Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that control a multitude of critical processes in mammalian cells. Increasing evidence has emerged that host miRNAs serve in animal cells to restrict viral infections. In turn, many viruses encode RNA silencing suppressors (RSS) which are employed to moderate the potency of the cell's miRNA selection against viral replication. Some viruses also encode viral miRNAs. In this review, we summarize findings from human immunodeficiency virus type 1 (HIV-1) and human T-cell leukemia virus type 1 (HTLV-1) that illustrate examples of host cell miRNAs that target the viruses, of RSS encoded by viruses, and of host cell miRNA profile changes that are seen in infected cells. This article is part of a Special Issue entitled: MicroRNAs in viral gene regulation.
Keywords: MicroRNA, Virus replication, Innate immunity, RNA silencing suppressor, Viral gene expression, Virus–host interaction
Research highlights
► Review of cellular miRNAs that have been described to interact with HIV-1 or HTLV-1. ► Review of RSS activities that have been described for HIV-1 and HTLV-1. ► Review of microRNA profile changes in cells after infection by HIV-1 or HTLV-1.
1. Introduction
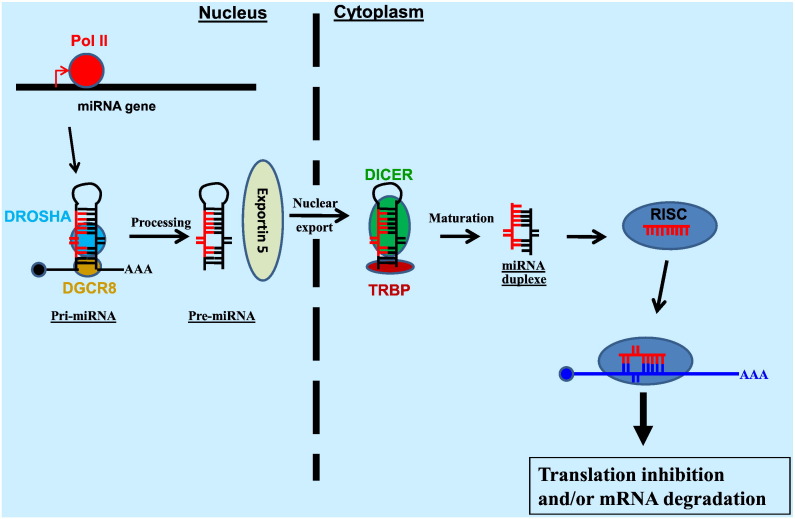
MicroRNAs (miRNAs) are small noncoding RNAs with approximate lengths of 21 to 22 nucleotides. The expression and biogenesis of miRNAs have been reviewed well elsewhere [1], [2] (Fig. 1 ). In brief, miRNAs are first transcribed as a long primary miRNA (pri-miRNA) by RNA polymerase II (Pol II). This pri-miRNA is processed sequentially by the RNase III enzymes Drosha in the nucleus and Dicer in the cytoplasm to generate an ~ 22 nucleotide duplex RNA that is incorporated into the RNA-induced silencing complex, RISC. The duplex miRNA is loaded into the RNA-induced silencing complex, RISC, where one strand will become the guide strand and will be incorporated to regulate the translation and/or degradation of target mRNA via incomplete base-pairing recognition [3]. The other passenger strand is discarded. The RISC complex is composed of a multitude of proteins including TAR RNA-binding protein (TRBP; [4], [5]) and TRBP-associated factors [6].
Fig. 1.
miRNA biogenesis and function. A miRNA gene is transcribed by RNA polymerase II (Pol II) to generate the primary miRNA (pri-miRNA). In the nucleus, the pri-miRNA is cleaved by the RNase III endonuclease Drosha to produce a ~ 70 nt precursor miRNA (pre-miRNA). Exportin-5 transports the pre-miRNA to the cytoplasm, where it is cleaved by another RNase III endonuclease, Dicer, together with TRBP, to produce the mature miRNA duplex. The miRNA duplex is then loaded on the Argonaute-containing RNA-induced silencing complex (RISC) where one strand is retained as a guide strand to regulate the translation and/or degradation of target mRNA via imperfect basepairing recognition while the other passenger strand is discarded.
Based on computational analyses of miRNAs and their putative mRNA targets, it has been speculated that mammalian miRNAs can regulate up to 30% of the protein-coding mRNAs. Accordingly, it is not surprising that functional studies on miRNAs have implicated their involvement in the regulation of almost all cellular processes and have documented changes in miRNA expression profiles in most human pathologies. The importance of miRNAs to vertebrate development is substantiated by the engineered loss of Dicer function resulting in profound developmental defects in both the zebrafish and the mouse [7], [8]. Additionally, the use of conditional Dicer knockout mice has verified the role of Dicer in limb morphogenesis [9], lung development [10], hair follicle generation [11], and T cell differentiation [12].
Dysregulated miRNAs are thought to be involved in the biology of cancers. The initial link between miRNAs and cancer arose from a study which showed that a chromosome deletion associated with chronic lymphocytic leukemia (CLL) led to the loss of miR-15a and miR-16-1 which were later shown to function as tumor suppressors [13]. Indeed, miRNA genes appear to be frequently perturbed in cancers [14], and increasing numbers of miRNAs that function as tumor suppressors or oncogenes are being described [15].
2. miRNAs and mammalian viruses
Given the importance of miRNAs in cellular development, metabolism and proliferation, what is the likelihood that miRNAs might also play a role in cellular regulation of pathogens? Although this area remains debated, many findings increasingly converge to suggest that the cell's RNAi-response, and in particular miRNAs, participates in host–virus interactions that can pivotally influence viral replication [16], [17], [18], [19], [20], [21] (Table 1 ). The first clue for this idea came from the observation that human miR-32 could limit the replication of primate foamy virus type 1 (PFV-1) [22] in cells. Next, the liver-specific miR-122 was unexpectedly found to enhance the replication of hepatitis C virus [23], while miR-199a-3p and miR-210 [24] and miR-125a-5p [25] were reported to suppress HBV replication. In other virus systems, miR-101 was shown to suppress HSV-1 propagation [26], and miR-323, miR-491, and miR-654 inhibited influenza virus [27].
Table 1.
Host miRNAs that have been experimentally demonstrated to target mammalian viruses.
If miRNAs can serve to restrict viral replication, then one might expect that viruses would evolve stratagems to counter this negative selection. Indeed, in response to the cell's RNAi mediated antiviral defense many mammalian viruses have developed viral proteins or RNA molecules that can function as RNA silencing suppressors (RSS) (Table 2 ). RSS in human viruses exert their activities using different mechanisms (reviewed in [28]). For example, the NS1 protein of influenza A virus [29] and the VP35 protein of Ebola virus [30] sequester small RNAs, while the HCV core [31], the HIV Tat proteins [32], and the HTLV-1 Rex protein [33] may interact with RNA and attenuate Dicer function; and the adenovirus VA 1 RNA, which is expressed at a very high amount during infection, exerts its RSS activity by saturating exportin-5 and Dicer [34]. Many RSS proteins encoded by viruses also suppress interferon-mediated antiviral defenses (e.g. the Tat protein also suppresses PKR [35], [36]) suggesting that viruses have evolved and maintain proteins that possess multifaceted host defense-neutralizing activities.
Table 2.
Mammalian viruses with identified RNA silencing suppressors.
| Virus | Viral suppressors | References |
|---|---|---|
| HIV-1 | Tat protein, TAR RNA | [30], [32], [39], [50], [51], [52] |
| HTLV-1 | Rex protein | [33] |
| Influenza | NS1 protein | [29], [109] |
| HCV | Core protein, Envelope E2 protein | [31], [107] |
| Ebola | VP35, VP30 and VP40 proteins | [30], [105] |
| Vaccinia | E3L protein | [109] |
| PFV-1 | Tas protein | [22] |
| LACV | NSs protein | [108] |
| Adenovirus | VA RNA | [34] |
| SARS-CoV | 7a protein | [106] |
3. HIV and miRNAs
3.1. Multiple HIV-1–host RNAi interactions
For human retroviruses, it is instructive to examine the multitude of interactions between miRNAs and HIV. A hint that an RNAi-related mechanism might be employed by cells to regulate HIV-1 came from an observation that the knockdown of Dicer and Drosha, the two RNases involved in RNA silencing, increased HIV expression in infected cells [37]. The mechanistic explanation for this finding is in part due to miRNA mediated silencing [38], as well as an increase in HIV-1 mRNA translation following Dicer knockdown [39]. That the cell's RNAi machinery negatively modulates the HIV-1 lifecycle is consistent with an earlier suggestion that the HIV-1 RRE RNA is susceptible to processing into a short non-coding RNA (ncRNA) [32] by cellular RNAse. Indeed, a subsequent report found that the related HTLV-1 RxRE RNA motif is similarly sensitive to cellular RNase III cleavage [33]. Moreover, using next generation pyrosequencing, Yeung et al. reported that multiple RNA regions of HIV-1 can be found as processed viral ncRNAs in infected cells [40]. While the implications of many of these viral ncRNAs remain unknown, a small PBSncRNA, which likely resulted from Dicer-mediated processing of an RNA hybrid formed between cellular Lys3tRNA and the viral PBS sequence, was found to possess an anti-HIV-1 property [40], [41].
Similar to approaches that have been applied to other viruses, using multiple target prediction software [42], Hariharan et al. were the first to deduce 5 miRNA target sites in the HIV-1 genome [43] (Table 3 ). Thereafter, other investigators, using computational miRNA target site prediction followed by experimental validation, have substantiated the concept that cellular miRNAs can indeed act to modulate HIV expression and replication (Table 4 ). Among the initially identified 5 target sites, a miR-29a targeted site in the HIV-1 nef gene was shown by two independent research groups to interfere functionally with HIV replication in human cells [44], [45]. Separately, Huang et al. demonstrated that 5 other miRNAs, miR-28, miR-125b, miR-150, miR-223 and miR-382, converge to target the 3′ ends of HIV messenger RNAs to confer proviral latency in resting CD4+ T cells [46]. These “anti-HIV” miRNAs are also likely involved in the differential susceptibility of monocytes and differentiated macrophages to HIV infection [47]. Accordingly, when the expression of 4 of these anti-HIV miRNAs (except miR-223) was inhibited, HIV infection of monocytes was enhanced [48].
Table 3.
Human miRNA target sites predicted in the HIV and HTLV genomes using computational analysis.
| Virus | Human miRNA target sites | References |
|---|---|---|
| HIV-1 | Plus strand: miR-29a, miR-29b, miR-149, miR-324-5p, miR-378 | [43] |
| HTLV-1 | Plus strand: miR-653, miR-648, miR-596, miR-644, miR-496, miR-431, miR-326 | [83] |
| Plus strand: miR-125b, miR- 432, miR-125a, miR-663, miR-939, miR-1538, miR-1908 Minus stranda: miR-150, miR-24, miR-20b, miR-337, miR-432, miR-125a, miR-324-3p, miR-766, miR-1913 |
[82] |
Minus strand corresponds to the antisense HBZ primary transcript.
Table 4.
Human miRNAs experimentally shown to be involved in the regulation of HIV-1 expression.
| Human miRNAs | Direct (viral position)/indirect (targeted factor) | Involved in | References |
|---|---|---|---|
| miR-29a | Direct (nef) | Decrease HIV infectivity | [44], [45] |
| miR-28, miR-125b, miR-150, miR-223 and miR-382 | Direct (3′ LTR) | Latency in primary resting CD4+ T cells Increase monocyte/macrophage infection by HIV-1 Induced by opioid |
[46] [47] [48] |
| miR-17-5p; miR-20a | Indirect (PCAF) | Decrease HIV infection | [37] |
| miR-198 | Indirect (Cyclin T1) | Restrict HIV-1 replication in monocytes | [49] |
Cellular miRNAs can influence HIV replication in two ways — direct targeting of HIV-1 RNA sequences or indirect targeting of a cellular factor involved in HIV replication (Table 4). In the latter category, Triboulet et al. described a role for the miR-17/92 cluster via the production of miR-17-5p and miR-20a that target the mRNA coding for PCAF. PCAF is a cellular transcriptional cofactor of the viral Tat protein. Because Tat is required for productive transcription from the HIV-1 long terminal repeat (LTR) promoter, miR-17-5p and miR-20a enforced repression of PCAF results in reduced viral transcription and replication [37]. Another example of a miRNA-regulated factor linked to HIV-1 replication was reported by Sung et al. They described a miR-198 restriction of HIV infection in monocytes via its repression of another Tat co-factor, Cyclin T1 [49].
3.2. Viral RNA silencing suppressors (RSS)
How does HIV-1 replicate in the face of the various RNAi-restrictions as outlined above? One idea is that the virus can apparently weaken RNAi-restriction by encoding RNA silencing suppressors (RSS). The HIV-1 RSS can come in two forms. First, its Tat protein, an avid RNA-binding moiety, has RSS activity [30], [32], [39], [50], [51]. Second, the virus' TAR RNA is capable of acting as a miRNA-decoy. Thus, an RSS activity has been described by Bennasser et al. for the HIV-1 TAR RNA which obstructs RNAi by binding and sequestering TRBP, an essential Dicer-cofactor [52]. There are other ways that the virus could suppress RNAi-attack. For example, the viral core proteins sheath the incoming viral RNA genome and make it inaccessible to cellular RNAi protecting the RNA during the early phase of the infection [53] and ensuring that HIV-1 can reach the strategic step of establishing a stably integrated provirus. Thus, by using Tat, TAR RNA and other means (reviewed in [18]), HIV-1 has evolved ways to evade RNAi-mediated restriction somewhat analogous to the manner that the virus maintains Vif and Vpu proteins to combat APOBEC- and tetherin-mediated restrictions [54].
How RSS proteins like Tat and Rex might work mechanistically is worth some discussion because while many studies have reported their activity [30], [32], [33], [39], [50] some investigators have measured weak to no effect [55]. Like many plant virus RNAi suppressors [56], [57], [58], both the viral Tat and Rex polypeptides are double stranded RNA-binding proteins. Double-stranded RNA-binding proteins as a general class of polypeptides have been characterized to be capable of acting as RNAi-suppressors that ameliorate cellular antiviral defenses [59]. A recent study has clarified that the mechanism of these RSS proteins is to bind reversibly siRNAs or miRNAs in multiple turnover reactions to outcompete cellular Dicer and RISC constituents [60]. Given that the mechanism of this class of RNAi suppressors is based on competitive binding with multiple turnovers, it is not surprising that in settings when siRNAs, miRNAs, Dicer and RISC constituents are in excess no measurable RSS activity can be observed from these viral RNAi-suppressors. Such results are compatible with the fundamental competitive mechanism of RNA-binding RSS proteins [60]. Indeed, the recent identification of a small poly-l-lysine peptide as a potent miRNA suppressor in an unbiased screen of a chemical library [61] is consistent with double-stranded RNA-binding [60] as an RSS mechanism for lysine- and arginine-rich proteins like Tat and Rex. It is also likely that the competitive binding of HIV-1 TAR RNA [52] or adenovirus VA RNA [62] to RNAi-pathway constituents explains their RSS activities. In such a mechanistic scenario, there will be settings where TAR and VA RNAs outcompete siRNAs and miRNAs for binding to Dicer and RISC, and other settings where they do not. However, in HIV-1 infection, one notes that TAR RNA is the single most abundant viral RNA motif in virus-infected cells since it is always present in two copies (at the 5′ end and at the 3′ end) in all HIV-1 transcripts [52], [63]. TAR RNA is also transcribed from the HIV-1 LTR promoter in large abundance as short abortive transcripts in infected cells [64].
3.3. Mutational escape from RNAi-restriction
An RSS-independent way for the virus to adapt to RNAi-restriction would be to mutate its nucleic acid sequence. Because RNAi is a basepairing-complementarity driven process, the high propensity for mutations could help HIV escape miRNA-mediated silencing. That HIV-1 can remedy RNAi-restriction through sequence mutation is supported by the observation that early attempts to repress HIV-1 replication using a single synthetic siRNA or a single shRNA-expressing vector rapidly selected for RNAi-escape genomes with changes within [65], [66] or without [67], [68] the targeted sequence. This finding suggests that constant ambient RNAi exerted by cellular miRNAs could provide a selective force that continuously shapes the nucleotide sequence evolution of the HIV-1 genome.
3.4. miRNA profile changes in HIV-1 infection
Several studies have reported changes in the profile of cellular miRNAs following viral gene expression [37], [69]. Using a high throughput microarray method, Yeung et al. described that the expression of transfected HIV-1 molecular clones in HeLa cells downregulated more than 43% of the 312 miRNAs which were assayed [69]. Triboulet et al. also observed altered miRNA expression profiles in HIV-1 infected cells and found that the observed downregulation of the polycistronic miRNA cluster mir-17/92 was responsible for an increase in PCAF expression. PCAF is an important cofactor for Tat in HIV-1 gene expression, and its upregulation promotes more efficient viral replication [37]. Separately, Houzet et al. compared the miRNA profiles of 36 HIV-1 positive and 12 non-infected individuals and documented changed expression in 62 miRNAs [70]. Currently, the processes that regulate miRNA profile changes after virus-infection are unknown. Perhaps some of the in vivo changes are consequences of activation of signal pathways by innate or adaptive immune responses to the virus; other changes may be triggered by viral factors that alter the cellular transcriptome after infection.
3.5. Is TAR RNA a viral miRNA?
Viral miRNAs were first discovered in EBV by Pfeffer et al. [71]. Currently, more than 200 viral miRNAs have been identified in herpesviruses which have large genomes while viral miRNAs have been rarely reported in DNA viruses with smaller genomes [72], [73]. In the case of RNA viruses, some have suggested that viral RNA genomes that contain a miRNA would be labile and that the processing of the miRNA would be incompatible with viral replication. However, when miRNAs were deliberately positioned into lentiviral vectors, they created only very minor reductions in viral titers, suggesting that miRNA-processing in the context of an RNA genome remains compatible with viral replication [74].
Early in silico analyses of HIV-1 genome sequence suggested the possible existence of several candidate viral miRNAs [75]. Subsequently, a miRNA derived from the nef gene at the 3′ end of the HIV-1 genome, miR-N367, was reported experimentally [76], [77]. The HIV-1 TAR element is a stem loop RNA present at the 5′ extremity of the HIV genome and was predicted as a potential viral miRNA [75]. Interestingly, the TRBP protein which is a Dicer cofactor for miRNA biogenesis and which serves generally to bind miRNAs, facilitating their loading into the RISC complex, was originally cloned and identified based on its strong association with TAR RNA [5]. Accordingly, it is perhaps not surprising that several investigators have described and/or cloned small non-coding RNAs containing TAR RNA sequence from HIV-1 infected human cells [40], [78], [79]. One group has reported that TAR could be bound and processed by Dicer in vitro, and that a TAR derived RNA can be detected in chronically infected T cells [79]. These investigators detected TAR miRNA in both latent and productively infected cells and showed that it is involved in the downregulation of viral gene expression via the recruitment of chromatin remodeling components to the LTR [79]. More recently, TAR miRNA has been suggested to downregulate cellular apoptotic genes, protecting HIV-1 infected cells from apoptosis [80].
4. HTLV-1 and non-coding RNAs
4.1. HTLV-1 and miRNAs
Like HIV-1, human T cell leukemia virus type 1 (HTLV-1) also infects CD4+ T cells [81]. HTLV-1 encodes two major regulatory proteins, Tax and Rex. As mentioned above, it was shown recently that the Rex protein of HTLV-1 has an RSS activity, and that Rex interacts with RNA and Dicer to suppress the latter's ribonuclease activity [33]. This finding suggests that HTLV-1 may also have biological interactions with the host cell's RNAi machinery (Table 3).
Compared to HIV-1, less is known about HTLV-1 interaction with host cell RNAi activities. Two computational studies have predicted several host miRNA target sites in the HTLV genome [82], [83] (Table 3). These predictions provide a biological rationale for why the viral Rex protein might have an RSS function in order to quell miRNA-mediated restriction. To further understand HTLV-1–host miRNA interactions, two miRNA-profiling studies have been performed in infected cell lines and ATL (adult T-cell leukemia) cells [84], [85]. The two studies showed many divergent results, but they did find two common miRNAs that are consistently downregulated in the context of HTLV-1 infection [82]. These are interesting initial findings which need more investigation in order to understand the biological relevance of the downregulation of these miRNAs by HTLV-1 infection.
4.2. HTLV-1 HBZ antisense RNA
Human cells have a surfeit of non-coding RNAs that cannot be classified into known small RNA families (e.g. siRNA, piRNA, miRNA) [86]. In vertebrate viruses, it is apparent that many small non-coding viral RNAs (ncvRNAs) that are not siRNAs nor miRNAs are increasingly being documented [40], [87]. Currently, while there is no evidence for HTLV-1 encoded ncvRNAs, studies on HTLV-1 have firmly established the existence of a long antisense transcript that codes for an HBZ protein [88]. This antisense transcript is fully complementary to viral sense transcripts that are transcribed from the 3′ end of the HTLV-1 genome. Conceivably, long double stranded viral RNAs could form between viral sense and antisense transcripts, providing a substrate for Dicer-mediated production of multiple non-coding RNAs. Preliminary analyses by next generation pyrosequencing of small RNAs in HTLV-1 cell lines have identified many such small viral non-coding transcripts (Yeung, Chen and Jeang, unpublished). The functions of these non-coding HTLV-1 small transcripts, like human cell “dark matter” RNAs [86], await further clarification.
4.3. HTLV-1 and oncogenic miRNAs (oncomiRs)
HTLV-1 transforms T cells in vivo and etiologically causes ATL [89]. The virus encodes a Tax oncoprotein which has pleiotropic effects on several signaling pathways [90], [91], [92], [93]. An emerging concept is that small miRNAs can function as tumor suppressors and oncogenes, and therefore these entities could be termed oncogenic miRNAs or oncomiRs [15], [94]. To date, whether oncomiRs are important cofactors for HTLV-1 transformation of cells is poorly understood. Two miRNAs that are upregulated in HTLV-1 infected/transformed cells, miR-93 and miR-130b, have recently been shown to be instrumental in repressing a cellular tumor suppressor protein, TP53INP1 [84]. These two miRNAs have been proposed as the first examples of host oncomiRs that could be cofactors for HTLV-1 cellular transformation.
Integrating extant knowledge, a plausible biological scenario could be that the virus employs Tax-mediated protein signaling pathways [95] and Tax-induced oncomiR changes [96] to create oncogene-addicted and oncomiR-addicted stages of HTLV-1 cellular transformation [97]. The existence of oncomiR-addicted cellular transformation in tissue culture was recently supported by the discovery through an unbiased screening of a chemical library of two small molecule compounds that can suppress miRNA activities; and the finding that these compounds can successfully treat miR-93- and miR-130b-dependent tumorigenesis in a tissue culture/animal model [61]. The notion of oncomiR-addiction has been recently extended in vivo by Medina et al. who showed that mice overexpressing miR-21 develop tumors that can regress completely when miR-21 is inactivated [98].
5. Concluding remarks
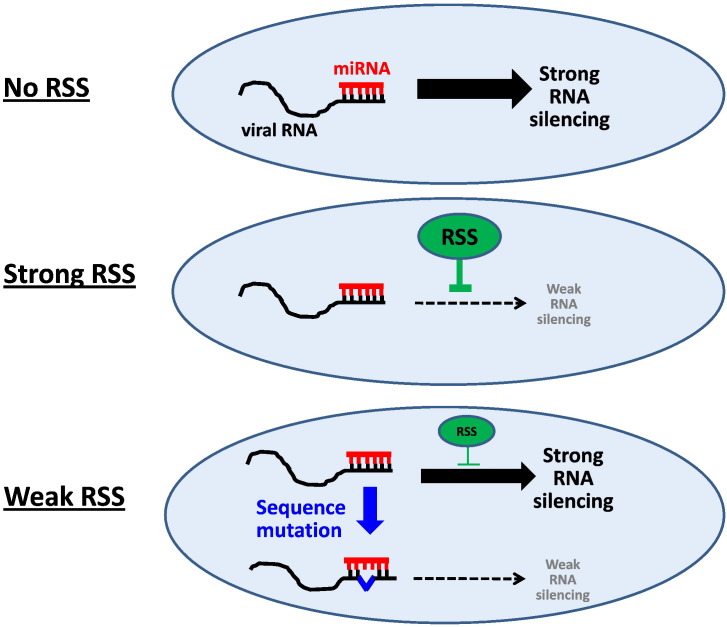
What are some general implications that one might learn from studying the interactions between human retroviruses and host cell RNAi? First, the existence of miRNAs that target human retroviral sequences can teach us about some of the selective pressures on the evolution of viral nucleotide sequences. For example, many of the miRNA–mRNA duplexes seen for HIV-1 [43], [44], [45], [46] and HTLV-1 [82], [83] are imperfect with three or more mispairings within a 21–22 stretch. Could it be that basepair complementarities at an earlier time were more perfectly matched? Could the current imperfect complementarities be RNAi-driven sequence “escapes” engineered by viruses to evade the otherwise more severe restriction enforced by perfect miRNA–mRNA basepairing? The biology of mammalian cells and their viruses may reflect an equilibrium whereby viral genomes change sequences that were originally perfect targets for miRNAs to sequences that are less perfectly complementary in order to ameliorate RNAi-based restriction. Viral genomes generally cannot change without some fitness cost; thus the currently maintained moderate miRNA–mRNA target pairings may represent a compromise between escape from RNAi and tolerable fitness loss from such nucleotide changes. Interestingly, unlike that seen in animal cells, plant cell miRNA–mRNA effector–target pairs are generally perfectly complementary without any mispairing [99]. It is unclear why this difference exists between animal and plant cells, but one explanation could be that RSS in animal cells may be less potent than their plant RSS counterparts. In a setting of less effective RSS function, RNAs that are targeted by highly complementary miRNAs may choose to mutate the sequence to avoid strong silencing (Fig. 2 ). On the other hand, when there is a strong RSS function, the selection pressure of highly complementary miRNA–mRNA basepairing is blunted; and thus there may be no need to evolve mRNA sequence changes in order to evade RNAi-restriction (Fig. 2).
Fig. 2.
Potential effects of RSS potency on the maintenance of miRNA-target site sequence complementarity. In the absence of RSS, miRNA–mRNA silencing is strong (No RSS). Strong RSS will moderate the RNAi-mediated silencing effect. Thus, in the setting of strong RSS, a perfect miRNA–mRNA complementarity could be tolerated without selecting for mRNA sequence changes (Strong RSS). In the case of weak RSS, the inhibition of the RNAi-mediated silencing by the RSS is modest and consequently will elicit a strong selection against a perfect miRNA–mRNA target complementarity (Weak RSS, top). In this setting, the viral mRNA target sequence may be selected for mutations in order to reduce base complementarity with the miRNA, thereby allowing the virus to evade an otherwise strong RNAi-mediated silencing (Weak RSS, bottom).
A second lesson from studying natural virus–miRNA interactions is that they could inform us on how to design better antiviral siRNAs. Conceptually, siRNA therapy is now well considered and is mediated using either synthetic siRNAs or vectors expressing small hairpin RNAs (shRNAs) which can be processed intracellularly to yield siRNAs. For HIV-1, one view is that siRNA-therapy could be an important adjunct to Highly Active Antiretroviral Therapy (HAART) which has documented toxicities and can quickly elicit the emergence of viral resistance. As mentioned above, HIV-1 rapidly escapes siRNA-restriction when the virus is targeted by only one siRNA [65], [66], [67], [68]. This viral escape can be countermanded when a combination of several discrete siRNAs is employed. ter Brake et al. have shown that a combination of four different shRNAs against highly conserved HIV-1 regions can minimize viral escape [100]. Similarly, a recent in silico modeling of HIV gene therapy using shRNA expressing vectors also illustrated that a combination of four shRNAs was required to suppress HIV-1, in order to prevent the development of resistance [101]. More recently, Schopman et al. showed that this viral escape could also be minimized by using second generation shRNAs which anticipate the viral escape by targeting the favored viral escape routes [102]. Whether synthetic siRNAs or vector-expressed shRNAs can be applied successfully toward future therapy remains unknown; however, a recent phase II gene therapy trial in HIV infected patients using the infusion of hematopoietic stem cells transduced with a plasmid expressing anti-HIV tat/vpr ribozyme does offer proof of principle that RNA-based gene therapy can be performed safely [103]. There remain major complexities to the appropriate in vivo delivery of RNAi effectors. Indeed, the challenges of these applications have prompted at least one major pharmaceutical firm to withdraw from RNAi research and development [104]. The quest for better understanding of the biology of retrovirus–RNAi interaction, therefore, holds many promises and many obstacles.
One of the initial suggestions that mammalian viruses encode RSS to combat functional RNAi-restriction was made for HIV-1 [32]. Many independent studies have now reported the finding of similar RSS in other mammalian viruses like the Ebola, hepatitis C, HTLV-1, SARS corona virus, and influenza viruses among others [30], [31], [33], [51], [62], [105], [106], [107], [108], [109] illustrating that the RSS strategy to counteract cellular RNAi may be common to many viruses. The finding of viral RSS proteins complements a multitude of reports that animal viruses are restricted by miRNAs in host cells [22], [23], [24], [25], [27], [44], [45], [46], [47], [48], [110], [111]. From a therapeutic perspective, future discoveries of small molecules that antagonize viral RSS activities could enhance natural host RNAi defenses against infection. The challenge in the coming years is to understand better how RNA-mediated surveillance integrates with protein-mediated and cell-mediated surveillances in defending humans against pathogens.
Acknowledgements
Work in our laboratory is supported in part by NIAID intramural fund and by the IATAP program from the Office of the Director, NIH. We thank members of our laboratory for critical readings of this manuscript.
Footnotes
This article is part of a Special Issue entitled: MicroRNAs in viral gene regulation.
Contributor Information
Laurent Houzet, Email: lhouzet@mail.nih.gov.
Kuan-Teh Jeang, Email: kj7e@nih.gov.
References
- 1.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 2.Chua J.H., Armugam A., Jeyaseelan K. MicroRNAs: biogenesis, function and applications. Curr. Opin. Mol. Ther. 2009;11:189–199. [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haase A.D., Jaskiewicz L., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gatignol A., Buckler-White A., Berkhout B., Jeang K.T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 6.Chi Y.H., Jeang K.T., Semmes O.J. A proteomic study of TAR-RNA binding protein (TRBP)-associated factors. Cell Biosci. 2011;1 doi: 10.1186/2045-3701-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wienholds E., Koudijs M.J., van Eeden F.J., Cuppen E., Plasterk R.H. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 9.Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris K.S., Zhang Z., McManus M.T., Harfe B.D., Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andl T., Murchison E.P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muljo S.A., Ansel K.M., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C.M. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C.M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lever A.M., Jeang K.T. Insights into cellular factors that regulate HIV-1 replication in human cells. Biochemistry. 2011;50:920–931. doi: 10.1021/bi101805f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umbach J.L., Cullen B.R. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassmann R., Jeang K.T. The roles of microRNAs in mammalian virus infection. Biochim. Biophys. Acta. 2008;1779:706–711. doi: 10.1016/j.bbagrm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung M.L., Benkirane M., Jeang K.T. Small non-coding RNAs, mammalian cells, and viruses: regulatory interactions? Retrovirology. 2007;4:74. doi: 10.1186/1742-4690-4-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe Y., Kishi A., Yachie N., Kanai A., Tomita M. Computational analysis of microRNA-mediated antiviral defense in humans. FEBS Lett. 2007;581:4603–4610. doi: 10.1016/j.febslet.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 21.Berkhout B., Jeang K.T. RISCy business: microRNAs, pathogenesis, and viruses. J. Biol. Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 22.Lecellier C.H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C., Saib A., Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 23.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 24.Zhang G.L., Li Y.X., Zheng S.Q., Liu M., Li X., Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Potenza N., Papa U., Mosca N., Zerbini F., Nobile V., Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. Feb 11 2011 doi: 10.1093/nar/gkr067. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng S.Q., Li Y.X., Zhang Y., Li X., Tang H. MiR-101 regulates HSV-1 replication by targeting ATP5B. Antiviral Res. 2011;89:219–226. doi: 10.1016/j.antiviral.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Song L., Liu H., Gao S., Jiang W., Huang W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010;84:8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bivalkar-Mehla S., Vakharia J., Mehla R., Abreha M., Kanwar J.R., Tikoo A., Chauhan A. Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res. 2011;155:1–9. doi: 10.1016/j.virusres.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delgadillo M.O., Saenz P., Salvador B., Garcia J.A., Simon-Mateo C. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J. Gen. Virol. 2004;85:993–999. doi: 10.1099/vir.0.19735-0. [DOI] [PubMed] [Google Scholar]
- 30.Haasnoot J., de Vries W., Geutjes E.J., Prins M., de Haan P., Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Kato N., Jazag A., Dharel N., Otsuka M., Taniguchi H., Kawabe T., Omata M. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology. 2006;130:883–892. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 32.Bennasser Y., Le S.Y., Benkirane M., Jeang K.T. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Abe M., Suzuki H., Nishitsuji H., Shida H., Takaku H. Interaction of human T-cell lymphotropic virus type I Rex protein with Dicer suppresses RNAi silencing. FEBS Lett. 2010;584:4313–4318. doi: 10.1016/j.febslet.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 34.Lu S., Cullen B.R. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai R., Carpick B., Chun R.F., Jeang K.T., Williams B.R. HIV-I Tat inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch. Biochem. Biophys. 2000;373:361–367. doi: 10.1006/abbi.1999.1583. [DOI] [PubMed] [Google Scholar]
- 36.McMillan N.A., Chun R.F., Siderovski D.P., Galabru J., Toone W.M., Samuel C.E., Mak T.W., Hovanessian A.G., Jeang K.T., Williams B.R. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 37.Triboulet R., Mari B., Lin Y.L., Chable-Bessia C., Bennasser Y., Lebrigand K., Cardinaud B., Maurin T., Barbry P., Baillat V., Reynes J., Corbeau P., Jeang K.T., Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 38.Chable-Bessia C., Meziane O., Latreille D., Triboulet R., Zamborlini A., Wagschal A., Jacquet J.M., Reynes J., Levy Y., Saib A., Bennasser Y., Benkirane M. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. 2009;6:26. doi: 10.1186/1742-4690-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian S., Zhong X., Yu L., Ding B., de Haan P., Boris-Lawrie K. HIV-1 Tat RNA silencing suppressor activity is conserved across kingdoms and counteracts translational repression of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 2009;106:605–610. doi: 10.1073/pnas.0806822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeung M.L., Bennasser Y., Watashi K., Le S.Y., Houzet L., Jeang K.T. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral–cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pederson T. Regulatory RNAs derived from transfer RNA? RNA. 2010;16:1865–1869. doi: 10.1261/rna.2266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindow M., Gorodkin J. Principles and limitations of computational microRNA gene and target finding. DNA Cell Biol. 2007;26:339–351. doi: 10.1089/dna.2006.0551. [DOI] [PubMed] [Google Scholar]
- 43.Hariharan M., Scaria V., Pillai B., Brahmachari S.K. Targets for human encoded microRNAs in HIV genes. Biochem. Biophys. Res. Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 44.Nathans R., Chu C.Y., Serquina A.K., Lu C.C., Cao H., Rana T.M. Cellular microRNA and P bodies modulate host–HIV-1 interactions. Mol. Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahluwalia J.K., Khan S.Z., Soni K., Rawat P., Gupta A., Hariharan M., Scaria V., Lalwani M., Pillai B., Mitra D., Brahmachari S.K. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang J., Wang F., Argyris E., Chen K., Liang Z., Tian H., Huang W., Squires K., Verlinghieri G., Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Ye L., Hou W., Zhou Y., Wang Y.J., Metzger D.S., Ho W.Z. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113:671–674. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., Ye L., Zhou Y., Liu M.Q., Zhou D.J., Ho W.Z. Inhibition of anti-HIV microRNA expression: a mechanism for opioid-mediated enhancement of HIV infection of monocytes. Am. J. Pathol. 2011;178:41–47. doi: 10.1016/j.ajpath.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sung T.L., Rice A.P. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of Cyclin T1. PLoS Pathog. 2009;5:e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnettler E., de Vries W., Hemmes H., Haasnoot J., Kormelink R., Goldbach R., Berkhout B. The NS3 protein of rice hoja blanca virus complements the RNAi suppressor function of HIV-1 Tat. EMBO Rep. 2009;10:258–263. doi: 10.1038/embor.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Vries W., Haasnoot J., Fouchier R., de Haan P., Berkhout B. Differential RNA silencing suppression activity of NS1 proteins from different influenza A virus strains. J. Gen. Virol. 2009;90:1916–1922. doi: 10.1099/vir.0.008284-0. [DOI] [PubMed] [Google Scholar]
- 52.Bennasser Y., Yeung M.L., Jeang K.T. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J. Biol. Chem. 2006;281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- 53.Westerhout E.M., ter Brake O., Berkhout B. The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference. Retrovirology. 2006;3:57. doi: 10.1186/1742-4690-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strebel K., Luban J., Jeang K.T. Human cellular restriction factors that target HIV-1 replication. BMC Med. 2009;7:48. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin J., Cullen B.R. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemmes H., Lakatos L., Goldbach R., Burgyan J., Prins M. The NS3 protein of rice hoja blanca tenuivirus suppresses RNA silencing in plant and insect hosts by efficiently binding both siRNAs and miRNAs. RNA. 2007;13:1079–1089. doi: 10.1261/rna.444007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merai Z., Kerenyi Z., Kertesz S., Magna M., Lakatos L., Silhavy D. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 2006;80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silhavy D., Molnar A., Lucioli A., Szittya G., Hornyik C., Tavazza M., Burgyan J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lichner Z., Silhavy D., Burgyan J. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 2003;84:975–980. doi: 10.1099/vir.0.18987-0. [DOI] [PubMed] [Google Scholar]
- 60.Rawlings R.A., Krishnan V., Walter N.G. Viral RNAi suppressor reversibly binds siRNA to outcompete Dicer and RISC via multiple turnover. J. Mol. Biol. Apr 29 2011;408(2):262–276. doi: 10.1016/j.jmb.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watashi K., Yeung M.L., Starost M.F., Hosmane R.S., Jeang K.T. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J. Biol. Chem. 2010;285:24707–24716. doi: 10.1074/jbc.M109.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andersson M.G., Haasnoot P.C., Xu N., Berenjian S., Berkhout B., Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berkhout B., Silverman R.H., Jeang K.T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 64.Kao S.Y., Calman A.F., Luciw P.A., Peterlin B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by Tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 65.Das A.T., Brummelkamp T.R., Westerhout E.M., Vink M., Madiredjo M., Bernards R., Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boden D., Pusch O., Lee F., Tucker L., Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leonard J.N., Shah P.S., Burnett J.C., Schaffer D.V. HIV evades RNA interference directed at TAR by an indirect compensatory mechanism. Cell Host Microbe. 2008;4:484–494. doi: 10.1016/j.chom.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Westerhout E.M., Ooms M., Vink M., Das A.T., Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeung M.L., Bennasser Y., Myers T.G., Jiang G., Benkirane M., Jeang K.T. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Houzet L., Yeung M.L., de Lame V., Desai D., Smith S.M., Jeang K.T. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5:118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfeffer S., Zavolan M., Grasser F.A., Chien M., Russo J.J., Ju J., John B., Enright A.J., Marks D., Sander C., Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 72.Grundhoff A., Sullivan C.S. Virus-encoded microRNAs. Virology. Mar 15 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boss I.W., Plaisance K.B., Renne R. Role of virus-encoded microRNAs in herpesvirus biology. Trends Microbiol. 2009;17:544–553. doi: 10.1016/j.tim.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y.P., Vink M.A., Westerink J.T., Ramirez de Arellano E., Konstantinova P., Ter Brake O., Berkhout B. Titers of lentiviral vectors encoding shRNAs and miRNAs are reduced by different mechanisms that require distinct repair strategies. RNA. 2010;16:1328–1339. doi: 10.1261/rna.1887910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bennasser Y., Le S.Y., Yeung M.L., Jeang K.T. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Omoto S., Fujii Y.R. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J. Gen. Virol. 2005;86:751–755. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- 77.Omoto S., Ito M., Tsutsumi Y., Ichikawa Y., Okuyama H., Brisibe E.A., Saksena N.K., Fujii Y.R. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ouellet D.L., Plante I., Landry P., Barat C., Janelle M.E., Flamand L., Tremblay M.J., Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klase Z., Kale P., Winograd R., Gupta M.V., Heydarian M., Berro R., McCaffrey T., Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klase Z., Winograd R., Davis J., Carpio L., Hildreth R., Heydarian M., Fu S., McCaffrey T., Meiri E., Ayash-Rashkovsky M., Gilad S., Bentwich Z., Kashanchi F. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology. 2009;6:18. doi: 10.1186/1742-4690-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsuoka M., Jeang K.T. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: viral infectivity, Tax, HBZ and therapy. Oncogene. Mar 24 2011;30(12):1379–1389. doi: 10.1038/onc.2010.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ruggero K., Corradin A., Zanovello P., Amadori A., Bronte V., Ciminale V., D'Agostino D.M. Role of microRNAs in HTLV-1 infection and transformation. Mol. Aspects Med. 2010;31:367–382. doi: 10.1016/j.mam.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 83.Hakim S.T., Alsayari M., McLean D.C., Saleem S., Addanki K.C., Aggarwal M., Mahalingam K., Bagasra O. A large number of the human microRNAs target lentiviruses, retroviruses, and endogenous retroviruses. Biochem. Biophys. Res. Commun. 2008;369:357–362. doi: 10.1016/j.bbrc.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 84.Yeung M.L., Yasunaga J., Bennasser Y., Dusetti N., Harris D., Ahmad N., Matsuoka M., Jeang K.T. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 2008;68:8976–8985. doi: 10.1158/0008-5472.CAN-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bellon M., Lepelletier Y., Hermine O., Nicot C. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood. 2009;113:4914–4917. doi: 10.1182/blood-2008-11-189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kapranov P., St Laurent G., Raz T., Ozsolak F., Reynolds C.P., Sorensen P.H., Reaman G., Milos P., Arceci R.J., Thompson J.F., Triche T.J. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parameswaran P., Sklan E., Wilkins C., Burgon T., Samuel M.A., Lu R., Ansel K.M., Heissmeyer V., Einav S., Jackson W., Doukas T., Paranjape S., Polacek C., dos Santos F.B., Jalili R., Babrzadeh F., Gharizadeh B., Grimm D., Kay M., Koike S., Sarnow P., Ronaghi M., Ding S.W., Harris E., Chow M., Diamond M.S., Kirkegaard K., Glenn J.S., Fire A.Z. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsuoka M., Green P.L. The HBZ gene, a key player in HTLV-1 pathogenesis. Retrovirology. 2009;6:71. doi: 10.1186/1742-4690-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matsuoka M., Jeang K.T. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer. 2007;7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 90.Peloponese J.M., Jr., Jeang K.T. Role for Akt/protein kinase B and activator protein-1 in cellular proliferation induced by the human T-cell leukemia virus type 1 Tax oncoprotein. J. Biol. Chem. 2006;281:8927–8938. doi: 10.1074/jbc.M510598200. [DOI] [PubMed] [Google Scholar]
- 91.Iha H., Kibler K.V., Yedavalli V.R., Peloponese J.M., Haller K., Miyazato A., Kasai T., Jeang K.T. Segregation of NF-kappaB activation through NEMO/IKKgamma by Tax and TNFalpha: implications for stimulus-specific interruption of oncogenic signaling. Oncogene. 2003;22:8912–8923. doi: 10.1038/sj.onc.1207058. [DOI] [PubMed] [Google Scholar]
- 92.Rosin O., Koch C., Schmitt I., Semmes O.J., Jeang K.T., Grassmann R. A human T-cell leukemia virus Tax variant incapable of activating NF-kappaB retains its immortalizing potential for primary T-lymphocytes. J. Biol. Chem. 1998;273:6698–6703. doi: 10.1074/jbc.273.12.6698. [DOI] [PubMed] [Google Scholar]
- 93.Jin D.Y., Teramoto H., Giam C.Z., Chun R.F., Gutkind J.S., Jeang K.T. A human suppressor of c-Jun N-terminal kinase 1 activation by tumor necrosis factor alpha. J. Biol. Chem. 1997;272:25816–25823. doi: 10.1074/jbc.272.41.25816. [DOI] [PubMed] [Google Scholar]
- 94.Esquela-Kerscher A., Slack F.J. Oncomirs — microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 95.Yasunaga J., Jeang K.T. Viral transformation and aneuploidy. Environ. Mol. Mutagen. 2009;50:733–740. doi: 10.1002/em.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jeang K.T. HTLV-1 and adult T-cell leukemia: insights into viral transformation of cells 30 years after virus discovery. J. Formos. Med. Assoc. 2010;109:688–693. doi: 10.1016/S0929-6646(10)60112-X. [DOI] [PubMed] [Google Scholar]
- 97.Jeang K.T. Human T cell leukemia virus type 1 (HTLV-1) and oncogene or oncomiR addiction? Oncotarget. 2010;1:453–456. doi: 10.18632/oncotarget.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Medina P.P., Nolde M., Slack F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 99.Millar A.A., Waterhouse P.M. Plant and animal microRNAs: similarities and differences. Funct. Integr. Genomics. 2005;5:129–135. doi: 10.1007/s10142-005-0145-2. [DOI] [PubMed] [Google Scholar]
- 100.ter Brake O., t Hooft K., Liu Y.P., Centlivre M., von Eije K.J., Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 101.Applegate T.L., Birkett D.J., McIntyre G.J., Jaramillo A.B., Symonds G., Murray J.M. In silico modeling indicates the development of HIV-1 resistance to multiple shRNA gene therapy differs to standard antiretroviral therapy. Retrovirology. 2010;7:83. doi: 10.1186/1742-4690-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schopman N.C., ter Brake O., Berkhout B. Anticipating and blocking HIV-1 escape by second generation antiviral shRNAs. Retrovirology. 2010;7:52. doi: 10.1186/1742-4690-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mitsuyasu R.T., Merigan T.C., Carr A., Zack J.A., Winters M.A., Workman C., Bloch M., Lalezari J., Becker S., Thornton L., Akil B., Khanlou H., Finlayson R., McFarlane R., Smith D.E., Garsia R., Ma D., Law M., Murray J.M., von Kalle C., Ely J.A., Patino S.M., Knop A.E., Wong P., Todd A.V., Haughton M., Fuery C., Macpherson J.L., Symonds G.P., Evans L.A., Pond S.M., Cooper D.A. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat. Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blow N.S. Our crystal ball. Biotechniques. 2011;50:77. doi: 10.2144/000113603. [DOI] [PubMed] [Google Scholar]
- 105.Fabozzi G., Nabel C.S., Dolan M.A., Sullivan N.J. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 2011;85:2512–2523. doi: 10.1128/JVI.01160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Karjee S., Minhas A., Sood V., Ponia S.S., Banerjea A.C., Chow V.T., Mukherjee S.K., Lal S.K. The 7a accessory protein of severe acute respiratory syndrome coronavirus acts as an RNA silencing suppressor. J. Virol. 2010;84:10395–10401. doi: 10.1128/JVI.00748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ji J., Glaser A., Wernli M., Berke J.M., Moradpour D., Erb P. Suppression of short interfering RNA-mediated gene silencing by the structural proteins of hepatitis C virus. J. Gen. Virol. 2008;89:2761–2766. doi: 10.1099/vir.0.2008/002923-0. [DOI] [PubMed] [Google Scholar]
- 108.Soldan S.S., Plassmeyer M.L., Matukonis M.K., Gonzalez-Scarano F. La Crosse virus nonstructural protein NSs counteracts the effects of short interfering RNA. J. Virol. 2005;79:234–244. doi: 10.1128/JVI.79.1.234-244.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li W.X., Li H., Lu R., Li F., Dus M., Atkinson P., Brydon E.W., Johnson K.L., Garcia-Sastre A., Ball L.A., Palese P., Ding S.W. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Otsuka M., Jing Q., Georgel P., New L., Chen J., Mols J., Kang Y.J., Jiang Z., Du X., Cook R., Das S.C., Pattnaik A.K., Beutler B., Han J. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 111.Pedersen I.M., Cheng G., Wieland S., Volinia S., Croce C.M., Chisari F.V., David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]