Abstract
The ability to modulate gene expression in response to sensory experience is critical to the normal development and function of the nervous system. Calcium is a key activator of the signal transduction cascades that mediate the process of translating a cellular stimulus into transcriptional changes. With the recent discovery that the mammalian Cav1.2 calcium channel can be cleaved, enter the nucleus and act as a transcription factor to control neuronal gene expression, a more direct role for the calcium channels themselves in regulating transcription has begun to be appreciated. Here we report the identification of a nuclear localization sequence (NLS) in the C. elegans transient receptor potential vanilloid (TRPV) cation channel OCR-2. TRPV channels have previously been implicated in transcriptional regulation of neuronal genes in the nematode, although the precise mechanism remains unclear. We show that the NLS in OCR-2 is functional, being able to direct nuclear accumulation of a synthetic cargo protein as well as the carboxy-terminal cytosolic tail of OCR-2 where it is endogenously found. Furthermore, we discovered that a carboxy-terminal portion of the full-length channel can localize to the nucleus of neuronal cells. These results suggest that the OCR-2 TRPV cation channel may have a direct nuclear function in neuronal cells that was not previously appreciated.
Introduction
The transient receptor potential (TRP) family of ion channels can be found in all eukaryotes from yeast to mammals [1], [2]. TRP channel proteins contain 6 transmembrane domains and are thought to assemble as homo- or hetero-tetramers to form cation selective channels [1], [3]. The commonly accepted nomenclature subdivides TRP proteins into 7 subfamilies: TRPA, TRPC, TRPM, TRPML, TRPN, TRPP and TRPV [1], [3], [4]. TRP channels are activated by a wide range of stimuli, including intra- and extracellular messengers, chemicals, mechanical forces and osmotic stress [1], [3] and they have been linked to many different sensory modalities including vision, taste, olfaction, hearing and touch [2], [3]. Although the different TRP subfamilies have diverse activation properties and functions, all TRP channels share the unifying characteristic of being permeable to calcium [4].
Mammalian TRP vanilloid family (TRPV) channels range from modestly to highly calcium permeable and contain 3–5 cytoplasmic amino-terminal ankyrin repeats as well as a long, unconserved carboxy-terminal cytoplasmic tail [1], [5]. TRPVs are widely expressed in mammals, but most thoroughly studied in sensory neurons [4]. While they are best known for their thermosensitivity, with 4 of the 6 members being activated by heat, TRPVs are also broadly involved in nociception and are activated by a variety of physiologically important cues including osmotic cell swelling, noxious chemicals, analgesic compounds, inflammatory cytokines, calcium store depletion and polyunsaturated fatty acids (PUFAs) [1], [3], [4]. Furthermore, most TRPVs are polymodally activated, allowing them to integrate signals from multiple cellular pathways [3].
The C. elegans genome encodes members of all seven TRP subfamilies, including 5 TRPVs (OSM-9, OCR-1, OCR-2, OCR-3 and OCR-4) [2], [5], [6]. Each ocr gene is only expressed in a subset of cells that express OSM-9, suggesting that the individual OCR channel subunits usually function together with OSM-9 [5]. C. elegans TRPV channels have been well studied for their role in olfaction and nociception [2], [5], [7], [8], [9], [10]. OSM-9 and OCR-2 are expressed together in the sensory cilia of AWA and ASH head sensory neurons, where it is believed that they promote each other's proper localization to the dendritic cilia to function in sensory transduction [5]. Loss of OSM-9 or OCR-2 results in mild to severe defects in AWA-mediated olfactory responses and ASH-mediated nociceptive avoidance behaviors [7], [8], [9], [10].
Beyond their roles in sensory signaling, OSM-9 and OCR-2 also regulate the transcription of sensory genes. osm-9 and ocr-2 mutant animals have reduced expression of ODR-10, a G protein-coupled receptor expressed in the AWA olfactory neurons [5], as well as the serotonin biosynthetic enzyme TPH-1 in the ADF chemosensory neurons [11]. While the precise molecular mechanism by which OSM-9/OCR-2 controls expression of these downstream targets is unknown, it has been proposed that OSM-9/OCR-2 function in activity-dependent gene expression pathways to regulate odr-10 and tph-1 expression [5], [11]. First discovered almost 3 decades ago, activity-dependent regulation of transcription is a cellular mechanism whereby a transient signaling event results in long-lasting changes in gene expression [12], [13], [14]. This process is particularly important in neuronal cells as it ensures the proper development and maturation of synaptic connections and allows for adaptations within the adult nervous system that underlie learning and memory [15], [16]. Mammalian genome-wide analysis has revealed over 300 genes that are transcriptionally regulated in response to neuronal activity [15], [16], [17].
Studies of mammalian activity-dependent genes have revealed that most transcriptional changes occur downstream of calcium entry into the cell either through plasma membrane channels or by release of calcium from internal stores [15], [18]. Typically, this influx activates a number of signal transduction cascades, such as Ca2+/calmodulin-dependent protein kinases and Ras/MAPK pathways, that converge to modify nuclear transcription factors, such as CREB, and regulate expression of activity-dependent genes [15], [16], [19], [20], [21], [22], [23]. However, a new mechanism directly linking calcium channels to transcriptional regulation was recently discovered. Gomez-Ospina et al. (2006) reported that a carboxy-terminal fragment of the mammalian Cav1.2 L-type voltage-gated calcium channel localizes to the nucleus, functions as a transcription factor, and directly regulates expression of a number of neuronal genes [24]. This new mode of regulation bypasses calcium-activated signaling cascades, previously thought to be the sole means connecting calcium channels to transcription, and was the first example of a calcium channel functioning in the nucleus as a transcription factor.
In this study, we report the discovery of a bipartite nuclear localization sequence (NLS) in the cytoplasmic carboxy-terminus of the C. elegans TRPV cation channel OCR-2. NLSs are stretches of amino acids that mediate the active transport of proteins into the nucleus [25]. We find that the OCR-2 NLS is functional, being both sufficient for the nuclear import of a GFP fusion in neuronal cells and able to direct nuclear accumulation of a carboxy-terminal cytoplasmic fragment of the OCR-2 channel. Key basic residues required for mammalian NLS function are also required for the OCR-2 NLS to direct nuclear localization. Furthermore, while our investigation into the subcellular localization of the full-length OCR-2 channel supports previous reports of expression in neuronal cell bodies and cilia, we also discovered that a portion of the full-length channel can localize to the nucleus in both head and tail neurons.
Results
The Carboxy-Terminal Tail of OCR-2 Contains a Bipartite Nuclear Localization Sequence
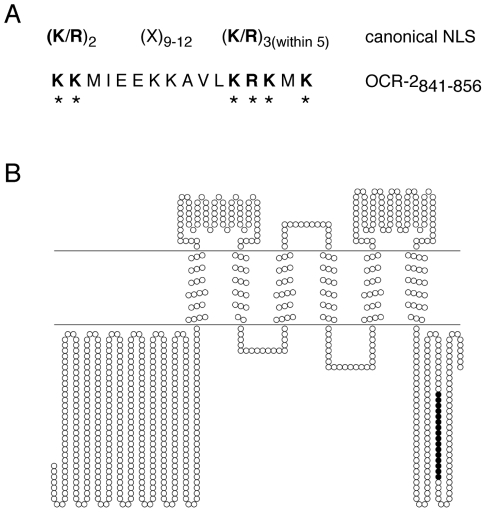
Canonical bipartite NLSs contain two clusters of basic residues, separated by a spacer region of 9–12 amino acids [25], [26]. Using the Expasy Prosite Protein Domain Database (http://ca.expasy.org/prosite/), we identified a putative bipartite NLS spanning residues 841–856 of the C. elegans TRPV channel OCR-2 that closely matches the canonical NLS sequence (Figure 1A). The NLS resides in the cytoplasmic carboxy-terminal tail of the channel (Figure 1B). To establish whether this putative NLS is able to direct nuclear transport of a cargo protein, we generated a GFP::LacZ fusion (using OCR-2 residues 841–856) to visualize subcellular localization in live animals. The inclusion of LacZ ensured that any nuclear accumulation observed was due to active nuclear import through the nuclear pore complex, rather than passive diffusion into the nucleus. Although GFP alone is small enough to passively diffuse into the nucleus, the fusion to LacZ creates a protein that greatly exceeds the 40–60 kDa passive diffusion limit of the nucleus [27], [28], [29]. We used the osm-10 promoter [30] to drive expression of the GFP::LacZ or OCR-2841–856::GFP::LacZ fusion proteins in the ASHs, a pair of head sensory neurons where the endogenous OCR-2 channel is expressed [5], and observed subcellular localization via GFP fluorescence. In the absence of the putative OCR-2 NLS, the GFP::LacZ fusion was excluded from the nucleus (Figure 2A). However, inclusion of the putative NLS resulted in dramatic nuclear accumulation of the fusion protein (Figure 2B). We conclude that the OCR-2 NLS is sufficient to direct nuclear import of a cargo protein.
Figure 1. A putative bipartite NLS in the TRPV channel OCR-2.
Amino acids 841–856 of OCR-2 contain two clusters of lysine/arginine residues, separated by 9 amino acids. (A) Alignment with the canonical bipartite nuclear localization sequence demonstrates that the OCR-2 sequence closely matches an NLS. (B) A diagram of the C. elegans OCR-2 channel, with the putative NLS in black, reveals that the sequence is located in the cytoplasmic carboxy-terminus of the channel. The cytoplasmic side is located below the horizontal lines that denote the plasma membrane. (Diagram was created using TOPO2, Transmembrane Protein Display software, http://www.sacs.ucsf.edu/TOPO2).
Figure 2. The putative NLS in OCR-2 is sufficient to drive nuclear localization of a GFP::LacZ fusion protein, and requires basic residues within the NLS.
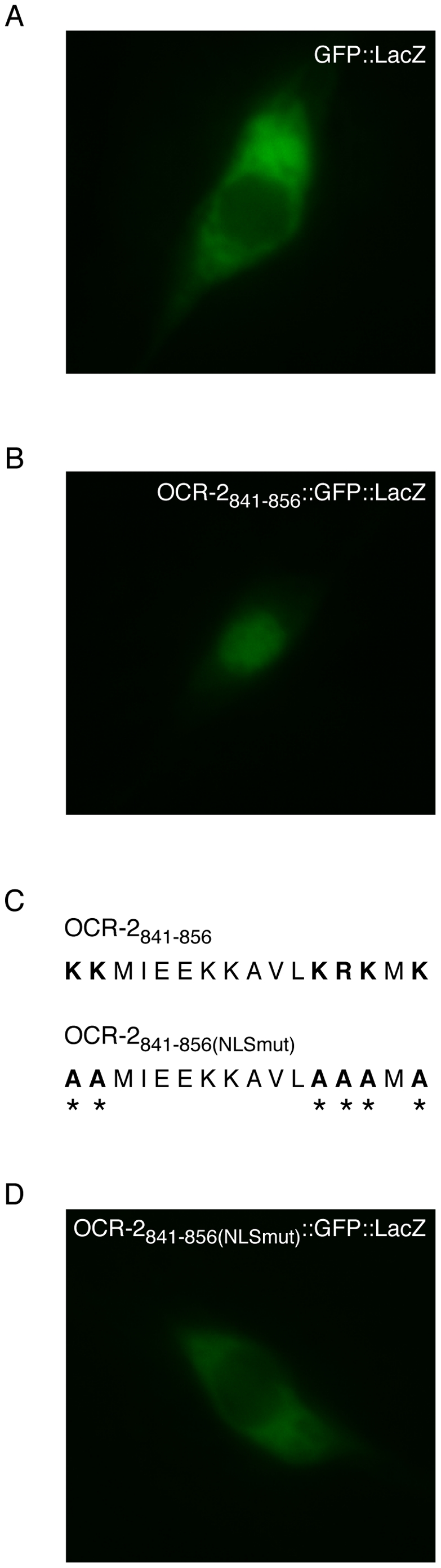
The osm-10 promoter [30] was used to express GFP::LacZ fusion proteins in the ASH neurons. (A) The GFP::LacZ fusion (osm-10p::gfp::lacZ) alone is excluded from the nucleus of the ASH neurons. GFP fluorescence was restricted to the cytoplasm in 78/78 animals assessed. (B) Inclusion of the putative NLS, upstream of and in frame with GFP::LacZ (osm-10p::ocr-2841–856::gfp::lacZ), resulted in robust nuclear translocation of GFP. The ASH neurons of 97/103 worms examined displayed pronounced nuclear accumulation of GFP. (C) Six of the basic lysine and arginine residues (indicated in bold) were replaced with the nonpolar amino acid alanine (indicated with an asterisk and in bold) and the mutated NLS was fused upstream of and in frame with GFP::LacZ (osm-10p::ocr-2841–856(NLSmut)::gfp::lacZ). (D) Mutating these basic clusters abolished nuclear localization. The OCR-2841–856(NLSmut)::GFP::LacZ fusion was restricted to the cytoplasm of ASH in 44/44 animals examined. Numbers represent the combined data of ≥3 independent transgenic lines for each construct.
Basic Residues within the OCR-2 NLS are Necessary for Nuclear Localization
The basic residues of canonical NLSs are critical for interactions with importin-α, an adaptor protein that mediates the first step of import through the nuclear pore complex [31], [32], [33]. The 2 clusters of basic residues in a bipartite NLS interact with the minor and major binding pockets of importin-α, which initiates the import of the NLS-containing protein into the nucleus [32]. To determine whether the 2 patches of basic residues within the putative OCR-2 NLS were required for the observed nuclear localization of OCR-2841–856::GFP::LacZ, we performed site directed mutagenesis, converting 6 basic residues within the NLS to alanine (Figure 2C). The resulting OCR-2841–856(NLSmut)::GFP::LacZ fusion failed to accumulate in the nucleus (Figure 2D). This suggests that, like canonical NLSs, the basic patches within the OCR-2 NLS are necessary for nuclear localization of associated proteins.
A Carboxy-Terminal Cytosolic Fragment of OCR-2 Localizes to the Nucleus and Requires the NLS for Nuclear Accumulation
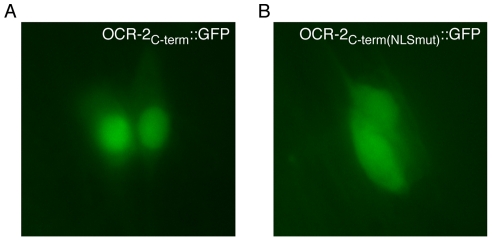
We next sought to determine whether the OCR-2 NLS could function in the context of the channel sequence. OCR-2 is a 6-pass transmembrane protein with the endogenous NLS located within the carboxy-terminal cytoplasmic tail (Figure 1B). To establish whether the carboxy-terminus of OCR-2 could localize to the nucleus, we fused GFP to the entire cytoplasmic fragment of the channel (“C-term”, residues 765–900) and expressed this fusion protein under the control of the ocr-2 promoter, which drives expression in the AWA, ASH, ADL and ADF head neurons, as well as the PHA and PHB tail neurons [5]. In all of these neurons we observed OCR-2C-term::GFP accumulating to high levels within the nucleus (Figure 3A). Additionally, fusing GFP to the OCR-2 cytoplasmic tail in which the 6 basic residues of the identified NLS had been mutated to alanine (OCR-2C-term(NLSmut)::GFP) abolished the nuclear accumulation of GFP, and the signal was distributed evenly throughout the cell body (Figure 3B). The low-level nuclear expression seen with this fusion protein was likely due to passive diffusion into the nucleus because these fusions are relatively small, approximately 40 kDa, and may not require active transport to enter the nucleus. Furthermore, the subcellular localization pattern seen when the NLS was mutated (Figure 3B) was similar to that seen when GFP alone was expressed under the control of the ocr-2 promoter (data not shown). As GFP lacks an endogenous NLS, this supports the notion that the low level nuclear accumulation of OCR-2C-term(NLSmut)::GFP observed in the absence of the functional NLS was due to diffusion. Taken together, our data indicates that the NLS can function to transport the cytoplasmic portion of the OCR-2 channel into the nucleus, and that nuclear accumulation requires the basic residues within the NLS.
Figure 3. The cytoplasmic carboxy-terminal fragment of OCR-2 can utilize the NLS to localize to the nucleus.
The entire carboxy-terminus (amino acids 765–900, “OCR-2C-term”) was fused upstream of and in frame with GFP and expressed under the control of the ocr-2 promoter (ocr-2p::ocr-2C-term::gfp). (A) The OCR-2C-term::GFP fusion was detected in the nucleus of ocr-2 expressing head and tail neurons in 91/91 transgenic animals examined. The PHA/PHB tail neurons are shown here. Furthermore, the basic residues of the NLS are required for the nuclear accumulation observed. (B) A GFP fusion to the carboxy-terminal tail in which the 6 previously identified essential lysine/arginine residues had been mutated to alanine (Figure 2C) (ocr-2p::ocr-2C-term(NLSmut)::gfp) resulted in a much less pronounced signal in the nucleus. The OCR-2C-term(NLSmut)::GFP signal was observed evenly distributed in the cytoplasm and nucleus of ocr-2 expressing neurons in 94/94 transgenic animals examined. The PHA/PHB tail neurons are shown here. Numbers represent the combined data of ≥3 independent transgenic lines for each construct.
The Carboxy-Terminus of the Intact OCR-2 Channel can Localize to the Nucleus
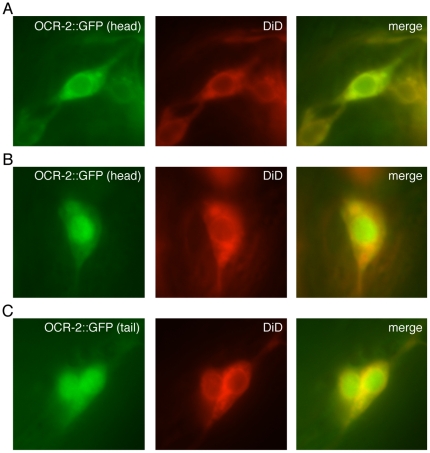
To determine whether a portion of the full-length OCR-2 channel can translocate into the nucleus, we fused GFP to the extreme carboxy-terminus of the full-length channel. Previous characterization of OCR-2 localization in live worms made use of a cytoplasmic amino-terminal GFP fusion, and expression in neuronal sensory cilia and cell bodies was seen [5]. Consistent with reported localization, we observed GFP expression in the sensory cilia and cell body (Figure 4A and data not shown). However, with GFP fused to the carboxy-terminus of the full-length channel, we also observed nuclear accumulation of GFP in at least one ocr-2 expressing neuron in 40% of transgenic animals (Figure 4B–C). While the majority of OCR-2 expression was seen in the cell body and the cilia, where it would fulfill its role in sensory transduction, the nuclear accumulation we observed suggests that the carboxy-terminal portion of the full-length channel is capable of translocating to the nucleus, albeit at a lower frequency.
Figure 4. A GFP fusion to the carboxy-terminus of the full-length OCR-2 channel can be detected in the nucleus.
GFP was fused downstream of and in frame with full-length OCR-2 and expressed under the control of the ocr-2 promoter (ocr-2p::ocr-2::gfp). Transgenic animals were incubated with the lipophilic dye DiD (shown in red), which is taken up by several of the OCR-2 expressing neurons and was used to visualize the neuronal cell body. (A) As previously reported for an amino-terminal GFP fusion [5], flourescence was detected in the cell body and cilia of the AWA, ASH, ADL and ADF head neurons and PHA and PHB tail neurons [5]. The cell body of a head neuron is shown. (B–C) In 82/203 (40%) transgenic animals examined, distinct nuclear accumulation of GFP was detected in at least one neuron. Head (B) and tail (C) neurons are shown. Numbers represent the combined data of 3 independent transgenic lines. To show the animal-to-animal variability in subcellular localization within a transgenic line, all three images shown here were obtained from a single line.
The OCR-2 NLS is not Necessary to Regulate tph-1 Expression in ADF or odr-10 Expression in AWA
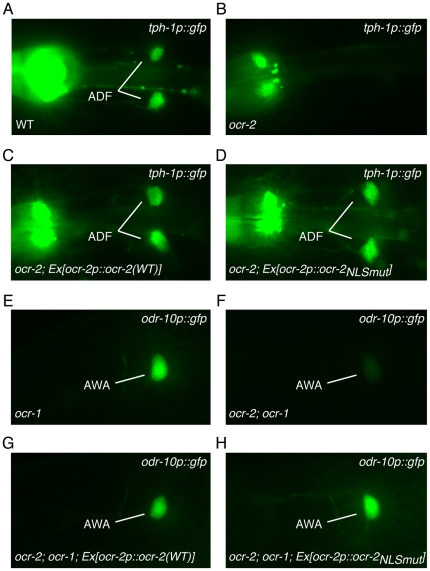
C. elegans TRPV channels have previously been shown to regulate transcription of select genes in the ADF and AWA head sensory neurons [5], [11]. Both OSM-9 and OCR-2 upregulate transcription of the serotonin biosynthetic enzyme TPH-1 in the ADF neurons [11]. To assess whether the NLS of OCR-2 was required to regulate expression of tph-1, we used a tph-1p::gfp transcriptional reporter expressed in the ADF and NSM head neurons [11] (Figure 5A). Expression of GFP in the ADF neurons is dramatically reduced in ocr-2 mutants (Figure 5B) [11]. We found that GFP expression was significantly restored in ADF by reintroducing either wild-type full-length OCR-2 (Figure 5C), or the full-length channel in which the basic residues of the NLS had been changed to alanine (OCR-2NLSmut) (Figure 5D).
Figure 5. OCR-2 regulates expression of tph-1 and odr-10 independently of the NLS.
(A–D) The OCR-2 NLS is not required for tph-1::gfp expression. (A) TPH-1 is a serotonin biosynthetic enzyme and, as previously described [11], a tph-1p::gfp transcriptional reporter is expressed in the ADF and NSM head neurons. The ADF neurons are indicated. The NSM neurons are seen in the left of each panel. (B) OCR-2 has been reported to modulate transcription of tph-1 [11], and ocr-2(ak47) null mutants have reduced expression of tph-1p::gfp in the ADF neurons. Using the ocr-2 promoter, either (C) the wild-type OCR-2 channel (ocr-2p::ocr-2), or (D) the channel in which the basic residues of the NLS had been mutated to alanine (ocr-2p::ocr-2NLSmut), was re-introduced into ocr-2(ak47) mutant animals. Both channels restored tph-1p::gfp expression in ADF. (E-H) The OCR-2 NLS is not required for odr-10::gfp expression. ODR-10 is a G protein-coupled receptor and an odr-10p::gfp transcriptional reporter is expressed solely in the AWA head neurons [5]. (E) Expression of odr-10p::gfp in ocr-1(ak46) null mutants is indistinguishable from wild-type animals (and [5]). (F) ocr-2(ak47);ocr-1(ak46) double mutants have drastically reduced expression of odr-10p::gfp (and [5]). Using the ocr-2 promoter, either (G) the wild-type OCR-2 channel (ocr-2p::ocr-2), or (H) the channel in which the basic residues of the NLS had been mutated to alanine (ocr-2p::ocr-2NLSmut), was re-introduced into ocr-2(ak47);ocr-1(ak46) mutant animals. Both channels restored odr-10p::gfp expression in AWA. ≥2 independent transgenic lines were examined for each construct, n≥40 transgenic animals.
The AWA neurons express an additional TRPV channel, OCR-1 [5]. OCR-1 and OCR-2 have overlapping functions in regulating expression of the diacetyl receptor, ODR-10, in the AWA neurons [5]. ocr-1 mutants have no change in expression of an odr-10p::gfp reporter [5] (and Figure 5E), while odr-10p::gfp expression is only slightly reduced in ocr-2 mutant animals [5]. However, ocr-1;ocr-2 double mutants have little to no expression [5] (and Figure 5F). To determine whether the NLS of OCR-2 was necessary to regulate expression of odr-10, we used the odr-10p::gfp transcriptional reporter expressed solely in AWA [5]. While ocr-1;ocr-2 double mutants have drastically reduced GFP expression (Figure 5G), we found that GFP expression was restored to levels comparable to the ocr-1 single mutant when we reintroduced either wild-type full-length OCR-2 (Figure 5H), or the full-length channel in which the NLS had been mutated (OCR-2NLSmut) (Figure 5G).
Given that the OCR-2 channel that lacked a functional carboxy-terminal NLS was able to restore tph-1p::gfp expression in ADF and odr-10p::gfp expression in AWA to levels comparable to the wild-type channel, we conclude that the NLS is not necessary to regulate transcription of tph-1 or odr-10. Additionally, expression of OCR-2C-term, under the control of the ocr-2 promoter (ocr-2p::ocr-2Cterm), was not sufficient to restore either tph-1p::gfp or odr-10p::gfp expression (data not shown), further supporting the conclusion that nuclear localization of the OCR-2 carboxy-terminal cytoplasmic tail does not contribute to transcriptional regulation of these 2 genes. However, as these are currently the only 2 genes known to be transcriptionally regulated by TRPV channels in C. elegans, we can not rule out the possibility that the OCR-2 NLS is required to regulate the transcription of other, as yet unidentified, genes.
Discussion
Transient changes in the environment can bring about long-lasting modifications in an animal's behavior by altering neural circuitry [16], [34]. This plasticity is due in part to the nervous system's ability to convert short-lived excitation into changes in gene expression, which have enduring effects. While it has long been appreciated that calcium plays a key role in coupling brief neuronal activity into long-term transcriptional changes [16], [18], [35], a direct role for the actual calcium channels involved has just begun to be appreciated [24]. The Cav1.2 channel was the first calcium channel described to encode a transcription factor that directly regulates expression of a wide variety of neuronal genes [24]. TRP channels are calcium permeable channels that are involved in a wide variety of cellular processes. While members of the nematode TRPV subfamily of channels have been implicated in modulating neuronal gene expression (a novel role for TRPV channels), their precise role in this process has yet to be elucidated.
Our results demonstrate that the OCR-2 TRPV channel contains an NLS that is functional and can direct nuclear accumulation of both synthetic cargo proteins as well as the cytoplasmic portion of the native channel where it is found. Furthermore, we report that despite being a 6-pass transmembrane protein, a carboxy-terminal portion of the full-length channel is capable of accumulating in the nucleus. Previous characterization of the OCR-2 channel by other labs may have failed to detect this localization due to GFP insertions into the amino-terminus of OCR-2 [5], fixative conditions used for FLAG epitope detection [11], or because of the relatively low level frequency with which nuclear accumulation occurs.
The mechanism by which a portion of the OCR-2 transmembrane channel would be released has yet to be established. One possibility is that OCR-2 may participate in regulated intramembrane proteolysis (RIP) to release its cytoplasmic tail. RIP is a cellular process whereby a portion of a transmembrane protein is cleaved, releasing a soluble fragment that can enter the nucleus and modify gene expression, bypassing the need to activate signaling cascades in the cytoplasm [36], [37]. Since its discovery, the process has been found to be conserved from bacteria to humans and controls a wide range of cellular processes [36], [38]. While RIP has been described for surface receptors and transmembrane proteins found in the endoplasmic reticulum [39], [40], [41], [42], [43], calcium channels have only recently been added to the list of transmembrane proteins identified to undergo this type of cleavage [24]. While protease cleavage predictor programs did not reveal any obvious candidate cleavage sites within the OCR-2 channel, RIP cleavage sites may exist. Prediction programs do not use an exhaustive list of cellular proteases, and the available data for most proteases is still very incomplete. Thus, legitimate cleavage sites may go undetected.
Although the OCR-2 NLS does not seem to be required to regulate expression of the two previously identified C. elegans targets, tph-1 in ADF or odr-10 in AWA, it is possible that the TRPV channels modulate expression of other neuronal genes and that the full repertoire of genes regulated by the TRPV channels has yet to be identified. Although it has been proposed that OCR-2 also regulates gene expression in ASH [8], specific targets remain to be identified in ASH, ADL and PHA/PHB, where OCR-2 is also expressed.
We suggest that the OCR-2 channel may transcriptionally regulate neuronal gene expression both indirectly, by activating traditional calcium-dependent cascades, and directly via NLS-mediated nuclear translocation. For example, tph-1 and odr-10 transcripts may be regulated by calcium-mediated signaling pathways. Consistent with this possibility, a gain-of-function mutation in the Ca2+/calmodulin-dependent protein kinase UNC-43 partially suppresses the tph-1p::gfp expression defects of ocr-2 mutants [11]. This suggests that traditional calcium-mediated signaling events downstream of OCR-2 are important, at least in part, for transcriptional regulation in the ADF neurons. While it is possible that the OCR-2 NLS has no biological role, an intriguing possibility is that the OCR-2 channel may also directly affect transcription, via nuclear localization of the carboxy-terminus, for other, yet unidentified targets in the head and tail sensory neurons. As TRP channels have already been recognized as integrators of multiple regulatory pathways, this could place TRPVs in the unique position of integrating various cellular signals and directly transducing this information to the nucleus to alter gene expression, and ultimately neuronal activity.
Materials and Methods
Strains
Strains were maintained under standard conditions on NGM agar plates seeded with OP50 E. coli bacteria [44]. Strains used in this study include: N2 Bristol wild-type, GE24 pha-1(e2123), GR1366 mgIs42[tph-1::GFP+pRF4(rol-6(su1006))], FG221 ocr-2(ak47);mgIs42[tph-1::GFP+pRF4(rol-6(su1006))], CX3260 kyIs37(odr-10p::gfp), FG231 ocr-1(ak46);kyIs37(odr-10p::gfp), FG240 ocr-2(ak47);ocr-1(ak46);kyIs37(odr-10p::gfp).
To create transgenic strains, germline transformations were performed as previously described [45]. For gfp::LacZ fusion lines, GE24 pha-1(e2123) animals were injected with 100 ng/ul of pFG31 osm-10p::gfp::lacZ, pFG36 osm-10p::ocr-2841–856::gfp::lacZ, or pFG41 osm-10p::ocr-2841–856(NLSmut)::gfp::lacZ, along with 150 ng/ul of pBX1 pha-1(+) [46] plasmid co-injection marker. For carboxy-terminal::gfp lines, GE24 pha-1(e2123) animals were injected with 50 ng/ul pFG42 ocr-2p::ocr-2C-term::gfp or pFG55 ocr-2p:: ocr-2C-term(NLSmut)::gfp, along with 75 ng/ul of pBX1 pha-1(+) [46] plasmid co-injection marker. For full length ocr-2::gfp lines, GE24 pha-1(e2123) animals were injected with 50 ng/ul of pFG43 ocr-2p::ocr-2::gfp, along with 75 ng/ul of pBX1 pha-1(+) [46] plasmid co-injection marker. For tph-1p::gfp rescue experiments FG221 ocr-2(ak47);mgIs42[tph-1::GFP+pRF4(rol-6(su1006))] animals were injected with 50 ng/ul of pAJ35 wild-type ocr-2p::ocr-2 (gift of Cori Bargmann) or pFG40 ocr-2p::ocr-2NLSmut, along with 75 ng/ul of pJM67 elt-2::gfp [47] co-injection marker. For odr-10p::gfp rescue experiments FG240 ocr-2(ak47);ocr-1(ak46);kyIs37(odr-10p::gfp) animals were injected with 50 ng/ul of pAJ35 wild-type ocr-2p::ocr-2 (gift of Cori Bargmann) or pFG40 ocr-2p::ocr-2NLSmut, along with 75 ng/ul of pJM67 [47] elt-2::gfp co-injection marker.
Plasmid Construction
pFG40 ocr-2p::ocr-2NLSmut: The 6 basic lysine/arginine residues in the NLS were mutated to alanine using site directed mutagenesis (QuikChange, Stratagene) of pAJ35 (ocr-2p::ocr-2, gift of Cori Bargmann). Residue changes were verified by sequencing. In addition the entire promoter and ocr-2 coding region was sequenced to ensure there were no unwanted base pair changes.
pFG31 osm-10p::gfp::lacZ: Site directed mutagenesis (QuikChange, Stratagene) was used to remove the SV40 NLS from pPD96.04 (Fire Lab C. elegans Vector Kit, Addgene), leaving gfp::lacZ and generating pFG24. The ∼900 bp osm-10 promoter was removed from CR142 [48] using PstI and BamHI and inserted into the PstI/BamHI sites of pFG24, upstream of gfp and lacZ.
pFG36 osm-10p::ocr-2841–856::gfp::lacZ: The putative NLS from OCR-2 (encompassing amino acids 841–856) was PCR amplified from pAJ35 (ocr-2p::ocr-2, gift of Cori Bargmann) with an ATG start codon added 5′, an additional GC added to the 3′ end to maintain correct frame usage, and AgeI sites added to both the 5′ and 3′ ends. The ocr-2841–856 PCR product and pFG31 were digested with AgeI and the NLS was inserted upstream of and in frame with gfp::lacZ. Correct orientation of the NLS was verified by sequencing.
pFG41 osm-10p::ocr-2841–856(NLSmut)::gfp::lacZ: The mutated NLS was PCR amplified from pFG40 (full length OCR-2 following site directed mutagenesis of the NLS) with an ATG start codon added 5′, an additional GC added to the 3′ end to maintain correct frame usage, and AgeI sites added to both the 5′ and 3′ ends. The ocr-2841–856(NLSmut) (mutated NLS) PCR product and pFG31 were digested with AgeI and the NLS was inserted upstream of and in frame with gfp::lacZ. Correct orientation of the NLS was verified by sequencing.
pFG38 ocr-2p::gfp: The ∼2.5 kb ocr-2 promoter was digested out of pAJ35 (ocr-2p::ocr-2, gift of Cori Bargmann) with SphI and XmaI, and inserted into the SphI/XmaI sites of pPD49.26 (Fire Lab C. elegans Vector Kit, Addgene) to generate pFG35. gfp was PCR amplified from pPD95.77 (Fire Lab C. elegans Vector Kit, Addgene) with KpnI and SacI sites added 5′ and 3′, respectively. gfp was inserted into the KpnI/SacI sites of pFG35, downstream of the ocr-2 promoter.
pFG42 ocr-2p::ocr-2C-term::gfp: The carboxy-terminus of ocr-2, starting with CGTACATATGAA, was PCR amplified from pAJ35 (ocr-2p::ocr-2, gift of Cori Bargmann). An ATG start codon was added 5′, the TGA stop codon was not included, and NheI and KpnI sites were added 5′ and 3′, respectively. pFG38 was digested with NheI and KpnI and the ocr-2 carboxy-terminus was inserted into these sites, upstream of and in frame with gfp.
pFG55 ocr-2p:: ocr-2C-term(NLSmut)::gfp: The carboxy-terminus of ocr-2 was PCR amplified from pFG40 (full length OCR-2 following site directed mutagenesis of the NLS). An ATG start codon was added 5′, the TGA stop codon was not included, and NheI and KpnI sites were added 5′ and 3′, respectively. pFG38 was digested with NheI and KpnI and the ocr-2 carboxy-terminus with the mutated NLS was inserted into those sites, upstream of and in frame with gfp.
pFG43 ocr-2p::ocr-2::gfp: Full length ocr-2 was PCR amplified from pAJ35 (ocr-2p::ocr-2, gift of Cori Bargmann). XbaI and KpnI sites were included 5′ and 3′, respectively, and the TGA stop codon was not included. pFG38 was digested with NheI and KpnI and ocr-2 was inserted upstream of and in frame with gfp.
pFG52 ocr-2p::ocr-2C-term: The carboxy-terminus of ocr-2, starting with CGTACATATGAA, was PCR amplified from pAJ35 (ocr-2p::ocr-2, gift of Cori Bargmann). An ATG start codon was added 5′ and NheI and KpnI sites were added 5′ and 3′, respectively, for insertion into these sites of pFG35 (see above).
All constructs were verified by sequencing when appropriate.
Imaging
To identify the cell body of dye-filling neurons, animals were incubated with the lipophilic dye DiD (Molecular Probes, Invitrogen), as previously described [49]. Images were obtained with a Zeiss Axio Imager Z1 microscope (using a 100× Plan-APO oil objective and epi-flourescence), high resolution AxioCam MRm digital camera and Zeiss AxioVision software.
Acknowledgments
We thank Michelle Krzyzanowski, Paul Kuehnert, Kerry Michelson, Benjamin Peterson, Nikhila Sundareswaran and Jordan Wood for feedback on this manuscript and Noelle L'Etoile for valuable discussions. We also thank Cori Bargmann, Andrew Fire, Ji Ying Sze, and the Caenorhabditis Genetics Center for reagents.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Science Foundation (http://www.nsf.gov, MCB-0917896 to DF). An award from the Julian Park Publication Fund (College of Arts and Sciences, University at Buffalo) helped to defray publication costs for this manuscript. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pedersen SF, Owsianik G, Nilius B. TRP channels: an overview. Cell Calcium. 2005;38:233–252. doi: 10.1016/j.ceca.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 2.Kahn-Kirby AH, Bargmann CI. TRP channels in C. elegans. Annu Rev Physiol. 2006;68:719–736. doi: 10.1146/annurev.physiol.68.040204.100715. [DOI] [PubMed] [Google Scholar]
- 3.Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 5.Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, et al. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron. 2002;35:307–318. doi: 10.1016/s0896-6273(02)00757-2. [DOI] [PubMed] [Google Scholar]
- 6.Xiao R, Xu XZ. Function and regulation of TRP family channels in C. elegans. Pflugers Arch. 2009;458:851–860. doi: 10.1007/s00424-009-0678-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezak MJ, Hong E, Chaparro-Garcia A, Ferkey DM. Caenorhabditis elegans TRPV channels function in a modality-specific pathway to regulate response to aberrant sensory signaling. Genetics. 2010;185:233–244. doi: 10.1534/genetics.110.115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilliard MA, Apicella AJ, Kerr R, Suzuki H, Bazzicalupo P, et al. In vivo imaging of C. elegans ASH neurons: cellular response and adaptation to chemical repellents. Embo J. 2005;24:63–72. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilliard MA, Bergamasco C, Arbucci S, Plasterk RH, Bazzicalupo P. Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. Embo J. 2004;23:1101–1111. doi: 10.1038/sj.emboj.7600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Sokolchik I, Blanco G, Sze JY. Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development. 2004;131:1629–1638. doi: 10.1242/dev.01047. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg ME, Greene LA, Ziff EB. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. J Biol Chem. 1985;260:14101–14110. [PubMed] [Google Scholar]
- 14.Curran T, Morgan JI. Superinduction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229:1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- 15.Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, et al. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dash PK, Karl KA, Colicos MA, Prywes R, Kandel ER. cAMP response element-binding protein is activated by Ca2+/calmodulin- as well as cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1991;88:5061–5065. doi: 10.1073/pnas.88.11.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu Z, Ghosh A. A brief history of neuronal gene expression: regulatory mechanisms and cellular consequences. Neuron. 2008;60:449–455. doi: 10.1016/j.neuron.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 21.Kapiloff MS, Mathis JM, Nelson CA, Lin CR, Rosenfeld MG. Calcium/calmodulin-dependent protein kinase mediates a pathway for transcriptional regulation. Proc Natl Acad Sci U S A. 1991;88:3710–3714. doi: 10.1073/pnas.88.9.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng M, Thompson MA, Greenberg ME. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 23.Cullen PJ, Lockyer PJ. Integration of calcium and Ras signalling. Nat Rev Mol Cell Biol. 2002;3:339–348. doi: 10.1038/nrm808. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 27.Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 28.Keminer O, Peters R. Permeability of single nuclear pores. Biophys J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart AC, Kass J, Shapiro JE, Kaplan JM. Distinct signaling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J Neurosci. 1999;19:1952–1958. doi: 10.1523/JNEUROSCI.19-06-01952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dingwall C, Laskey RA. Nuclear targeting sequences–a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 32.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 33.Gorlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 34.Peckol EL, Troemel ER, Bargmann CI. Sensory experience and sensory activity regulate chemosensory receptor gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:11032–11038. doi: 10.1073/pnas.191352498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- 36.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 37.Ebinu JO, Yankner BA. A RIP tide in neuronal signal transduction. Neuron. 2002;34:499–502. doi: 10.1016/s0896-6273(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 38.Urban S, Freeman M. Intramembrane proteolysis controls diverse signalling pathways throughout evolution. Curr Opin Genet Dev. 2002;12:512–518. doi: 10.1016/s0959-437x(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 39.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 40.Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 41.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niwa M, Sidrauski C, Kaufman RJ, Walter P. A role for presenilin-1 in nuclear accumulation of Ire1 fragments and induction of the mammalian unfolded protein response. Cell. 1999;99:691–702. doi: 10.1016/s0092-8674(00)81667-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 44.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- 48.Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- 49.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]