et al., 2011). The small size of Musa genomes, i.e. 500– 600 Mbp (1C) (Dolezel et al., 1994; Lysak et al., 1999; Bartos et al., 2005) and thus of Musa chromosomes (1·5–3·5 µm) makes conventional karyotypic analysis difficult (Isobe and Hashimoto, 1994; Osuji et al., 1996a, b). However, many conventional cytogenetic studies have been performed in Musa, in particular to study chromosome pairing during meiosis (Wilson, 1945; Simmonds, 1962; Dessauw, 1987; Fauré et al., 1993; Shepherd, 1999; Therdsak et al., 2010). They suggested that M. accuminata subspecies differed in their chromosome structure due to rearrangements, particularly translocations (Dodds, 1943; Fauré et al., 1993; Shepherd, 1999). These variations in chromosome structures disrupt meiosis in the hybrids and are believed to contribute to their sterility. This character as well as parthenocarpy was selected by early farmers for the production of edible fruits.
Different production areas currently suffer from new emerging diseases and have to face an ever-increasing range of pests and diseases (Pennisi, 2010). In the absence of locally adapted resistant varieties, the crop requires extensive use of pesticides, which threatens the sustainability of the crop and environment. Despite its economic importance, the banana export industry has been affected, as it relies on monoculture of genetically extremely closely related clones of the Cavendish subgroup (sterile triploids, AAA). There is thus an urgent need for a wider diversity of genetically improved banana cultivars with more robust disease resistance, increased productivity and better adaptability to a wide large range of growing conditions. However, breeding programmes are facing serious difficulties, mainly due to the low level of fertility, structural heterozygosity and triploidy of cultivated banana and to the absence of knowledge on the genetic factors involved in important agricultural traits. Musa balbisiana has important potential for breeding, such as conferring good ratooning ability, a strong root system and more generally resistance to biotic and abiotic stresses (Bakry et al., 2009). However, there is a serious lack of information regarding chromosome segregation in interspecific hybrids between M. accuminata and M. balbisiana and the possibility of chromosome exchange between these two genomes. This is due to the fact that chromosomes from these two species cannot be differentiated based on their size and/or morphology.
Genomic in situ hybridization (GISH) is a powerful tool to differentiate chromosomes from parental species in interspecific hybrids. It was developed by Schwarzacher et al. (1989) since when it has been applied to mitotic chromosomes from many plants resulting from interspecific hybridization (Jiang and Gill, 1994; D'Hont et al., 1996; Lashermes et al., 1999; Tanguy et al., 2005; Konnan et al., 2009; Mason et al., 2010). Likewise, GISH was successfully applied to differentiate the A and B chromosome of Musa on mitotic chromosome spreads prepared from root tips (Osuji et al., 1997; D'Hont et al., 2000). However, application of GISH on meiotic chromosomes is challenging and has been reported in just a few species, such as Solanum (Zhong et al., 1996), Hordeum (Anamthawat-Jonsson et al., 1993), Medicago tranculata (Kulikova et al., 2001), Beta vulgaris (Desel et al., 2001), Arabidopsis thaliana (Armstrong et al., 1998), Alstroemeria hybrids (Kamstra et al., 2004), Brassica (Armstrong et al., 1998; Kun et al., 2006; Nicolas et al., 2007), Lilium (Zhou et al., 2008), Lolium (Kopecky et al., 2008) and Brachiaria (Souza-Kaneshima et al., 2010). In Musa, a first report of fluorescence in situ hybridization (FISH) on meiotic chromosomes at pachytene stage was published recently (De Capdeville et al., 2009) but no report exists on GISH on meiotic chromosomes.
Here we describe a protocol developed to perform GISH on meiosis preparations at metaphase I from pollen mother cells and detail the technical challenges faced. We applied this protocol to study chromosome pairing in two interspecific triploid cultivated clones. The results demonstrated for the first time that pairing between chromosomes A and B occurs and may be frequent. Finally, we discuss the potential of these findings on Musa cultivar evolution and for banana breeding.
MATERIALS AND METHODS
Plant material
Two interspecific triploid banana clones involving Musa acuminata and M. balbisiana and belonging to two different banana groups (AAB and ABB) were analysed: ‘Figue Pomme’ (AAB, 2n = 33) of the ‘Silk’ subgroup and ‘Praha’ (ABB, 2n = 33) of the ‘Pisang Awak’ subgroup. These two clones were provided from the African Centre for Research on Banana and Plantain in Cameroon.
Meiotic chromosome preparation
Young anthers containing meiotic chromosomes were selected according to Fauré et al. (1993). The stage of development was determined via a acetocarmine squash preparation using a single anther from a flower. If at metaphase I, the remaining anthers were fixed directly in ethanol/acetic acid (3 : 1) and could be stored in 70 % ethanol, 4 °C for a few months. Flowers were harvested from 45-d-old inflorescences and the appropriate stages were found within buds in flower clusters (=‘hand’) number 30 for ‘Figue Pomme’ and number 35 for ‘Praha’ (flower size 0·5–1 cm). Flower cluster number 0 corresponds to the latest mature flowers that are located under the latest opened bract. Fixed anthers were rinsed twice in deionized water, then in citrate buffer (10 mm, pH 4·5) and then incubated for 6 h at 37 °C in a mixture of pectolytic enzymes containing 0·3 % (w/v) cytohelicase (Duchefa, www.duchefa.com), 0·3 % cellulase ‘Onozuka’ RS (Duchefa), 0·3 % pectolyase Y-23 (Duchefa) completed with variable concentrations of macerozyme R-10 (Duchefa) depending on cultivars in 10 mm citrate buffer, pH 4·5. For ‘Figue Pomme’ and ‘Praha’ accessions, best results were obtained with 6 and 7 % macerozyme, respectively. After washes in deionized water, the pollen mother cells were dissected out of the anthers into a watch glass using fine-mounted needles, taking care to remove as much as possible of the supporting tissues.
A 3 μL droplet of the cell suspension was then carefully transferred into grease-free slides, 15–30 µL of 60 % acetic acid was added and the pollen mother cells were left for 3 min on a hot plate at 50 °C for ‘Figue Pomme’ or 65 °C for ‘Praha’. A ring of freshly prepared ice-cold fixative (3 : 1) was added around the droplet containing the meiotic cells. Shortly after the fixative had mixed with the cell suspension, the cells were spread on the slide.
The quality of the slides was controlled by microscopic observation under phase contrast optics. Slides were used directly for in situ hybridization or were stored at –20 °C until needed.
Genomic in-situ hybridization
Before hybridization, the slides were fixed for 1 h in an oven at 65 °C, treated with RNase (Sigma, www.sigmaaldrich.com; 1 µg mL−1) in a moist chamber at 37 °C for 45 min and washed twice with 2× sodium saline citrate (SSC). The hybridization mixture (30 µL per slide) consisted of 50 % formamide, 10 % dextran sulphate, 2× SSC, 0·66 % sodium dodecyl sulphate and 6 ng μL−1 of each parental total genomic DNA probe. Genomic DNA of ‘Pisang Klutuk Wulung’ (M. balbisiana, B genome) and ‘Pahang’ (M. acuminata, A genome) were labelled by random priming with biotin-14-dUTP (Invitrogen Life Technology, www.invitrogen.com) and digoxigenin-11-dUTP (High Prime DNA Labeling Kit, Roche, www.roche.com), respectively. The hybridization mixture was denatured for 10 min in boiling water and dispensed on the slides. The chromosomes (with the hybridization mixture) were then denatured in a moist chamber placed on a water bath equilibrated at 80 °C for 2 min. Hybridization were performed overnight in a moist chamber at 37 °C. After hybridization, slides were successively washed for 10 min in 2 × , 0·5× and 0·1× SSC at 42 °C. Biotinylated probes were immunodetected with Texas-Red-conjugated avidin antibodies (ABCYS, www.abcysonline.com). Digoxigenin-labelled probes were detected with digoxigenin antibody conjugated with fluorescein isothiocyanate (FITC) (ABCYS). The chromosomes were mounted in Vectashield antifade solution (Vector Laboratories, www.vectorlabs.com) containing 2·5 µg mL−1 4′,6-diamidino-2-phenylindole (DAPI) as counterstaining. Fluorescence images were captured separately using a cooled high-resolution black and white CCD camera (Orca Hamamatsu, www.hamamatsucameras.com) and a Leica (www.leica-microsystems.com) DMRAX2 fluorescence microscope. The camera was interfaced to a PC running Volocity software (Perkin Elmer, www.perkinelmer.com).
RESULTS AND DISCUSSION
Development of GISH on Musa meiotic chromosomes from pollen mother cells
GISH on mitotic chromosomes obtained from root tips has been reported on many species, including Musa (Osuji et al., 1997; D'Hont et al., 2000), with quite similar protocols. By contrast, GISH on meiotic chromosomes in plants is quite challenging and the protocols used are complex and highly variable depending on the species. Accordingly, we encountered a series of technical challenges in Musa for preparing meiotic chromosome spreads suitable for GISH. The main challenge was the hardness of the cell wall and the density of the microsporocyte's cytoplasm, which hamper the accessibility of the probes to the chromosomes and generate higher levels of background, as already reported for Musa by De Capdeville et al. (2009).
We compared and tested the digestion protocols used in several species which vary in particular for the composition and concentration of the following enzyme cocktail: cellulase, pectinase, cytohelicase and pectolyase (Armstrong et al., 1998; Desel et al., 2001; Kulikova et al., 2001; Kun et al., 2006; De Capdeville et al., 2009). However, in our hands and with our plant material they did not provide acceptable results. We performed staining experiments on cells with calcofluor, propidium iodide and ruthenium red, which revealed that chromosomes were still embedded in an outer crust of pectin. Finally, we managed to digest those pectins by adding macerozyme to 0·3 % cytohelicase, pectolyase and cellulase. Macerozyme is used for protoplast production in various plants such as banana (Assani et al., 2001).
The addition of macerozyme in the enzyme mix, the duration of digestion and the temperature of the hot plate during the spreading phase were the key parameters in the success of our protocol. Note that these three parameters have to be adapted slightly for each clone and stage of meiosis. With this protocol, we were able to obtain meiotic chromosome spreads suitable for FISH for various accessions of AA, AB and AABB genomic constitution in addition to those reported in the present paper and from different stages of meiosis (prophase I, metaphase I and anaphase I) (data not shown). This protocol requires fresh material (fixed for no more than a few months). For both triploid clones, ‘Figue Pomme’ and ‘Praha’, the development of microsporocytes was not synchronized. This phenomena was more important in ‘Figue Pomme’, for which we observed on the same slide (from a single anther) early meiotic stages still in prophase I together with microsporocytes that had already reached the tetrad stage. Clones with highly asynchronous microsporocyte development required much more effort to get a reasonable rate of suitable chromosome spreads.
We then optimized our classical GISH protocol (D'Hont et al., 2000) by including the following changes: probes were labelled by random priming, slides with chromosome preparation were fixed for 1 h at 65 °C, chromosome denaturation was performed on a floating moist chamber over a water bath (Leflon et al., 2006) and the washes were softened.
Homoeologous chromosome pairing between the A and B genome
This protocol was used to perform GISH on meiotic metaphases of interspecific hybrids involving M. acuminata and M. balbisiana with genomic DNA of these species. The differential labelling obtained for A versus B chromosomes was excellent, so it was possible for the first time to attribute an A or B origin to the chromosomes involved in various pairing configuration (Figs 1 and 2). Labelling was essentially located on the centromeric and pericentromeric part of the chromosome leaving most of the chromosome arms unlabelled. This partial labelling was observed by D'Hont et al. (2000) when analysing Musa mitotic chromosomes but is much clearer on meiotic chromosomes. This uneven labelling observed on species with small chromosomes (Barre et al., 1998; Lashermes et al., 1999; Ollitrault et al., 2000; Ali Hoda et al., 2004) most probably results from the lower repeated sequence content in these genomes (D'Hont, 2005) and to the fact that the repeated sequences are typically more abundant in centromeric and pericentromeric parts of the chromosomes.
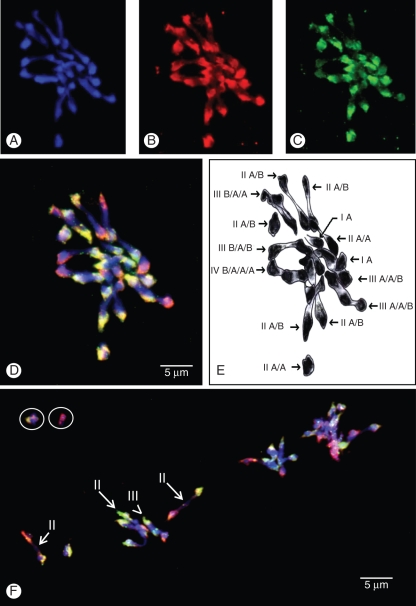
Fig. 1.
GISH on meiotic metaphase I of interspecific triploid Musa cultivar ‘Figue pomme’ (genome AAB). (A) DAPI, (B) B genomic DNA revealed in red (Texas Red), (C) A genomic DNA revealed in green (FITC) and (D, F) after superimposition of the three colours. (E) Schematic interpretation of (D). Circles indicate chromosomes that were oustside of the presented field. Arrows indicate homoeologous bivalents and arrowsheads indicate homoeologous multivalents.
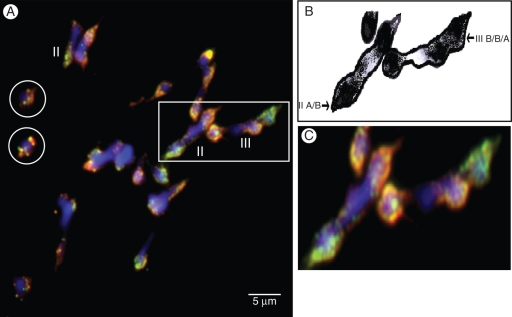
Fig. 2.
(A) GISH on meiotic metaphase I of interspecific triploid Musa cultivar ‘Praha’ (genome ABB) after superimposition of DAPI, Texas Red and FITC (cf. Fig. 1). (B) Schematic interpretation of part of (A). (C) Focus on part of (A). Circle indicates chromosomes that were oustside of the presented field. Arrows indicates homoeologous bivalents and arrowsheads indicate homoeologous multivalents.
It is also interesting to note that the labelling obtained with the B genomic DNA was relatively homogeneous on all centromeric regions of all chromosomes (Fig. 1B) in contrast to the labelling obtained with the A genomic DNA, which was variable depending on chromosomes (Fig. 1C). This may be related to the fact that B genomes are slightly smaller than A genomes (Dolezel et al., 1994; Lysak et al., 1999; Bartos et al., 2005) and thus species-specific repeated sequences appeared more numerous in M. acuminata than in M. balbisiana (Baurens, 1997).
Two triploid interspecific accessions were analysed involving a different balance of A and B genomes: accession ‘Figue Pomme’ with an AAB genomic constitution (Fig. 1) and accession ‘Praha’ with an ABB genomic constitution (Fig. 2). Classical cytogeneticists have reported that it is particularly difficult to obtain good spreading at meiosis metaphase for interspecific triploid Musa clones (Wilson, 1945; Simmonds, 1962). Accordingly, for most of the cell only part of the pairing configuration could be analysed, the other chromosomes being not sufficiently spread. We observed that chromosome associations were quite variable between the different metaphases of the same clone. This has been noted in interspecific hybrids from Musa (reviewed by Shepherd, 1999) and in other genera such as Lilium (Zhou et al., 2008).
For both genotypes, we identified univalents, bivalents and multivalents (Figs 1 and 2) in accordance with previous results based on acetocarmin staining (Shepherd, 1999). In addition, for the first time, we were able to visualize homoeologous pairing. Homoeologous bivalents were observed in all analysed cells with two to five homoeologous bivalents for ‘Figue Pomme’ (Fig. 1) and one to four for ‘Praha’ (Fig. 2). Moreover, all multivalents (trivalents and tetravalents) observed involved homoeologous chromosomes (Figs 1 and 2).
Information regarding the possibility of homoeologous chromosome pairing have tentatively been inferred from classical chromosome pairing studies. In diploid AB interspecific hybrids, bivalents were observed in variable proportions from three to 11, suggesting partial homoeologous chromosome pairing (Dodds and Simmonds, 1946; Shepherd, 1999). In polyploid interspecific hybrids (AAB, ABB), interpretations were also based on the numbers of bivalents and multivalents observed. However, as multivalents are observed in monospecific clones (AA and AAA) (reviewed by Shepherd 1999), these studies were largely speculative and sometimes contradictory; for example, Shepherd (1999) concluded that there was low affinity between the chromosomes of the two species whereas Dessauw (1987) concluded the opposite. GISH on mitotic chromosomes was unable to resolve this issue as the partial labelling obtained prevented the detection of interspecific recombinant chromosomes (Osuji et al., 1997; D'Hont et al., 2000).
Here for the first time, we clearly demonstrated that interspecific recombination between A and B chromosomes does exist and may be frequent. This information has important implications for the domestication and improvement of interspecific cultivars through breeding.
Implications for Musa cultivar evolution and for banana breeding
Interspecific triploid cultivars derived from M. acuminata and M. balbisiana are believed to result from one or two steps of combination between the parental species, featuring 2n gametes derived from AA and AB genotypes (Cheesman, 1947; Simmonds and Shepherd, 1955; Perrier et al., 2009). De Langhe et al. (2010) emphasized that additional steps of combination may have occurred. The possibility of A/B pairing revealed in our study implies that chromosome re-assortments and exchanges of chromosome segments may have occurred between the two genomes, leading to unbalanced genome transmission with respect to the parental species. The 8A + 25B chromosome constitution of the triploid interspecific cultivar ‘Pelipita’ revealed by GISH (D'Hont et al., 2000) constitutes the first accurately documented example of important unbalanced genome constitution; note that this reveals essentially the origin of the centromeric regions and may overlook chromosome arm recombinants. Further work to understand the impact of interspecific chromosome pairing on cultivar chromosome constitution will require the analysis of interspecific cultivars and/or progenies with numerous A- and B-specific molecular marker alleles. The ongoing Musa genome sequencing project will further facilitate development of markers for future high-throughput characterizations (http://www.genoscope.cns.fr/spip/September-8th-2009-Banana-genome.html).
Production of edible banana requires sterile clones or clones with only residual fertility; consequently, developing new cultivars requires complex breeding strategies. The main strategy used so far to create new interspecific cultivars is based on exploitation of residual fertility of triploid cultivars in combination with fertile diploid accessions (Menendez and Shepherd, 1975; Bakry and Horry, 1992; Rowe and Rosales, 1993; Vuylsteke et al., 1993). The tetraploid progeny resulting from un-reduced 2n gametes from the triploid parent and n gametes from the diploid parent are then selected. We are currently testing in Guadeloupe (French West Indies) another strategy which consists in doubling AB hybrids and crossing them with AA or BB diploids (Bakry et al., 2009). The occurrence of interspecific chromosome pairing demonstrated in the present study opens new perspectives for this latter strategy as it implies that a much wider range of gametic genotypes can be obtained from an AABB genotype compared with a pure disomic inheritance. In addition, the possibility of introgressing only fragments of B chromosomes opens new perspectives to exploit the B genomes for improving resistance and rusticity of monospecific acuminata cultivars. This should allow us to introgress only B chromosome segments bearing a target character and avoid introgression of B chromosome fragments harboring endogenous banana streak virus (eBSV) sequences. eBSV sequences are able to release infectious virions and such activations are known to be favoured in an intergenomic context (Lheureux et al., 2003; Gayral et al., 2008). Recent results suggested that eBSV sequences are present at only a few loci in the B genome (Iskra-Caruana et al., 2010).
Finally, homoeologous pairing and thus interspecific chromosome exchanges will most probably vary depending on genotypes, through genic control (Griffiths et al., 2006; Kopecky et al., 2008; Nicolas et al., 2009), and environment. The possibility of testing the extent of homoeologous pairing, demonstrated in this paper using GISH, in various types of parental materials and crosses should help to define new efficient breeding strategies.
ACKNOWLEDGEMENTS
We thank Raphaël Mercier and Liudmila Chelysheva for advice. We thank the ‘Ministère de l'Enseignement Supérieur de la Recherche Scientifique et de la Technologie’ of Tunisia, the European AVERROES programme and CIRAD for financing assistance.
LITERATURE CITED
- Ali Hoda BM, Lysak MA, Schubert I. Genomic in situ hybridization in plants with small genomes is feasible and elucidates the chromosomal parentage in interspecific Arabidopsis hybrids. Genome. 2004;47:954–960. doi: 10.1139/g04-041. [DOI] [PubMed] [Google Scholar]
- Anamthawat-Jonsson K, Schwarzacher T, Heslop-Harrison JS. Behavior of parental genomes in the hybrid Hordeum vulgare × H. bulbosum. The Journal of Heredity. 1993;84:78–82. [Google Scholar]
- Armstrong SJ, Paul F, David FM, Gareth HJ. Physical mapping of DNA repetitive sequences to mitotic and meiotic chromosomes of Brassica oleracea var. alboglabra by fluorescence in situ hybridization. Heredity. 1998;81:666–673. [Google Scholar]
- Assani A, Haicour R, Wenzel G, et al. Plant regeneration from protoplasts of dessert banana cv. Grande Naine (Musa spp., Cavendish sub-group AAA) via somatic embryogenesis. Plant Cell Reports. 2001;20:482–488. [Google Scholar]
- Bakry F, Horry JP. Tetraploid hybrids from interploid 3 × /2 × crosses in cooking bananas. Fruits. 1992;47:641–647. [Google Scholar]
- Bakry F, Carreel F, Jenny C, Horry JP. Genetic improvement of banana. In: Mohan Jain Shri, PM Priyadarshan., editors. Breeding plantation tree crops: tropical species. New York: Springer; 2009. pp. 3–50. [Google Scholar]
- Barre P, Layssac M, D'Hont A, et al. Relationship between parental chromosomic contribution and nuclear DNA content in the coffea interspecific hybrid: C. pseudozanguebariae × C. liberica var. dewevrei. Theoretical and Applied Genetics. 1998;96:301–305. [Google Scholar]
- Bartos J, Alkhimova O, Dolezelova M, De Langhe E, Dolezel J. Nuclear genome size and genomic distribution of ribosomal DNA in Musa and Ensete (Musaceae): taxonomic implications. Cytogenetics and Genome Research. 2005;109:50–57. doi: 10.1159/000082381. [DOI] [PubMed] [Google Scholar]
- Baurens FC. Identification par PCR des espèces impliquées dans la composition génomique des cultivars de bananier, à l'aide de séquences répétées = PCR identification of the species involved in the genomic composition of the banana cultivar using repeated sequences. 1997. Thèse de doctorat, Université de Toulouse 3, Toulouse, France. [Google Scholar]
- Carreel F, Gonzalez de Leon D, Lagoda P, et al. Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome. 2002;45:679–692. doi: 10.1139/g02-033. [DOI] [PubMed] [Google Scholar]
- Chalmin P, editor. Cyclope: les marchés mondiaux, 2009. 2009. [Google Scholar]
- Cheesman EE. The classification of the bananas II. The genus Musa L. Kew Bulletin. 1947;2:106–117. [Google Scholar]
- Creste S, Neto AT, Vencovsky R, De Oliveira Silva S, Figueira A. Genetic diversity of Musa diploid and triploid accessions from the Brazilian banana breeding program estimated by microsatellite markers. Genetic Resources and Crop Evolution. 2004;51:723–733. [Google Scholar]
- De Capdeville G, Manoel TSJ, Dora S, et al. The potential of high-resolution BAC-FISH in banana breeding. Euphytica. 2009;166:431–443. [Google Scholar]
- De Langhe E, Hribova E, Carpentier S, Dolezel J, Swennen R. Did backcrossing contribute to the origin of hybrid edible bananas? Annals of Botany. 2010;106:849–857. doi: 10.1093/aob/mcq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desel C, Christian J, Daguang C, Michael K, Thomas S. High-resolution mapping of YACs and the single-copy gene Hs1pro-1 on Beta vulgaris chromosomes by multi-colour fluorescence in situ hybridization. Plant Molecular Biology. 2001;45:113–122. doi: 10.1023/a:1006405911442. [DOI] [PubMed] [Google Scholar]
- Dessauw D. Etude des facteurs de la stérilité du bananier (Musa spp) et des relations cytotaxinomiques entre M. acuminata Colla et M. balbisiana Colla. 1987 PhD thesis, University of Paris-Sud Centre d'Orsay. [Google Scholar]
- D'Hont A. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenetics and Genome Research. 2005;109:27–33. doi: 10.1159/000082378. [DOI] [PubMed] [Google Scholar]
- D'Hont A, Grivet L, Feldmann P, Glaszmann JC, Rao S, Berding N. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Molecular and General Genetics. 1996;250:405–413. doi: 10.1007/BF02174028. [DOI] [PubMed] [Google Scholar]
- D'Hont A, Paget-Goy A, Escoute J, Carreel F. The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theoretical and Applied Genetics. 2000;100:177–183. [Google Scholar]
- Dodds KS. Genetical and cytological studies of Musa. V. Certain edible diploids. Journal of Genetics. 1943;45:113–138. [Google Scholar]
- Dodds KS, Simmonds NW. Genetical and cytological studies of Musa. VIII. The formation of polyploid spores. Journal of Genetics. 1946;47:223–241. doi: 10.1007/BF02986248. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Dolezelova M, Novak FJ. Flow cytometric estimation of nuclear DNA amounts in diploid bananas (Musa acuminate and M. balbisiana) Biologia Plantarum. 1994;36:351–357. [Google Scholar]
- Fauré S, Bakry F, Gonzâlez LD. Cytogenetic studies of diploid bananas. In: Ganry J, editor. Breeding banana and plantain for resistance to diseases and pests. Montpellier: CIRAD-FLHOR; 1993. pp. 77–92. International symposium on genetic improvement of bananas for resistance to diseases and pests, 1992-09-07/1992-09-09, Montpellier, France. [Google Scholar]
- Gayral P, Noa-Carrazana JC, Lescot M, et al. A single Banana Streak Virus integration event in the banana genome as the origin of infectious endogenous Pararetrovirus. Journal of Virology. 2008;82:6697–6710. doi: 10.1128/JVI.00212-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, et al. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- Hřibová E, Čížková J, Christelová P, Taudien S, de Langhe E, Doležel J. The ITS1-5·8S-ITS2 sequence region in the Musaceae: structure, diversity and use in molecular phylogeny. PLoS ONE. 2011;6:e17863. doi: 10.1371/journal.pone.0017863. doi:10.1371/journal.pone.0017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskra-Caruana ML, Baurens FC, Gayral P, Chabannes M. A four-partner plant–virus interaction: enemies can also come from within. Molecular Plant-Microbe Interactions. 2010;23:1394–1402. doi: 10.1094/MPMI-05-10-0107. [DOI] [PubMed] [Google Scholar]
- Isobe M, Hashimoto K. The chromosome count of nine taxa in Musa and its allied genus Musella. Bulletin of the Hiroshima Botanical Garden. 1994;15:7–11. [Google Scholar]
- Jiang J, Gill BS. Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome. 1994;37:717–725. doi: 10.1139/g94-102. [DOI] [PubMed] [Google Scholar]
- Kamstra SA, De Jong HJ, Jacobsen E, Ramanna MS, Kuipers AGJ. Meiotic behaviour of individual chromosomes in allotriploid Alstroemeria hybrids. Heredity. 2004;93:15–21. doi: 10.1038/sj.hdy.6800465. [DOI] [PubMed] [Google Scholar]
- Konnan O, Baudoin JP, D'Hont A, Mergeai G. Bridging classical and molecular cytogenetics of Gossypium. In: Paterson Andrew H, editor. Genetics and genomics of cotton. New York: Springer; 2009. pp. 257–281. [Google Scholar]
- Kopecky D, Lukaszewski AJ, Dolezel J. Meiotic behaviour of individual chromosomes of Festuca pratensis in tetraploid Lolium multiflorum. Chromosome Research. 2008;16:987–998. doi: 10.1007/s10577-008-1256-0. [DOI] [PubMed] [Google Scholar]
- Kulikova O, Gualtieri G, Geurts R, et al. Integration of the FISH pachytene and genetic maps of Medicago truncatula. The Plant Journal. 2001;27:49–58. doi: 10.1046/j.1365-313x.2001.01057.x. [DOI] [PubMed] [Google Scholar]
- Kun Y, Hong-Yan QI, Zhu LQ, Wang XJ. Localization of S genes on extended DNA fibers (EDFs) in Brassica oleracea by high-resolution FISH. Actu Geneticu Sinicu. 2006;33:277–284. doi: 10.1016/S0379-4172(06)60051-6. [DOI] [PubMed] [Google Scholar]
- Lashermes P, Combes MC, Robert J, et al. Molecular characterisation and origin of the Coffea arabica L. genome. Molecular and General Genetics. 1999;261:259–266. doi: 10.1007/s004380050965. [DOI] [PubMed] [Google Scholar]
- Lassois L, Busogoro JP, Jijakli H. La banane: de son origine à sa commercialisation. Biotechnology, Agronomy, Society and Environment. 2009;13:575–586. [Google Scholar]
- Leflon M, Eber F, Letanneur JC, et al. Pairing and recombination at meiosis of Brassica rapa (AA) Brassica napus (AACC) hybrids. Theoretical and Applied Genetics. 2006;113:1467–1480. doi: 10.1007/s00122-006-0393-0. [DOI] [PubMed] [Google Scholar]
- Lheureux F, Carreel F, Jenny C, Lockhart BEL, Iskra-Caruana ML. Identification of genetic markers linked to banana streak disease expression in inter-specific Musa hybrids. Theoretical and Applied Genetics. 2003;106:594–598. doi: 10.1007/s00122-002-1077-z. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Dolezelova M, Horry HJP, Swennen R, Dolezel J. Flow cytometric analysis of nuclear DNA content in Musa. Theoretical and Applied Genetics. 1999;98:1344–1350. [Google Scholar]
- Mason AS, Huteau V, Eber F, et al. Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chromosome Research. 2010;18:655–666. doi: 10.1007/s10577-010-9140-0. [DOI] [PubMed] [Google Scholar]
- Menendez T, Shepherd K. Breeding new bananas. 1975:104–112. World CropsMay/June. [Google Scholar]
- Nicolas SD, Le Mignon G, Eber F, et al. Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics. 2007;175:487–503. doi: 10.1534/genetics.106.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas SD, Leflon M, Monod H, et al. Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. The Plant Cell. 2009;21:373–385. doi: 10.1105/tpc.108.062273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollitrault O, Dambier D, Froelicher Y, et al. Apport de l'hybridation somatique pour l'exploitation des ressources génétiques des agrumes. Cahiers Agricultures. 2000;9:223–236. [Google Scholar]
- Osuji JO, Okoli BE, Ortiz R. An improved procedure for mitotic studies of the Eumusa section of the genus Musa L. (Musaceae) Infomusa. 1996a;5:12–14. [Google Scholar]
- Osuji JO, Okoli BE, Ortiz R. Relevance of cytogenetics to Musa research. 1996b:24–28. Proceedings of the NARS-IITA meeting on Musa research in West and Central Africa, IITA High Rainfall Station, Onne, Rivers State Nigeria. September 1995, 79. [Google Scholar]
- Osuji JO, Crouch J, Harrison G, Heslop-Harrison JS. Identification of the genomic constitution of Musa L. lines (bananas, plantains and hybrids) using molecular cytogenetics. Annals of Botany. 1997;80:787–793. [Google Scholar]
- Pennisi E. Armed and dangerous. Science. 2010;327:804–805. doi: 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- Perrier X, Bakry F, Carreel F, et al. Combining biological approaches to highlight the evolution towards edible bananas. Ethnobotany Research & Applications. 2009;7:199–216. [Google Scholar]
- Rowe P, Rosales F. Diploid breeding at FHIA and the development of Goldfinger. InfoMusa. 1993;2:9–11. [Google Scholar]
- Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. In situ localization of parental genomes in a wide hybrid. Annals of Botany. 1989;64:315–324. [Google Scholar]
- Shepherd K. Cytogenetics of the genus Musa. Montpellier: International Network for the Improvement of Banana and Plantain; 1999. [Google Scholar]
- Simmonds NW. The evolution of the bananas. London: Longman; 1962. [Google Scholar]
- Simmonds NW, Shepherd K. The taxonomy and origins of the cultivated bananas. Botanical Journal of the Linnean Society. 1955;55:302–312. [Google Scholar]
- Souza-Kaneshima AMD, Simioni C, Felismino MF, et al. Meiotic behaviour in the first interspecific hybrids between Brachiaria brizantha and Brachiaria decumbens. Plant Breeding. 2010;129:186–191. [Google Scholar]
- Tanguy AM, Coriton O, Abélard P, Dedryver F, Jahier J. Structure of Aegilops ventricosa chromosome 6Nv, the donor of wheat genes Yr17, Lr37, Sr38, and Cre5. Genome. 2005;48:541–546. doi: 10.1139/g05-001. [DOI] [PubMed] [Google Scholar]
- Therdsak T, Benchamas S, Yingyong P, Pradit P. Meiotic behavior in microsporocytes of some bananas in Thailand. Kasetsart Journal (Natural Science) 2010;44:536–543. [Google Scholar]
- Ude G, Pillay M, Nwakanma D, Tenkouano A. Genetic diversity in Musa acuminata Colla and Musa balbisiana Colla and some of their natural hybrids using AFLP Markers. Theoretical and Applied Genetics. 2002;104:1246–1252. doi: 10.1007/s00122-002-0914-4. [DOI] [PubMed] [Google Scholar]
- Vuylsteke D, Swennen R, Ortiz R. Registration of 14 improved tropical Musa plantain hybrids with Black Sigatoka resistance. HortScience. 1993;28:957–959. [Google Scholar]
- Wilson GB. Cytological studies in the Musa. I. Meiosis in some triploid clones. Genetics. 1945;31:241–258. [PubMed] [Google Scholar]
- Zhong XB, De Jong HJ, Zabel P. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH) Chromosome Research. 1996;4:24–28. doi: 10.1007/BF02254940. [DOI] [PubMed] [Google Scholar]
- Zhou S, Ramanna MS, Visser RGF, Jaap MVTJ. Analysis of the meiosis in the F1 hybrids of Longiflorum × Asiatic (LA) of lilies (Lilium) using genomic in situ hybridization. Journal of Genetics and Genomics. 2008;35:687–695. doi: 10.1016/S1673-8527(08)60091-0. [DOI] [PubMed] [Google Scholar]