Conspectus
RNA owes its role as the central conduit for gene expression to its ability to encode information at two levels: in its linear sequence and in its ability to fold back on itself to form complex higher-order structures. Understanding the global structure-function interrelationships mediated by RNA remains a great challenge in molecular and structural biology. We describe evolving work in our laboratory focused creating facile, generic, quantitative, accurate, and highly informative approaches for understanding RNA structure at nucleotide resolution for RNAs of wide ranges of size s and complexities in biological environments.
The core innovation derives from our discovery that the nucleophilic reactivity of the ribose 2′-hydroxyl in RNA is gated by local nucleotide flexibility. The 2′-hydroxyl is reactive at conformationally flexible positions but is unreactive at nucleotides constrained by base pairing. Sites of modification in RNA can be efficiently detected using either primer extension or protection from exoribonucleolytic degradation. This technology is now called SHAPE, for selective 2′-hydroxyl acylation analyzed by primer extension (or protection from exoribonuclease). SHAPE reactivities are largely independent of nucleotide identity but correlate closely with model-free measurements of molecular order. The simple SHAPE reaction is thus a robust, nucleotide-resolution, biophysical measurement of RNA structure. SHAPE can be used to provide an experimental correction to RNA folding algorithms and, in favorable cases, yield kilobase-scale secondary structure predictions with high accuracies.
SHAPE chemistry is based on very simple reactive carbonyl centers that can be varied to yield slow- and fast-reacting reagents. Differential SHAPE reactivities can be used to detect specific RNA positions with slow local nucleotide dynamics. These positions, which are often in the C2′-endo conformation, have the potential to function as molecular timers that regulate RNA folding and function. In addition, fast-reacting SHAPE reagents can be used to visualize RNA structural biogenesis and RNA-protein assembly reactions in 1 second snapshots via very straightforward experiments.
Application of SHAPE to challenging problems in biology has revealed surprises in well-studied systems and has indentified new regions likely to have critical functional roles based on their high levels of RNA structure. For example, SHAPE analysis of large RNAs like authentic viral RNA genomes suggests that RNA structure organizes regulatory motifs and regulates splicing, protein folding, genome recombination, and ribonucleoprotein assembly. SHAPE has also revealed the limitations of the hierarchical model for RNA folding. Continued development and application of SHAPE technologies will advance our understanding of the many ways in which the genetic code is expressed through the underlying structure of RNA.
Introduction
The central role of RNA in biology reflects its unique abilities to encode genetic information in its linear sequence and to fold into structures that contribute to diverse biological functions. RNA structure may selectively present or conceal access to the information stored in a linear RNA sequence. Alternatively, the structures themselves may be important; these structures can be as simple as the small hairpins processed to become miRNAs or as sophisticated as ligand binding switches, complex protein binding elements, and catalytic active sites.
The structure of an individual RNA is determined by the complex pattern of interactions among nucleotide bases at both secondary and tertiary structure levels. The secondary structure consists of base-pairing interactions that form helices and define loops in individual structural elements. Tertiary interactions are longer-range and bring individual structural elements together to create higher-order structures. There are strong hints that transcription initiation codes1, splicing codes2,4, translation initiation codes3, and protein folding codes4,5 are all expressed in the form of RNA structure. Thus, the raw genetic information encoded in the primary sequence of RNA is communicated by additional information encoded in the RNA secondary and tertiary structure.
We have only begun to glimpse these RNA codes because the structures of the vast majority of potential regulatory and functional elements in RNA are unknown. The current situation resembles playing on a complex jungle gym with one’s eyes closed. Experience in our laboratory emphasizes the importance of the following principles. First, each RNA has many possible secondary structures, but only a few are highly populated. Second, biologically important differences between structures can be subtle. Thus, identifying populated states of an RNA requires structural information for every nucleotide. Third, short fragments of long RNAs often do not fully recapitulate the structure that exists in the authentic RNA. Moreover, folding is subject to very strong “end effects” such that either omitting an outlying element or adding flanking sequences may induce misfolding. Thus, full-length, biologically active RNAs should be studied. Fourth, the fine details and dynamics that contribute to RNA folding are important. Thus, sequence specificities, time dependencies, and other idiosyncrasies of experiments designed to probe the RNA structure must be well understood. In this review, we discuss the applications of selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), a collection of technologies capable of measuring local nucleotide flexibility at single-nucleotide resolution for RNAs of arbitrary size in diverse biological environments6.
Simple, Generic, Covalent Chemistry at the RNA 2′-Hydroxyl
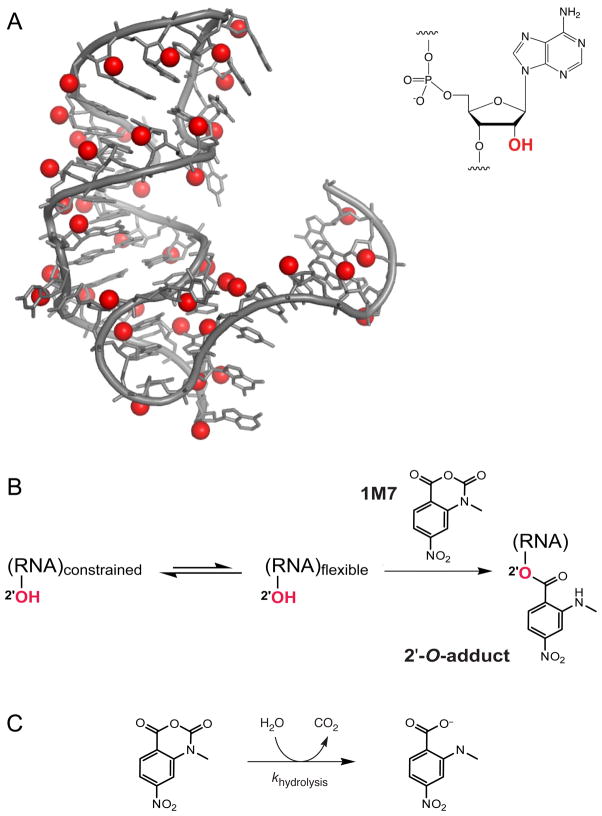
Our initial vision was to find a simple, generic approach to probe simultaneously the structure of every nucleotide within an RNA6. Early work from our lab showed that chemical reactivity at the 2′-ribose position (Figure 1A) is exquisitely sensitive to the local nucleotide environment7–10. We discovered that a wide variety of electrophilic reagents, especially anhydrides and acyl cyanides, react at the 2′-hydroxyl group, and that reactivities are strongly gated by local nucleotide flexibility (Figure 1B). Flexible nucleotides sample multiple conformations, a few of which strongly facilitate nucleophilic attack by the 2′-hydroxyl. The reactivity of individual nucleotides is determined by electrostatic communication between the 2′-OH and adjacent 3′-phosphate diester6,9 and other features intrinsic to favorable ribose conformations(McGinnis and Weeks, unpublished results).
Figure 1. The RNA 2′-hydroxyl group and SHAPE chemistry.
(A) Position of the 2′-hydroxyl group. (B) Mechanism of the SHAPE reaction to form 2′-O-adducts at flexible nucleotides. (C) Mechanism of the parallel hydrolysis reaction.
Electrophilic reagents capable of reacting with RNA 2′-hydroxyl groups also undergo simultaneous inactivation by hydrolysis with water (Figure 1C). This competing hydrolysis reaction imparts three advantages to SHAPE experiments. First, the hydrolysis reaction makes RNA probing extremely straightforward to perform. The hydroxyl-selective electrophile either reacts with RNA or is inactivated by hydrolysis: No explicit quench step is required. Second, although reaction with RNA is the experimental focus of SHAPE, the solution lifetime of a given SHAPE reagent is determined primarily by reagent hydrolysis with ~55 M water. Thus, SHAPE modification of RNA is insensitive to both the RNA concentration and to complications due to additional cross-reacting components, including buffer components, small molecules, or proteins. Third, as will be outlined below, hydrolysis rates for distinct SHAPE reagents vary over a large range and by comparing reactivities of different reagents it is possible to examine RNA folding kinetics and dynamics in very straightforward experiments.
The Basic SHAPE Experiment
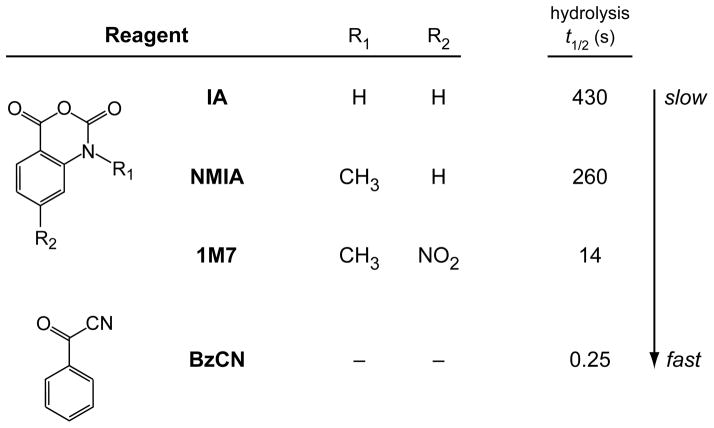
RNA structure analysis involves treatment of an RNA with a SHAPE reagent, such as isatoic anhydride (IA), N-methyl isatoic anhydride (NMIA), 1-methyl-7-nitro-isatoic anhydride (1M7), or benzoyl cyanide (BzCN) (Scheme 1)6,11,12. Reaction results in the selective formation of covalent adducts at the 2′-ribose position of flexible nucleotides. Reaction conditions are controlled to yield sparse modification of the RNA, typically modification of 1 in 300 nucleotides. The detected signal thus reflects uncorrelated adduct-forming events on unmodified RNA or in unmodified regions of a long RNA. The reaction is allowed to proceed until the reagent is consumed by either reaction with RNA or by hydrolysis with water. For the general analysis of RNA structure, the 1M7 SHAPE reagent11(Figure 1B) is easy to handle, reacts in ~1 minute, and reports RNA secondary and tertiary interactions accurately.
Scheme 1.
A toolbox of useful SHAPE reagents.
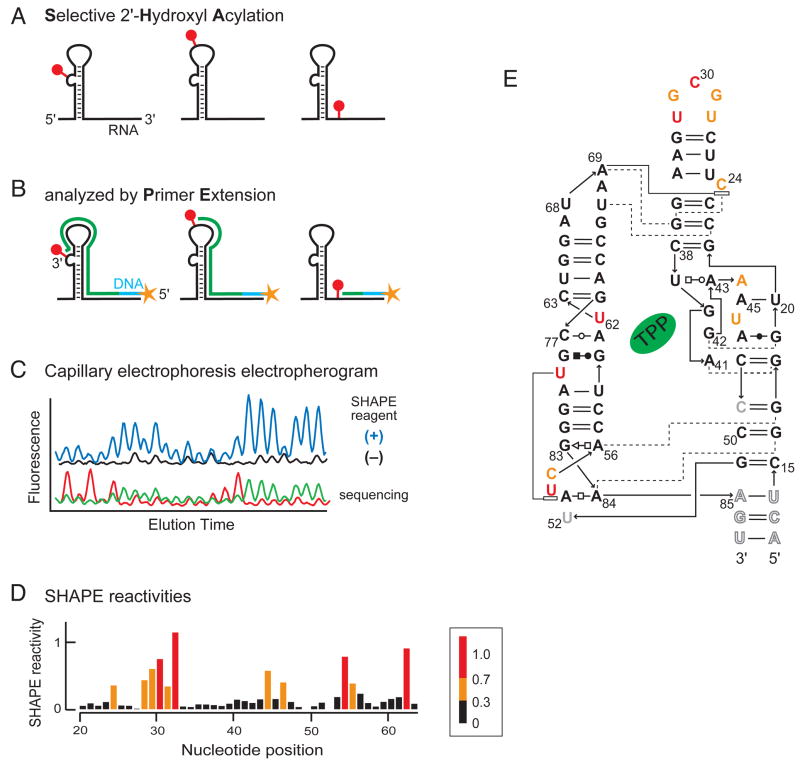
Following selective 2′-hydroxyl acylation (Figure 2A), covalent SHAPE adducts are detected by reverse transcriptase-mediated primer extension6,13. DNA synthesis by reverse transcriptase stops one nucleotide prior to the position of an adduct (Figure 2B)6,10. The length of each cDNA reports the site of a SHAPE modification in the original RNA. A control primer extension reaction is performed in parallel using unmodified RNA to locate sites of natural reverse transcriptase pausing and pre-existing RNA degradation. Peaks in both the modified and no-reagent control lanes are assigned by comparison with sequencing ladders(Figure 2C). Subtraction of the intensity of modified RNA peaks from intensities of no-reagent control peaks yields a reactivity profile (Figure 2D)13,14. Nucleotides constrained by base-pairing and tertiary interactions have low SHAPE reactivities whereas single-stranded and unconstrained nucleotides have higher reactivities, as illustrated for the TPP riboswitch aptamer domain(Figure 2E).
Figure 2. Overview of the SHAPE Experiment.
(A) RNA is selectively modified (red dots) at flexible nucleotides in an RNA. (B) Positions of adduct formation are detected by primer extension. (C) Primer extension products from the experimental, no-reagent control, and sequencing markers are resolved by capillary electrophoresis. (D) Electropherograms are computationally deconvoluted to yield normalized SHAPE reactivities(see scale). (E) Superposition of SHAPE reactivities on a secondary structure model for the TPP riboswitch aptamer domain16.
High-throughput SHAPE can be performed using fluorescently labeled primers and capillary electrophoresis using a commercial DNA sequencing instrument15 and analyzed using custom software14. A single high-throughput SHAPE experiment typically provides data on several hundred RNA nucleotides and is particularly well suited for analyzing the structure of large RNAs15. 2′-O-adducts can also be detected by their ability to protect an RNA from degradation by a 3′ → 5′ exoribonuclease (RNase-detected SHAPE)16. RNase-detected SHAPE allows analysis of short RNAs and nucleotides very close to the ends of an RNA.
SHAPE as a Quantitative Measure of Nucleotide Flexibility
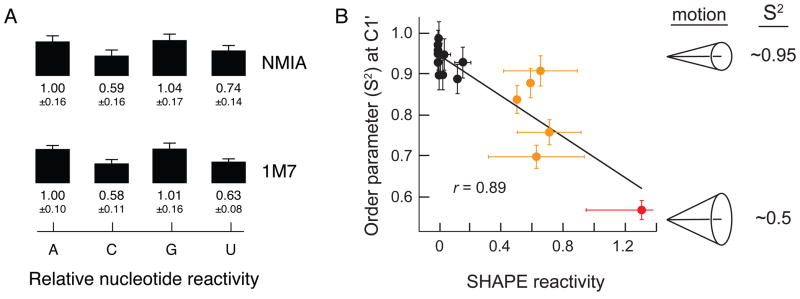
Our laboratory has pursued three classes of experiments to place SHAPE chemistry on a rigorous biophysical foundation. First, analysis of nucleotide reactivity preferences for NMIA and 1M7 shows that adenosine and guanine react approximately 1.6-fold more rapidly than do cytosine and uridine (Figure 3A). These small, but statistically significant, differences appear to have no effect on the ability of SHAPE to measure quantitative conformational features at all nucleotides in an RNA. Variations in nucleotide reactivity for SHAPE reagents are much smaller than the biases found for other chemical probes that form covalent adducts with RNA 17.
Figure 3. SHAPE reactivities are independent of nucleotide identity and correlate quantitatively with local nucleotide flexibility.
(A) Intrinsic nucleotide-specific reactivities for NMIA and 1M717. (B) Correlation between the model-free generalized order parameter, S2, and SHAPE reactivity in the U1A-binding RNA element20 Low, medium, and high SHAPE reactivities are emphasized in black, yellow and red, respectively.
Second, nucleotide-resolution dynamics in RNA can be measured by NMR using the model-free generalized order parameter, S2,18,19, which varies from 0.0 (fully disordered) to 1.0 (fully ordered). Nucleotides that are highly ordered have large S2 values (~0.95), whereas nucleotides that sample a larger conformational space have much lower S2 values (~0.5) 19. SHAPE and NMR analyses were performed on three well characterized RNAs20. Nucleotide resolution SHAPE reactivities and the NMR-derived S2 parameter correlate strongly. For example, for the RNA regulatory element that binds the U1A protein, r is 0.89 (Figure 3B). This correlation is impressive given that S2 measurement are derived primarily from NMR measurements on the picosecond to nanosecond timescale, whereas SHAPE is likely influenced by these rapid motions and by motions on a slower time scale. From these experiments, we conclude that SHAPE measures spatial disorder at single nucleotide resolution.
Third, it was formally possible that SHAPE reactivity might be modulated by the physical accessibility of the 2′-hydroxyl group. Although this group is solvent accessible in an isolated helix, it is often buried in folded RNA structures (Figure 1A). Multiple experiments on simple duplexes and on RNAs with complex tertiary structures have shown there is essentially no relationship between SHAPE reactivities and solvent accessibility4,6,20–22. Critically, many fully buried, but unconstrained, nucleotides react readily with SHAPE reagents. SHAPE thus probes the local nucleotide flexibility of RNA nucleotides regardless of solvent accessibility.
Accurate SHAPE-directed RNA Secondary Structure Prediction
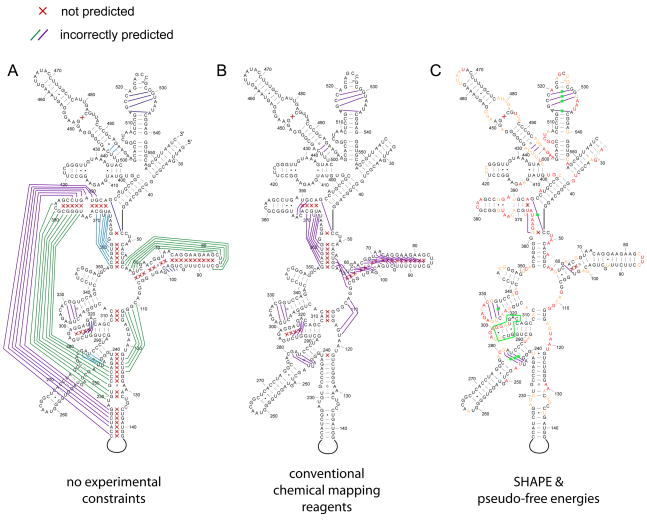
Most computational methods predict RNA secondary structure by approximating the free energy of structural motifs as the sum of individual base-pairing energies within these motifs (reviewed in 23,24). This works well for simple structures, especially hairpins, but is not accurate for large RNAs. We lack thermodynamic parameters for all possible sequence elements, do not understand the contributions of higher-order interactions to free energy, and cannot predict the influences of RNA folding kinetics or interactions with proteins or small molecules. For example, when the secondary structure of the E. coli 16S rRNA is predicted based on sequence alone using one of the most accurate programs currently available, RNA structure25,26, only 50% of accepted base pairs are predicted correctly (Figure 4A)27. Accuracies can be improved by incorporation of experimental information into the prediction algorithm28. In general, data obtained from small chemical probes improves prediction accuracy to a much greater extent than data generated by RNase enzymes29,30. Prediction of 16S rRNA structure improves to 72% upon incorporation of data from traditional chemical probing reagents (Figure 4B)27. However, this structure still contains serious errors.
Figure 4. Comparison of RNA secondary structure predictions for the 5′ domain of E. coli 16S rRNA.
Using (A) no experimental constraints, (B) conventional chemical probing reagents, or (C) SHAPE reactivity-based pseudo free energy change terms (ΔGSHAPE) as constraints27. Base pairs in the accepted structure that are missing in the prediction (×) and base pairs that are predicted incorrectly (green and blue lines) are indicated. Green boxes and dots indicate areas in the co-variation structure where SHAPE reactivities support formation of alternate base pairs.
As outlined above, SHAPE yields a true biophysical measurement of RNA dynamics. We thus converted SHAPE reactivities into a pseudo-free energy change term (ΔGSHAPE) and used this as an experimental correction to an RNA folding algorithm. Using SHAPE pseudo-free energies derived from reaction with the 1M7 reagent, the structures of 16S rRNA (Figure 4C) and a set of smaller RNAs ranging in size from 75–155 nt are predicted with accuracies of ~95% 27. In a few areas (Figure 4C), SHAPE analysis suggests that the 16S rRNA may adopt alternative conformations not found in the accepted model. In general, SHAPE-directed RNA secondary structure prediction has proven highly accurate; additional innovations will be required to predict structures in which only a few nucleotides are unconstrained and thus reactive11,27.
Structural Surprises
The nucleotide-resolution data produced by SHAPE has consistently yielded structural surprises. Before showing examples in which SHAPE supports alternative models of secondary structure and folding, we do want to emphasize that SHAPE cannot prove that any of the proposed RNA structures are correct. However, any proposed model must be substantially compatible with SHAPE analyses.
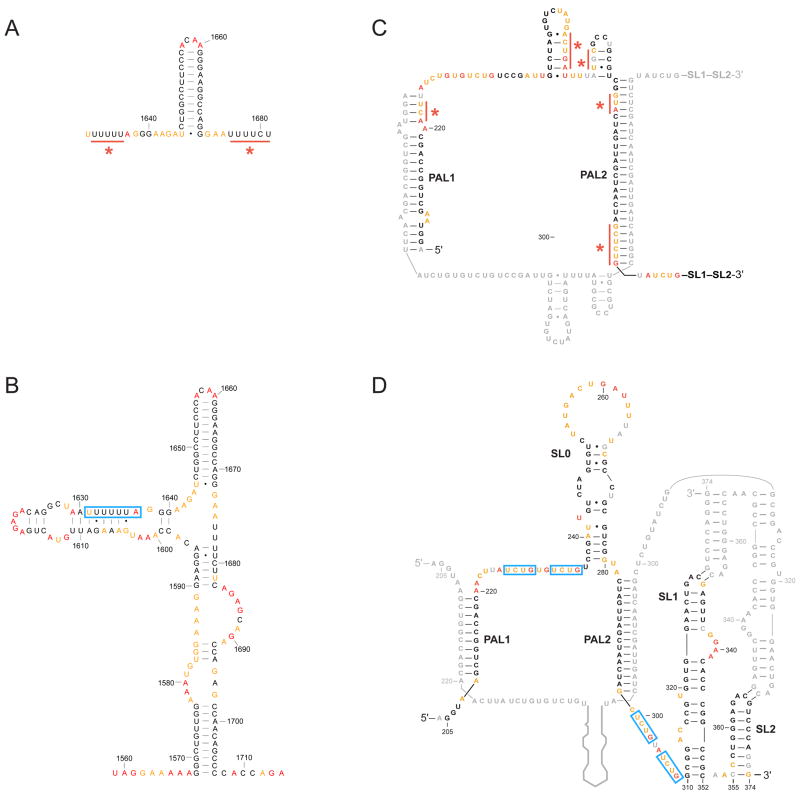
The frameshift element in the HIV-1 RNA genome causes a -1 frameshift to enable the translation of the HIV-1 Pol gene31. This element has traditionally been modeled as a simple stem-loop downstream of a single-stranded, poly-U slippery sequence (Figure 5A). SHAPE analysis of an intact, full-length HIV-1 genomic RNA revealed that several regions proposed to be single stranded actually had low SHAPE reactivities (Figure 5A, red bars)4. SHAPE-derived pseudo-free energies suggest that the slippery sequence base pairs to an upstream sequence and the complete regulatory signal forms a large branched RNA domain (Figure 5B)4. The revised model is consistent with previous findings that the HIV-1 frameshift element required additional sequences downstream of the simple stem-loop for full activity32,33. Phylogenetic analysis revealed that the structure proposed based on SHAPE data is conserved among divergent HIV-1 isolates4.
Figure 5. SHAPE reactivities support revised RNA secondary structure models.
(A) Original model for the HIV-1 frameshift element31 and (B) revised model based on S HAPE data4. (C) Initial model for the viral dimerization and packaging domain of MuLV34 and (D) SHAPE-supported model37. Red bars and asterisks highlight areas where the SHAPE data are not consistent with a secondary structure model. Notable functional elements in the SHAPE-supported structures are highlighted by blue boxes.
In a second example, the RNA genome of Murine Leukemia Virus (MuLV) forms a dimer during a critical step in virion packaging. The dimer structure is minimally composed of two palindromic sequences (PAL1 and PAL2) that form intermolecular duplexes and two stem loops (SL1 and SL2) that form loop-loop interactions (reviewed in 34,35). SHAPE analysis of the MuLV RNA domain using authentic genomic RNA revealed several regions in which nucleotides proposed to base pair actually had high SHAPE reactivities (Figure 5C)35–37. The SHAPE-directed model of the dimerization domain suggests that the PAL1 and PAL2 duplexes are significantly shorter than traditionally drawn34 and that many of the nucleotides between PAL1 and PAL2 form a large, bulged stem-loop structure, SL0 (Figure 5D)37. Subsequent work showed that the two sets of UCUG sequences, which are exposed and flanked by helices in the SHAPE-directed model but partially obscured in the prior model, constitute the viral packaging signal38.
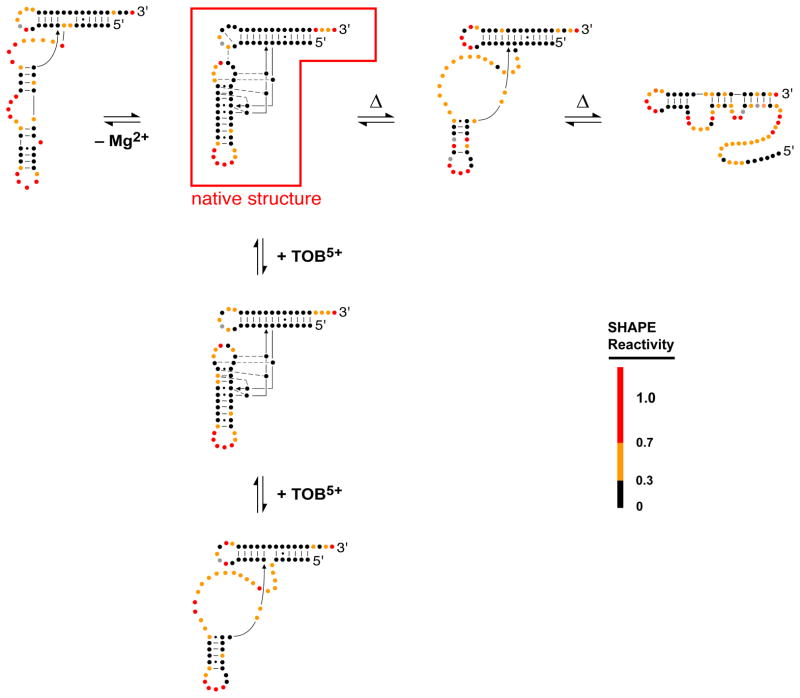
In a third example, we monitored the unfolding of tRNAAsp transcripts as a function of three distinct perturbations: reduced Mg2+ concentration, increased temperature, and addition of the cationic antibiotic tobramycin39,40. Although presumed to be simple, the SHAPE data revealed that the energy landscape for the folding of this molecule contains many structurally diverse states: Each of these three conditions denatured the tRNA in a unique way and yielded distinct end structures (Figure 6). These studies reveal that the folding of even a small RNA molecule can be very complex and non-hierarchical.
Figure 6. Analysis of complex RNA structural transitions by SHAPE.
Structural changes in tRNAAsp induced by varying the Mg 2+concentration, upon heating (Δ), or upon binding by tobramycin are shown39,40.
These three examples illustrate principles that apply broadly to the analysis of RNA structure using SHAPE. First, many important elements of prior models remain key features in the SHAPE-directed analysis, for example, the 1660 hairpin in HIV-131, the UCUG elements in the dimerization domain of MuLV41, and the sensitivity of tRNAAsp structure to ligand binding. Second, SHAPE reveals structural details that may result in revision of accepted models. The revised structural models are substantially consistent with prior functional data, but these data are interpreted in a different way. Third, elements within SHAPE-directed RNA structure models sometimes do not correlate perfectly with the SHAPE data. This reflects, in part, that multiple structures can be formed at equilibrium and that a single model may be an over simplification37,40,42. In addition, pseudoknots and other tertiary interactions need to be better accounted for in our algorithms.
Analysis of RNA Dynamics using SHAPE
In aqueous solution, SHAPE reagents react with RNA and also undergo inactivation by hydrolysis (Figure 1C). The rate of reaction with RNA is proportional to the rate of hydrolysis. Our laboratory has created a tool kit of SHAPE reagents (Scheme 1) that makes it possible to study the local folding dynamics of individual RNA nucleotides. There are two general approaches for using SHAPE to obtain time-resolved information regarding RNA folding: by differential reactivity and by using fast-reacting reagents.
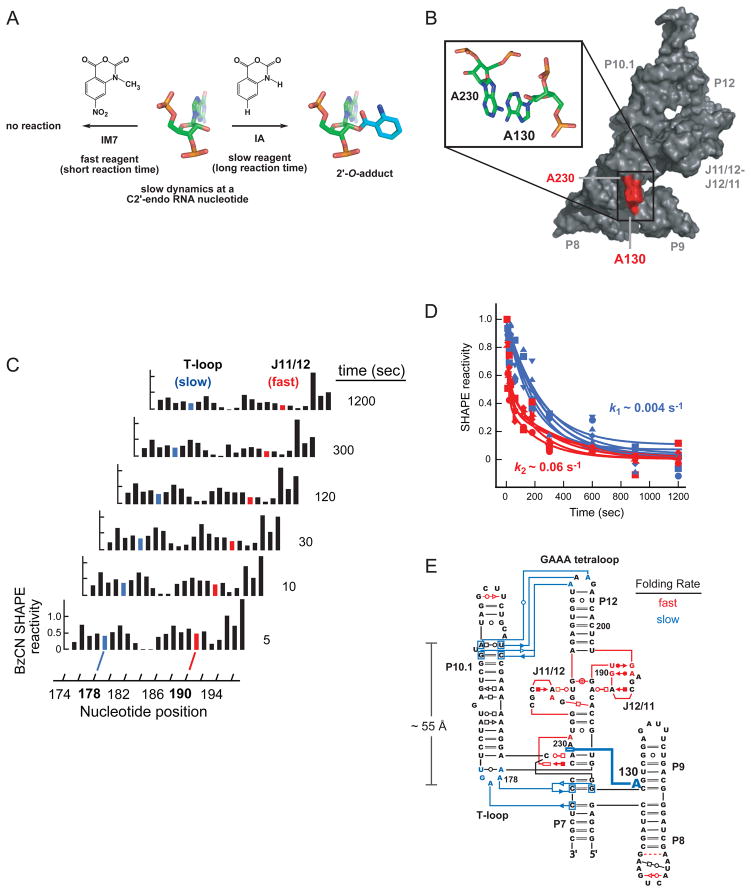
When differential reactivities of slow- versus fast-reacting SHAPE reagents, such as IA versus 1M7 (Scheme 1) are compared, most nucleotides have identical reactivities43,44. However, a critical subset is reactive towards the slow reagent but unreactive towards the fast reagent (Figure 7A). These nucleotides exhibit very slow local nucleotide dynamics with half-lives on the order of minutes and many adopt the C2′-endo conformation. Nucleotides with a C2′-endo conformation are much less prevalent than C3′-endo nucleotides but are highly over-represented in regions of RNA molecules with complex tertiary structures44,45. One such nucleotide is A130 in the specificity domain of the B. subtilis RNase P ribozyme. A130 shows strong differential SHAPE reactivity, adopts the C2′-endo conformation, and mediates a critical tertiary interaction by stacking with nucleotide A230 (Figure 7B). These two nucleotides play a critical role in recognition of the tRNA substrate46,47.
Figure 7. Measuring RNA dynamics by SHAPE.
(A) Use of fast (1M7) and slow (IA) reagents to detect nucleotides with slow conformational dynamics43. (B) Structural context of A130, which exhibits slow dynamics and functions as a molecular timer for folding of an RNase P specificity domain44. (C) Time-resolved SHAPE reactivities and (D) representative time–progress curves. (E) Model for time-resolved tertiary folding of the B. subtillis RNase P specificity domain12. Nucleotides that fold in the fast verses slow phase are emphasized in red and blue, respectively, in panels C–E.
The time-frame of many RNA folding events is on the order of a few seconds to minutes. Benzyl cyanide (BzCN, Scheme 1) reacts completely in 1 second and is used in time-resolved SHAPE12,48 to take snap-shots of RNA structure (Figure 7C). Time-resolved SHAPE showed that the tertiary folding of the RNase P domain proceeds in two distinct, kinetically significant steps (Figure 7D)12. Local tertiary structures involving the J11/12 and J12/11 loops fold at ~0.06 sec−1. In contrast, inter-domain tertiary interactions, like that involving A130 and which span 55 Å, fold much more slowly (~0.004 sec−1; Figure 7E). Strikingly, deleting A130 sped up folding of the whole domain by an order of magnitude44. Together, differential and time-resolved SHAPE support the general model that certain C2′-endo nucleotides function as molecular timers to control RNA folding kinetics.
RNA-Protein Interactions
SHAPE is a robust tool for analyzing RNA structure in biologically important RNA-protein complexes (RNPs). If a protein binds to a simple, single-stranded element, SHAPE can identify the precise binding site directly. Conversely, protein binding can promote large-scale structural rearrangement and thereby change SHAPE reactivity in many regions of an RNA.
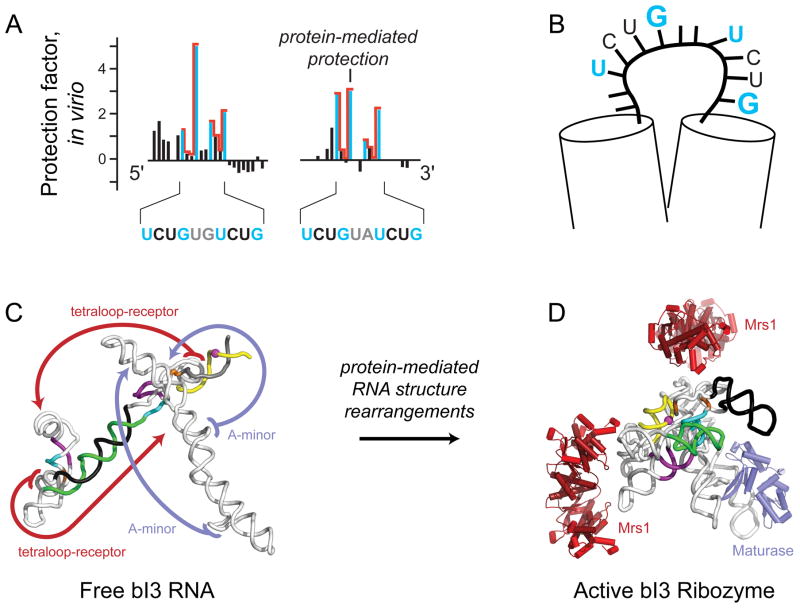
As an example of detection of direct binding, we studied packaging of the retrovirus, MuLV. Retroviruses have evolved conserved pathways to recognize genomic RNA dimers and to specifically exclude monomeric genomes and cellular RNAs. Packaging of RNA genomes into nascent virions is mediated by interactions with the nucleocapsid domain of the Gag protein. SHAPE reagents readily penetrate viral particles and were used to detect differences between protein-bound RNA inside virions and protein-free RNA that had been gently extracted from virions15,38. In addition, protein-RNA interactions were probed inside viral particle in reverse-footprinting experiments in which nucleocapsid-RNA interactions were weakened by treatment with a reagent that disrupted the fold of the nucleocapsid domain. SHAPE analysis indicated that the nucleocapsid domain of Gag binds and constrains the first and last nucleotides within a tandemly repeated UCUG motif (in blue, Figure 8A), flanked by base paired regions (Figure 8B)38. This work demonstrates the ability of SHAPE to precisely detect protein-RNA interactions at single-nucleotide resolution, to reveal the overall architecture of a binding site, and to do so in a complex virus particle environment. Analogous studies identified key components of the packaging signal for HIV-115.
Figure 8. SHAPE analysis of RNA-protein interactions.
(A) Binding of nucleocapsid to the MuLV genomic RNA results in protection of a small number of nucleotides from SHAPE modification. (B) Model for the nucleocapsid RNA binding site. Nucleotides most strongly protected are emphasized in blue; flanking helices are shown as cylinders38. (C) The bI3 group I intron RNA exists in a mis- and unfolded state prior to protein binding52. (D) Binding by the maturase and Mrs1 proteins stabilize long-range tertiary interactions (blue and red arrows) and induce widespread structural rearrangements as shown by changes in SHAPE reactivities21,49.
Protein-facilitated folding of the bI3 group I intron ribozyme illustrates the ability of SHAPE to visualize large-scale, protein-induced conformational changes in RNA21,49. To achieve its enzymatically active splicing conformation, the bI3 intron binds one copy of a maturase protein and two copies of the Mrs1 protein50,51. SHAPE analysis of the free bI3 RNA revealed that much of the RNA forms base pairs that are not consistent with the active conformation (see green-black and yellow-gray helices, Figure 8C)52. The complex alternative secondary for the free RNA could be specifically tested by the SHAPE-analysis-of-point-mutations approach42,52, which readily detects long-range RNA interactions mediated by structures separated by hundreds of nucleotides. SHAPE and hydroxyl radical probing were then used to identify the binding sites for the bI3 maturase and Mrs1 proteins and to determine the complex RNA structural rearrangements induced by each protein21,49. Strikingly, protein binding induced large-scale structural rearrangements in the RNA that ultimately stabilized specific long-distance tertiary interactions (Figure 8C, red and blue arrows). Protein-mediated stabilization of tertiary interactions appears to pull the secondary structure into an energetically disfavored, but functional conformation (Figure 8D)49,53.
Experimental Frontiers in RNA Structure Analysis
It is likely that RNA structure governs numerous elements of gene expression. However, until recently, it was not possible to obtain an experimentally-based understanding of the relationship between RNA structure and gene function on a global scale. This general problem is by no means solved, but a number of groups have begun to explore hypotheses based on structural information collected on RNA genomes or transcriptomes4,54,55. SHAPE was recently used to probe the structure of 99% of the 9,173 nucleotides in the genome of the NL4-3 strain of HIV-14, providing the first data-guided model for the structure of an entire RNA genome. When data collected on the HIV-1 genome were smoothed over a 75-nucleotide window, we observed that regions known to be functionally important – including elements in the 5′ region of the genome, at the frameshift element that allows translation of Pol, and at the Rev responsive element (RRE) – tended to have low SHAPE reactivities and were thus highly structured4. The SHAPE analysis also showed that there were numerous additional highly structured regions that had not previously been associated with a biological role.
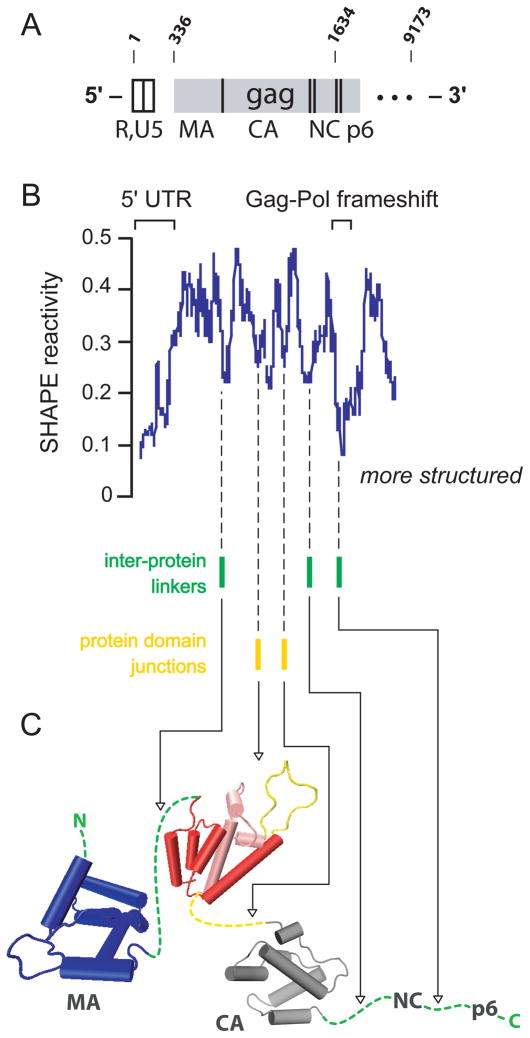
Intriguingly, there is a strong correlation between RNA structure and the domain structure of the encoded protein4. HIV-1 genome regions with low SHAPE reactivities correspond to loops between domains in the encoded protein (Figure 9). This observation is consistent with the hypothesis that RNA structure regulates the rate of ribosome processivity to facilitate protein folding. This model is supported by ribosome mapping experiments that show that pausing occurs preferentially in regions of high levels of RNA structure4 and with a study that found that yeast mRNAs tend to have a greater degree of RNA structure within their coding sequences than in their untranslated regions54.
Figure 9. SHAPE reveals a relationship between RNA structure and protein domain structure in the HIV-1 genomic RNA.
(A) Organization of the HIV-1 genome, emphasizing the Gag coding region. (B) Median SHAPE reactivities over a moving 75 nucleotide window. (C) Relationship between structured RNA regions and the peptide loops that link independent protein domains in Gag4.
Genome-wide SHAPE data was also used to generate a working model for the secondary structure of an entire authentic HIV-1 genome. This model includes proposals for new regulatory motifs in the genome, including the ribosome frameshifting element (Figure 6B), for structures that comprise protein domain junctions, for a domain that encodes the signal peptide in the Env protein, and for regions that have a conserved lack of structure, including both hypervariable sequences in Env and splice site acceptors4. Subsequent independent work has found that there is a strong correlation between HIV-1 RNA genome structure and recombination56.
Perspective
The first analysis of an entire RNA genome revealed that the HIV-1 genomic RNA is densely populated with highly structured regions, many of which have clearly identifiable functional roles in HIV-1 replication4. Moreover, it has become possible to predict RNA structures on the kilobase scale with good accuracy27. The extent of structure in intact RNAs appears to be pervasive, to correlate extensively with key elements of gene expression, and to plausibly constitute a second level of the genetic code. The available information suggests that we have just scratched the surface regarding how biological information is encoded in the structure of RNA.
The ability of SHAPE to probe the local nucleotide environment of RNA inside viral membranes15,38, opens the door to new and exciting experiments. There appear to be no significant limitations to using SHAPE in even more complex environments, including cells. RNA modification by SHAPE reagents leaves stable adducts that capture an imprint of the RNA structure during the short time the reagents are active. Subsequent purification of the RNA does not obscure this imprint and SHAPE is likely to prove broadly useful for exploring numerous structural states of RNA in vivo.
Biographies
Kevin M. Weeks grew up in Nashville, Tennessee where he caught the teaching bug during many years working as a camp counselor and as a tour guide at the Country Music Hall of Fame. He began his research career and earned a B.A. at the College of Wooster in Wooster, Ohio. He spent a year in Göttingen, Germany as a Fulbright Scholar; earned his Ph.D. with Donald Crothers at Yale; and did postdoctoral studies with Thomas Cech at the University of Colorado. Dr. Weeks is currently Professor of Chemistry at the University of North Carolina. His laboratory focuses on creating new chemical microscopes for understanding RNA structure and on applying these technologies to challenging problems in biology. The National Science Foundation and National Institutes of Health support work in his laboratory.
David M. Mauger was raised in Pottstown, Pennsylvania, where he first developed an interest in science. He earned a B.S. at the University of Pittsburgh, majoring in both chemistry and biology, and conducted research in yeast genetics in the lab of Karen Arndt. He earned his Ph.D. with Mariano Garcia-Blanco at Duke University where he studied regulation of alternative splicing in mammalian mRNAs. Dr. Mauger is currently an American Cancer Society Postdoctoral Fellow in the Weeks laboratory at the University of North Carolina at Chapel Hill. His research focuses on the development of new experimental approaches to analyze the role of RNA structure in viral replication.
References
- 1.Muesing MA, Smith DH, Capon DJ. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–13. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 4.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–6. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–31. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlin SI, Weeks KM. Mapping local nucleotide flexibility by selective acylation of 2′-amine substituted RNA. J Am Chem Soc. 2000;122:216–224. [Google Scholar]
- 8.John DM, Merino EJ, Weeks KM. Mechanics of DNA flexibility visualized by selective 2′-amine acylation at nucleotide bulges. J Mol Biol. 2004;337:611–9. doi: 10.1016/j.jmb.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlin SI, Merino EJ, Weeks KM. Catalysis of amide synthesis by RNA phosphodiester and hydroxyl groups. Proc Natl Acad Sci U S A. 2002;99:14688–93. doi: 10.1073/pnas.212527799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamberlin SI, Weeks KM. Differential helix stabilities and sites pre-organized for tertiary interactions revealed by monitoring local nucleotide flexibility in the bI5 group I intron RNA. Biochemistry. 2003;42:901–9. doi: 10.1021/bi026817h. [DOI] [PubMed] [Google Scholar]
- 11.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–5. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 12.Mortimer SA, Weeks KM. Time-Resolved RNA SHAPE Chemistry. J Am Chem Soc. 2008;130:16178–80. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–6. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 14.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–90. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steen KA, Malhotra A, Weeks KM. Selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease. J Am Chem Soc. 2010;132:9940–3. doi: 10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–21. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules.1. Theory and Range of Validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 19.Shajani Z, Varani G. 13C NMR relaxation studies of RNA base and ribose nuclei reveal a complex pattern of motions in the RNA binding site for human U1A protein. J Mol Biol. 2005;349:699–715. doi: 10.1016/j.jmb.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008;130:12244–5. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan CD, Weeks KM. The Mrs1 splicing factor binds the bI3 group I intron at each of two tetraloop-receptor motifs. PLoS One. 2010;5:e8983. doi: 10.1371/journal.pone.0008983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortimer SA, Johnson JS, Weeks KM. Quantitative analysis of RNA solvent accessibility by N-silylation of guanosine. Biochemistry. 2009;48:2109–14. doi: 10.1021/bi801939g. [DOI] [PubMed] [Google Scholar]
- 23.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol. 2006;16:270–8. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–8. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowell RD, Eddy SR. Evaluation of several lightweight stochastic context-free grammars for RNA secondary structure prediction. BMC Bioinformatics. 2004;5:71. doi: 10.1186/1471-2105-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathews DH. RNA secondary structure analysis using RNAstructure. In: Baxevanis AD, Davidson DB, Page RD, Petsko GA, Stein LD, Stormo GD, editors. Curr Protoc Bioinformatics. John Wiley & Sons, Inc; Hoboken, NJ: 2006. pp. 12.6.1–12.6.14. [Google Scholar]
- 27.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 2004;101:7287–92. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–40. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 30.Mauger DM, Weeks KM. Toward global RNA structure analysis. Nat Biotechnol. 2010;28:1178–9. doi: 10.1038/nbt1110-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–3. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 32.Dulude D, Baril M, Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed -1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30:5094–102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinman JD, Richter S, Plant EP, Taylor RC, Hammell AB, Rana TM. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc Natl Acad Sci U S A. 2002;99:5331–6. doi: 10.1073/pnas.082102199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3:643–55. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 35.Badorrek CS, Weeks KM. Architecture of a gamma retroviral genomic RNA dimer. Biochemistry. 2006;45:12664–72. doi: 10.1021/bi060521k. [DOI] [PubMed] [Google Scholar]
- 36.Gherghe C, Weeks KM. The SL1-SL2 (stem-loop) domain is the primary determinant for stability of the gamma retroviral genomic RNA dimer. J Biol Chem. 2006;281:37952–61. doi: 10.1074/jbc.M607380200. [DOI] [PubMed] [Google Scholar]
- 37.Gherghe C, Leonard CW, Gorelick RJ, Weeks KM. Secondary Structure of the Mature Ex Virio Moloney Murine Leukemia Virus Genomic RNA Dimerization Domain. J Virol. 2010;84:898–906. doi: 10.1128/JVI.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW, Jr, Gorelick RJ, Rein A, Weeks KM. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc Natl Acad Sci U S A. 2010;107:19248–53. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. J Am Chem Soc. 2005;127:4659–67. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 40.Wang B, Wilkinson KA, Weeks KM. Complex ligand-induced conformational changes in tRNA(Asp) revealed by single-nucleotide resolution SHAPE chemistry. Biochemistry. 2008;47:3454–61. doi: 10.1021/bi702372x. [DOI] [PubMed] [Google Scholar]
- 41.D’Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–90. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 42.Badorrek CS, Weeks KM. RNA flexibility in the dimerization domain of a gamma retrovirus. Nat Chem Biol. 2005;1:104–11. doi: 10.1038/nchembio712. [DOI] [PubMed] [Google Scholar]
- 43.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008;130:8884–5. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mortimer SA, Weeks KM. C2′-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc Natl Acad Sci U S A. 2009;106:15622–7. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Julien KR, Sumita M, Chen PH, Laird-Offringa IA, Hoogstraten CG. Conformationally restricted nucleotides as a probe of structure-function relationships in RNA. RNA. 2008;14:1632–43. doi: 10.1261/rna.866408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–4. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 47.Loria A, Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–25. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- 48.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc. 2009;4:1413–21. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duncan CD, Weeks KM. Nonhierarchical ribonucleoprotein assembly suggests a strain-propagation model for protein-facilitated RNA folding. Biochemistry. 2010;49:5418–25. doi: 10.1021/bi100267g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bassi GS, de Oliveira DM, White MF, Weeks KM. Recruitment of intron-encoded and co-opted proteins in splicing of the bI3 group I intron RNA. Proc Natl Acad Sci U S A. 2002;99:128–33. doi: 10.1073/pnas.012579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bassi GS, Weeks KM. Kinetic and thermodynamic framework for assembly of the six-component bI3 group I intron ribonucleoprotein catalyst. Biochemistry. 2003;42:9980–8. doi: 10.1021/bi0346906. [DOI] [PubMed] [Google Scholar]
- 52.Duncan CD, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–13. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGinnis JL, Duncan CD, Weeks KM. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol. 2009;468:67–89. doi: 10.1016/S0076-6879(09)68004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–7. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon-Loriere E, Martin DP, Weeks KM, Negroni M. RNA structures facilitate recombination-mediated gene swapping in HIV-1. J Virol. 2010;84:12675–82. doi: 10.1128/JVI.01302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]