Abstract
Discussion on the intricate regulation of C/EBPA transcriptional activity and the delicate balance between normal myelopoiesis and transformation to AML.
Keywords: granulocytes, macrophages, monocytes
Lineage determination during blood cell development, known as hematopoiesis, is regulated primarily by transcription factors and their target genes. Several of these transcription factors have been identified as being necessary for differentiation of hematopoietic stem cells into myeloid cells, including monocytes and granulocytes. The question is how specific transcription factors can confer lineage specificity when they associate with other proteins to regulate gene expression. The paper by Hong et al. [1] describes the interaction of c-Jun or c-Fos to the C/EBPA and that heterodimerization of C/EBPA with the AP-1 leads to preferential differentiation toward the monocyte lineage.
AP-1 proteins belong to a family of transcription factors that heterodimerize with c-Fos, c-Jun, activating transcription factor, and Jun dimerization protein. These proteins regulate gene expression in response to a variety of extracellular stimuli, including growth factors, cytokines, stress, and infections. AP-1 proteins also regulate important cellular decisions, such as proliferation, differentiation, and apoptosis. C/EBPA is critical for granulocyte differentiation and is a member of a family of transcription factors that contains a bZIP region [2, 3], which facilitates dimerization with other bZIP-containing transcription factors and is required for DNA binding.
In terms of the structure and function of C/EPBA, the gene is located on chromosome 19q13.1 and lacks introns [2]. Two isoforms result, with the shorter p30 isoform lacking 117 aa of the N-terminal region [2, 4]. The transactivation domains that interact with the transcriptional machinery are in the p42 form, whereas the chromatin-remodeling complex and the region responsible for interaction with other transcription factors are present in both [2, 5]. C/EBPA is expressed in a variety of tissues, including adipose, liver, airway epithelial cells, and myeloid cells [2, 6, 7]. Therefore, the specificity of direct target genes in a tissue-restricted manner is dependent on the heterodimeric complexes of C/EBPA. The relative contribution of the various partners that interact with C/EBPA in myeloid progenitor cells is necessary for our understanding of the molecular regulation of myeloid differentiation.
The role of C/EBPA has been elucidated through various mouse models, in which the gene has been knocked out. C/EBPA−/− mice die at birth as a result of severe hypoglycemia and abnormal liver development [2, 8, 9]. Nonconditional targeted disruption of C/EBPA in mice results in a block in early granulocyte maturation [2, 10]. C/EBPA knockout mice also have lung abnormalities, specifically hyperproliferation of type II pneumocytes [2]. In terms of the hematopoietic effects, there was notable absence of granulocytes and accumulation of early myeloid cells, suggestive of a maturational arrest [2, 3]. Therefore, C/EBPA clearly has pleiotropic effects on numerous tissues.
Previous reports have suggested that C/EBPA not only induces myeloid differentiation but also growth arrest [3]. Disruption of C/EBPA affects cells by inhibiting cell cycle regulation and granulocyte differentiation, indicating the role of this protein as a tumor suppressor [3]. Hence, mutations in C/EBPA appear to contribute to myeloid leukemias. Among important target genes of C/EBPA are myeloid growth factor receptors and microRNA miR233, which controls granulocyte differentiation [2, 3]. Up-regulation of p21Cip1 by C/EBPA leads to cell cycle arrest [2]. Down-regulation of c-myc may also play a role in blocking cell proliferation and inducing differentiation [2]. One of the primary reasons for this diversity of functions is the fact that C/EBPA can interact with other transcription factors, in addition to post-transcriptional and post-translational modifications, as well as high order changes, such as chromatin remodeling.
The importance of C/EBPA in myeloid differentiation is highlighted by several groups identifying mutations in myeloid leukemia cells from patients with AML [2, 3]. The frequency of C/EBPA mutations has been reported between 5% and 14% [10]. In AML, there are N-terminal frameshift mutations that result in premature termination, whereas the 30-kDa form acts in a dominant-negative manner for the remaining, normal C/EBPA. The C-terminal in-frame mutations disrupt the bZIP, thereby interfering with dimerization with other family members and subsequent DNA transcription [10]. Most studies so far suggest that AML patients with C/EBPA mutations have improved survival [10]. The reasons for this are not well understood. Interestingly, the majority (72%) of AML patients with C/EBPA mutations has more than one type of mutation rather than a single mutation. Approximately 41% of patients have the N-terminal frameshift mutation, whereas 33% have the C-terminal insertion/deletion mutation. The remaining 26% have one N-terminal mutation and one C-terminal frameshift mutation. Other studies have reported over 90% of patients having both mutations. Therefore, it is difficult to assess which mutation is responsible for improved prognosis [10].
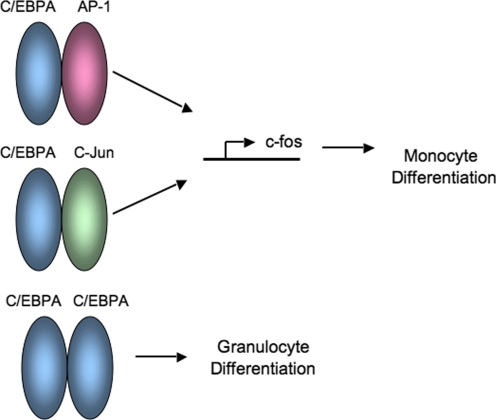
Very little is known about the molecular regulation of C/EBPA and normal granulocyte and monocyte differentiation. Interestingly, C/EBPA has been reported to be expressed in myeloid progenitors and granulocytes, but not macrophages [3]. This is interesting, given the fact that the paper by Hong et al. [1] suggests that association of C/EBPA with c-Fos preferentially drives myeloid progenitors to monocyte differentiation. This may be a result of the fact that experiments were performed with cell lines that are immortalized or transformed. The significance of the paper by Hong et al. [1] is the demonstration of the diversity of transcription factors based on their binding partners. Depending on its partners, the C/EBPA homo- or heterodimers recognize different DNA sequences in promoters of target genes. Subsequently, this leads to differential gene expression that results in granulocyte or monocyte differentiation (Fig. 1). Interestingly, C/EBPA interacts with c-Jun or c-Fos and binds to a hybrid consensus DNA-binding site. In addition, C/EBPA or c-Jun alone binds weakly to a consensus promoter sequence compared with the C/EBPA:c-Jun heterodimer, which binds to this sequence with higher affinity. Furthermore, similar to in vitro experiments, the stoichiometry of these transcription factors was also found in myeloid cells. Previous work by this same group demonstrated that C/EBPA and NF-κB cooperate in inducing genes that are involved with the inflammatory response [11]. The C/EBPA basic region was found to interact with the p50, not p65, subunit of NF-κB to induce expression of bcl-2, which in turn increases myeloid proliferation and survival [11]. In the future, additional leucine zipper transcription factors, such as CREB, could modulate C/EBP function in normal myelopoiesis and AML. In conclusion, the intricate regulation of C/EBPA transcriptional activity reminds us of the enormous complexity of myeloid proliferation and differentiation and the delicate balance between normal myelopoiesis and transformation to AML.
Figure 1. Myeloid differentiation and C/EBPA partners.
ACKNOWLEDGMENTS
K.M.S. is supported by NIH grants HL75826 and HL83077 and an American Society of Hematology Alternative Training Pathway Award in Bone Marrow Failure Syndromes and is a scholar of the Leukemia and Lymphoma Society.
SEE CORRESPONDING ARTICLE ON PAGE 643
- bZIP
- basic leucine zipper
REFERENCES
- 1. Hong S., Skaist A. M., Wheelan S. J, Friedman A. D. (2011) AP-1 protein induction during monopoiesis favors C/EBP:AP-1 heterodimers over C/EBP homodimerization and stimulates FosB transcription. J. Leukoc. Biol. 90, 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koschmieder S., Halmos B., Levantini E., Tenen D. G. (2009) Dysregulation of the C/EBPα differentiation pathway in human cancer. J. Clin. Oncol. 27, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reckzeh K., Cammenga J. (2010) Molecular mechanisms underlying deregulation of C/EBPα in acute myeloid leukemia. Int. J. Hematol. 91, 557–568 [DOI] [PubMed] [Google Scholar]
- 4. Lin F. T., MacDougald O. A., Diehl A. M., Lane M. D. (1993) A 30-kDa alternative translation product of the CCAAT/enhancer binding protein α message: transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. USA 90, 9606–9610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Friedman A. D., McKnight S. L. (1990) Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 4, 1416–1426 [DOI] [PubMed] [Google Scholar]
- 6. Birkenmeier E. H., Gwynn B., Howard S., Jerry J., Gordon J. I., Landschulz W. H., McKnight S. L. (1989) Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 3, 1146–1156 [DOI] [PubMed] [Google Scholar]
- 7. Antonson P., Xanthopoulos K. G. (1995) Molecular cloning, sequence, and expression patterns of the human gene encoding CCAAT/enhancer binding protein α (C/EBP α). Biochem. Biophys. Res. Commun. 215, 106–113 [DOI] [PubMed] [Google Scholar]
- 8. Flodby P., Barlow C., Kylefjord H., Ahrlund-Richter L., Xanthopoulos K. G. (1996) Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein α. J. Biol. Chem. 271, 24753–24760 [DOI] [PubMed] [Google Scholar]
- 9. Sugahara K., Iyama K. I., Kimura T., Sano K., Darlington G. J., Akiba T., Takiguchi M. (2001) Mice lacking CCAAt/enhancer-binding protein-α show hyperproliferation of alveolar type II cells and increased surfactant protein mRNAs. Cell Tissue Res. 306, 57–63 [DOI] [PubMed] [Google Scholar]
- 10. Pabst T., Mueller B. U. (2007) Transcriptional dysregulation during myeloid transformation in AML. Oncogene 26, 6829–6837 [DOI] [PubMed] [Google Scholar]
- 11. Friedman A. D. (2007) C/EBPα induces PU.1 and interacts with AP-1 and NF-κB to regulate myeloid development. Blood Cells Mol. Dis. 39, 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]