Abstract
Immunoassays for detection of bacterial pathogens rely on the selectivity and stability of bio-recognition elements such as antibodies tethered to sensor surfaces. The search for novel surfaces that improve the stability of biomolecules and assay performance has been pursued for a long time. However, the anticipated improvements in stability have not been realized in practice under physiological conditions because the surface functionalization layers on commonly used substrates, silica and gold, are themselves unstable on time scales of days. In this paper, we show that covalent linking of antibodies to diamond surfaces leads to substantial improvements in biological activity of proteins as measured by the ability to selectively capture cells of the pathogenic bacterium Escherichia coli O157:H7 even after exposure to buffer solutions at 37 °C for extended periods of time, approaching 2 weeks. Our results from ELISA, XPS, fluorescence microscopy, and MD simulations suggest that by using highly stable surface chemistry and controlling the nanoscale organization of the antibodies on the surface, it is possible to achieve significant improvements in biological activity and stability. Our findings can be easily extended to functionalization of micro and nanodimensional sensors and structures of biomedical diagnostic and therapeutic interest.
1. Introduction
Food and water-borne pathogens can pose serious long-term health risks, and in severe cases, can be fatal. In 2008, there were 3.5 million cases reported in the United States for infection with five major pathogens Escherichia coli O157:H7, non-O157 shiga toxin-producing E.coli, Salmonella, Listeria monocytogenes, and Campylobacter.[1] The losses associated with these cases can be reduced by monitoring food and water supplies using continuous, real-time pathogen sensors. While many miniaturized pathogen sensors have been developed based on antibody, DNA, or biochemical-based methods in the last decade,[2] the need for highly selective and sensitive sensors that can monitor for extended periods of time still remains unfulfilled. Such sensors could ideally be envisioned to continuously sense the presence of a target pathogen until it is captured, at which time the sensor could be replaced. The key to the realization of such extended use sensors would be to achieve extended stability of biorecognition elements like antibodies immobilized on sensing surfaces. These sensors could include label free approaches such as quartz crystal microbalance, surface plasmon resonance based sensors, photonic crystals sensors, field-effect based sensors, or resonance-based micro-cantilever sensors.
Previous studies have shown that improvements in stability of immobilized proteins can be achieved by reducing the protein-surface interactions.[3,4] These interactions depend on the water molecule density at the interface, the surface functionalization chemistry and the charge distribution on the protein and the surface. Silicon dioxide (SiO2) and gold (Au) are currently the most commonly used substrates for anchoring proteins and are known to denature proteins upon direct interaction with bare surface[5,6]. Such protein-denaturing protein-surface interactions can be reduced by co-immobilization of polyethylene glycol (PEG) molecules[4,6] or lipid bilayers[7] via thiol chemistry on Au or silane chemistry on SiO2, improving the stability of the surface immobilized proteins. However, PEG or lipid bilayer surface chemistries lack the robustness to withstand physiological conditions for more than two days.[8] The search for novel surfaces and surface chemistries that retain the stability of antibodies and assay performance has been pursued for a long time. One such surface is ultrananocrystalline diamond (UNCD). UNCD films are attractive to construct biosensors due to its high chemical stability,[9] biocompatibility,[10] large electrochemical potential window,[11,12] and ease of integration with microfluidic architectures.[13] Ultra-nanocrystalline diamond (UNCD) films utilized in this research comprise of 2–5 nm diameter, phase-pure diamond grains. UNCD films have 10% (by volume) grain boundaries, which consist of carbon in several different bonding states including sp2 (graphite).[9] UNCD films of 1 μm were grown on n-type Si (100) at Advanced Diamond Technologies in a hot-filament chemical vapor deposition reactor at about 700 °C using CH4/H2 chemistry at a pressure of about 5 Torr. Figure 1 shows the atomic force microscopy (AFM), scanning electron microscopy (SEM), and X-ray photoelectron spectroscopy (XPS) data characterizing the surface structure of the as-deposited UNCD film and verifying its hydrogen-termination. Previously we have shown that UV-based grafting of alkenes on UNCD surfaces provides a surface chemistry that is hydrolytically more stable when compared to silane chemistry on silica, thiol chemistry on gold, UV-based alkene grafting chemistry on silicon, or glassy carbon surfaces.[14] Since then UV-alkene chemistry has been used to attach DNA,[15] antibodies[16] and enzymes[11] to diamond surfaces, and biological activities on diamond have been demonstrated. However, no reports suggest the impact of diamond surface and its surface chemistry on the protein stability and function especially under physiological conditions, which is important for application of diamond surfaces for biomedical diagnostics.
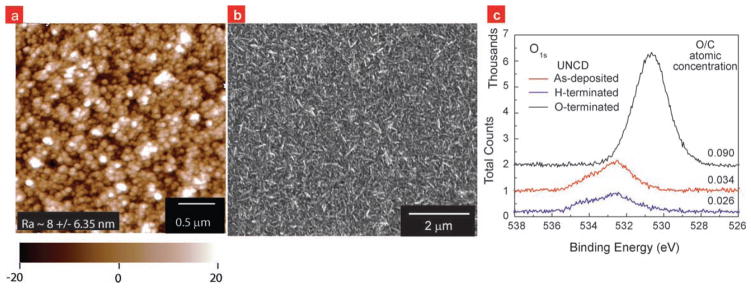
Figure 1.
UNCD surface characterized via microscopy. a) AFM and b) SEM image of ultra nanocrystalline diamond films deposited via hot-filament chemical vapor deposition. AFM image shows that the average roughness is ~8 nm and largest peak-to-valley height is ~40 nm. SEM image shows the continuity in the diamond film at micron-scale. c) O1s peaks obtained during XPS analysis of O-terminated, as-deposited and H-terminated UNCD films (top to bottom) show that the as-deposited UNCD films were more H-terminated than O-terminated. XPS data were obtained using an ultrahigh vacuum (P < 7 × 10−10 Torr) XPS system with a monochromatized Mg Kα (1253.6 eV) source (350 W, 14.0 kV) and a hemispherical analyzer equipped with a multichannel detector. H-terminated sample was produced by exposing the as-deposited samples to a 13.56 MHz inductively coupled hydrogen plasma (15 torr) for 20 minutes at 800 °C. O-terminated sample was produced by exposing the as-deposited samples to UV-lamp (254 nm, 10 mW/cm2) in air.
In this paper we provide detailed characterization of antibody-tethered diamond surfaces as a biosensor, compare it to antibody-tethered glass surfaces, and more importantly examine the stability of antibodies attached to a diamond surfaces under physiological conditions as a function of time (0.137 M NaCl, phosphate-buffered pH 7.4, 37°C). Due to the growing concern of detecting and eliminating food and water borne pathogens as mentioned above, we chose to work with anti-bacterial antibodies. Scheme 1 shows the reaction steps used to create and use antibody functionalized UNCD surface. We used the UV-alkene grafting chemistry to attach antibodies to UNCD films. We have shown that H-terminated diamond surfaces on exposure to 254 nm UV light, photoemit electrons into adjacent liquid layer,[17] allowing to graft alkene monolayers to the diamond surface. However, the complete picture of the grafting mechanism remains unclear including, the origin of sub-bandgap photoemission from diamond,[18] and how excitations in the diamond and in the adjacent fluid may impact the monolayer growth. To compare the performance of antibody functionalized UNCD surfaces with an existing standard and well characterized surface, we prepared antibody functionalized Corning GAPS II amine-terminated glass (GAPSG) slides. GAPSG surfaces were covalently liked to antibodies via glutaraldehyde the same way as amine-terminated UNCD surfaces.
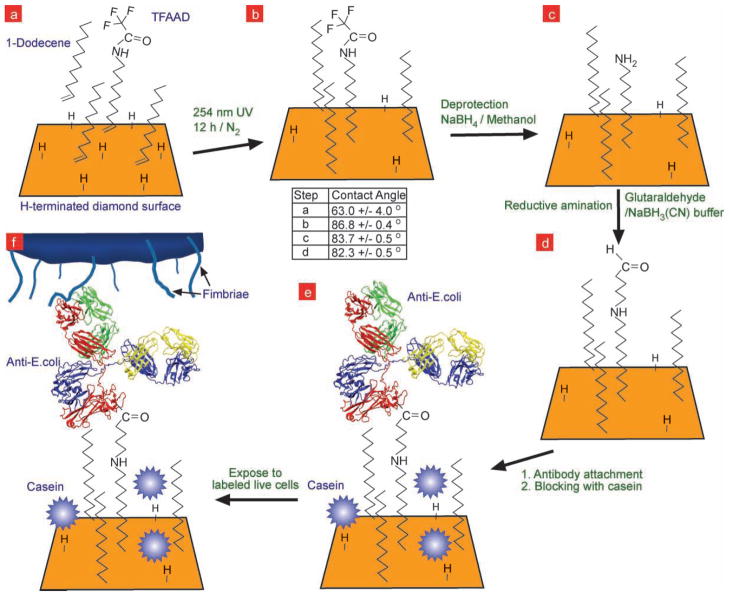
Scheme 1.
Reaction steps used to attach antibody to hydrogen-terminated UNCD surface, followed by its use to capture fluorescently-labeled bacteria E.coli. a) The as-deposited hydrogen terminated UNCD film surface was photochemically grafted with trifluoroacetamide protected 10-aminodec-1-ene (TFAAD) and 1-dodecene. After attaching TFAAD to the surface (b), the amide group was deprotected in sodium borohydride (NaBH4)-methanol solution, leaving a primary amine termination on the surface (c). The deprotected amines were further reacted with glutaraldehyde via reductive amination in sodium cyanoborohydride buffer to yield an aldehyde termination on the surface (d), which was then used to attach antibodies to the surface (e) and capture fluorescently-labeled bacteria (f).The table shows the contact angles measured on UNCD surfaces during steps (a), (b), (c), and (d).
2. Results and Discussion
Figure S1 (Supporting Information) shows XPS spectra for C1s, N1s, O1s, and F1s peaks obtained for the H-terminated, TFAAD-grafted, amine- and aldehyde- terminated surfaces confirming the UNCD surface chemical composition in steps (a), (b), (c), and (d) of Scheme 1. Sessile drop contact angle (θc) measurements were carried out at the latter steps of Scheme 1 by taking six readings over three 3 μl droplets. We found that the amine-terminated UNCD was hydrophobic compared to GAPSG surface (θc~45°). Water contact angle measurements show an increase from 63 ± 4° to 86.8 ± 4° on grafting TFAAD/Dodecene to UNCD, and a decrease to 83.7° and 82.3° on amine and aldehyde termination of the UNCD surfaces, respectively.
2.1. Antibody Activity on Freshly Prepared Surfaces
We attached FITC-conjugated anti-E.coli to UNCD surface as per Scheme 1. Figure 2a shows higher fluorescence on antibody-conjugated UNCD compared to the low fluorescence from the control surface. This confirms the tethering of antibodies to UNCD surfaces. To examine the activity of immobilized antibody we used UNCD and GAPSG surfaces functionalized with O157:H7 and K12 antibody to selectively capture E. coli O157:H7 and E.coli K12 bacteria respectively from live isolated cultures (Figure 2b–c). We found that the anti-E.coli O157:H7 functionalized UNCD and GAPSG surfaces did not show a significant difference in capture efficiency; ~270 cells mm−2 captured on UNCD versus ~250 cells mm−2 on GAPSG. Capture of E. coli O157:H7 was quantified by averaging results from seven experimental repeats. Also, exposure of anti-E.coli O157:H7 tethered UNCD and GAPSG films to E.coli K12 culture showed minimal non-specific capture of E.coli K12 (~20 cells/mm2) compared to specific capture of E. coli O157:H7 (~270 cells mm−2). As an ever more stringent test of selectivity, E.coli O157:H7 cells were captured from live co-cultures of E. coli O157:H7 (~5 × 106 cfu/ml, SYTO84 dye) and L. monocytogenes V7 (~5 × 106 cfu/ml, Hoescht 33342 dye) (Figure 2d). L.monocytogenes was chosen for selectivity experiments due to its known notoriety in non-specific binding. UNCD and GAPSG immunosurfaces showed similar capture selectivity (~4) during co-culture experiments. During this test we also evaluated the effectiveness in blocking of non-specific binding by three blocking agents, casein, bovine serum albumin (BSA), and protein-free block (Pierce Scientific). As shown in Figure 2d the use of different blocking agent did not affect the specific bacteria capture count on UNCD or GAPSG; however we did see a difference in effectiveness of the three blocking agents to reduce non-specific binding on UNCD surfaces. We found casein to be more effective on UNCD immunosurfaces, while BSA blocking was more effective on silica surfaces. More work needs to be done to optimize surface functionalization scheme leading to reduced non-specific binding of L.mono. on UNCD. One solution may be to replace dodecene with PEG-terminated alkenes as spacer molecules, as shown by Lasseter et al. [19]
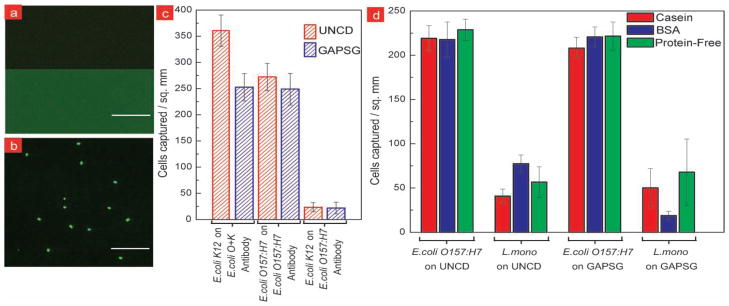
Figure 2.
Selective capture of E.coli on antibody functionalized UNCD thin films. a) Fluorescence image of FITC-labeled antibody tethered to UNCD surface showing higher fluorescence compared to control UNCD surface. b) Fluorescence image showing labeled E.coli O157:H7 captured on antibody-UNCD surface. c) Bacteria capture density on antibody functionalized UNCD and GAPSG substrates from live isolated cultures. Fluorescently-labeled E.coli O157:H7 and K12 cells (106 cfu ml−1 each) were exposed to anti-O157:H7 and anti-O+K functionalized surfaces, respectively. Anti-O157:H7 functionalized surfaces were also exposed to E.coli K12 cells (106 cfu ml−1) to check cross-serotype reactivity. Results show similar capture and cross-serotype reactivity with antibody-tethered UNCD and GAPSG surfaces. d) Bacteria capture density for targeted and non-targeted bacteria obtained by exposing the anti-O157:H7 functionalized UNCD and GAPSG surfaces to live co-cultures of E.coli O157:H7 and Listeria monocytogenes (5 × 106 cfu ml−1 each). Three different blocking agents viz. casein, bovine serum albumin (BSA), and protein-free block (Pierce Scientific) were used to evaluate the effectiveness in blocking non-specific binding. Experiments in (c) and (d) were performed in triplicates.
2.2. Regeneration of Immobilized Antibody and Spatial Distribution of Capture Events
Stability and reusability are also key factors in designing bio-sensors. The ideal situation would be to integrate a reversible antibody–antigen reaction in a sensor that is able to maintain the same activity through a high number of assays. After primary binding of antigen and antibody has occurred through hydrophobic and electrostatic interactions, the epitope (antibody Fab region) and the parotope (protein or carbohydrate expression on bacteria surface) will be close enough to allow Van der Waals and hydrogen bonds to become operative.[20] In order to dissociate the antigen–antibody complexes, the strength of these forces may be reduced by changing the pH, ionic strength, or temperature; or through the addition of dehydrating agents or organics.[21] We found that antibody renaturation on UNCD or GAPSG surfaces is not affected during capture-regeneration cycling (Figure 3), where the DiOC6(3)-labeled heat-killed E.coli O157:H7 cells (107 cfu ml−1) were captured on anti-O157:H7 functionalized surface, imaged for fluorescence, and released from the surface bound antibodies using 0.1M glycine buffer (pH 2.1, 22 °C, 20 min). Non-specific binding sites, which may have been created in the process of regeneration, were blocked using casein blocking solution (20 min, 22 °C). Figure 3 shows the cell capture activity on UNCD and silica remains within the same range (400–700 cells/mm2) for 7 capture cycles. The regeneration experiments were carried out in quadruplicate wells. Control surfaces (without antibodies) showed minimal non-specific capture or false positives.
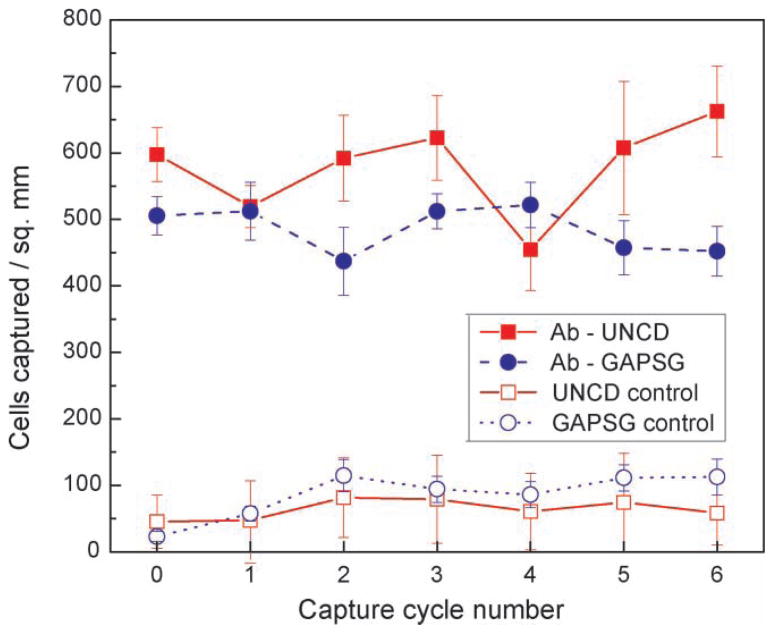
Figure 3.
Plot of E.coli O157:H7 cells captured during six regeneration and capture cycles on functionalized UNCD and GAPSG surfaces. Regeneration was performed using 0.1M glycine-HCl buffer (pH 2.1). Bacteria count after regeneration was found to coincide with readings from controls. Experiments were performed in quadruplicates.
We also used the data from the first five capture-regeneration cycles on freshly-prepared immunosurfaces (within 16 hours) to statistically quantify the temporal and spatial randomness in bacteria capture events. UNCD and GAPSG surfaces showed similar spatial and temporal randomness in bacterial capture. Temporal randomness was compared between substrates using the histogram of overlaid capture-step images. Figure S2 (Supporting Information) shows a sample sum intensity projection of bacteria-capture images taken during the first five capture steps of the capture-regeneration cycle (at same spot). If there were patches on the surface that ended up capturing bacteria in all the cycles, we would see a sharp peak in the histogram of the projection image at a higher pixel value. Histogram of the projection image confirms temporal homogeneity in capture events on UNCD and silica immunosurfaces. Second-order spatial analysis was carried out to check for spatial randomness in capture events during each cycle and was evaluated by computing the corrected neighbourhood density fuction (NDF),[22] which is the relative mean density (observed density/event set density) at distance x from all bacteria; under complete spatial randomness the corrected NDF ~ 1. The bacterial positions in the images were calculated using ImageJ and the NDF values were calculated using SpPack.[23] Figure S3 (Supporting Information) shows plots of NDF calculated on four samples as a function of distance from an event. The calculated value for NDF reaches ~1 at a distance of 25 pixels in case of UNCD and silica (bacteria size: ~5 × 3 pixel2). This shows equal spatial randomness in bacteria capture events on UNCD and GAPSG immunosurfaces.
2.3. Antibody Stability Study
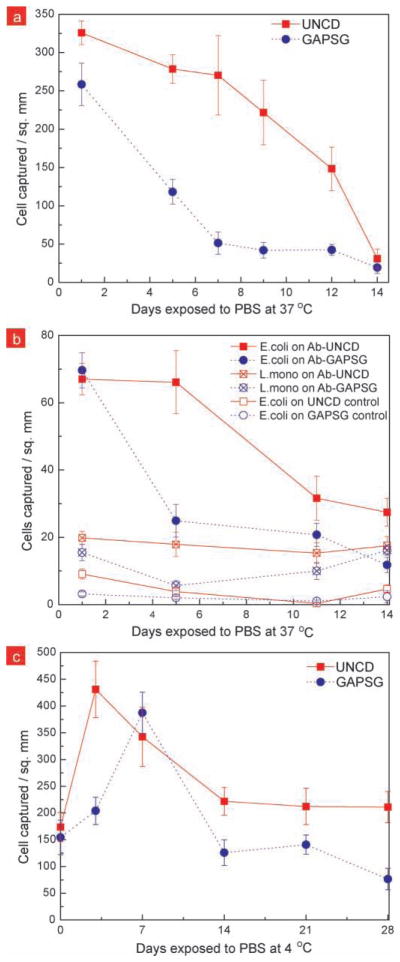
In order to test the stability of immobilized-antibodies, functionalized surfaces were exposed to phosphate buffered saline (PBS) at 37 °C or 4 °C for extended time and the stability of the immobilized antibodies was measured by performing bacterial capture from isolated or co- cultures. It was important to make sure that the blocker protein does not desorb during these stability tests. This was confirmed by using control surfaces with no antibody, as well as by looking at the non-specific binding of FITC-labeled goat antibodies on exposed surfaces (see Figure S4 (Supporting Information). Half-life of antibody activity was calculated to quantitatively compare protein stability on surfaces. As shown in Figure 4 during the 37 °C stability testing, we found that the antibodies on UNCD surfaces were active for a longer period of time compared to when on GAPSG surfaces. Figure 5a, Figure 5b quantifies the E.coli O157:H7 capture from live isolated culture and co-cultures with Listeria monocytogenes, respectively as a function of exposure time to physiological conditions. The half-life of antibody activity was measured to be ~4 days on GAPSG surfaces, while it was ~11 days on UNCD surface. This indicates that either there is a loss of proteins from the GAPSG surfaces at physiological conditions, or the surface-protein interactions on GAPSG are strong enough to deactivate the Fab sites of the antibody or make them inaccessible within a few days, while the functionalized UNCD surface reduces such surface-protein interactions. During the 4 °C stability tests (Figure 5c), we found that antibodies on UNCD and GAPSG surfaces exhibited sustained activity for the first 14 days, however past that the antibodies on GAPSG showed a decrease in antibody activity. The antibody activity half-life at 4 °C was found to be ~27 days on GAPSG, but we could not measure it on UNCD within the duration of our testing (28 days).
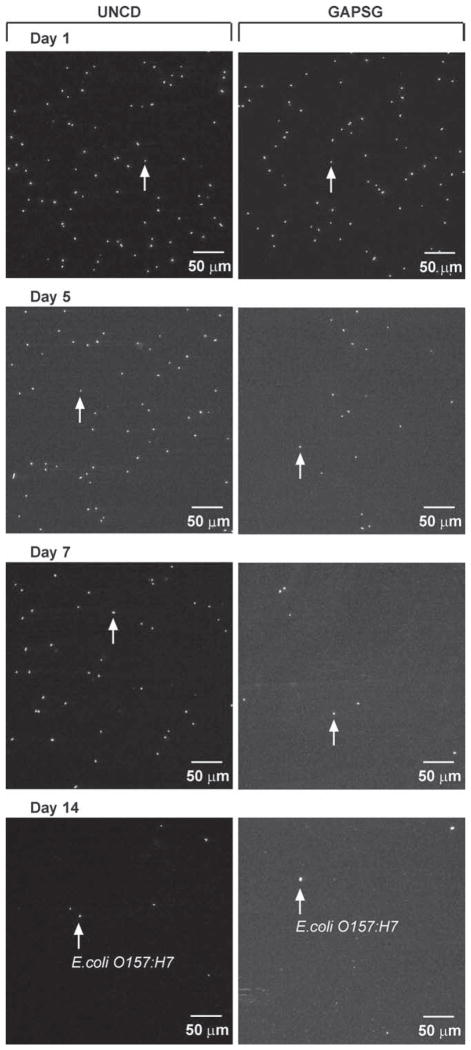
Figure 4.
E.coli O157:H7 capture on antibody-tethered UNCD and GAPSG substrates exposed to PBS at 37 °C for upto 14 days. Fluorescent images show E.coli O157:H7 captured on anti-O157:H7 functionalized UNCD and GAPSG surfaces, which were stored for 1, 5, 7, and 14 days in PBS at 37 °C. The images show the relative decrease in antibody activity on GAPSG by day 5, which was gradually also seen in case of UNCD after day 7.
Figure 5.
Plot of bacteria capture density on UNCD and GAPSG surfaces a) from live E.coli O157:H7 culture (106 cfu ml−1) versus the number of days the antibody-tethered surfaces were exposed to PBS at 37 °C; b) from live co-culture of E.coli O157:H7 and Listeria monocytogenes (5 × 105 cfu ml−1, total) against the number of days the functionalized surfaces were stored in PBS at 37 °C; and c) from live E.coli O157:H7 culture (106 cfu ml−1) versus the number of days the antibody-tethered surfaces were exposed to PBS at 4 °C. The unusual increase in antibody activity on UNCD and GAPSG at 4 °C on day 5 and day 7, respectively, may be attributed to the favorable rearrangement of antibodies at the surface. Experiments in a, b and c were performed in triplicates.
2.4. Elisa and XPS Study to Detect Change in Antibody Content from Surface
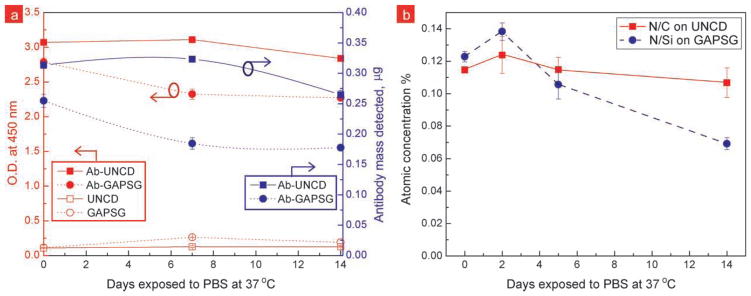
In order to detemine if the loss of proteins was completely responsible for the rapid decrease in antibody activity, we performed ELISA and XPS studies. ELISA was used to quantify the O157:H7 antibody bound to the surface at days 1, 7, and 14 of the 37 °C stability tests (Figure 6a). We were able to correlate the ELISA optical readings to antibody mass with the help of a calibration curve generated using MaxiSorp® surface (Figure S5, Supporting Information). ELISA readings show that there was ~24% decrease in antibody mass from GAPSG surfaces in the first 7 days, however this loss is inadequate to explain the complete loss in antibody activity on GAPSG surfaces. ELISA readings show that there was no change in antibody mass on UNCD during the first 7 days; however there was ~13% decrease during the later 7 days. This decrease may be due to the loss of small antibody fragments from the surface or variation in antibody content between chips. Control surfaces for UNCD and GAPSG containing no antibody confirm that the contribution to ELISA readings from non-specifically bound HRP-antibody was negligible. To corroborate our ELISA findings we monitored the total nitrogen content using XPS on the antibody modified surfaces that were subjected to 37 °C stability tests (Figure 6b). N/C and N/Si atomic concentration ratios on the UNCD and GAPSG surfaces, respectively, show that antibody content is not completely lost from the GAPSG surface (see Figure S6, Supporting Information for XPS spectra). ELISA and XPS studies suggest that the unstable silane chemistry on GAPSG surface may not be responsible for the complete loss in antibody activity seen on GAPSG; however, the question remains whether protein-surface interactions were responsible for the early loss of antibody activity on GAPSG and delayed activity loss on UNCD.
Figure 6.
a) ELISA and b) XPS results confirm presence of antibody on the UNCD and GAPSG surfaces during pro-longed exposure to PBS at 37 °C. a) Plot of optical density readings at 450 nm for quenched enzyme substrate solutions in ELISA to detect goat anti-O157:H7 tethered to UNCD and GAPSG surfaces. UNCD and GAPSG control surfaces were created by skipping the antibody functionalization step. Controls demonstrate the minimal non-specific binding during ELISA. b) Plot of N/C and N/Si atomic concentration ratios obtained via XPS on functionalized UNCD and GAPSG surfaces during the 37 °C stability tests. Nitrogen content is directly proportional to the antibody mass immobilized on the surface, assuming the nitrogen content from the self assembled layer, substrate, and contamination is negligible.
2.5. Molecular Dynamics Simulations
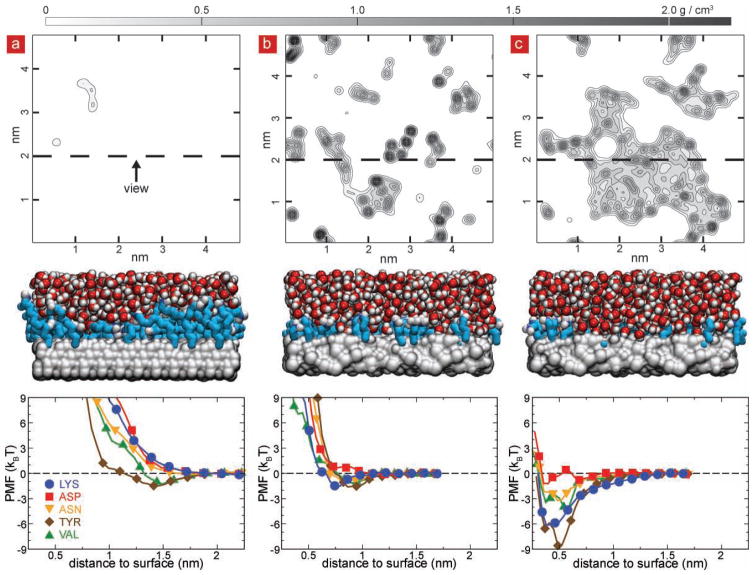
We believe that the interactions between the surface and the proteins through water molecules played the major role, causing the antibody to lose its configuration and its activity. It is known that without a protective coating of aminopropylsilane on silica or aminodecane on diamond surfaces, the antibodies can bind nonspecifically to both surfaces,[19,24] losing their conformation and specific affinity to antigens. To gain insight into the interactions of antibodies with the functionalized forms of such surfaces, we performed molecular dynamics (MD) simulations of aminopropylsilane-terminated SiO2 (4.0 × 1014 molecules cm−2) and aminodecane-terminated diamond surfaces (2.6 × 1014 molecules cm−2) exposed to saline solution. An amorphous silica slab was tiled in a two-by-two grid to create a 5.0 × 5.0 × 2.6 nm3 silica membrane with two exposed faces consisting of four identical patches of silica. Further details of silica membrane construction are given in the experimental section. To construct the diamond membrane, a diamond unit cell was replicated and cut along the (1 1 1) face to create a 4.8 × 4.8 × 2.5 nm3 block, periodic in the plane orthogonal to the (1 1 1) face. The exposed carbon atoms were terminated with hydrogen atoms. To model the functionalized surfaces, each membrane was coated with a covalently attached layer of aminopropylsilane (on silica) or aminodecane (on diamond) as described by Luan and Aksimentiev.[25] The resulting functionalized surfaces were submerged in a 0.138 M NaCl solution consisting of 5,194 water molecules, 13 Na+ and 13 Cl- ions and each system was simulated in the NPT ensemble for at least 40 ns. As shown in Figure 7a, a contour plot of the water density within 0.5 nm of the diamond surface, averaged over a representative 1 ns period of the 40 ns MD trajectory indicates that the aminodecane layer of the diamond surface effectively blocks water access to the diamond substrate, whereas as shown in Figure 7b small pockets of water were observed in the aminopropyl layer of the SiO2 surface. Supplementary Movie 1 shows water molecules within 0.5 nm of the diamond surface during representative 3-ns fragment of the 40-ns MD trajectory. Water molecules can be seen to spontaneously approach the diamond surface, however stable water pocket formation on the diamond surface was not observed. Similarly Supplementary Movie 2 shows the water molecules within 0.5 nm of the silica surface. The silica surface can be seen to have constant pockets of water in addition to those spontaneously forming during this representative 3-ns fragment of the 40-ns MD trajectory. To model effect of variations in functionalization layer density, we have simulated diamond and SiO2 substrates with a range of functional group densities. Supplementary Figure S7 shows the average density of water versus the distance from the surface for each substrate for all functionalization group densities considered.
Figure 7.
Water pockets on the immunosurfaces revealed by molecular dynamics (MD). a) H-terminated diamond coated with 2.6 × 1014 molecules cm−2 aminodecane. The top panel shows a contour plot of the water density within 0.5 nm of the diamond surface, averaged over a representative 1 ns period of the 40 ns MD trajectory. The center panel shows a cut-away view of the same surface; the location of the cut away surface is indicated at the top panel as a dashed line, diamond is shown as a grey molecular surface, aminodecane as teal chains with nitrogen in blue and water as vdw spheres (oxygen in red and hydrogen in white). The bottom panel shows the potential of mean force (PMF) for bringing different types of amino acids from bulk solution to the surface. b) Water density (top), a cut away view (center) and the PMF profiles (bottom) for the silica surface coated with a 4.0 × 1014 molecules cm−2 layer of aminopropylsilane. Silica is shown as a gray molecular surface and other molecules are shown as in panel (a). c) Water density (top), a cut away view (center) and the PMF profiles (bottom) for partially degraded the silica surface (see text). The degradation of the functionalization layer on silica creates pockets of water within the functionalization layer, which facilitates binding of amino acids as revealed by the PMF plots.
To quantify the affinity of the two surfaces to the antibody proteins, we computed the potential of mean force (PMF) for bringing representative types of amino acids from bulk solution to the respective surfaces (Figure 7 bottom panels). The potential of mean force (PMF) of an amino acid as a function of its position relative to the substrate surface was determined from a set of umbrella sampling simulations, which were analyzed using the weighted histogram analysis method (WHAM)[26]. The umbrella sampling simulations were performed by restraining the alpha-carbon atom of an amino acid to a specified distance z from the substrate surface using the following potential energy function: wi(z) = 1/2 kz (z-zi)2, where zi was the center of sampling window i and kz = 4 kcal mol−1 Å−2 was the spring constant. The location of the amino acids within the plane of the substrate was harmonically restrained using a spring constant of 0.8 kcal mol−1 Å−2. The sampling windows were placed every 0.06 nm for either 1.5 < z < 2.82 nm (silica) or 1.5 < z < 3.42 nm (diamond), for a total of 23 or 33 windows, respectively. Following 200 steps of energy minimization, the system in each window was simulated at fixed volume for at least 6 ns, with the first 0.2 ns of each simulation excluded from the WHAM PMF calculation. For tyrosin and valine, the adhesion energy to functionalized diamond did not exceed 2 kBT, whereas charged (Lys, Asp) and polar (Asn) amino acids repelled from the surface. Similar PMF profiles were obtained in the case of the functionalized silica, indicating that both surfaces are unlikely to bind proteins.
We know that instability of silane layers on silica is quite pronounced in saline solutions[25,26], especially for short-chain aminosilanes where the positively charged primary amine can coordinate to the silicon center and catalyze hydrolysis via the formation of a stable five-membered ring[27]. To see if loss of dangling aminosilanes could explain the quick loss in activity over GAPSG we created a partially degraded SiO2 surface (Figure 7c) that was obtained by iteratively removing 25% of the aminosilane groups (1.0 × 1014 molecules cm−2) from the surface characterized in Figure 7b (4.0 × 1014 molecules cm−2). To model the degradation we performed an eight-step molecular dynamics simulation starting from the coordinates obtained from the simulations of silica system with surface functionalization density of 4.0 × 1014 molecules cm−2. We assumed that the probability of losing a functional group is directly proportional to the total exposure of the Si-C bond to water. Hence, to choose the functionalization groups for removal, a 1-ns fragment of the simulation trajectory was used to compute the average number of water molecules located within 3 Å of each aminopropylsilane group and 0.5 nm within the silica substrate. All aminopropylsilane groups were ranked according to the average number of contacts with this water; the top 10% (rounding down) were removed. Following 2000 steps of energy minimization, the resulting system was simulated for 2.5 ns. This procedure was repeated 8 times, until the density of aminopropylsilane molecules fell to 75% of the original density, or 3.0 × 1014 molecules cm−2. Supplementary Figure S8 illustrates the results of these simulations. Dramatically different results were obtained for the partially degraded SiO2 surface. The water density plot (Fig 5c top) revealed large pockets of water near the degraded surface, whereas the PMF for bringing amino acids from the bulk solution to the substrate through one of such pockets (Figure 5c bottom) indicated adhesion energies exceeding 6 kBT. Thus, we find that degradation of the functional layer on SiO2 exposes patches of unprotected substrate to the solution, which considerably increases affinity of the surfaces to proteins, facilitating reduction of the antibody activity.
3. Conclusions
We have shown that proteins can be tethered to UNCD surfaces to achieve extended stability at physiological conditions compared to glass surfaces, which makes UNCD surfaces attractive for producing extended use biosensors. We find that the loss of activity on glass surface can be explained mainly by the loss of the antibody stability through increased interactions with the surface via pockets of water molecules. Whereas, on the UNCD surface, the desorption of antibody from the surface is minimized due to the strong chemical bond, while the activity is maintained due to reduced water density and reduced antibody-surface interactions. We believe that our experimental and modeling approaches may provide a foundation for improving the stability of immobilized biomolecules and enhancing the performance of protein-tethered surfaces for a range of biomedical applications.
4. Experimental Section
Materials
All stock solutions were prepared using deionized water from a Millipore water purification system to obtain a minimum resistivity of 18.0 MΩ-cm. Casein blocking solution, 1-dodecene, sodium borohydride, 3,3′-Dihexyloxacarbocyanine iodide stain (DiOC6(3)), phosphate-buffered saline (PBS), PBS with Tween 20, aluminum sealing film and glycine was obtained from Sigma Aldrich. Methanol, Ethanol and HCl was obtained from Fisher Scientific. Corning GAPS II slides were obtained from Corning Life Sciences. House nitrogen was used after filtering it through a 0.22 micron teflon filter. Stains Hoechst 33342 and SYTO 84 were bought from Invitrogen. BBL Brain heart infusion broth (BHI) was obtained from Becton, Dickinson and Company. Antibodies, BSA diluent and SureBlue Reserve (HRP substrate) were bought from Kirkegaard & Perry Laboratories.
Preparation of Antibody Functionalized Ultra-nanocrystalline Diamond (UNCD) Films
UNCD-coated silicon wafer was diced into 1 cm × 3 cm pieces, rinsed with deionized water (DI), sonicated in ethanol, and dried with nitrogen. Trifluoroacetamide protected 10-aminodec-1-ene (TFAAD) mixed with 1-dodecene in 2:1 ratio (v/v) was used as functionalization mix. The 1-dodecene provides space between the TFAAD molecules and increases the efficiency of deprotection, which to be performed later. Photochemical attachment was carried out in a nitrogen-purged reaction chamber by applying 2 μl cm−2 of TFAAD-Dodecene liquid mixture uniformly between the sample surface and a piranha-cleaned quartz slide (1 mm thick), and illuminating with a 254 nm low-pressure mercury lamp for 12 h (~16 mW cm−2 measured at sample surface)[16]. Excess reaction mix from the samples surface was removed by soaking the functionalized UNCD surfaces in chloroform and ethanol for 15 min each. TFAAD group attached to the UNCD were deprotected in a tightly-sealed scintillation vial containing 65 mM sodium borohydride (NaBH4)-anhydrous methanol solution (65 °C, ≥6 h) creating a primary amine group termination.[27] Sample surfaces were rinsed with methanol and DI, and dried with nitrogen. The primary amines on the deprotected UNCD surfaces and Corning GAPS II slides were reacted with glutaraldehyde via reductive amination in sodium cyanoborohydride buffer (4 h, 22 °C) to yield an aldehyde termination on the surface.[16] The aldehyde-terminated surfaces were rinsed with DI, dried with nitrogen and in an oven at 70 °C for 30 min. Home-made polycarbonate wells with a silicone gasket (4.7 mm well diameter) were attached to the aldehyde-terminated surfaces using stainless steel clips to carry out further reactions. Functionalized surfaces were incubated overnight (18–22 h, 4 °C) with antibody solution (10 μg ml−1). Wells were sealed with a sterile aluminum adhesive film. Next day, wells were washed to remove non-specifically bound antibody. Washing routine used in our experiments consisted of rinsing with PBS-Tween20 (0.05%) twice followed by PBS once. Non-specific binding sites on surfaces were blocked using casein blocking solution (1 h, 22 °C), and wells were further washed to remove excess blocker protein.
Fluorescent Labeling of Bacteria
Isolated cultures of Escherichia coli K12, Escherichia coli O157:H7 EDL933, and Listeria monocytogenes V7 were preparted in BHI broth at 37 °C with 2% inoculum and incubation time of 12 h. Based on optical density measurements at 600 nm, all bacteria were found to reach stationary phase of growth within 12 h. This was also confirmed by plating cultures at 10th, 12th and 15th hour. Live cultures were used for work in this paper, except heat-killed culture (80 °C for 15 min) was used for capture-regeneration cycling experiment. Heat-killed culture was agar plated for 48 h and showed no bacterial viability. Fresh cultures (1 ml in 1.5 ml centrifuge tubes) were washed with phosphate saline buffer (PBS, 0.9 ml, pH 7.4 at 25 °C) prior to use by pelletizing cells at 12k rpm (2 min), removing supernatant (0.9 ml), and resuspending cells in PBS. Blue fluorescent Hoechst 33342 stain, orange fluorescent SYTO 84 stain, and green fluorescent DiOC6(3) were used for labeling at concentrations, 10 μM, 10 nM, and 17 nM, respectively. Nuclear stains Hoechst 33342 and SYTO 84 were used to stain live cultures. Membrane stain DiOC6(3) was used to stain dead cultures. Labeling was performed at 37 °C for 20 min with constant shaking. Excess labeling dye was removed by washing the labeled culture with PBS thrice using above mentioned culture-washing routine. The labeled cells were resuspended in 1 ml PBS. Concentration of labeled-bacteria was estimated using Petroff-Hausser counting chamber and agar plating of 10−4 and 10−5 dilutions of the labeled cell suspension. On average, the concentration in the labeled cell suspension was found to be 109 colony forming units per ml (cfu ml−1). Safety measures were exercised to handle E.coli O157:H7 and L. monocytogenes cultures.
Bacteria Capture Assay from Isolated and Co-cultures
In cases where detection from isolated cultures was demonstrated, solutions of labeled-bacteria (E.coli K12 or O157:H7) were diluted 10−3 times and deposited on antibody-functionalized surfaces. In case of capture from co-cultures, O157:H7 and L. monocytogenes cultures were diluted 10−3 times, mixed in 1:1 ratio, and then transferred onto antibody-functionalized surfaces; so concentrations of O157:H7 in co-cultures was half of that in isolated cultures. The surfaces exposed to bacterial solutions were then placed on a shaker at 37 °C for 1 h. Evaporation from wells was minimized using adhesive aluminum film. At the end of capture step, bacterial solution was removed and wells were washed with PBS (thrice) to remove non-specifically bound bacteria. In case of O157:H7, captured cells were further labeled with FITC-labeled anti-O157:H7 (5 μg ml−1 bovine serum albumin diluent, 30 min, 22 °C). To remove the excess FITC-labeled anti-O157:H7, surfaces were further washed with PBS twice. The surface captured bacteria were imaged with an Olympus BX51WI fluorescence microscope equipped with SpotFlex camera and DAPI/FITC/TRITC filter sets. Five or more images were taken per well. Experiments were performed in triplicate wells each time. Bacterial enumeration in fluorescence images was performed using an image analysis routine in ImageJ (Wayne Rasband, NIH, Bethesda, MD).
Testing the Stability of Antibodies on Surfaces
Wells on antibody-functionalized surfaces were filled with PBS and sealed with adhesive aluminum sheet. When testing stability at 37 °C sealed wells were immersed in a PBS container, which was incubated at 37 °C. At the end of 1, 3, 7, 11, and 14 days, functionalized surfaces were retrieved, PBS was removed from the wells, and bacteria capture was performed using the treated surfaces. When testing for stability at 4 °C sealed wells were stored in a 4 °C fridge, samples were retrieved at the end of 3, 7, 14, 21, and 28 days, and tested for bacteria capture capability. In all above cases, the tested surfaces were blocked with casein blocking solution before and after exposure to stability test conditions. Studies have shown that blocker protein plays an essential role in stabilization of surface attached antibodies, and so it was necessary to block the surfaces prior to stability tests. During the stability test non-specific binding sights might have been created due to desorption of blocking agent, which may lead to non-specific binding of bacteria and result in higher bacteria capture activity; this may not be a true indicator of the antibody stability.
Quantitative Enzyme-Linked Immunoassay to Detect Antibody Loss from Functionalized Surfaces
HRP-labeled rabbit anti-goat was used to quantify the amount of goat anti-O157:H7 bound to the immunosurfaces obtained from 37 °C stability test. PBS from wells was removed and wells were blocked with casein blocking solution (30 min, 22 °C). Excess blocking agent was rinsed from surfaces using PBS-Tween20 twice and PBS once and surfaces were incubated with HRP-antibody solution (50 ng ml−1) for 30 min at 22 °C. Non-specifically bound HRP-antibody was removed using the washing routine. Surfaces were exposed to 50 μl SureBlue Reserve (HRP substrate) for 5 min; the reacted substrate solution was transferred to a 384 well plate, quenched using equal amount of hydrochloric acid (1N), and measured for optical density at 450 nm. The amount of surface bound antibody was quantified using a calibration curve given in Supplementary Figure S5.
X-ray Photoelectron Spectroscopy (XPS) to Setect Antibody Loss from Functionalized Surfaces
XPS analyses were performed with a Kratos Axis Ultra spectrometer (Kratos Analytical, Manchester, UK) equipped with a monochromatized aluminum X-ray source (powered at 10 mA and 13 kV). Instrument was calibrated to Cu2p3/2 (932.7 eV) and Au4f7/2 (84 eV). The samples were attached on conductive multi-specimen holder using copper-beryllium clips and screws, in order to avoid differential charging. The pressure in the analysis chamber was about 10−7 Pa. The angle ϕ between the normal to the sample surface and the direction of photoelectrons collection was 0°. Analysis was performed in the hybrid lens mode with the slot aperture; the analyzed area was 700 μm × 300 μm. The pass energy was set at 160 eV for the wide scan and 40 eV for narrow scans. Charge stabilization was achieved by using the Kratos Axis device. The following sequence of spectra was recorded: survey spectrum, C1s, N1s, O1s, and Si2p. The C1s peak of carbon was fixed to 284.5 eV to set the binding energy scale. Data treatment was performed with the Kratos Vision software. Atomic concentration ratios were calculated using peak areas normalized on the basis of acquisition parameters after a linear background subtraction, experimental sensitivity factors and transmission factors provided by the manufacturer. Antibody functionalized UNCD and GAPSG surfaces were prepared as previously stated. The surface was analyzed as freshly prepared and after completing the XPS analysis the surfaces were stored in PBS at 37 °C.
Molecular Dynamics Simulations of Diamond and Silica Immunosurfaces
All molecular dynamics (MD) simulations were performed using the program NAMD,[28] particle mesh Ewald (PME) electrostatics,[29] periodic boundary conditions, and a multiple time step integration scheme consisting of a 2-fs time step for bonded and non-bonded calculations and a 6-fs time step for long-range electrostatics. Water bonds and angles were kept rigid with SETTLE,[30] while all other covalent bonds with hydrogen were kept rigid with RATTLE.[31] Simulations in the NPT ensemble (constant number of particles N, pressure P and temperature T) were performed using a Langevin thermostat[32] and Nosé-Hoover Langevin piston pressure control[33] set at 310.15 K (37 °C) and 1 atm, respectively; the area of the unit cell within the plane coincident with the plane of the surface was kept constant. The damping coefficient of the Langevin thermostat was 1 ps−1; the thermostat was applied only to the atoms of the substrate (diamond or silica). A smooth (1.0–1.2 nm) cutoff was used to compute the van der Waals forces. All simulations were performed using the CHARMM[34] force field for diamond, aminopropyl and aminodecane, the TIP3P model for water,[35] the CHARMM-compatible force field for ions[36] and a custom CHARMM-compatible force field for silica.[37] The CHARMM fore field has been shown to properly model both short- and long-range non-bonded interactions for diamond carbons at ambient temperatures.[34] The silica membrane was generated using the BKS potential.[38,39]
To produce the silica membrane used in this study, we created a 2.5 × 2.5 × 3.5 nm3 block of crystalline silica containing 500 silicon and 1000 oxygen atoms by replicating a unit cell of SiO2. The membrane was annealed through NVT simulations in a 2.5 × 2.5 × 5.5 nm3 periodic cell lasting 20 ps at 7000 K, 20 ps at 5000 K, 50 ps at 2000 K, 100 ps at 1000 K and finally 50 ps at 300 K. The silica annealing was performed using the BKS potential,[38] modified at small distances to prevent spurious behavior at high temperature,[39] a 0.55 nm cutoff for the evaluation of the van der Waals forces, and a 5 ps−1 Langevin damping coefficient. To prevent atoms from evaporating into the vacuum region, external forces were applied with grid-steered molecular dynamics.[40]
To study the effect of coating density, we used two simulation systems for each substrate material. For silica surfaces, each face of the substrate was coated with a layer of 25 or 62 (system 1), or 100 or 151 (system 2) aminopropylsilane molecules, corresponding to a density of 1.0, 2.5, 4.0 or 6.0 × 1014 molecules cm−2, respectively. Similarly, a pair of simulation systems containing a diamond membrane was coated with a layer of 25 or 60 (system 1), or 35 or 46 (system 2) aminodecane molecules, corresponding to a density of 1.0, 2.6, 1.5 or 2.0 × 1014 molecules cm−2, respectively. The resulting functionalized surfaces were submerged in a 0.138 M NaCl solution consisting of 5,194 water molecules, 13 Na+ and 13 Cl- ions, producing final systems of 22,652 (silica 1), 24,620 (silica 2), 29,196 (UNCD 1), and 29,068 (UNCD 2) atoms. Prior to energy minimization, all water molecules located within 0.3 nm of the substrate or coating molecules were moved into the bulk solution. Following 2000 steps of energy minimization using the conjugate gradient method, each system was subjected to an initial equilibration for 5 ns with all the heavy atoms of the aminopropylsilane or aminodecane molecules harmonically restrained (with spring constants of 1.0 kcal mol−1 Å−2) to their initial conformations, which allowed the water and ions to approach thermal equilibrium. Following that, the harmonic restraints on the aminopropylsilane or aminodecane molecules were released, and each system was simulated in the NPT ensemble for at least 40 ns. For all simulations, all substrate atoms (silica or UNCD carbon atoms) were harmonically restrained with spring constants of 20.0 kcal mol−1 Å−2.
To study the interaction between amino acids and the immunosurfaces, we created 15 systems consisting of a 0.138 M aqueous solution of NaCl, one copy of an amino acid and one of the following three functionalized substrates: The 1.0/2.6 × 1014 molecules cm−2 aminodecane diamond substrate; the 4.0/6.0 × 1014 molecules cm−2 aminopropylsilane silica substrate; or the 3.0/6.0 × 1014 molecules cm−2 aminopropylsilane substrate created through the degradation process described above. For each functionalized substrate, five systems were created corresponding to the five types of amino acids that we used to model the interactions of surfaces with the antibody protein: Lysine (charged, positive); aspartic acid (charged, negative); asparagine (polar); valine (nonpolar); and tyrosine (aromatic). The systems containing charged amino acids were made electrically neutral by adding an extra chloride or sodium counterion. Prior to PMF calculations, each system was subject to 2000 steps of energy minimization followed by a 3-ns equilibration in the NPT ensemble.
Supplementary Material
Acknowledgments
This research was supported primarily by the Defense Threat Reduction Agency (DTRA) under contract HDTRA1–09-C-0007. We also acknowledge the support of the Center for Food Safety Engineering at Purdue University, a cooperative agreement with US Department of Agriculture Agricultural Research Service (Project number 1935–4200-035). R.C. and A.A. were supported by grants from the National Institutes of Health (PHS 5 P41-RR05969), National Science Foundation (PHY0822613), and the Petroleum Research Fund (48352-G6). AFM and XPS were carried out in part in the Frederick Seitz Materials Research Laboratory Central Facilities, University of Illinois, which are partially supported by the U.S. Department of Energy under grants DE-FG02–07ER46453 and DE-FG02–07ER46471.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Adarsh D. Radadia, Micro and Nanotechnology Laboratory, University of Illinois, 208 North Wright Street, Urbana, Illinois 61801, USA
Courtney J. Stavis, Department of Chemistry, University of Wisconsin, Madison, WI 53706, USA
Rogan Carr, Department of Physics, University of Illinois, Urbana, IL 61801, USA.
Hongjun Zeng, Advanced Diamond Technologies, Inc, Romeoville, IL 60446, USA.
Prof. William P. King, Department of Mechanical Science and Engineering, Micro and Nanotechnology Laboratory, University of Illinois, 208 North Wright Street, Urbana, Illinois 61801, USA
Dr. John A. Carlisle, Advanced Diamond Technologies, Inc Romeoville, IL 60446, USA
Prof. Aleksei Aksimentiev, Department of Physics, University of Illinois, Urbana, IL 61801, USA
Prof. Robert J. Hamers, Email: rjhamers@wisc.edu, Department of Chemistry, University of Wisconsin, Madison, WI 53706, USA
Prof. Rashid Bashir, Email: rbashir@illinois.edu, Micro and Nanotechnology Laboratory, University of Illinois, 208 North Wright Street, Urbana, Illinois 61801, USA
References
- 1.USDA/ERS. Foodborne Illness Cost Calculator. ( http://www.ers.usda.gov/Data/FoodBorneIllness/)
- 2.Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E. Biosensors Bio-electron. 1999;14:599. doi: 10.1016/s0956-5663(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 3.Haynes CA, Norde W. Colloids Surfaces B. 1994;2:517. [Google Scholar]
- 4.Jeon SI, Lee JH, Andrade JD, Degennes PG. J Colloid Interface Sci. 1991;142:149. [Google Scholar]
- 5.Giacomelli CE, Bremer MGEG, Norde W. J Colloid Interface Sci. 1999;220:13. doi: 10.1006/jcis.1999.6479. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimoto K, Nishio M, Sugasawa H, Nagasaki Y. J Am Chem Soc. 2010;132:7982. doi: 10.1021/ja910372e. [DOI] [PubMed] [Google Scholar]
- 7.Sackmann E. Science. 1996;271:43. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 8.Dekeyser CM, Buron CC, Mc Evoy K, Dupont-Gillain CC, Marchand-Brynaert J, Jonas AM, Rouxhet PG. J Colloid Interface Sci. 2008;324:118. doi: 10.1016/j.jcis.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Gruen DM. Ann Rev Mater Sci. 1999;29:211. [Google Scholar]
- 10.Bajaj P, Akin D, Gupta A, Sherman D, Shi B, Auciello O, Bashir R. Biomed Microdevices. 2007;9:787. doi: 10.1007/s10544-007-9090-2. [DOI] [PubMed] [Google Scholar]; Chong KF, Loh KP, Vedula SRK, Lim CT, Sternschulte H, Steinmüller D, Sheu FS, Zhong YL. Langmuir. 2007;23:5615. doi: 10.1021/la070037y. [DOI] [PubMed] [Google Scholar]; Nebel CE, Shin D, Rezek B, Tokuda N, Uetsuka H, Watanabe H. J Royal Soc Interface. 2007;4:439. doi: 10.1098/rsif.2006.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Härtl A, Schmich E, Garrido JA, Hernando J, Catharino SCR, Walter S, Feulner P, Kromka A, Steinmüller D, Stutzmann M. Nat Mater. 2004;3:736. doi: 10.1038/nmat1204. [DOI] [PubMed] [Google Scholar]
- 12.Hupert M. Diamond Related Mater. 2003;12:1940. [Google Scholar]
- 13.a) Yang N, Uetsuka H, Osawa E, Nebel CE. Angew Chem Int Ed. 2008;47:5183. doi: 10.1002/anie.200801706. [DOI] [PubMed] [Google Scholar]; b) Yang W, Baker SE, Butler JE, Lee C-s, Russell, John N, Shang L, Sun B, Hamers RJ. Chem Mater. 2005;17:938. [Google Scholar]; c) Yang W, Butler JE, Russell JN, Hamers RJ. Analyst. 2007;132:296. doi: 10.1039/b612201a. [DOI] [PubMed] [Google Scholar]; d) Yang W, Hamers RJ. Appl Phys Lett. 2004;85:3626. [Google Scholar]
- 14.Stavis C, Clare TL, Butler JE, Radadia AD, Carr R, Zeng H, King WP, Carlisle JA, Aksimentiev A, Bashir R, Hamers RJ. Proc Nat Acad Sci. 2011 doi: 10.1073/pnas.1006660107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang W, Auciello O, Butler JE, Cai W, Carlisle JA, Gerbi JE, Gruen DM, Knickerbocker T, Lasseter TL, Russell JN, Smith LM, Hamers RJ. Nature Mater. 2002;1:253. doi: 10.1038/nmat779. [DOI] [PubMed] [Google Scholar]
- 15.Lu M, Knickerbocker T, Cai W, Yang W, Hamers RJ, Smith LM. Biopolymers. 2004;73:606. doi: 10.1002/bip.20007. [DOI] [PubMed] [Google Scholar]
- 16.Yang WS, Butler JE, Russell JN, Hamers RJ. Analyst. 2007;132:296. doi: 10.1039/b612201a. [DOI] [PubMed] [Google Scholar]
- 17.a) Nichols BM, Butler JE, Russell JN, Hamers RJ. J Phys Chem B. 2005;109:20938. doi: 10.1021/jp0545389. [DOI] [PubMed] [Google Scholar]; b) Nichols BM, Metz KM, Tse KY, Butler JE, Russell JN, Hamers RJ. J Phys Chem B. 2006;110:16535. doi: 10.1021/jp062443g. [DOI] [PubMed] [Google Scholar]
- 18.Bandis C, Pate B. Phys Rev B. 1995;52:12056. doi: 10.1103/physrevb.52.12056. [DOI] [PubMed] [Google Scholar]; b) Takeuchi D, Ri S, Kato H, Nebel C, Yamasaki S. Diamond Related Mater. 2006;15:698. [Google Scholar]
- 19.Lasseter TL, Clare BH, Abbott NL, Hamers RJ. J Am Chem Soc. 2004;126:10220. doi: 10.1021/ja047642x. [DOI] [PubMed] [Google Scholar]
- 20.Van Oss CJ. Mol Immunol. 1995;32:199. doi: 10.1016/0161-5890(94)00124-j. [DOI] [PubMed] [Google Scholar]
- 21.Van Oss CJ, Good RJ, Chaudhury MK. J Chromatogr– Biomed Appl. 1986;376:111. [PubMed] [Google Scholar]
- 22.Condit R. Science. 2000;288:1414. doi: 10.1126/science.288.5470.1414. [DOI] [PubMed] [Google Scholar]
- 23.Perry G. Environ Modelling Software. 2004;19:559. [Google Scholar]
- 24.Carr R, Comer J, Ginsberg MD, Aksimentiev A. IEEE Trans Nanotechnol. 2010 doi: 10.1109/tnano.2010.2062530. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luan B, Aksimentiev A. Soft Matter. 2010;6:243. doi: 10.1039/b917973a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux B. Comput Phys Commun. 1995;91:275. [Google Scholar]
- 27.Sun B, Baker SE, Butler JE, Kim H, Russell JN, Jr, Shang L, Tse KY, Yang W, Hamers RJ. Diamond Related Mater. 2007;16:1608. [Google Scholar]
- 28.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. J Comput Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batcho PF, Case DA, Schlick T. J Chem Phys. 2001;115:4003. [Google Scholar]
- 30.Miyamoto S, Kollman PA. J Comput Chem. 1992;13:952. [Google Scholar]
- 31.Andersen HC. J Comput Phys. 1983;52:24. [Google Scholar]
- 32.Brünger AT. The Howard Hughes Medical Institute and Department of Molecular Biophysics and Biochemistry. Yale University; CT: 1992. X-PLOR, Version 3.1: A System for X-ray Crystallography and NMR. [Google Scholar]
- 33.Martyna G, Tobias D, Klein M. J Chem Phys. 1994;101:4177. [Google Scholar]
- 34.Deb B, Hu W, Song K, Hase WL. Phys Chem Chem Phys. 2008;10:4565. doi: 10.1039/b803155j. [DOI] [PubMed] [Google Scholar]
- 35.Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M. J Chem Phys. 1983;79:926. [Google Scholar]
- 36.Joung I, Cheatham T. J Phys Chem B. 2008;112:9020. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruz-Chu ER, Aksimentiev A, Schulten K. J Phys Chem B. 2006;110:21497. doi: 10.1021/jp063896o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Beest BWH, Kramer GJ, van Santen RA. Phys Rev Lett. 1990;64:1955. doi: 10.1103/PhysRevLett.64.1955. [DOI] [PubMed] [Google Scholar]
- 39.Vollmayr K, Kob W, Binder K. Phys Rev B. 1996;54:15808. doi: 10.1103/physrevb.54.15808. [DOI] [PubMed] [Google Scholar]
- 40.Wells DB, Abramkina V, Aksimentiev A. J Chem Phys. 2007:127. doi: 10.1063/1.2770738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.