Abstract
The molecular chaperone GroEL is required for bacterial growth under all conditions, mediating folding assistance, via its central cavity, to a diverse set of cytosolic proteins; yet the subcellular localization of GroEL remains unresolved. An earlier study, using antibody probing of fixed Escherichia coli cells, indicated colocalization with the cell division protein FtsZ at the cleavage furrow, while a second E. coli study of fixed cells indicated more even distribution throughout the cytoplasm. Here, for the first time, we have examined the spatial distribution of GroEL in living cells using incorporation of a fluorescent unnatural amino acid into the chaperone. Fluorescence microscopy indicated that GroEL is diffusely distributed, both under normal and stress conditions. Importantly, the present procedure uses a small, fluorescent unnatural amino acid to visualize GroEL in vivo, avoiding the steric demands of a fluorescent protein fusion, which compromises proper GroEL assembly. Further, this unnatural amino acid incorporation avoids artifacts that can occur with fixation and antibody staining.
Keywords: GroEL, chaperonin, chaperone, protein localization, unnatural amino acid, tRNA
The chaperonin (large, ring-shaped chaperones are called chaperonins) GroEL with its co-chaperonin lid GroES is required for growth at all temperatures,1 unlike the other bacterial chaperone systems, which are required only at elevated temperatures.2,3 GroEL is a double ring structure of identical subunits that assists a large set of other cytosolic proteins to achieve their final active state in the crowded cellular cytosol. This chaperone function can take place both with and without forming a privileged GroES covered, hydrophilic folding chamber, termed the cis cavity.4 Because of the central role played by this machine in many different organisms, a great deal of effort has been dedicated to understanding the structural, biochemical, and physiological details of GroEL function.5 However, a fundamental question concerns GroEL cellular localization both under normal growth conditions and in cells that have been subjected to stress (heat, oxidation, pH change, toxicity, or hyperosmotic shock). It is known that GroEL is upregulated during cellular stress, but whether or not this induces a change in subcellular localization remains controversial. A study using antibody immunofluorescence of fixed E. coli cells indicated that GroEL colocalizes with the tubulin homologue, FtsZ, involved in cell division.6 A later study, also using fixation and antibody staining of E. coli cells, however, suggested a diffuse localization.7
Examining GroEL in vivo could further address the issue of localization. However, this has been complicated by the difficulties associated with fluorescently labeling intact GroEL in vivo. Both the N- and C-termini are located inside the GroEL barrel, rendering attachment of a bulky, fluorescent protein (e.g. green fluorescent protein contains 238 amino acids with M.W. = 27 kDa) unfeasible without disassembly of the chaperonin. Attempts have been made to extend the GroEL C-termini outside of the cavity, a technique employed for other purposes (e.g. the C-termini were fused to GroES subunits without impairing function of the machine)8. But when a fluorescent protein was fused to extended C-termini to allow for a fluorescent signal produced from outside of the cis cavity, this failed to produce a functional tetradecameric assembly and led to large, terminal, fluorescent polar inclusion bodies excluded from the nucleoid (Supplemental Fig. 1). In an effort to find a viable system for the study of GroEL in vivo, we have employed the technique of Schultz and coworkers using an evolved tRNA/tRNA synthetase pair from Methanococcus jannaschii to introduce a fluorescent unnatural amino acid at the position of an inserted amber (UAG) nonsense mutation.9
The technique of unnatural amino acid incorporation, here a fluorescent coumarin amino acid, uses a nonsense (amber) mutation inserted into a gene of interest, here groEL, and an orthogonal tRNA/tRNA synthetase pair that activates the unnatural amino acid and enables its specific incorporation at the amber position during translation. With this system, a potential side product of the reaction is a truncated form of the protein of interest, with the amount of the side product inversely dependent on the efficiency of suppression. To test incorporation of the coumarin amino acid into GroEL, DH10B cells were transformed with two plasmids, pBKCouRS, containing a constitutively expressed copy of the evolved tRNA synthetase that activates the coumarin amino acid, and pBADJYgroELTAG, containing both a constitutively expressed copy of the evolved tRNA that accepts the coumarin amino acid and an arabinose inducible copy of the groE operon with groES and an amber-containing groEL mutant. An initial test was carried out to insert the coumarin amino acid in place of GroEL aspartate 473, whose side chain points out from the external surface of the equatorial domain, using an amber codon in the groEL gene. Position 473 has been observed to be innocuous with respect to cysteine substitution and with respect to subsequent fluorophore attachment.10 Here, however, following induction of groEL473TAG in the presence of the coumarin amino acid, a large amount of truncation product was observed (~70%) and a relatively small amount of full-length GroEL (~30%) was detected. The truncation product was observed to co-assemble with full-length GroEL, resulting in cell growth impairment (not shown). Therefore, additional positions more N-terminal to 473 were tested with amber suppression. When position 129, also situated at the outer aspect of the equatorial domain,11 was tested, arabinose induction in the presence of coumarin amino acid produced a full-length GroEL in the absence of any observable truncation product (Fig. 1a). When over-expressed, the full-length GroEL species was readily detected by Coomassie staining and the species was fluorescent, reflecting the incorporation of the coumarin amino acid (Fig. 1a). Notably, in the absence of coumarin amino acid supplied to the medium, induction of GroEL synthesis failed to produce either a fluorescent GroEL species or a Coomassie-stained GroEL species (Fig. 1a). This indicates that coumarin amino acid was specifically and efficiently incorporated into GroEL using position 129.
Figure 1. GroEL129CouAA biochemical characterization.
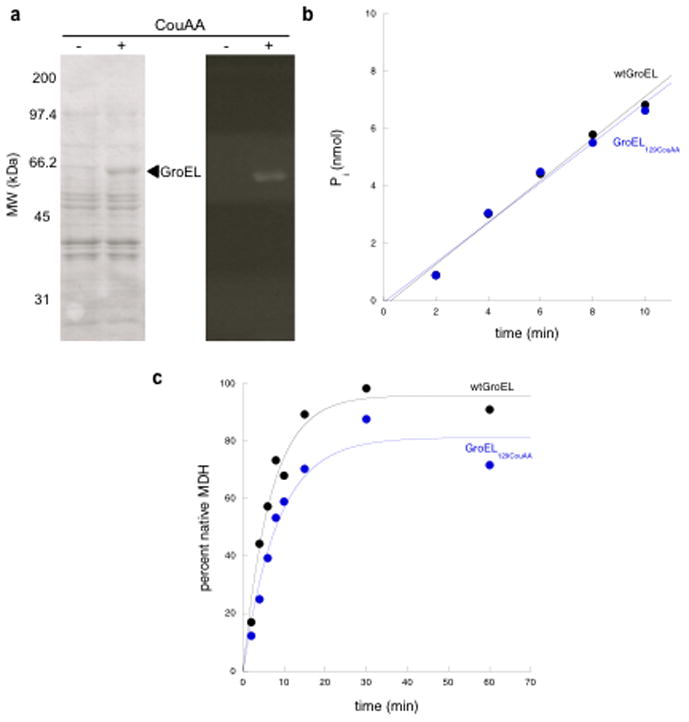
a) The efficiency of amber suppression was tested by growing cells harboring plasmids for GroEL129CouAA, MjtRNA, and CouRS expression in the absence or presence of coumarin amino acid, and by analyzing the whole cell lysates by SDS PAGE. The left panel shows the Coomassie- stained gel. The right panel shows GroEL129CouAA excited using a 305 nm transilluminator. b) The rate of ATP hydrolysis was measured using a standard malachite green-based assay using 1 μM chaperonin tetradecamer and 1 mM ATP. Displayed is a plot of the amount of inorganic phosphate released as a function of time. c) The ability to refold MDH, a stringent GroEL substrate, was measured. The percentage of MDH activity relative to a native MDH control is plotted as a function of time.
Next, to demonstrate biochemical function of the GroEL129CouAA, DH10B cells transformed with pBKcouRS and pBADJYgroEL129TAG were used for over-expression and purification. The standard GroEL purification protocol was employed, involving anion exchange chromatography, followed by hydrophobic interaction chromatography, and then Affigel Blue incubation (to remove GroEL-bound proteins).11 Light was excluded at each step to avoid photobleaching of the CouAA. A fluorescent protein of the expected mass was detected by mass spectrometry (51,313.8 Da calculated vs. 51,314 Da observed). The function of the GroEL129CouAA variant was assessed by measuring ATP turnover using a malachite green assay and by measuring refolding proficiency of a stringent GroEL substrate (i.e. requiring GroEL, GroES, and ATP), malate dehydrogenase (MDH), by measuring MDH activity recovered after incubation with the chaperonin system for measured time periods. For the ATPase assay, the amount of inorganic phosphate released was calculated from a standard curve and plotted as a function of time (Fig. 1b).12 The MDH refolding data were plotted by calculating the percent MDH activity relative to a non-denatured control as a function of time (Fig. 1c).13 Both ATPase and MDH refolding activities of the fluorescent variant were found to be effectively equivalent to wt GroEL.
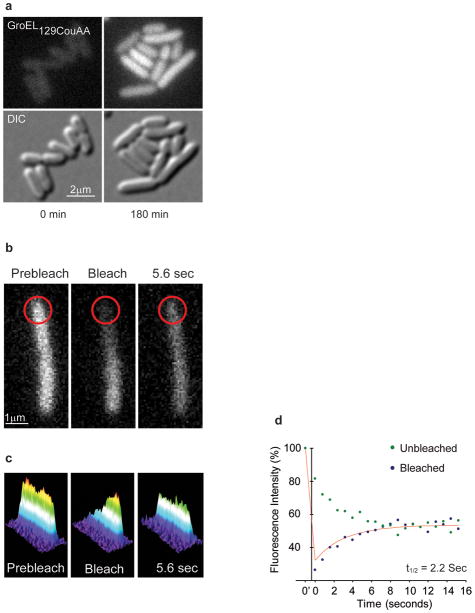
To study GroEL localization in living cells, MC1000 cells (a wild-type E. coli strain) transformed with pBKcouRS and pBADJYgroEL129TAG were employed and grown in either LB media (as shown in Fig. 2) or M9 minimal media. Prior to microscopy, it was necessary to remove unincorporated coumarin amino acid by washing the cells with minimal media. We observed that in cells grown in the presence of the coumarin amino acid but without induction of GroEL129CouAA synthesis, no fluorescence over background obtained (not shown). Also, cells imaged by fluorescence and DIC microscopy within 1 minute after arabinose induction of GroEL129CouAA and washing showed no fluorescence over background, as expected (Fig. 2a). In contrast, after 180 minutes of GroEL129CouAA induction, fluorescence images displayed a diffuse fluorescent pattern (Fig. 2a). These data demonstrate that the orthogonal suppression system produced fluorescent GroEL and no other fluorescent proteins.
Figure 2. GroEL129CouAA fluorescence characterization and fluorescence recovery after photobleaching.
a) Incorporation of the coumarin amino acid was analyzed using fluorescence microscopy. The upper panels show fluorescence using a DAPI filter set and the lower panels show DIC images. The times post-induction are shown beneath the images. b)-d) The dynamics of GroEL were measured using FRAP. b) Single cell FRAP images pre-bleaching, immediately post-bleaching, and 5.6 seconds post-bleaching (5.6). c) A plot of cellular fluorescence intensity corrected for photobleaching during acquisition of images. d) Fluorescence intensity was plotted as a function of time and fit to a single exponential curve to calculate the rate of fluorescence recovery (t1/2 = 2.2 sec).
To study GroEL diffusion dynamics, fluorescence recovery after photo bleaching (FRAP) was employed. Cells were grown as above in M9 minimal medium and GroEL129CouAA synthesis was induced for 180 minutes in the presence of 1 mM coumarin amino acid. The cells were then washed with M9, and placed on an agarose pad for visualization. Cephalexin was added to the cell culture to halt cell division for more reproducible measurements of diffusion dynamics.14 A small area of individual cells was then photo bleached using a Photonic Instruments Micropoint Laser at 364 nm and the rate of fluorescence recovery measured. A representative FRAP experiment is shown with the bleached area indicated by a red circle (Fig. 2b). The intensity of the fluorescent signal as a function of cell position was also determined and plotted (Fig. 2c). A plot of relative fluorescence density as a function of time was fit to a single exponential decay to yield a half time of diffusion of 2.0 ± 1.1 (mean ± SD) seconds (n = 5 cells) for ~6 μm long cells (Fig. 2d). Using the protocol of Cone and coworkers, an apparent diffusion constant can be calculated, Dapp = 0.16 ± 0.15 μm2sec−1 (n = 17 cells).15 The rate of diffusion of several GFP polymers (from 1–5 GFPs linked end to end) linked to either cytosolic or periplasmic proteins has been used to estimate size dependence of apparent diffusion rates.16 The apparent diffusion constant of GroEL was slower than the largest cytoplasmic test cases reported (138 kDa; Dapp = 2.8 ± 1.9 μm2sec−1) reflecting either the greater size of the 800 kDa assembled chaperonin or the binding dynamics of GroEL to polypeptide substrates.17 The latter is unlikely to have a large effect, however, as the substrates are mostly small (<60 kDa), cytosolic proteins4,18,19,20.
Inquiries concerning cellular localization of the GroEL-GroES chaperonin system in response to environmental stress have been complicated by the inability to generate a functional, labeled GroEL variant. Therefore, we have utilized GroEL129CouAA to study stress effects. We first examined the effects of severe heat shock. Cells were grown and GroEL129CouAA synthesis was induced as above. However, prior to visualization cells were washed, heated to 45°C for 5 minutes, placed on an agarose pad, and visualized on a microscope stage heated to 45°C. Under these stressful conditions, the diffuse cellular localization of the GroEL signal was observed to remain despite apparent stress-induced effects, such as cell elongation and slow cell division rate (Fig. 3a and supplemental movie 1). Next, the effects of other stress conditions were examined including toxic stress by a one hour treatment with ampicillin in a hypertonic solution (Fig. 3b), and osmotic stress by treatment with a hypertonic solution (Fig. 3c).6 These cellular stress conditions, coupled with fixation and antibody staining, were used by Ogino et al. who observed colocalization of GroEL with the cell division protein FtsZ. Further studies by Winkler et al., also using fixation and antibody staining, however, indicated GroEL remains diffuse under heat shock.7 In each of the present studies, the stressed cells displayed an elongated morphology, but showed the same diffuse GroEL129CouAA localization observed under normal conditions.
Figure 3. GroEL remains diffuse after all tested insults.
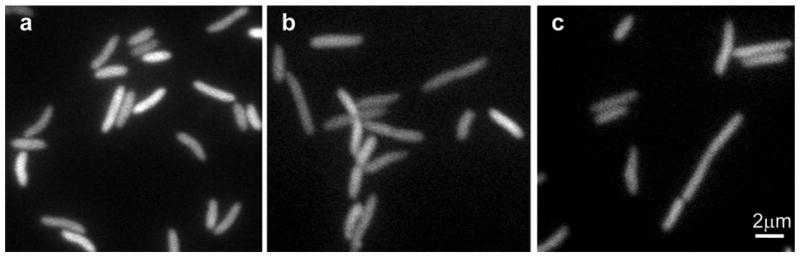
Fluorescence images of cells growing at 45°C (a), in the presence of 100 μg/ml ampicillin and 0.5 M NaCl (b), and in hypertonic solution (0.5 M NaCl) (c).
Genetic and proteomic evidence indicate that a major function of GroEL is to assist newly translated, cytosolic proteins to achieve their functional form.4,18,19,20 This activity takes place along with the assistance of other chaperone systems, but it is known that GroEL-GroES over-expression can rescue cells deficient in these other chaperones, at otherwise non-permissive temperatures.21 However, once stress-induced aggregation obtains, the ClpB/DnaK/DnaJ/GrpE system has been shown to be the primary machinery for disentangling these aggregated proteins. As such, this latter chaperone system was shown to colocalize with aggregated proteins at the cell poles.7 In contrast, GroEL remains diffusely distributed under the same aggregation-promoting conditions. In addition to newly tranlated proteins, GroEL also assists pre-existent proteins to maintain native form, for instance, under heat shock conditions. As such, GroEL seems to be needed throughout the cytosolic compartment including the nucleoid. More specifically, localization in the nucleoid suggests a role in assisting proteins involved in chromosomal maintenance and replication, in agreement with the discovery of proteins of this class in proteomic studies.4,19,20
Supplementary Material
Acknowledgments
This work was supported by NIH RO1 GM086225. G.C. was supported by the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement n°PIRG05-GA-2009-247241. J. W. was supported by the National Science Foundation of China (30870592 and 90913022) and the Major State Basic Research Program of China (2010CB912301). This work was partially supported by NIH (GM076698 and GM065835 to C.J.-W.) and by NIH (RO1 GM62159 to P.G.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fayet O, Ziegelhoffer T, Georgopoulos C. J Bacteriol. 1989;171:1379. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Nature. 1999;400:693. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 3.Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. Cell. 1999;97:755. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 4.Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, Pypaert M, Horwich AL. Proc Natl Acad Sci USA. 2006;103:15800. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horwich AL, Fenton WA, Chapman E, Farr GW. Annu Rev Cell Dev Biol. 2007;23:115. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 6.Ogino H, Wachi M, Ishii A, Iwai N, Nishida T, Yamada S, Nagai K, Sugai M. Genes Cells. 2004;9:765. doi: 10.1111/j.1365-2443.2004.00770.x. [DOI] [PubMed] [Google Scholar]
- 7.Winkler J, Seybert A, König L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B. EMBO J. 2010;29:910. doi: 10.1038/emboj.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farr GW, Fenton WA, Chaudhuri TK, Clare DK, Saibil HR, Horwich AL. EMBO J. 2003;22:3220. doi: 10.1093/emboj/cdg313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Xie J, Schultz PG. J Am Chem Soc. 2006;128:8738. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhuri TK, Farr GW, Fenton WA, Rospert S, Horwich AL. Cell. 2001;107:235. doi: 10.1016/s0092-8674(01)00523-2. [DOI] [PubMed] [Google Scholar]
- 11.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. Nature. 1994;371:578. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 12.Chapman E, Farr GW, Fenton WA, Johnson SM, Horwich AL. Proc Natl Acad Sci USA. 2008;105:19205. doi: 10.1073/pnas.0810657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rye HS, Burston SG, Fenton WA, Beechem JM, Xu Z, Sigler PB, Horwich AL. Nature. 1997;388:792. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 14.Elowitz MB, Surette MG, Wolf PE, Stock JB, Leibler S. J Bacteriol. 1999;181:197. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wey CL, Cone RA, Edidin MA. Biophys J. 1981;33:225. doi: 10.1016/S0006-3495(81)84883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nenninger A, Mastroianni G, Mullineaux CW. J Bacteriol. 2010;192:4535. doi: 10.1128/JB.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sprague BL, McNally JG. Trends Cell Biol. 2005;15:84. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara K, Ishihama Y, Nakahigashi K, Soga T, Taguchi H. EMBO J. 2010;29:1552. doi: 10.1038/emboj.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Nature. 1999;402:147. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 20.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, Hartl FU. Cell. 2005;122:209. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 21.Vorderwülbecke S, Kramer G, Merz F, Kurz TA, Rauch T, Zachmann-Brand B, Bukau B, Deuerling E. FEBS Lett. 2004;559:181. doi: 10.1016/S0014-5793(04)00052-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.