Abstract
G protein-coupled receptors (GPCRs) transmit signals by forming active-state complexes with heterotrimeric G proteins. It has been suggested that some GPCRs also assemble with G proteins prior to ligand-induced activation, and that inactive-state preassembly facilitates rapid and specific G protein activation. However, no mechanism of preassembly has been described, and no functional consequences of preassembly have been demonstrated. Here we show that M3 muscarinic acetylcholine receptors (M3R) form inactive-state complexes with Gq heterotrimers in intact cells. The M3R C terminus is sufficient, and a six amino-acid polybasic sequence distal to helix 8 (565KKKRRK570) is necessary for preassembly with Gq. Replacing this sequence with six alanine residues prevents preassembly, slows the rate of Gq activation, and decreases steady-state agonist sensitivity. Other Gq-coupled receptors possess similar polybasic regions and also preassemble with Gq, suggesting that these GPCRs may utilize a common preassembly mechanism to facilitate activation of Gq heterotrimers.
Hormones, neurotransmitters and other ligands often exert their physiological actions by binding to G protein-coupled receptors (GPCRs)1. Ligand binding to GPCRs promotes an active receptor state that binds to heterotrimeric (Gαβγ) G proteins and promotes the release of GDP from Gα subunits. Nucleotide-free G proteins, in turn, stabilize the ligand-bound active state of GPCRs2. These active-state ligand-GPCR-G protein ternary complexes can be observed when guanine nucleotides are absent, but are short-lived intermediates in intact cells because GTP and GDP are abundant2. No high-resolution structure of an active-state GPCR-G protein complex is presently available, although many GPCR-G protein interaction sites have been determined using biochemical and biophysical methods3,4. It is expected that the structure of an active-state complex will shed considerable light on the mechanism whereby GPCR activation is coupled to nucleotide release5.
In addition to active-state GPCR-G protein complexes it has been suggested that some receptors and G proteins associate prior to receptor activation, by mechanisms that do not depend on the conformational state of either protein. Evidence for “precoupled” or preassembled inactive-state complexes has come from ligand-binding and biochemical studies6–8, and more recently from biophysical studies in live cells9–11. In contrast to active-state GPCR-G protein complexes, almost nothing is known about inactive-state complexes. Mutagenesis studies have not identified common structural elements necessary for GPCR-G protein preassembly, and have not provided mutant receptors or G proteins that could be used to test the impact of preassembly on signaling. Signaling by preassembled GPCR-G protein complexes would differ from the collision coupling model first proposed for GPCR signaling12, in that random collision of receptors and G proteins would not be necessary. Therefore, preassembly could increase the rate of G protein activation by obviating this potentially rate-limiting step. However, this possibility has not been tested, in part because the tools necessary to do so are not available.
In this study we investigated preassembly of M3 muscarinic acetylcholine receptors (M3R) and Gq heterotrimers using fluorescence recovery after photobleaching in live cells. We found that immobile M3R decreased the lateral mobility of Gq at the plasma membrane in a manner consistent with specific preassembly of inactive-state M3R-Gq complexes. We identified a polybasic motif in the C terminus of M3R that is necessary for this interaction, and used this information to test the involvement of preassembly in signaling. Our results are consistent with the idea that preassembly of inactive-state complexes with Gq increases the sensitivity and accelerates the onset of signaling by M3 muscarinic acetylcholine receptors.
RESULTS
FRAP indicates preassembly of M3R and Gq heterotrimers
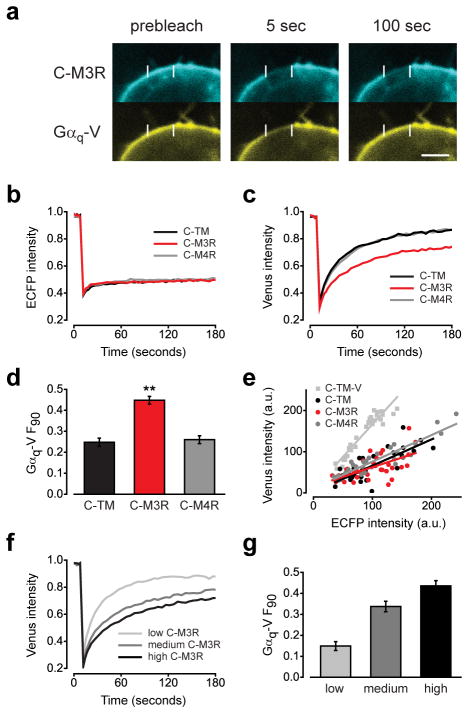
The M3R signals by activating Gq heterotrimers. To determine if M3R associates with Gq prior to receptor activation, we immobilized M3R labeled at its N terminus with ECFP (C-M3R) on the plasma membrane (see Methods), and then measured the lateral mobility of venus-labeled Gαq (Gαq-V). If C-M3R and Gαq-V assembled in inactive-state complexes we expected that immobilizing C-M3R would decrease the lateral mobility of Gαq-V, which would be evident by a slowing of fluorescence recovery after photobleaching (FRAP). A protein consisting of an extracellular ECFP and a single transmembrane domain (C-TM) served as a negative control.
When HEK 293 cells were transiently transfected with C-M3R and Gαq-V (together with unlabeled Gβ1 and Gγ2 subunits) both proteins were located at the plasma membrane (Fig. 1a). Cell surface crosslinking effectively immobilized C-M3R and C-TM, as ECFP fluorescence did not recover after photobleaching (Fig. 1b). Immobilization did not affect the ability of C-M3R to bind to the antagonist [3H] N-methylscopolamine ([3H]NMS), or to activate Gq heterotrimers in response to acetylcholine (Supplementary Fig. 1; Supplementary Results), thus the transmembrane regions of C-M3R were not compromised by this procedure. Immobile C-M3R significantly decreased the lateral mobility of Gαq-V on the plasma membranes of intact cells (Fig. 1c). The fraction of Gαq-V fluorescence that was unrecovered 90 seconds after photobleaching (F90) was 0.45 ± 0.01 (mean ± s.e.m.) in cells with immobile C-M3R, as compared to 0.25 ± 0.01 in cells with immobile C-TM (Fig. 1d). Five minutes after photobleaching the unrecovered fractions were similar for C-M3R and C-TM (0.23 ± 0.01 and 0.17 ± 0.01, respectively), suggesting that immobile C-M3R slowed but did not completely immobilize Gαq-V (Supplementary Fig. 2). This effect of immobile C-M3R was specific, as immobile ECFP-labeled M4 muscarinic receptors (C-M4R), which activate Gi and Go heterotrimers but not Gq heterotrimers, were unable to decrease Gαq-V mobility (F90=0.26 ± 0.01) (Fig. 1c,d). Similarly, immobile C-M3R did not significantly decrease the mobility of venus-labeled GαoA (GαoA-V), a G protein that is not activated by M3R (Supplementary Fig. 3). Immobile C-M4R also had no effect on GαoA-V mobility, suggesting that this receptor does not detectably preassemble with its cognate G protein (Supplementary Fig. 4)13.
Figure 1. Immobile C-M3R decreases Gαq-V mobility.
(a) HEK293 cells transiently expressing C-M3R, Gαq-V, Gβ1 and Gγ2 were biotin-avidin crosslinked and imaged before and after photobleaching of a 4 μm segment of the plasma membrane; scale bar=4 μm. Recovery of ECFP (b) and venus (c) fluorescence after photobleaching in cells expressing immobile C-TM (n=31), C-M3R (n=32) or C-M4R (n=33); traces are averages of all cells. (d) Unrecovered fluorescence 90 seconds after photobleaching (F90) for cells expressing C-TM, C-M3R and C-M4R; **, P<0.001 compared to C-TM. (e) ECFP and venus intensity at the plasma membrane for the same cells as panels b-d, and for cells expressing C-TM-V. C-TM, C-M3R and C-M4R were expressed in excess of Gαq-V. (f) Gαq-V fluorescence recovery after photobleaching in cells expressing low (n=16), medium (n=23) and high (n=17) levels of C-M3R and constant levels of Gαq-V. Average C-M3R fluorescence intensity in these three groups was 58 ± 3, 106 ± 3, and 150 ± 6 arbitrary units, respectively. Average Gαq-V fluorescence intensity was 85 ± 5, 86 ± 7, and 92 ± 8 a.u., respectively. (g) Unrecovered fluorescence in the same cells as panel f. Data represent mean values ± s.e.m..
In order for this FRAP assay to detect transient protein interactions the immobile binding partner (in this case C-M3R) must be relatively abundant, ideally in excess of the mobile partner. Accordingly, C-M3R was expressed at a high density (24.3 pmol mg-1) in our experiments, and the intensity of ECFP fluorescence in the cells used for FRAP experiments suggested that C-TM, C-M3R and C-M4R were expressed at comparable levels. The ratio of ECFP to venus fluorescence in these cells was compared to that of a construct with a 1:1 ECFP:venus stoichiometry (C-TM-V), and this comparison indicated that the immobile proteins were present in excess of Gαq-V (Fig. 1e). In order to determine the relationship between Gαq-V mobility and immobile C-M3R abundance we performed experiments in cells expressing a range of C-M3R levels, as determined by ECFP fluorescence intensity, but constant Gαq-V levels. Cells were categorized as having low (ECFP intensity 0–74, arbitrary units (a.u.)), medium (75–124 a.u.) and high (>124 a.u.) C-M3R expression. The C-M3R:Gαq-V ratios (calculated by comparison to C-TM-V) in these three groups of cells were 1.2, 2.3 and 3.1, respectively. The effect of immobile C-M3R on Gαq-V mobility increased as C-M3R expression increased, and was detectable only when C-M3R outnumbered Gαq-V (Fig. 1f, g).
Although Gαq-V was expressed with unlabeled Gβ1 and Gγ2 in these experiments, it was possible that immobile C-M3R decreased the mobility of free Gαq-V subunits as opposed to Gq heterotrimers (i.e. Gαq-Vβ1γ2). To test this possibility we labeled Gβ1γ2 dimers with bimolecular fluorescence complementation of venus (see Methods), and expressed Gβ1γ2-V with unlabeled Gαq subunits. Gβ1γ2-V mobility was significantly lower with immobile C-M3R (F90=0.48 ± 0.02) than with immobile C-TM (F90=0.32 ± 0.03) (Supplementary Fig. 5). These results suggest that immobile C-M3R interacts (directly or indirectly) with Gq heterotrimers, and that the complexes formed are sufficiently stable to decrease the apparent rate of Gq lateral diffusion.
Inactive- and active-state M3R-Gq complexes are distinct
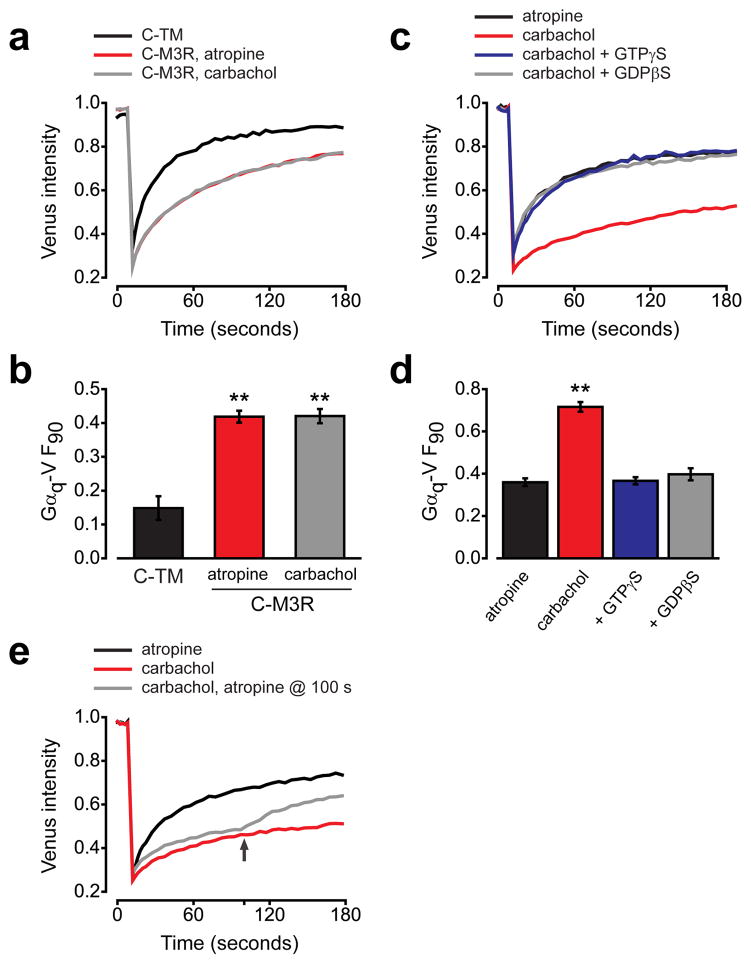
GPCRs and G protein heterotrimers signal by forming active-state complexes in which agonist-bound GPCRs promote the exchange of GDP for GTP on Gα subunits1. In order to determine whether or not the interaction between M3R and Gq that we observed also depended on the activation state of M3R, we repeated our experiments in the presence of the M3R agonist carbachol or the M3R inverse agonist atropine (Fig. 2a). Gαq-V mobility was indistinguishable in the presence of these two ligands (Fig. 2b), suggesting that receptor activation did not stabilize or disrupt M3R-Gq complexes, and that preassembly was not dependent on constitutive receptor activity.
Figure 2. Receptor activity does not affect Gαq-V mobility in intact cells.
(a) Gαq-V fluorescence recovery after photobleaching in cells expressing C-TM (n=22), C-M3R in the presence of 10 μM atropine (n=44) or C-M3R in the presence of 50 μM carbachol (n=42). (b) Unrecovered fluorescence for the same cells as panel a; **, P<0.001 compared to C-TM. (c) Gαq-V fluorescence recovery in permeabilized, nucleotide-depleted cells expressing C-M3R in the presence of atropine (n=15), carbachol (n=15), or in the presence of carbachol with 0.3 mM GTPγS (n=15) or 0.3 mM GDPβS (n=24). (d) Unrecovered fluorescence for the same cells as panel c; **, P<0.001. (e) Gαq-V fluorescence recovery in permeabilized, nucleotide-depleted cells expressing C-M3R in the presence of atropine (n=20), carbachol (n=20), or in the presence of carbachol with atropine added at 100 seconds (arrow; n=20). Data represent mean values ± s.e.m..
The inability of carbachol to decrease Gq mobility in these experiments suggested that active-state complexes were not sufficiently stable to further decrease Gαq-V mobility in intact cells. One possible explanation for this finding is that active-state complexes dissociate quickly after Gα subunits bind GTP, which is abundant in intact cells. In order to test this idea we performed similar experiments in cells that were depleted of guanine nucleotides (see Methods). In contrast to what was observed in intact cells, Gαq-V mobility was dramatically slowed by carbachol in nucleotide-depleted cells expressing immobile C-M3R (Fig. 2c), suggesting that under these conditions Gαq-V subunits remained “empty” for longer periods of time, thus prolonging the lifetime of active-state complexes. In contrast, when atropine was applied to nucleotide-depleted cells to promote the inactive state of immobile C-M3R Gαq-V mobility was similar to that observed in intact cells. The activity-dependent decrease in Gαq-V mobility in nucleotide-depleted cells was reversed if either GTPγS or GDPβS was present, suggesting that stable active-state complexes did, in fact, require nucleotide-empty Gαq-V (Fig. 2d). Switching from carbachol to atropine after photobleaching produced an abrupt increase in the rate of Gαq-V fluorescence recovery, consistent with rapid destabilization of active-state complexes after receptor deactivation (Fig. 2e). These results demonstrate that C-M3R and Gαq-V-containing heterotrimers were capable of forming active-state ligand-GPCR-G protein ternary complexes. Furthermore, they show that inactive M3R-Gq complexes can be distinguished from active-state M3R-Gq complexes. In order to emphasize the distinction between the two types of M3R-Gq complexes, we refer to the formation of inactive-state complexes as preassembly, and the formation of active-state complexes as coupling.
The M3R C terminus mediates preassembly with Gq
We next set out to identify the regions of M3R that participate in preassembly with Gq. Because C-M4R did not detectably decrease the mobility of Gq we constructed a series of chimeras where intracellular portions of M3R were replaced with the analogous segments of M4R. Because of its large size (>200 amino acids) we initially focused on the third intracellular loop (i3) of M3R. However, we found that a C-M3R(M4i3) chimera with the i3 loop of M4R decreased the mobility of Gαq-V as effectively as M3R (Supplementary Fig. 6), suggesting the i3 loop of M3R was not necessary for preassembly. We were unable to test C-M3R chimeras that contained the first and second intracellular loops (i1 and i2) of M4R, as these constructs expressed poorly at the plasma membrane. In contrast, a C-M3R(M4ct) chimera bearing the C terminus of M4R (including helix 8) expressed well but lost the ability to decrease Gαq-V mobility (Fig. 3a,b), as did a chimera containing both i3 and the C terminus from M4R (Supplementary Fig. 6). Conversely, a C-M4R(M3ct) chimera bearing the C terminus of M3R gained the ability to decrease Gαq-V mobility (Fig. 3c,d). These results indicate that the C terminus of M3R is necessary and sufficient for preassembly of muscarinic receptors with Gq. Interestingly, we found that fusing ECFP to the C terminus of M3R also prevented preassembly with Gq (F90=0.18 ± 0.02), suggesting that a free C terminus may be necessary.
Figure 3. The M3R C terminus is necessary and sufficient for preassembly with Gq.
(a) Gαq-V fluorescence recovery in cells expressing C-TM (n=30), C-M3R (n=30), a chimeric C-M3R with the C terminus of M4R (C-M3R(M4ct); n=30) or C-M4R (n=24). (b) Unrecovered fluorescence for the same cells as panel a; **, P<0.001 compared to C-TM. (c) Gαq-V fluorescence recovery in cells expressing C-TM (n=16), C-M3R (n=18), a chimeric C-M4R with the C terminus of M3R (C-M4R(M3ct); n=37) or C-M4R (n=30). (d) Unrecovered fluorescence for the same cells as panel c; **, P<0.001 compared to C-TM. Unrecovered fluorescence for C-M3R and C-M4R(M3ct) were not significantly different (P=0.14). Data represent mean values ± s.e.m..
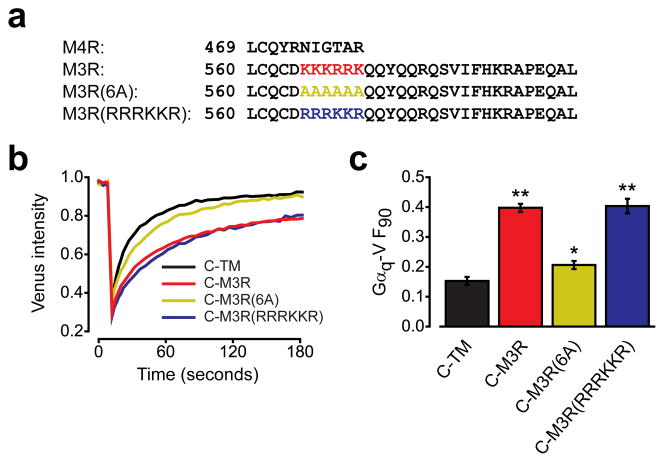
A polybasic region is necessary for preassembly with Gq
The 46 amino acids immediately following the NPxxY motif in TM7 of M3R were sufficient to allow M4R to decrease the mobility of Gαq-V. Roughly the first 16 of these amino acids are predicted to form helix 8, and are conserved in M3R and M4R, as is a cysteine residue (C561 in the human M3R) that is palmitoylated in many GPCRs. A prominent feature of the distal M3R C terminus that is not found in the shorter M4R C terminus is a series of six consecutive basic residues, 565KKKRRK570 (Fig. 4a). We also noticed that similar polybasic regions were present in other Gq-coupled muscarinic acetylcholine receptors and adrenergic receptors (see below). To test the involvement of this polybasic region in preassembly with Gq, we mutated these residues to alanines. The resulting C-M3R(6A) mutant expressed well at the cell surface (see below), but decreased Gαq-V mobility only slightly (P=0.047 compared to C-TM), significantly less than the wild-type C-M3R (Fig. 4b). In order to determine whether the specific order of amino acids or the net charge of the M3R C-terminal polybasic region was important for preassembly with Gq, we constructed a mutant with an altered sequence but the same net charge in this region by exchanging lysine and arginine residues (565RRRKKR570). This C-M3R(RRRKKR) mutant decreased Gαq-V mobility to the same extent as wild-type C-M3R (Fig. 4c), suggesting the positive net charge of the C-terminal polybasic region is critical for preassembly. When a polybasic region (KKKRRR) was added to the C terminus of the C-M4R, the immobilized C-M4R(RKKKRRR) significantly decreased Gαq-V mobility (F90=0.30 ± 0.02), but not to the same extent as immobile C-M3R (F90=0.44 ± 0.03; Supplementary Fig. 7). This result suggests that a polybasic region alone is not sufficient to fully support preassembly, and that other elements in the M3R C terminus are important.
Figure 4. A polybasic region in the M3R C terminus is necessary for preassembly with Gq.
(a) Alignment of the M4R and M3R C termini, together with mutants where amino acids 565–570 are replaced with alanines (6A) or have lysines and arginines exchanged for each other (RRRKKR). (b) Gαq-V fluorescence recovery in cells expressing C-TM (n=43), C-M3R (n=43), C-M3R(6A) (n=43) or C-M3R(RRRKKR) (n=20). (c) Unrecovered fluorescence for the same cells as panel b; **, P<0.001; *, P=0.047 compared to C-TM. Data represent mean values ± s.e.m..
We then tested the ability of the C-M3R(6A) mutant to participate in active-state complexes. In nucleotide-depleted cells immobile C-M3R and C-M3R(6A) inhibited empty Gαq-V mobility to the same extent when the agonist carbachol was present, suggesting that the C-M3R(6A) mutant retained its ability to form active-state ternary complexes (Supplementary Fig. 8). In nucleotide-depleted cells in the presence of atropine Gαq-V mobility was greater in cells expressing immobile C-M3R(6A) than in cells expressing immobile C-M3R, consistent with defective inactive-state preassembly of C-M3R(6A) (Supplementary Fig. 8).
An electrostatic interaction participates in preassembly
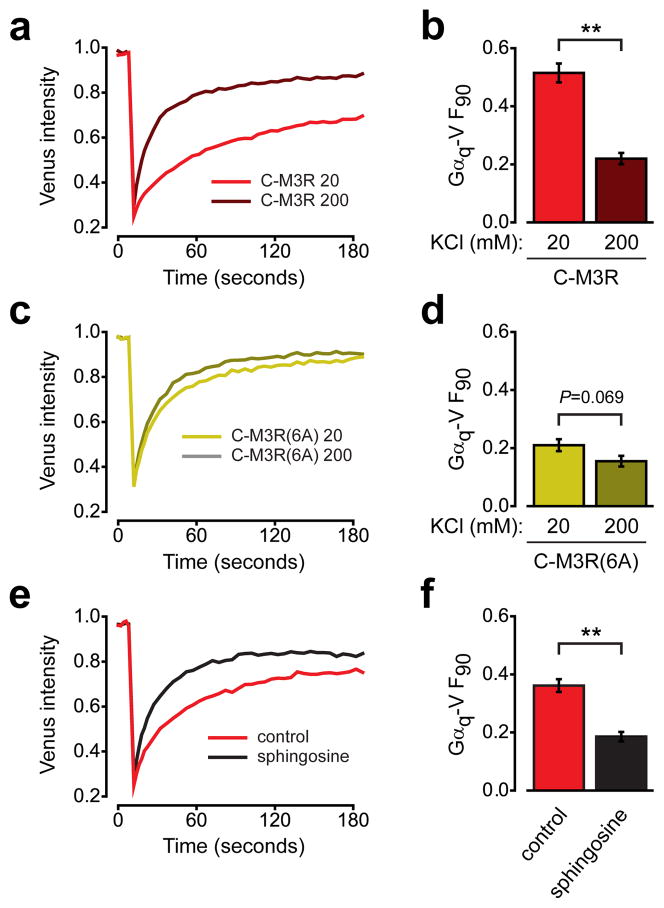
The importance of positively-charged residues suggested that an electrostatic mechanism might be involved in preassembly of M3R and Gq. To test this idea we manipulated the ionic strength of the intracellular environment in permeabilized cells. Electrostatic interactions are shielded (weakened) in high ionic strength conditions, and are strengthened in low ionic strength conditions. Increasing the ionic strength essentially abolished the slowing of Gαq-V mobility by immobile C-M3R (F90=0.22 ± 0.02). In contrast, decreasing the ionic strength enhanced the slowing of Gαq-V mobility (F90=0.51 ± 0.03) (Fig. 5a,b). These results suggest that an electrostatic interaction participates in preassembly of M3R and Gq heterotrimers. The effects of changing ionic strength on Gαq-V mobility were largely absent with the C-M3R(6A) mutant (Fig. 5c,d), consistent with the specific involvement of the C-terminal polybasic region in this interaction.
Figure 5. Preassembly involves an electrostatic interaction and requires an electronegative plasma membrane.
(a) Gαq-V fluorescence recovery in permeabilized cells expressing immobile C-M3R bathed in 20 mM KCl (n=19) or in 200 mM KCl (n=20). (b) Unrecovered fluorescence for the same cells as panel a; **, P<0.001. (c) Gαq-V fluorescence recovery in permeabilized cells expressing immobile C-M3R(6A) bathed in 20 mM KCl (n=21) or in 200 mM KCl (n=19). (d) Unrecovered fluorescence for the same cells as panel c. (e) Gαq-V fluorescence recovery in intact cells expressing immobile C-M3R in the absence (n=19) and presence (n=20) of 50 μM sphingosine. (f) Unrecovered fluorescence for the same cells as panel e; **, P<0.001. Data represent mean values ± s.e.m..
Unstructured clusters of basic amino acids often serve to anchor peripheral membrane proteins to the plasma membrane, which is negatively charged due to the abundance of phosphoinositides and phosphatidylserine14,15. To test the possibility that a similar interaction participated in preassembly of M3R and Gq, we neutralized the negative charge of the plasma membrane by applying sphingosine (50 μM). Sphingosine is a weak base that partitions into the plasma membrane and neutralizes the charge of acidic phospholipids, thus decreasing electrostatic attraction of polybasic clusters to the inner plasma membrane surface15. Sphingosine effectively prevented the slowing of Gαq-V mobility by immobile C-M3R (Fig. 5e,f). This result suggests that preassembly requires an electronegative plasma membrane, in addition to a positively charged M3R C terminus.
Polybasic regions are present in several GPCRs
Because polybasic regions similar to that found in M3R were present in other Gq-coupled muscarinic and adrenergic receptors, we tested the ability of two of these receptors to decrease the mobility of Gq-V. Both M5 muscarinic receptors (C-M5R) and α1b adrenergic receptors (C-α1bAR) decreased Gq-V mobility to a similar extent as C-M3R (Supplementary Fig. 9). F90 for Gαq-V was 0.42 ± 0.01 with immobile C-M5R, and 0.40 ± 0.01 with immobile C-α1bAR, suggesting that preassembly is not limited to M3R. We then asked if polybasic regions similar to that found in M3R are a common feature of GPCRs. We searched the sequences of 286 non-olfactory human GPCRs for polybasic regions that contained at least five basic amino acids (R, K or H), each separated from the nearest basic amino acid by no more than 3 intervening residues. We narrowed the search to the C terminus distal to a conserved cysteine residue at the end of helix 8, 10–27 residues after the NPxxY motif (see Methods). In many GPCRs this cysteine residue is covalently modified by addition of a palmitoyl group16. This search identified 21 GPCRs with polybasic regions matching these criteria, all belonging to the class A, rhodopsin-like subfamily (Supplementary Fig. 10). Of these, 13 activate only Gq, 3 couple to both Gq and some other G protein isoform, 3 couple to Gi/o, 1 couples to Gs and Gi/o, and 1 couples to Gs. While pronounced polybasic regions are found in GPCRs that do not couple to Gq, the relatively frequent occurrence of such regions in Gq-coupled receptors suggests that these regions may mediate preassembly with Gq in more cases than those studied here.
The C-terminal polybasic region facilitates M3R signaling
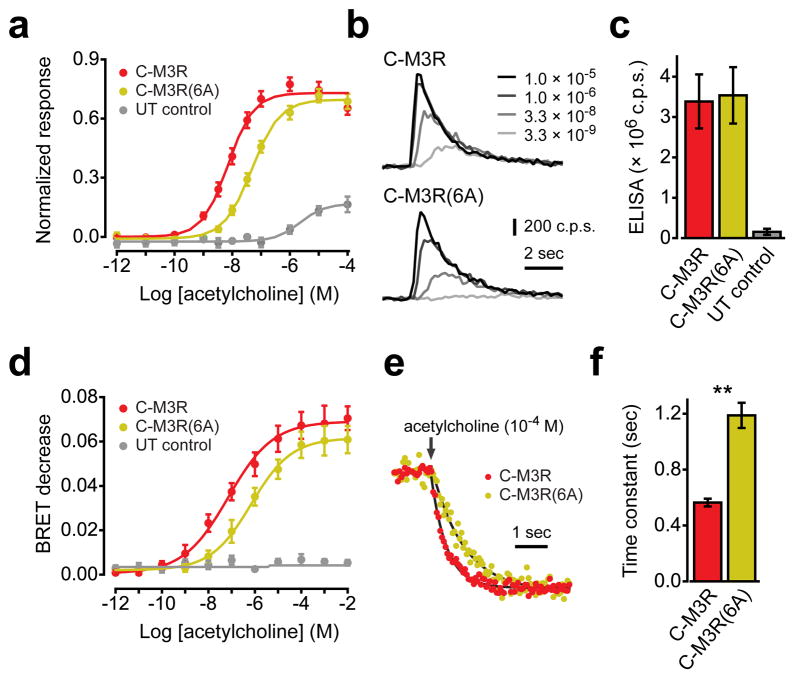
Preassembly of GPCRs and G proteins is often proposed as a mechanism to facilitate rapid activation of G proteins and downstream signaling pathways, in which case disrupting preassembly would be expected to inhibit signaling. In order to test this prediction for M3R and Gq we compared intracellular calcium responses mediated by C-M3R and C-M3R(6A) and endogenous Gq heterotrimers. Intracellular calcium transients were monitored using the calcium-sensitive photoprotein aequorin. Activation of either C-M3R or C-M3R(6A) with acetylcholine (Ach) induced transient concentration-dependent increases in intracellular calcium, as indicated by transient increases in aequorin luminescence (Fig. 6a,b). Introduction of the 6A mutation decreased sensitivity to Ach; the average EC50 for Ach-induced calcium transients was 7 nM in cells expressing C-M3R, and 47 nM in cells expressing C-M3R(6A) (n=5 experiments performed in triplicate; Fig. 6a). Maximal calcium signals mediated by the two receptors were similar. In cells expressing only endogenous Ach receptors the EC50 for Ach-induced calcium transients was >2000 nM, and maximum responses reached only ~20% of those mediated by exogenous receptors, suggesting that calcium responses in cells expressing C-M3R or C-M3R(6A) were largely mediated by exogenous receptors (Fig. 6a). The observed difference in agonist sensitivity was not due to differences in expression of C-M3R and C-M3R(6A) at the cell surface (Fig. 6c). An ELISA assay using an antibody directed against the extracellular ECFP moiety in intact cells indicated that the average ratio of C-M3R(6A) to C-M3R surface expression was 1.05 ± 0.03 (n=12), and varied between 0.83 and 1.27 in individual experiments.
Figure 6. A polybasic region in the M3R C terminus is necessary for efficient activation of Gq.
(a) Normalized aequorin luminescence (calcium) responses from cells expressing C-M3R (n=5), C-M3R(6A) (n=5) or untransfected control cells (UT control; n=4). (b) Exemplary luminescence traces from cells expressing C-M3R or C-M3R(6A) and exposed to a range of acetylcholine concentrations; c.p.s.=photon counts per second. (c) Cell surface expression of C-M3R (n=12) and C-M3R(6) (n=12) quantified by ELISA in intact cells, and compared to signals from untransfected controls (n=7). (d) Decreases in BRET between Gαq-Rluc8 and Gβ1γ2-V in cells expressing C-M3R (n=4), C-M3R(6A) (n=4) or untransfected control cells (n=4). (e) Normalized BRET responses from cells expressing C-M3R or C-M3R(6A) and exposed to 100 μM acetylcholine (arrow). Smooth lines are least squared fits to a single exponential function. (f) Fitted onset time constants from cells expressing C-M3R or C-M3R(6A) (n=4 each, 24 replicates per experiment); **, P<0.001. Data represent mean values ± s.e.m..
The decreased agonist sensitivity of C-M3R(6A)-mediated calcium responses was consistent with the notion that preassembly facilitates activation of G proteins. To more directly assess G protein activation we compared steady-state activation of Gq by C-M3R and C-M3R(6A) by measuring bioluminescence resonance energy transfer (BRET) between Renilla luciferase-labeled Gαq (Gαq-Rluc8) and Gβ1γ2-V in intact cells. Changes in energy transfer between labeled G protein subunits reliably reflect the conformational and/or associational changes that follow heterotrimer activation10,17. Activation of either C-M3R or C-M3R(6A) with Ach induced a concentration-dependent decrease in BRET between Gαq-Rluc8 and Gβ1γ2-V. As was the case for calcium responses, introduction of the 6A mutation decreased sensitivity to Ach in this BRET assay. The average EC50 for the Ach-induced BRET decrease was 71 nM in cells expressing C-M3R, and 696 nM in cells expressing C-M3R(6A) (n=4 experiments performed in quadruplicate; Fig. 6d). Similarly, the average EC50 for the Ach-induced BRET decrease was 36 nM in cells expressing C-M3R, and 282 nM in cells expressing C-M3R(M4ct) (n=2 experiments performed in quadruplicate). No Ach-induced BRET changes were observed in cells transfected only with Gαq-Rluc8 and Gβ1γ2-V, suggesting that endogenous muscarinic receptors were incapable of activating a significant fraction of Gαq-Rluc8:Gβ1γ2-V heterotrimers (Fig. 6d). The decrease in agonist sensitivity in cells expressing C-M3R(6A) is consistent with the suggestion that preassembly facilitates steady-state activation of Gq.
We then took advantage of the high temporal resolution of the BRET assay to investigate the possibility that preassembly increases the rate of Gq activation. We measured BRET between Gαq-Rluc8 and Gβ1γ2-V every 60 ms, before and after addition of a saturating concentration of Ach (100 μM). The low signal-to-noise ratio at this sampling interval meant that substantial signal averaging was required to adequately resolve the time course of Gq activation. The BRET signal approached a new (lower) steady-state after Ach application with a time course that could be fitted well with a single exponential time constant (Fig. 6e). The average time constant for Gq activation was 0.52 ± 0.02 seconds in cells expressing C-M3R, and 1.08 ± 0.06 seconds in cells expressing C-M3R(6A) (n=4 experiments, 24 replicates each) (Fig. 6f). This result is consistent with the hypothesis that preassembly with M3R increases the rate of Gq activation.
DISCUSSION
The steps involved in the formation of productive, coupled complexes between active GPCRs and their cognate heterotrimeric G proteins are beginning to be elucidated3,5,18. Several complementary studies have indicated that receptor activation exposes a pocket in the transmembrane core that provides a binding site for the Gα subunit C terminus19–21. This interaction is blocked by posttranslational modification of the Gα C terminus, or by inverse agonists that promote the inactive state of the receptor22.
This mechanism does not preclude the possibility that inactive GPCRs also interact with G proteins to form distinct inactive-state complexes, prior to productive coupling. Evidence for the existence of “precoupled” or preassembled complexes has been provided by a variety of methods, including several resonance energy transfer studies in live cells9–11. However, similar studies have failed to find evidence of preassembly13,23,24, and in no case is much known about the properties of putative inactive-state complexes. Mutagenesis studies have not identified mutant receptors or G proteins that are defective for preassembly, making it difficult to clearly distinguish inactive-state preassembly from active-state coupling. More importantly, the functional significance of inactive-state preassembly is uncertain, as tools that can prevent this type of interaction have not been available.
In the present study we used FRAP to detect the formation of inactive-state complexes containing M3R and Gq heterotrimers. This method is relatively insensitive to artifacts caused by overexpression, as proteins that do not interact have no detectable effect on each others’ mobility (e.g. M4R and Gq). We found that immobile M3R specifically slowed the lateral mobility of fluorescently-labeled Gq. This slowing was evident if either the Gαq subunit or the Gβγ dimer was labeled, and was not different if the agonist carbachol or the inverse agonist atropine was bound to the receptor. These properties are consistent with the formation of inactive-state (activity-independent) complexes containing M3R and Gq heterotrimers. Our results do not allow us to distinguish between a direct interaction between M3R and Gq heterotrimers and an indirect interaction mediated by an intermediate protein or a membrane microdomain. However, a recent crosslinking study detected several crosslinks between M3R and Gq that persisted in the presence of atropine, supporting the possibility of a direct interaction between inactive M3R and Gq25.
The interaction that we observed was insensitive to the activity state of the M3R, suggesting that inactive-state complexes are mechanistically distinct from active-state complexes. However, this does not rule out the possibility that common regions of the two proteins are involved in both types of interaction. Indeed, the fact that preassembly was specific for a receptor (the M3R) and G protein (Gq) that also form active-state complexes suggests that some properties may be shared. We found that the C terminus of M3R is necessary and sufficient for inactive-state preassembly. The N-terminal portion of the C terminus, including helix 8, has been implicated in active-state coupling of several GPCRs and G proteins26,27, including the M3R4,25. Therefore, it is possible that this region is involved both in inactive-state preassembly and active-state coupling. We found that a polybasic region distal to helix 8 was itself necessary for preassembly, with positive charge being more important than sidechain identity. It is possible that these residues are directly involved in an electrostatic interaction with Gq. Analogous electrostatic steering mechanisms have been shown to be important for rapid assembly of other protein complexes28. However, it is also possible that this positively-charged region interacts with negatively-charged phospholipids to impose a conformation necessary for inactive-state complex formation. In many GPCRs helix 8 is terminated by one or two cysteine residues that are S-palmitoylated, creating a fourth intracellular loop16. In the receptors that we studied these cysteines are followed immediately by a cluster of basic amino acids. Unstructured polybasic regions are frequently found adjacant to covalent lipid modifications, where they are important for targeting and stable attachment of peripheral membrane proteins to the plasma membrane. As examples, this combination is found in many small GTPases, growth associated protein 43 (GAP-43), and G protein α subunits14,29–31. Thus it is conceivable that polybasic regions that follow helix 8 serve to firmly anchor the C terminus to the plasma membrane, and that this function is necessary for preassembly of inactive-state receptor-G protein complexes. In support of this possibility, we found that preassembly required an electronegative plasma membrane, as it was prevented by neutralization of negatively charged phospholipids with sphingosine. However, because sphingosine may also have disrupted other important electrostatic interactions with the plasma membrane31, further studies will be required to determine the precise mechanism of specific M3R-Gq preassembly.
Our data suggest that inactive-state M3R-Gq complexes are transient, as immobile M3R did not completely immobilize Gq heterotrimers, and active-state complexes were demonstrably more stable when guanine nucleotides were depleted. The observation that M3R activation did not change Gq mobility in intact cells is consistent with the expectation that active-state complexes are transient intermediates when guanine nucleotides are available. This observation is also consistent with the suggestion that receptor activation does not trigger dissociation of preassembled receptor-G protein complexes in intact cells10. Further studies will be required to quantitatively estimate the lifetimes of inactive- and active-state M3R-Gq complexes under physiological conditions. It is important to emphasize that we observed slowing of Gq mobility by immobile M3R that were heterologously overexpressed, and that Gq may be more mobile in cells expressing lower levels of M3R. Indeed, we only detected preassembly when C-M3R was more abundant than Gαq-V, whereas native Gα subunits outnumber receptors by a wide margin in most cells32. However, since dissociation of a protein from a complex does not depend on concentration, this caveat does not affect the conclusion that inactive-state M3R-Gq complexes are as stable or more stable than active-state complexes in intact cells, or that M3R preassembles with Gq whereas M4R does not.
Although in this study we focused primarily on the M3R, it seems likely that several other Gq-coupled receptors with polybasic regions distal to helix 8 employ a similar preassembly mechanism. For example, we found that immobile M5 muscarinic receptors and α1b adrenergic receptors, both of which possess pronounced polybasic regions, decrease Gq mobility in a manner similar to immobile M3R. It will be interesting to study receptors that possess polybasic regions and do not activate Gq, to see if these receptors preassemble with their cognate G protein isoforms using a similar mechanism.
The functional significance of GPCR-G protein preassembly has been a longstanding subject of speculation6,7,33. Interest in the organization of G protein signaling components has been fueled by the possibility that preassembly via direct interaction or in discrete domains could impose receptor-G protein specificity, and speed the rate of G protein activation by eliminating (or shortening) the time required for GPCRs and G proteins to collide. In the present study we show that a mutant M3R that does not preassemble with Gq heterotrimers is functionally impaired in a manner consistent with slowed activation of Gq. These findings are consistent with the proposal that preassembly facilitates signaling by obviating or accelerating the collision step. However, we emphasize that alternative explanations can not be ruled out. For example, the 6A mutation that eliminated preassembly may also have affected the formation of active-state complexes, or even the conformational changes that occur during receptor activation. Indeed, if inactive-state complexes represent an intermediate step in the chain of events from ligand binding to G protein activation, it may not be possible to selectively disrupt preassembly without indirectly altering other steps.
If preassembly accelerates G protein activation, the ultimate physiological significance of this affect will depend on which step is rate-limiting for a particular signal transduction pathway in a particular cell. Preassembly will be less important in situations where collision of receptors and G proteins is fast, such as in cells that express an abundance of these proteins. Consequently, the functional importance of preassembly may be greater in native cells than what we observed in cells overexpressing M3R and Gq. Similarly, responses that are rate-limited by much slower downstream effector steps will not be accelerated by GPCR-G protein preassembly34, although steady-state agonist sensitivity may still be increased. Predicting the precise impact of preassembly on signaling will require detailed knowledge of the absolute abundance of several signaling components and their rates of activation, deactivation, association and dissociation35. Alternatively, the results of the present study may facilitate the development of tools that can be used to empirically test the functional significance of GPCR-G protein preassembly in a native system.
METHODS
Plasmid DNA constructs
C-M3R subunits consisted of (starting at the amino terminus) a cleavable signal peptide from human growth hormone, ECFP, and the human M3 muscarinic acetylcholine receptor. C-M4R, C-M5R, and C-α1bAR were constructed in an analogous fashion. Gαq-V and Gαq-Rluc8 were modified from Gαq-citrine, which was provided by Dr. Catherine Berlot (Geisinger Clinic, Danville, PA) 36. The C-TM control construct, C-TM-V and Gβ1γ2-V have been described previously24,37. CD8-aequorin was provided by Dr. David Fulton (Georgia Health Sciences University, Augusta, GA)38. All constructs were made using an adaptation of the QuikChange (Stratagene) mutagenesis protocol and were verified by automated sequencing.
Cell culture and transfection
HEK 293 cells (ATCC) were propagated in plastic flasks and on polylysine-coated glass coverslips according to the supplier’s protocol. Cells were transfected using linear polyethyleneimine and were used for experiments 12–48 hours later. In all cases complementary unlabeled G protein subunits were cotransfected with labeled subunits.
Crosslinking and permeabilization
For avidin-mediated crosslinking cells were rinsed 3 times in buffer containing 150 mM NaCl, 2.5 mM KCl, 10 mM HEPES, 12 mM glucose, 0.5 mM CaCl2, and 0.5 mM MgCl2 (pH 8.0), and incubated at room temperature for 15 minutes in 0.5 mg ml−1 NHS-sulfo-LC-LC-biotin (Thermo Scientific). Cells were washed and incubated for 15 minutes in 0.1 mg ml−1 avidin. For antibody-mediated crosslinking (used for permeabilized cells) cells were rinsed in buffer containing 140 mM potassium gluconate, 5 mM KCl, 10 mM HEPES, 1 mM EGTA, 0.3 mM CaCl2, and 1 mM MgCl2 (pH 7.2), and incubated at room temperature for 5 minutes in a 1:200 dilution of polyclonal rabbit anti-GFP antibody (Invitrogen A11122). Cells were washed and incubated for 5 minutes in a 1:1000 dilution of goat anti-rabbit antibody (Invitrogen B2770). Cells were permeabilized with 1000 U ml−1 α-hemolysin (Sigma H9395). For nucleotide depletion permeabilized cells were incubated in 5 mM KCN for at least 10 minutes before imaging. Changes in ionic strength were accomplished by substituting the indicated concentration of KCl for K gluconate. Low ionic strength (20 mM KCl) buffer was supplemented with mannitol to maintain an osmolarity equivalent to that of high ionic strength (200 mM KCl) buffer.
FRAP
Coverslips were transferred to the stage of a Leica (Wetzlar, Germany) SP2 scanning confocal microscope and imaged using a 63X, 1.4 NA objective. Cells were excited using 458 nm (for CFP), or 512 nm (for venus) laser lines. Low intensity illumination was used during a control (prebleach) period, after which a 4 μm segment of the plasma membrane was photobleached by increasing the laser intensity to 100%. Fluorescence recovery was monitored for 3 minutes using low intensity illumination. Average pixel intensity in the bleached region was corrected for photobleaching, normalized and plotted versus time. The unrecovered fluorescence at 90 seconds (F90) was calculated as F90=(Ipre−I90)/(Ipre−Ipost), where Ipre is fluorescence intensity immediately prior to bleaching, Ipost is intensity immediately after bleaching, and I90 is intensity 90 seconds after bleaching.
Intact cell radioligand binding
After transfection with C-M3R and biotin-avidin crosslinking, intact cells were incubated in DMEM containing [3H] N-methylscopolamine at concentrations ranging from 0.3125 nM to 20 nM for 90 minutes at room temperature. Cells were washed twice with PBS, and surface-bound ligand was extracted with 1 M NaOH for 2 hours. Radioactivity was counted by liquid scintillation spectrometry in 4 ml of Ecoscint A. Nonspecific binding was determined in the presence of 20 μM atropine.
Analysis of GPCR sequences to detect polybasic regions
Uniprot IDs were copied from the IUPHAR GPCR database (http://www.iuphar-db.org/DATABASE/GPCRListForward) and scanned using Scanprosite (http://us.expasy.org/tools/scanprosite/) using the search criterion [NDT]-P-x-x-Y-x(10,27)-C-x(0,10)-[HKR]-x(0,3)-[HKR]-x(0,3)-[HKR]-x(0,3)-[HKR]-x(0,3)-[HKR]. Regions matching this criterion (total of 24) were manually aligned around the cysteine residue. One match (CML1) was discarded because the region identified was not located in the C terminus. Two additional matches were discarded because the identified polybasic regions started within helix 8 (GASR and NK1R, both Gq-coupled).
ELISA
Cells were washed gently with PBS and incubated for 20 minutes in 1:500 or 1:1000 anti-GFP antibody. Cells were harvested, washed by centrifugation, and incubated for 20 minutes in 1:1000 peroxidase-conjugated goat anti-rabbit antibody (Thermo Scientific 32460). Cells were washed (3X) by centrifugation, resuspended in PBS, and mixed with chemiluminscent substrate.
Aequorin calcium release assay
Cells transfected with CD8-aequorin were washed with PBS and incubated in DMEM containing 0.1% (w/v) BSA and 10 μM coelenterazine h (Nanolight) in the dark for 2 hours. Cells were harvested and resuspended in DMEM with 0.1% BSA and kept in the dark. Suspended cells (0.1 ml) were injected into wells containing 0.1 ml of PBS with twice the final concentration of acetylcholine during 20 second luminescence measurements.
Background-subtracted luminescence was normalized to maximal signals produced by cell lysis according to normalized response=(Lx−Lback)/(Lmax−Lback), where Lx is integrated luminescence after injection into drug-containing PBS, Lback is integrated luminescence after injection into drug-free PBS, and Lmax is integrated luminescence after injection into triton X-containing PBS.
BRET
Cells were washed with PBS and harvested by trituration. For steady-state measurements cells were transferred to black 96-well plates containing PBS and acetylcholine. Coelenterazine h (5 μM) was added to all wells immediately prior to making measurements. Steady-state luminescence measurements were made using a Mithras LB940 photon-counting plate reader (Berthold Technologies). Raw BRET signals were calculated as the emission intensity at 520–545 nm divided by the emission intensity at 475–495 nm. For kinetic measurements cells were transferred to 96-well plates containing only PBS and coelenterazine h, and concentrated acetylcholine was injected during 6 second luminescence measurements. Donor and acceptor luminescence were acquired simultaneously every 60 ms using a BMG Polarstar plate reader (BMG Labtech).
Statistical analysis
Data are presented as mean ± s.e.m. for the indicated number of experiments. When more than two groups were compared hypothesis testing was done using one-way analysis of variance (ANOVA), followed by Bonferroni means comparison; reported values of P reflect the Bonferroni comparison to the control (C-TM) unless otherwise indicated. When two groups were compared hypothesis testing was done using the unpaired Student’s t-test.
Supplementary Material
Acknowledgments
We thank Greg Digby and Sudha Kuravi for invaluable assistance. This work was supported by NIH grants GM076167 (G.W.), GM096762 and GM078319 (N.A.L.).
Footnotes
Author contributions
K.Q., designed and performed experiments and analyzed data; C.D., performed experiments; G.W., designed experiments and analyzed data; N.A.L., designed experiments, analyzed data and wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 2.De Lean A, Stadel JM, Lefkowitz RJ. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J Biol Chem. 1980;255:7108–7117. [PubMed] [Google Scholar]
- 3.Oldham WM, Hamm HE. Structural basis of function in heterotrimeric G proteins. Q Rev Biophys. 2006;39:117–166. doi: 10.1017/S0033583506004306. [DOI] [PubMed] [Google Scholar]
- 4.Wess J, et al. Structural basis of receptor/G protein coupling selectivity studied with muscarinic receptors as model systems. Life Sci. 1997;60:1007–1014. doi: 10.1016/s0024-3205(97)00041-6. [DOI] [PubMed] [Google Scholar]
- 5.Tesmer JJ. The quest to understand heterotrimeric G protein signaling. Nat Struct Mol Biol. 2010;17:650–652. doi: 10.1038/nsmb0610-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neubig RR. Membrane organization in G-protein mechanisms. FASEB J. 1994;8:939–946. doi: 10.1096/fasebj.8.12.8088459. [DOI] [PubMed] [Google Scholar]
- 7.Hein P, Bunemann M. Coupling mode of receptors and G proteins. Naunyn Schmiedebergs Arch Pharmacol. 2009;379:435–443. doi: 10.1007/s00210-008-0383-7. [DOI] [PubMed] [Google Scholar]
- 8.Rebois RV, Hebert TE. Protein complexes involved in heptahelical receptor-mediated signal transduction. Receptors Channels. 2003;9:169–194. [PubMed] [Google Scholar]
- 9.Gales C, et al. Real-time monitoring of receptor and G-protein interactions in living cells. Nat Methods. 2005;2:177–184. doi: 10.1038/nmeth743. [DOI] [PubMed] [Google Scholar]
- 10.Gales C, et al. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat Struct Mol Biol. 2006;13:777–786. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
- 11.Nobles M, Benians A, Tinker A. Heterotrimeric G proteins precouple with G protein-coupled receptors in living cells. Proc Natl Acad Sci U S A. 2005;102:18706–18711. doi: 10.1073/pnas.0504778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolkovsky AM, Levitzki A. Mode of coupling between the beta-adrenergic receptor and adenylate cyclase in turkey erythrocytes. Biochemistry. 1978;17:3795. doi: 10.1021/bi00611a020. [DOI] [PubMed] [Google Scholar]
- 13.Kuravi S, Lan TH, Barik A, Lambert NA. Third-party bioluminescence resonance energy transfer indicates constitutive association of membrane proteins: application to class A G-protein-coupled receptors and G-proteins. Biophys J. 2010;98:2391–2399. doi: 10.1016/j.bpj.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heo WD, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 16.Torrecilla I, Tobin AB. Co-ordinated covalent modification of G-protein coupled receptors. Curr Pharm Des. 2006;12:1797–1808. doi: 10.2174/138161206776873716. [DOI] [PubMed] [Google Scholar]
- 17.Lohse MJ, Hoffmann C, Nikolaev VO, Vilardaga JP, Bunemann M. Kinetic analysis of G protein-coupled receptor signaling using fluorescence resonance energy transfer in living cells. Adv Protein Chem. 2007;74:167–188. doi: 10.1016/S0065-3233(07)74005-6. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann KP, et al. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci U S A. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 21.Ye S, et al. Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature. 2010;464:1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- 22.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hein P, Frank M, Hoffmann C, Lohse MJ, Bunemann M. Dynamics of receptor/G protein coupling in living cells. EMBO J. 2005;24:4106–4114. doi: 10.1038/sj.emboj.7600870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin K, Sethi PR, Lambert NA. Abundance and stability of complexes containing inactive G protein-coupled receptors and G proteins. FASEB J. 2008;22:2920–2927. doi: 10.1096/fj.08-105775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, et al. Structural basis of G protein-coupled receptor-G protein interactions. Nat Chem Biol. 2010;6:541–548. doi: 10.1038/nchembio.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst OP, et al. Mutation of the fourth cytoplasmic loop of rhodopsin affects binding of transducin and peptides derived from the carboxyl-terminal sequences of transducin alpha and gamma subunits. J Biol Chem. 2000;275:1937–1943. doi: 10.1074/jbc.275.3.1937. [DOI] [PubMed] [Google Scholar]
- 27.Marin EP, et al. The amino terminus of the fourth cytoplasmic loop of rhodopsin modulates rhodopsin-transducin interaction. J Biol Chem. 2000;275:1930–1936. doi: 10.1074/jbc.275.3.1930. [DOI] [PubMed] [Google Scholar]
- 28.Hemsath L, Dvorsky R, Fiegen D, Carlier MF, Ahmadian MR. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol Cell. 2005;20:313–324. doi: 10.1016/j.molcel.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Pedone KH, Hepler JR. The importance of N-terminal polycysteine and polybasic sequences for G14alpha and G16alpha palmitoylation, plasma membrane localization, and signaling function. J Biol Chem. 2007;282:25199–25212. doi: 10.1074/jbc.M610297200. [DOI] [PubMed] [Google Scholar]
- 30.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 31.Crouthamel M, Thiyagarajan MM, Evanko DS, Wedegaertner PB. N-terminal polybasic motifs are required for plasma membrane localization of Galpha(s) and Galpha(q) Cell Signal. 2008;20:1900–1910. doi: 10.1016/j.cellsig.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrom RS, Post SR, Insel PA. Stoichiometry and compartmentation in G protein-coupled receptor signaling: implications for therapeutic interventions involving G(s) J Pharmacol Exp Ther. 2000;294:407–412. [PubMed] [Google Scholar]
- 33.Hille B. G protein-coupled mechanisms and nervous signaling. Neuron. 1992;9:187–195. doi: 10.1016/0896-6273(92)90158-a. [DOI] [PubMed] [Google Scholar]
- 34.Jensen JB, Lyssand JS, Hague C, Hille B. Fluorescence changes reveal kinetic steps of muscarinic receptor-mediated modulation of phosphoinositides and Kv7.2/7.3 K+ channels. J Gen Physiol. 2009;133:347–359. doi: 10.1085/jgp.200810075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falkenburger BH, Jensen JB, Hille B. Kinetics of M1 muscarinic receptor and G protein signaling to phospholipase C in living cells. J Gen Physiol. 2010;135:81–97. doi: 10.1085/jgp.200910344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes TE, Zhang H, Logothetis DE, Berlot CH. Visualization of a functional Galpha q-green fluorescent protein fusion in living cells. Association with the plasma membrane is disrupted by mutational activation and by elimination of palmitoylation sites, but not be activation mediated by receptors or AlF4. J Biol Chem. 2001;276:4227–4235. doi: 10.1074/jbc.M007608200. [DOI] [PubMed] [Google Scholar]
- 37.Hynes TR, et al. Visualization of G protein betagamma dimers using bimolecular fluorescence complementation demonstrates roles for both beta and gamma in subcellular targeting. J Biol Chem. 2004;279:30279–30286. doi: 10.1074/jbc.M401432200. [DOI] [PubMed] [Google Scholar]
- 38.Church JE, Fulton D. Differences in eNOS activity because of subcellular localization are dictated by phosphorylation state rather than the local calcium environment. J Biol Chem. 2006;281:1477–1488. doi: 10.1074/jbc.M505968200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.