Abstract
The lysyl-tRNA synthetase paralog PoxA modifies elongation factor P (EF-P) with α-lysine at low efficiency. Cell-free extracts contained non-α-lysine substrates of PoxA that modified EF-P by a change in mass consistent with β–lysine, a substrate also predicted by genomic analyses. EF-P was efficiently, functionally, modified with (R)-β-lysine but not (S)-β-lysine or genetically encoded α-amino acids, indicating that PoxA has evolved an activity orthogonal to that of the canonical aminoacyl-tRNA synthetases.
The aminoacyl-tRNA synthetases family (aaRSs) encompasses 20 canonical enzymes that match amino acids with their corresponding tRNAs during translation1. The aaRSs are divided between classes I and II, each of which is characterized by a core catalytic domain responsible for ATP-dependent acyl-adenylate synthesis2. These core domains recognize and activate specific amino acids, which are transferred to the 3' end of tRNA. AaRSs are highly modular and contain domains appended to the catalytic core that assist in RNA binding and recognition, and in proofreading of incorrect aminoacylation products3. In addition to the canonical enzymes required for translation, the aaRS superfamily contains paralogs that recapitulate either the core or the appended domains of aaRSs4. AaRS paralogs generally retain some degree of specificity for canonical protein synthesis substrates, amino acid or tRNA, whether or not they function inside or outside translation5. One notable exception is PoxA, a paralog of lysyl-tRNA synthetase (LysRS), which does not recognize tRNA but instead post-translationally modifies elongation factor P (EF-P), a protein that mimics tRNA in both shape and size6–8. EF-P modification leads to specific alterations in the proteome and is required for virulence in Salmonella, but exactly how PoxA modifies EF-P to elicit these changes is unknown7.
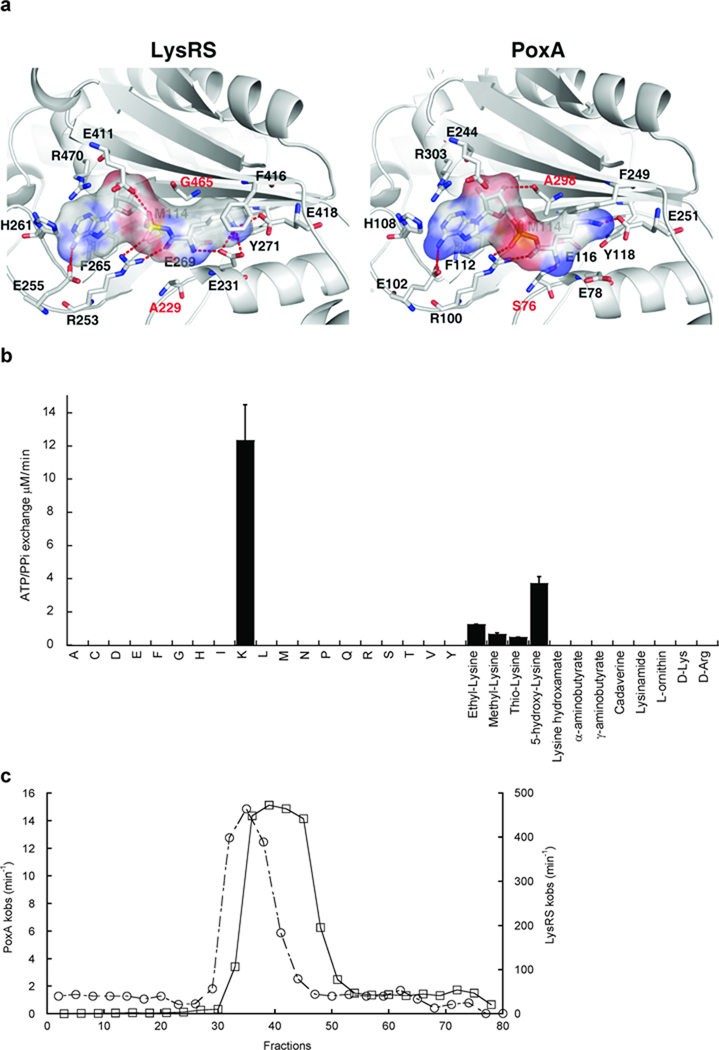
Structural studies suggest PoxA recognizes EF-P in a similar manner to that by which LysRS binds tRNALys 7,8. Aminoacylation assays show the KM of PoxA for EF-P (3.2 µM) is similar to values determined for bacterial class II-type LysRS with tRNALys (1–2 µM9,10). PoxA cannot aminoacylate tRNALys, indicating that while it is structurally similar to LysRS its activity is different from that of a canonical aaRS11. The amino acid binding pockets of PoxA and LysRS also appear highly similar and super positioning of the α-carbons within the active sites shows an RMSD of 1.1 Å (Fig. 1a). Using the LysRS co-crystal structure, lysyl-adenylate can be docked into the active site of PoxA, consistent with previous findings that lysine is a substrate for EF-P modification in vitro7. While lysine was a substrate for PoxA, the reaction was extremely inefficient compared to amino acid activation by LysRS (Supplementary Methods, Table 1). Characterization of amino acid activation by PoxA under a variety of conditions did not lead to significant improvements in the efficiency of the reaction (Supplementary Results, Supplementary Fig. 1), suggesting that lysine may not be the cognate substrate. This was supported by comparing the PoxA and LysRS active sites, which indicated divergence of two conserved residues predicted to impact substrate specificity, Ser76 and Ala294 (Fig. 1a; Escherichia coli PoxA numbering). The effect of reverting these PoxA residues to their conserved LysRS identities was investigated by comparing the apparent amino acid binding affinities of the S76A and A294G variants to wild-type. The S76A mutation abolished all activity, while A294G showed a 25-fold decrease in the KM for lysine compared to wild-type (Table 1). These data indicate PoxA is not optimized to use lysine as its preferred substrate, but has instead evolved amino acid specificity divergent from that of LysRS.
Figure 1. Amino acid recognition by PoxA.
(a) The 20 amino acids forming the lysyl-adenylate binding pocket of PoxA and LysRS are identical, apart from the two positions indicated in red. For clarity, only 14 residues are represented. Crystal structures are LysRS from Bacillus stearothermophilus24 and PoxA from Salmonella enterica Typhimurium7. The Van der Waals surface of the lysyl-adenylate is represented. S. enterica Ala298 corresponds to E. coli Ala294. (b) PoxA-catalyzed ATP/PPi exchange in the presence of non-cognate amino acids (single letter code) and lysine analogs. The concentration of each amino acid was 20 mM. Errors correspond to the standard deviation from three independent experiments. (c) Metabolite fractionation by LC on silica gel. Fractions were separately tested for stimulation of ATP/PPi exchange catalyzed by PoxA (○) or LysRS (□).
Table 1.
Steady-state kinetics of amino acid activation by E. coli LysRS and PoxA.
| KM (µM) | kcat (min−1) | kcat / KM (min−1µM−1) | |
|---|---|---|---|
| L-α-lysine | |||
| LysRS1 | 43 | 3000 | 70 |
| PoxA | 8600 ± 1200 | 15 ± 1.3 | 17 × 10−4 |
| PoxA A294G | 344 ± 44 | 3.8 ± 0.6 | 111 × 10−4 |
| (R)-β-lysine | |||
| PoxA | 213 ± 31 | 36 ± 3.3 | 1690 ×10 −4 |
| PoxA A294G | 414 ± 63 | 17 ± 0.5 | 410 × 10−4 |
| (S)-β-lysine | |||
| PoxA | 6950 ± 1894 | 2.8 ± 0 | 4 × 10−4 |
Values previously determined for the lysS-encoded protein25. Errors correspond to the standard deviation from three independent experiments.
Previous studies show different LysRSs preferentially recognize particular non-cognate amino acids12, and the specificity of PoxA was similarly tested (Fig. 1b). PoxA did not activate either other non-cognate genetically encoded amino acids or various commercially available analogs more efficiently than lysine. This raised the possibility that another amino acid was the preferred substrate of PoxA in vivo. E. coli cell-free extracts showed significantly higher stimulation of ATP / PPi exchange activity than expected based solely upon their lysine content, suggesting the presence of a second, more efficient, substrate (Supplementary Fig. 2a). Cell-free extracts were fractionated and the resulting samples tested for ATP / PPi exchange activity using LysRS and PoxA (Supplementary Fig. 2b). Optimal activities for LysRS and PoxA did not co-fractionate during purification, supporting the idea that E. coli cell-free extracts contained a substrate for EF-P modification distinct from lysine (Fig. 1c). The identity of the amino acid attached by PoxA in vivo was investigated by mass spectroscopy of both affinity purified EF-P, and of EF-P modified in vitro using enriched cell-free extracts. In both cases EF-P was site-specifically modified at Lys34 by a mass consistent with the attachment of lysine (Supplementary Fig. 3). These data showed that while the genetically encoded amino acid α-lysine was not the optimal substrate for PoxA, EF-P is nevertheless modified by a compound with the mass of lysine, consistent with previous reports8,13.
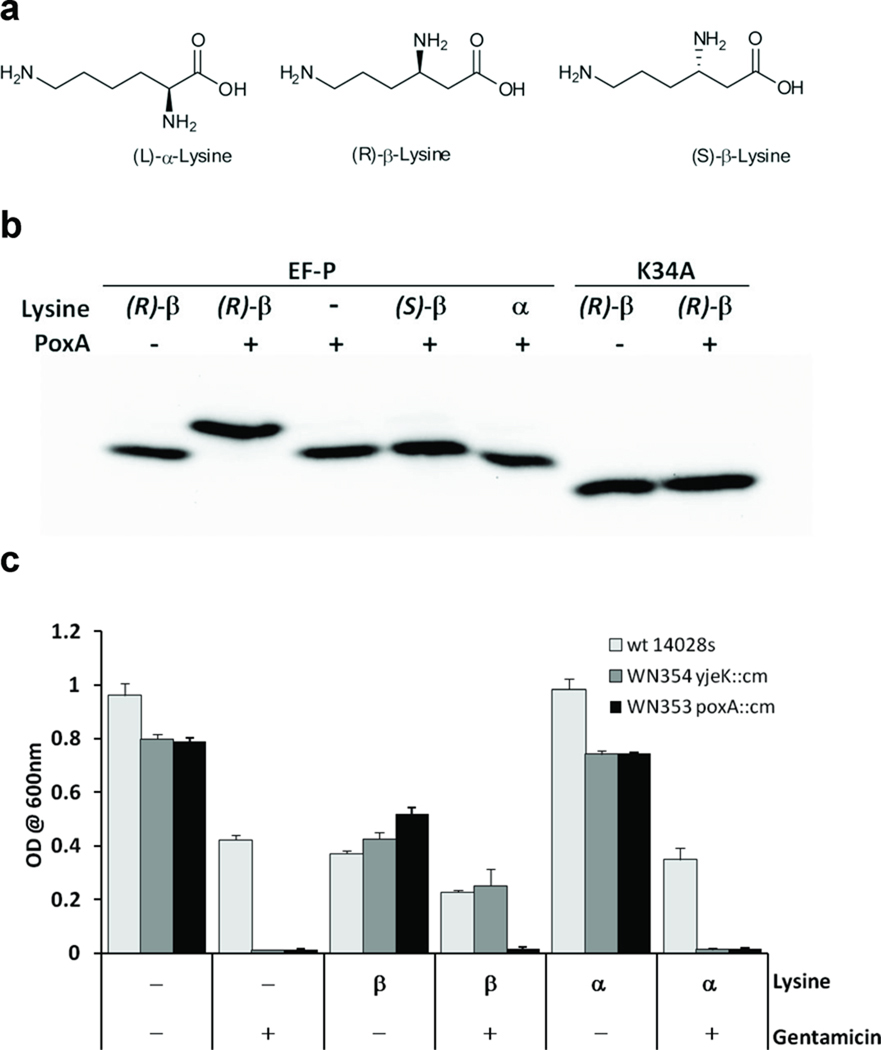
Genomic and phylogenetic studies predict that β-lysine might be the natural substrate for PoxA6,7, consistent with the mass observed for modified EF-P and with the detection in vivo of a substrate distinct from α-lysine. (R)-β-lysine was 100-fold more efficient as a substrate than either (S)-β-lysine or α-lysine (Fig. 2a), suggesting that (R)-β-lysine is the cognate substrate for EF-P modification (Table 1). In contrast, (R)-β-lysine was a poor substrate compared to α-lysine for LysRS (Supplementary Fig. 4a) indicating that the amino acid substrate specificity of PoxA is different to that of the ancestral aaRS from which it evolved. The ability of PoxA to post-translationally modify EF-P with β-lysine was examined in vitro and found to be considerably more efficient than modification with α-lysine (Fig. 2b). β-lysine was also a potent inhibitor of α-lysine modification of EF-P, consistent with this reaction being orthogonal to tRNA aminoacylation by LysRS (Supplementary Fig. 4b).
Figure 2. Modification of EF-P with β-lysine.
(a) Lysine structures (b) PoxA catalyzed aminoacylation of EF-P with (R)-β-lysine. Aminoacylation of EF-P or the inactive K34A variant was assayed in the presence or absence of 20µM (R)-β-lysine, (S)-β-lysine, or α-lysine. Modified EF-P containing an additional –NH3+ group was separated from the unmodified EF-P by 1D isoelectric focusing and detected by western blotting. (c) Complementation of Salmonella ΔyjeK strain with (R)-β-lysine. Growth in liquid LB with (+) or without (−) 8 µg/ml gentamicin for 24 hours at 37 °C. Growth was monitored in the presence of either 0.8 mM (R)-β-lysine, 0.8 mM α-lysine, or no lysine (−). Optical densities of replicate cultures are plotted. Growth rates were consistently slower on β-lysine than on α-lysine, suggesting a modest inhibitory effect at the concentrations used, the reason for which is presently unclear.
Previous studies established that EF-P, PoxA and the 2,3-β-lysine aminomutase encoded by yjeK8,14 act together in vivo to establish virulence in Salmonella, and are important for maintenance of stress resistance phenotypes including resistance to gentamicin and growth on AB2 media7,15. The latter phenotype was used to assess how modification with β-lysine affects the functional activity of EF-P in vivo. Complementation of a poxA yjeK double knockout strain showed that neither wild-type PoxA nor the A298G variant could restore normal growth on AB2 media, suggesting that lysylation alone (as opposed to β-lysylation) is not sufficient to create functional EF-P (Supplementary Fig. 5). To more directly investigate the role of β-lysylation of EF-P in vivo, we attempted to restore the growth of the yjeK mutant on gentamicin by supplementation with different forms of lysine. β-lysine, but not α-lysine, was able to restore growth of the yjeK mutant in the presence of gentamicin (Fig. 2c). These data show that functional EF-P is dependent on post-translational modification by PoxA specifically with β-lysine, and that synthesis of β-lysine by YjeK occurs prior to addition rather than by a post-addition rearrangement of α-lysine.
Perturbation of the post-translational modification of EF-P by PoxA gives rise to changes in translation that attenuate Salmonella virulence7,15,16. The chemical nature of this novel modification is a critical determinant of EF-P activity; while both α- and β-lysine are substrates for PoxA, only the latter amino acid generates the biologically active form of EF-P. What remains unclear is how attachment of β-lysine changes the activity of EF-P. One possibility is that β-lysine acts as a recognition element for ribosome binding, although this may not be a universal role as unmodified isoforms of EF-P can also bind to bacterial ribosomes17 and a number of bacteria encode EF-P but not PoxA or YjeK6. Comparisons to the eukaryotic homolog eIF5a18 suggest that addition of β-lysine may contribute to the ability of EF-P to promote translation initiation and /or elongation13,19. The modification of eIF5a with hypusine, by an unrelated pathway, is required for its stimulatory effect on translation20, and the availability of a defined in vitro modification system will now allow similar effects to be tested for with EF-P. The availability of a defined modification system in vivo will in addition facilitate investigation of the proposal that EF-P stimulates translation of specific messages7,16, a role also proposed for eIF5a but for which there is no known mechanism18.
Post-translational modification of EF-P is based on molecular mimicry of a pathway hijacked from the translation machinery. To achieve the substrate specificity necessary to modify EF-P with β-lysine, PoxA evolved two specificities orthogonal to those of the protein from which it is derived, the class II-type LysRS. Elimination of the amino-terminal anticodon-binding domain of LysRS would have substantially reduced affinity for tRNA10, and allowed PoxA to evolve the protein-protein interactions that permit it to specifically recognize the tRNA-mimic EF-P8 but not tRNALys. The evolution of amino acid specificity in PoxA appears to have been primarily driven by selection for β-lysine as a substrate, and may have been facilitated by the lack of selection to maintain a high turnover rate. The biological role of LysRS, providing Lys-tRNALys for protein synthesis, requires a high rate of product synthesis whereas PoxA would be required to have a far lower turnover rate. As a result, only very few active site replacements compared to LysRS are sufficient to provide PoxA with useful β-lysine specificity, since any accompanying loss in turnover number is still compatible with a biologically relevant rate of EF-P modification.
Many aaRS paralogs have been sequenced and annotated, but to date functions have only been assigned to a small fraction4,21,22. The relative ease with which PoxA apparently acquired new substrate specificities suggests that alternative mechanisms of post-translational modification, and amino acid-dependent transformations, may be associated with other aaRS paralogs. The substrate specificity of PoxA also opens up the possibility that aaRS paralogs could provide the basis for new tools for protein design and engineering. The orthogonality of PoxA to the translation machinery mirrors that of systems developed for co-translational insertion of unnatural amino acids23, suggesting that similar approaches could be developed in the future to evolve new post-translational protein modification activities.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Michael Thomas (University of Wisconsin) for the generous gift of (S)-β-lysine. This work was supported by the National Institutes of Health (GM65183, M.I.), the Canada Institutes for Health Research (MOP-86683 and MSH-87729, W.W.N.), and the Natural Sciences and Engineering Research Council of Canada (RGPIN 386286-10, W.W.N.; Vanier Graduate Scholarship, S.B.Z). We also acknowledge The Ohio State University for partial financial support.
Footnotes
AUTHOR CONTRIBUTIONS
H.R., S.B.Z., T.B., B.W. and M.G. performed experiments and analyzed the resulting data; H.R., C.F., W.W.N. and M.I. designed experiments and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
None of the authors declare any competing financial interests with respect to this study.
REFERENCES
- 1.Ibba M, Söll D. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 3.Alexander RW, Schimmel P. Prog. Nucleic Acid Res. Mol. Biol. 2001;69:317–349. doi: 10.1016/s0079-6603(01)69050-0. [DOI] [PubMed] [Google Scholar]
- 4.Francklyn C. In: Aminoacyl-tRNA Synthetases. Francklyn C, Ibba M, Cusack S, editors. Georgetown, TX: Landes Bioscience; 2005. pp. 285–297. [Google Scholar]
- 5.Vetting MW, Hegde SS, Blanchard JS. Nat. Chem. Biol. 2010;6:797–799. doi: 10.1038/nchembio.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailly M, de Crécy-Lagard V. Biol. Direct. 2010;5:3. doi: 10.1186/1745-6150-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navarre WW, et al. Mol. Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. Nat. Struct. Mol. Biol. 2010;17:1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- 9.Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M. Nucleic Acids Res. 1992;20:2335–2339. doi: 10.1093/nar/20.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Commans S, Lazard M, Delort F, Blanquet S, Plateau P. J. Mol. Biol. 1998;278:801–813. doi: 10.1006/jmbi.1998.1711. [DOI] [PubMed] [Google Scholar]
- 11.Ambrogelly A, O'Donoghue P, Söll D, Moses S. FEBS Lett. 2010;584:3055–3060. doi: 10.1016/j.febslet.2010.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levengood J, Ataide SF, Roy H, Ibba M. J. Biol. Chem. 2004;279:17707–17714. doi: 10.1074/jbc.M313665200. [DOI] [PubMed] [Google Scholar]
- 13.Aoki H, et al. FEBS J. 2008;275:671–681. doi: 10.1111/j.1742-4658.2007.06228.x. [DOI] [PubMed] [Google Scholar]
- 14.Behshad E, et al. Biochemistry. 2006;45:12639–12646. doi: 10.1021/bi061328t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaniga K, Compton MS, Curtiss R, 3rd, Sundaram P. Infect. Immun. 1998;66:5599–5606. doi: 10.1128/iai.66.12.5599-5606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bearson SM, Bearson BL, Brunelle BW, Sharma VK, Lee IS. Foodborne Pathog. Dis. 2011;8:725–732. doi: 10.1089/fpd.2010.0796. [DOI] [PubMed] [Google Scholar]
- 17.Blaha G, Stanley RE, Steitz TA. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park MH, Nishimura K, Zanelli CF, Valentini SR. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glick BR, Chladek S, Ganoza MC. Eur. J. Biochem. 1979;97:23–28. doi: 10.1111/j.1432-1033.1979.tb13081.x. [DOI] [PubMed] [Google Scholar]
- 20.Saini P, Eyler DE, Green R, Dever TE. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schimmel P, Ribas De Pouplana L. Trends Biochem. Sci. 2000;25:207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L, de Souza RF, Iyer LM. Biol. Direct. 2010;5:48. doi: 10.1186/1745-6150-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CC, Schultz PG. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 24.Sakurama H, et al. J. Biochem. 2009;145:555–563. doi: 10.1093/jb/mvp014. [DOI] [PubMed] [Google Scholar]
- 25.Ataide SF, Ibba M. Biochemistry. 2004;43:11836–11841. doi: 10.1021/bi0490542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.