Abstract
The natural ligands for family B G protein-coupled receptors are moderate length linear peptides having diffuse pharmacophores. The amino-terminal regions of these ligands are critical for biological activity, with their amino-terminal truncation leading to production of orthosteric antagonists. The carboxyl-terminal regions of these peptides are thought to occupy a ligand-binding cleft within the disulfide-bonded amino-terminal domains of these receptors, with the peptides in amphipathic helical conformations. In the current work, we have characterized the binding and activity of a series of 11 truncated and lactam-constrained secretin(5-27) analogues at the prototypic member of this family, the secretin receptor. One peptide in this series with lactam connecting residues 16 and 20 (c[E16,K20][Y10]sec(5-27)) improved the binding affinity of its unconstrained parental peptide 22-fold, while retaining absence of endogenous biological activity and competitive antagonist characteristics. Homology modeling with molecular mechanics and molecular dynamics simulations established that this constrained peptide occupies the ligand-binding cleft in orientation similar to natural full-length secretin, and provided insights into why this peptide was more effective than other truncated conformationally-constrained peptides in the series. This lactam bridge is believed to stabilize an extended α-helical conformation of this peptide while in solution and to not interfere with critical residue-residue approximations while docked to the receptor.
Keywords: Secretin, secretin receptor, family B G protein-coupled receptor, antagonist, lactam bridge, ligand binding
The secretin receptor is a prototypic member of family B G protein-coupled receptors (GPCRs) that contains a number of important drug targets. Members in this family include receptors for glucagon-like peptide-1 (GLP-1), glucagon, glucose-dependent insulinotropic polypeptide, parathyroid hormone (PTH), calcitonin, corticotropin releasing factor (CRF), vasoactive intestinal polypeptide (VIP), and pituitary adenylate cyclase-activating peptide (PACAP), and have potential roles in the management of many human diseases, such as diabetes, bone disease, neuropsychiatric, and even neoplastic disorders 1, 2. Understanding of the molecular basis for the binding and action of natural ligands for these receptors can provide insights useful in the development of drugs acting at their orthosteric binding sites with the capacity to produce a spectrum of biological effects. An important component in understanding receptor activation is the proposed two-site model of ligand interaction in which the peptide carboxyl terminus is thought to bind to the folded, disulfide-constrained receptor amino terminus and the peptide amino terminus is then directed to interact with the receptor helical bundle, thus initiating the conformational change in the receptor that results in biological activity 3-5.
All natural ligands for family B GPCRs are moderate length peptides that contain diffuse pharmacophores extending throughout the length of these ligands 2, 6. Secretin is quite typical, being a 27-residue carboxyamidated linear peptide that is produced in the small intestinal mucosa and secreted in response to luminal acid, resulting in the stimulation of biliary and pancreatic ductular bicarbonate and water secretion, and thereby regulating the pH of the duodenal contents 7, 8. Like all of these peptides, the carboxyl-terminal portion is most critical for high affinity binding, while the amino-terminal portion is most critical for receptor selectivity and activation 9. Residues throughout the length of secretin have been shown to contribute to receptor binding and biological activity 10. In a hydrophobic environment similar to the membrane, secretin has been shown to adopt a conformation consisting of two helical segments involving residues 7-13 and 17-25 11-14.
Family B GPCRs possess a characteristic amino-terminal structure established by three intradomain disulfide bonds, with two antiparallel β-sheets and several loops 15-24. This structure provides a long peptide-binding cleft that has been shown in NMR and crystal structures of isolated receptor amino-terminal domains and interacting peptides to accommodate α-helical portions of the peptides 15-24. Indeed, in intact receptor mutagenesis, ligand structure-activity, and photoaffinity labeling studies, the carboxyl-terminal regions of the natural ligands have been shown to interact with this domain 25-34. The receptor docking of the amino-terminal ends of the natural peptide ligands has been more difficult to define. This portion of the peptide ligands seems to be directed toward the receptor helical bundle and loop regions 3-5, but no consistent site of docking has yet been established. Even modeling such an interaction has been problematic, due to the absence of a three-dimensional structure for the intact receptors in this family and due to data suggesting that even the helical bundle may be structurally distinct from that characteristic of family A GPCRs 35-37. Recently, we have proposed a ligand-bound intact secretin receptor model that accommodates all existing experimental data, including all structure-activity observations, as well as spatial approximation constraints provided by 11 sets of photoaffinity labeling data and 16 sets of fluorescence resonance energy transfer distances 38.
It has long been recognized that amino-terminal truncation of natural peptide ligands for family B GPCRs results in competitive antagonists 39-41. However, because the peptide amino terminus also contributes receptor-binding determinants, truncation often leads to reduced binding affinity 39-44. Secretin is typical of this theme 10, 40, 42-44. It is necessary to truncate the amino-terminal four residues to eliminate all endogenous agonist activity from secretin, but the resulting peptide has a binding affinity more than two orders of magnitude lower than that of the natural ligand 40, 42-44. This likely reflects the absence of some critical binding determinants as well as the possibility that the helical conformation of this peptide is less stable due to loss of an effective helix N-capping motif 45.
A goal of this work was to utilize lactam bonds to constrain secretin analogues and thereby provide insights into the conformations of secretin that might be accommodated within the normal peptide-binding cleft within the receptor amino terminus. Utilizing these constraints within truncated secretin peptides had the additional potential benefit of helping to create more effective receptor antagonists. For this, we prepared a series of truncated secretin(5-27) analogues incorporating a lactam bridge constraint in various positions. Of the 11 such analogues, one with a lactam bridge linking residues in positions 16 and 20 showed significant improvement of binding affinity and inhibition of secretin-stimulated biological activity. Additionally, full length secretin analogues incorporating the lactam bridge in the same position maintained the same binding affinity and biological activity as natural secretin.
MATERIALS AND METHODS
Materials
Fmoc amino acids used for peptide synthesis were purchased from Advanced ChemTech (Louisville, KY) and PAL resin was from Sigma-Aldrich (St. Louis, MO). Ham's F-12 medium and soybean trypsin inhibitor were from Invitrogen (Carlsbad, CA). Bovine serum albumin was from Serologicals Corp. (Norcross, GA). 3-isobutyl-1-methylxanthine was from Sigma. Fetal clone II culture medium supplement was from Hyclone laboratories (Logan, UT). The solid-phase oxidant, N-chlorobenzenesulfonamide (IODO-BEADs) was purchased from Pierce Chemical Co (Rockford, IL). The secretin-like radioligand, [Tyr10]rat secretin(1-27), was prepared as we previously described 46. All other reagents were of analytical grade.
Peptide Synthesis
The 15 secretin analogues prepared for the current study are illustrated in Figure 1. These include [Tyr10]human secretin(1-27) ([Y10]sec(1-27)) (1), [Y10]human secretin(5-27) ([Y10]sec(5-27)) (2), and a series of [Tyr10]sec(5-27) analogues that contain a lactam bridge between lysine and glutamic acid residues present and spaced three to four residues apart within the peptide (3-13). Additionally, two lactam-constrained full length secretin analogues, cyclic [Glu16,Lys20][Tyr10]human secretin(1-27) (c[E16,K20][Y10]sec(1-27)) and cyclic [Glu16,Lys20]human secretin(1-27) (c[E16,K20]sec(1-27)) were synthesized (14,15). Each of the peptides (1-14) in the series incorporated a tyrosine in position 10 to replace the natural leucine in that position for accurate determination of peptide concentrations by UV absorption spectrometry and for potential use for radioiodination. This modification has previously been shown to be well tolerated, not interfering with peptide binding or biological activity 47, 48.
Figure 1.
Primary structures of secretin analogues used in this study. Shown are the amino acid sequences of natural human secretin (sec(1-27), [Y10]sec(1-27) (1), [Y10]sec(5-27) (2), and lactam-constrained truncated (3-13) and full length (14 and 15) secretin analogues. Natural residues are illustrated in grey, while modified residues are illustrated in black. Lactam bridges linking the side chains of Lys and Glu residues three to four positions apart are illustrated with solid lines, and identified as cyclo (c) analogues.
Each of the peptides described in Figure 1 was synthesized using standard solid-phase techniques with 0.25 mmol PAL resin (substitution, 0.6 mmol/gm) and 1 mmol of each amino acid in its Fmoc-protected form, using double coupling as necessary based on testing each cycle of synthesis for completion, as we have described previously 49. N-α-Fmoc-N-ε-4-methyltrityl-L-lysine and N-α-Fmoc-L-glutamic acid-γ-2-phenylisopropyl ester were incorporated into the peptides in the positions of the lactam bridges (Fig. 1) during synthesis. After the addition of all residues for each peptide, while the peptide was still on the resin, the methytrityl and phenylisopropyl protection groups were removed using 10 ml of 1.8% trifluoroacetic acid in CH2CL2 per gm of resin (3 min × 10) until the ninhydrin test became positive 50. The resin-bound peptide was then washed with dimethylformamide, and N,N,N′,N′-tetra-methyl-O-(1H-benzotriazol-1-yl)-uronium hexafluoro-phosphate (1.2 mmol) and N,N-diisopropylethylamine (2 mmol) were added to allow the formation of the lactam bridge through the ε-amino group of lysine and δ-carboxyl group of glutamic acid. This typically was allowed to react for two h until the ninhydrin test became negative. The peptide was then washed with dimethylformamide and the α-amino group Fmoc protection was removed with 20% piperidine in dimethylformamide. The peptide was then cleaved from the resin using a solution of 6.25% (wt/vol) phenol, 2% (vol/vol) triisopropylsilane, 4% (vol/vol) thioanisole, 4% (vol/vol) distilled water, and 83% (vol/vol) trifluoroacetic acid. This also removed all side chain-protecting groups. The peptide was then precipitated in ether and lyophilized. The yield of crude peptide was typically in the range of 10-15 percent of that theoretically possible if all resin sites were fully utilized. All the peptides were dissolved in 10% acetonitrile and purified to homogeneity by reversed-phase HPLC using an octadecylsilane column running a 10 to 60% acetonitrile concentration gradient in the presence of 0.1% trifluoroacetic acid. Purified peptide represented 3-8 percent of the crude peptide collected (typical profiles in Supplementary Material). Retention times and molecular masses, as determined by matrix-assisted laser desorption/ionization-time of flight mass spectrometry, are shown in Table 1.
Table 1.
Characterization of secretin analogues by mass spectrometry and HPLC. Molecular masses of the synthetic products were determined by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Purified peptides were analyzed by reversed-phase HPLC on an octadecylsilane column running a 10 to 60% acetonitrile gradient with a background of 0.1% trifluoroacetic acid.
| No. | Secretin analogues | Calculated mass (Da) | Measured mass (Da) | Retention time (min) |
|---|---|---|---|---|
| 1 | [Y10]sec(l-27) | 3089.0 | 3087.4 | 27.4 |
| 2 | [Y10]sec(5-27) | 2693.4 | 2694.2 | 27.9 |
| 3 | c[K5,E9][Y10]sec(5-27) | 2702.5 | 2702.5 | 27.8 |
| 4 | c[K6,E9][Y10]sec(5-27) | 2648.4 | 2648.8 | 27.0 |
| 5 | c[E11,K14][Y10]sec(5-27) | 2689.4 | 2689.7 | 29.5 |
| 6 | c[K11,E15][Y10]sec(5-27) | 2716.5 | 2714.5 | 28.1 |
| 7 | c[E15,K18][Y10]sec(5-27) | 2647.4 | 2647.6 | 31.0 |
| 8 | c[El6,K20][Yl0]sec(5-27) | 2747.5 | 2747.6 | 29.4 |
| 9 | c[E17,K20][Y10]sec(5-27) | 2733.5 | 2733.7 | 29.4 |
| 10 | c[E17,K21][Y10]sec(5-27) | 2705.4 | 2706.0 | 29.9 |
| 11 | c[E18,K21][Y10]sec(5-27) | 2620.3 | 2620.4 | 31.5 |
| 12 | c[E21,K24][Y10]sec(5-27) | 2648.4 | 2648.8 | 30.3 |
| 13 | c[E21,K25][Y10]sec(5-27) | 2719.4 | 2719.8 | 32.6 |
| 14 | c[E16,K20][Y10]sec(1-27) | 3144.0 | 3145.3 | 29.3 |
| 15 | c[E16,K20]sec(1-27) | 3094.0 | 3094.0 | 30.0 |
Radioiodination
The secretin radioligand was prepared by the oxidative radioiodination of [Tyr10]rat secretin(1-27) using 1 mCi Na125I and exposure to the IODO-BEAD solid-phase oxidant for 15 s in 0.1 M borate buffer (pH 9), as we have previously described 49. The product was purified using reversed-phase HPLC to yield a specific radioactivity of approximately 2,000 Ci/mmol 49.
Cell Line and Membrane Preparation
The previously established Chinese hamster ovary cell line expressing the human secretin receptor (CHO-SecR cells) 51 was used as a source of receptor for the current study. Cells were cultured at 37 °C in an environment containing 5% CO2 on tissue culture plasticware in Ham's F-12 medium supplemented with 5% Fetal Clone II and were passaged approximately twice a week. Enriched plasma membranes from CHO-SecR cells were prepared using discontinuous sucrose gradient centrifugation 52 and were stored in aliquots in Krebs-Ringers/HEPES (KRH) medium (25 mM HEPES, pH 7.4, 104 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM KH2PO4, 1.2 mM MgSO4) containing 0.01% soybean trypsin inhibitor and 1 mM phenylmethylsulfonyl fluoride at -80 °C until use.
Receptor Binding Assays
The receptor binding characteristics of each of the secretin analogues listed in Table 1 were determined using receptor-bearing membranes and a radioligand competition-binding assay. In brief, approximately 5-10 μg of CHO-SecR cell membranes were incubated with a constant amount of secretin radioligand (approximately 10,000 cpm, representing 1-3 pM) and increasing concentrations of [Tyr10]sec(1-27) (1), [Tyr10]sec(5-27) (2), or the lactam-constrained [Tyr10]sec(5-27) or [Tyr10]sec(1-27) analogues (3-15) (from 0 to 1 μM) in KRH medium containing 0.01% soybean trypsin inhibitor, 1 mM phenylmethylsulfonyl fluoride and 0.2% bovine serum albumin for 1 h at room temperature (reaction volume, 100 μl) in a 96-well plate. Separation of bound from free radioligand was performed using a UniFilter-96 in a FilterMate harvester (PerkinElmer), with bound radioactivity quantified using a TopCount spectrometer (PerkinElmer). Non-saturable binding, determined in the presence of 1 M [Tyr10]sec(1-27), represented less than 20% of total radioligand bound. Binding data were expressed as percentages of saturable binding with the nonsaturable portion subtracted.
cAMP Assays
The biological activity of each of the full length secretin analogues (1, 14 and 15) was determined by examining their ability to stimulate cAMP responses in CHOSecR cells. The truncated secretin(5-27) analogues are known to represent antagonists 40, 43, 53. The ability of selected members of this series ([Tyr10]sec(5-27) (2) and c[E16,K20][Y10]sec(5-27) (8) to antagonize the biological activity stimulated by [Tyr10]sec(1-27) (1) was determined by evaluating cAMP responses in CHO-SecR cells. In brief, after cells (~8,000 cells per well in 96-well plates) were grown for two days, they were washed with phosphate-buffered saline (PBS) and stimulated with various concentrations of secretin, its analogues, and various combinations of these ligands in KRH medium containing 0.01% soybean trypsin inhibitor, 0.2% bovine serum albumin, 0.1% bacitracin, and 1 mM 3-isobutyl-1-methylxanthine for 30 min at 37 °C. After cells were lysed with 6% ice-cold perchloric acid for 15 min with vigorous shaking, the lysates were adjusted to pH 6 with 30% NaHCO3. The cAMP levels in the lysates were determined in a 384-well white Optiplate using a LANCE kit from PerkinElmer (Boston, MA).
Computational Methods
Molecular mechanics simulations of the peptides
Molecular mechanics simulations of the isolated full-length secretin peptide [Y10]sec(1-27) (1), the truncated secretin peptide [Y10]sec(5-27) (2), and lactam analogues of the truncated peptide (3-13) (Table 1) were performed using the Internal Coordinate Mechanics (ICM) 54 program (version 3.6, Molsoft LLC). The lactam bridges were built by mutating the appropriate residues to Glu and Lys and creating the peptide bond between the side chains of those residues. Starting from the extended peptide chain, peptide folding simulations were performed by sampling the conformational space of the peptides in three independent runs of biased-probability Monte Carlo (BPMC) 55 simulations with 2×108 function calls at T = 600 K. The BMPC simulations minimize an energy function that is a sum of ECEPP/3 energy 56-58, side chain entropy 55, and implicit solvation 59 terms. This reflects the average properties of bulk water molecules, with their dielectric constant, rather than the explicit use of water molecules in the simulation.
Molecular dynamics simulations of the complex
Initial complexes of full length secretin, truncated secretin, and the lactam analogues of truncated secretin in which the lactam might directly modify peptide docking with the receptor amino-terminal domain were built based on the crystal structure of the complex of GLP-1 and its receptor amino-terminal domain (PDB: 3IOL) 24. ICM was then used for the homology modeling, wherein the sequences of the peptide and receptor were aligned and tethered to the GLP-1 peptide-receptor structure. It should be emphasized that this approach was designed to optimize the likelihood that each of the peptides would assume the normal helical conformation after docking, even if they did not assume such conformations while in solution. The secretin peptide-receptor structures were annealed to the tethers and the conformational space of the complex was sampled using 5×106 function calls of BPMC 55 simulations. The conformation with the best energy from the Monte Carlo simulation was used to provide the initial coordinates for the molecular dynamics (MD) simulations.
MD simulations in explicit solvent (including water molecules) were performed using the CHARMM force field 60 with CMAP 61 in the GROMACS software package (version 4.5.3) 62, 63. Each complex was solvated in TIP3P water model 64 and the system was made neutral by adding the appropriate number of counter ions. All systems were equilibrated under a canonical (NVT) ensemble for 200 ps using the thermostat described by Bussi et al. 65, with a relaxation time of 0.1 ps for temperature control. The systems were further equilibrated in an isothermal-isobaric (NPT) ensemble for another 500 ps using the Parrinello-Rahman barostat 66 with a relaxation time of 2.0 ps. The reference temperature and pressure for all systems were 300 K and 0.1 MPa, respectively, approximately corresponding to physiological conditions. Position restraints (with spring constant of 1000 kJ/mol·nm2) were applied to all heavy atoms of the protein complex during the equilibration. Following equilibration, 20 ns production MD using 2-fs time steps and the linear constraint solver method 67 to constrain all bond lengths was conducted per system using an NPT ensemble. The Lennard-Jones interactions were switched off between 10 Å and 12 Å and the neighbor list was updated every 10 fs. Electrostatic interactions were treated with particle mesh Ewald method 68 with fourth-order spline interpolation and 1.6 Å grid spacing and a short-range cut-off of 13 Å. Coordinates were saved every 1 ps for analysis using the built-in analysis tools in GROMACS. The analyses were performed for the last 10 ns of the simulation to ensure that the complex had adequate time to diverge from its initial structure and to sample local (atomic fluctuation and side chain motion) and medium-scale (loop motion) motions to gain insights into the peptide docking flexibility.
Since the length of the MD simulations does not allow for global motions, such as peptide dissociation, the energy components of the complexes were analyzed using ICM. Coordinates were extracted every 25 ps for the last 10 ns of the MD simulations. Monte Carlo side-chain optimizations were performed with ~15,000 functional calls for each structure. The energy components were calculated between the peptide region extending from residue 15 to residue 25 and all receptor residues with atoms within 5.0 Å of the peptide. In addition, the surface energy, defined as the product of the total solvent-accessible area and the surface tension parameter (0.020 kcal/mol·Å2) 59, was calculated for the complex and for each of its components.
Statistical Analysis
All biological assays were performed in duplicate in a minimum of three independent experiments and are expressed as the means ± S.E.M. Receptor binding and cAMP concentration-response curves were analyzed and plotted using the non-linear regression analysis program in the Prism software suite v3.0 (GraphPad Software, San Diego, CA). Binding kinetics were determined by analysis with the LIGAND program of Munson and Rodbard 69. Two-tailed P value tests were performed to determine the significance of data differences using InStat3 (GraphPad Software, San Diego, CA).
Computational analyses were presented as means ± S.D. for the data from three independent molecular mechanics simulations and for the data representing every 1 ps during the last 10 ns of the molecular dynamics simulations.
RESULTS
Peptides
Fifteen human secretin analogues, 13 of which contained a lactam bridge (Fig. 1), were synthesized by solid phase techniques and were purified by reversed-phase HPLC to exceed purities of 92 percent. The chemical identities of the purified products were verified by mass spectrometry. Table 1 shows the calculated and measured masses as well as the retention times for these peptides.
Binding Affinity of the Lactam-Constrained Secretin Analogues
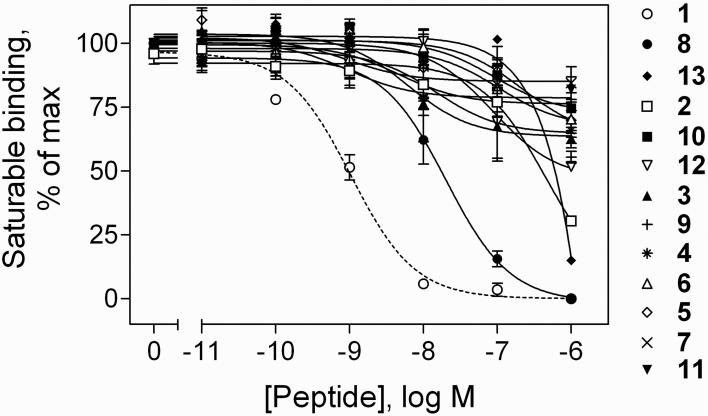
Figure 2 illustrates the receptor binding characteristics of each of the secretin analogues. Of all the truncated peptides tested (2-13), only the c[E16,K20][Y10]sec(5-27) (8) was able to fully compete for all saturable binding of the secretin radioligand to CHO-SecR membranes. Although c[E16,K20][Y10]sec(5-27) (8) had a lower affinity than that of the full length secretin peptide, [Y10]sec(1-27) (1) (Ki = 1.0 ± 0.2 nM), its affinity was significantly higher than that of [Y10]sec(5-27) (2). The peptide with the lactam constraint linking residues 16 and 20, c[E16,K20][Y10]sec(5-27) (8), had an IC50 value (concentration of this peptide competing for one half of the saturable binding of the radioligand) of 19 ± 6 (Ki =18 ± 5 nM), while its parental peptide, [Y10]sec(5-27) (2), had an IC50 of 410 ± 59 nM and did not fully displace all saturable radioligand binding even at 1 M (Fig. 2). The only other peptide that reached 50% of inhibition of saturable radioligand binding was c[E21,K25][Y10]sec(5-27) (13), with an IC50 of 690 ± 78 nM (Fig. 2). All other lactam-constrained peptides (2-7 and 9-12) had affinities greater than 1 M.
Figure 2.
Binding affinities of the lactam-constrained [Y10]sec(5-27) analogues. Shown are curves reflecting the ability of increasing concentrations of [Y10]sec(1-27) (1), [Y10]sec(5-27) (2), or lactam-constrained [Y10]sec(5-27) analogues (3-13) to compete for binding of the secretin radioligand, [Y10]rat sec(1-27), to CHO-SecR cell membranes. Values illustrated represent percentages of saturable binding, expressed as the means ± S.E.M. of duplicate values from a minimum of three independent experiments.
Antagonist Activity of the Lactam-Constrained Secretin Analogues
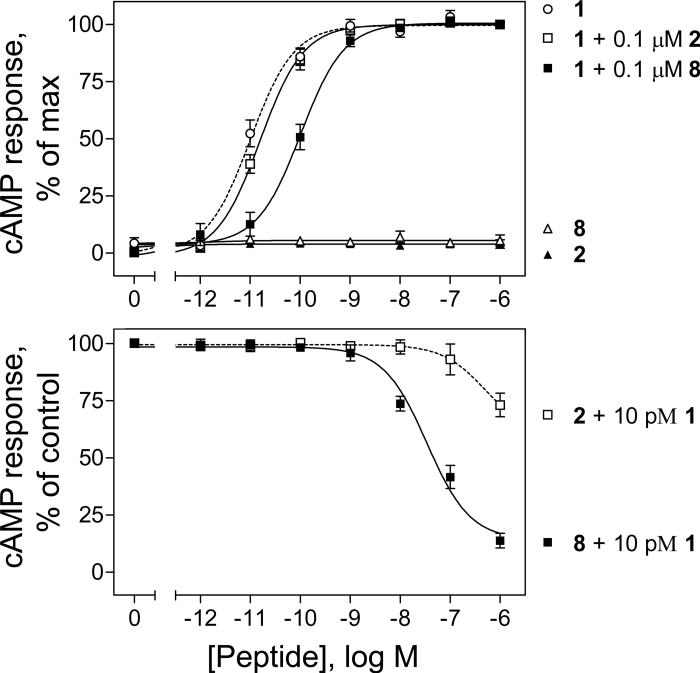
Figure 3 shows that c[E16,K20][Y10]sec(5-27) (8) was a secretin antagonist, having no endogenous activity to stimulate cAMP in CHO-SecR cells at a concentration as high as 1 M. The [Y10]sec(1-27) (1)-stimulated concentration-dependent cAMP responses in CHO-SecR cells were shifted to the right by 0.1 M c[E16,K20][Y10]sec(5-27) (8). The EC50 value for the agonist in the presence of this antagonist was 103 ± 19 pM, significantly different from that of stimulation by [Y10]sec(1-27) (1) alone (10 ± 2 pM). In addition, c[E16,K20][Y10]sec(5-27) (8) inhibited 10 pM [Y10]sec(1-27) (1)-stimulated cAMP responses in a concentration-dependent manner (IC50 = 34 ± 8 nM) (Fig. 3). In contrast, the parental peptide [Y10]sec(5-27) (2), previously reported as a secretin antagonist 40, 42-44, was much less effective in inhibiting [Y10]sec(1-27) (1)-induced cAMP responses in CHO-SecR cells (Fig. 3). The EC50 value for concentration-dependent cAMP responses in CHO-SecR cells stimulated by [Y10]sec(1-27) (1) in the presence of [Y10]sec(5-27) (2) was 16 ± 4 pM, not significantly different from that stimulated by [Y10]sec(1-27) (1) alone. In addition, [Y10]sec(5-27) (2) was only able to inhibit 10 pM [Y10]sec(1-27) (1)-stimulated cAMP responses by 27% at a concentration of 1 μM.
Figure 3.
Antagonist activities of [Y10]sec(5-27) (2) and its lactam-constrained [E16,K20][Y10]sec(5-27) (8) analogue. Top, intracellular cAMP responses to increasing concentrations of [Y10]sec(1-27) (1) or [Y10]sec(5-27) (2) or c[E16,K20][Y10]sec(5-27) (8) or [Y10]sec(1-27) (1) in the presence of 0.1 M [Y10]sec(5-27) (1 + 2) or c[E16,K20][Y10]sec(5-27) (1 + 8) in CHO-SecR cells. Bottom, intracellular cAMP responses in CHO-SecR cells by 10 pM [Y10]sec(1-27) in the presence of increasing concentrations of [Y10]sec(5-27) (2) or c[E16,K20][Y10]sec(5-27) (8). Data points represent the means ± S.E.M. of three independent experiments performed in duplicate, normalized relative to the maximal responses in these cells to [Y10]sec(1-27) (1). Basal and maximal cAMP levels stimulated by [Y10]sec(1-27) (1) were 4.0 ± 0.9 and 197 ± 51 pmol/million cells, respectively.
Binding and Biological Activities of Lactam-Constrained Full Length Secretin Analogues
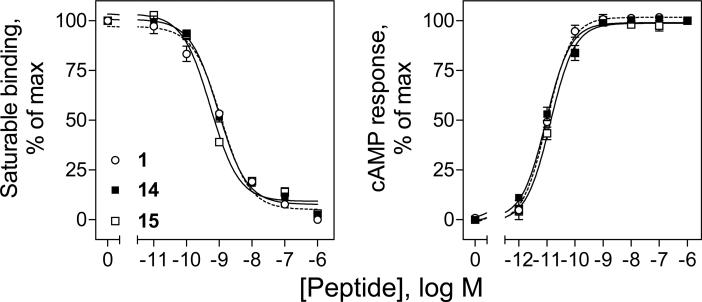
As described above, the best position for incorporation of a lactam bond was between residues 16 and 20 of secretin. Therefore, we prepared two full length secretin analogues incorporating a lactam bond in this position with and without a tyrosine in position 10, i.e. c[E16,K20][Y10]sec(1-27) (14) and c[E16,K20]sec(1-27) (15). Figure 4 shows that each peptide exhibited similar abilities to compete for secretin radioligand binding, reflecting affinities similar to the parental [Y10]sec(1-27) peptide (1). These full length secretin analogues represented full agonists, stimulating cAMP responses in CHOSecR cells that were not significantly different from that elicited by [Y10]sec(1-27) (1).
Figure 4.
Binding and biological activities of lactam-constrained full length [Y10]sec(1-27) analogues. Left, curves reflecting the ability of increasing concentrations of [Y10]sec(1-27) (1) or c[E16,K20][Y10]sec(1-27) (14) or c[E16,K20]sec(1-27) (15) to compete for binding of the secretin radioligand, [Y10]rat sec(1-27) to CHO-SecR cell membranes. Values illustrated represent percentages of saturable binding, expressed as the means ± S.E.M. of duplicate values from a minimum of three independent experiments. Right, intracellular cAMP responses in CHO-SecR cells by [Y10]sec(1-27) (1) or c[E16,K20][Y10]sec(1-27) (14) or c[E16,K20]sec(1-27) (15). Data points represent the means ± S.E.M. of three independent experiments performed in duplicate, normalized relative to the maximal responses in these cells to [Y10]sec(1-27) (1).
Molecular Mechanics Simulations of the Secretin Peptides in Solution
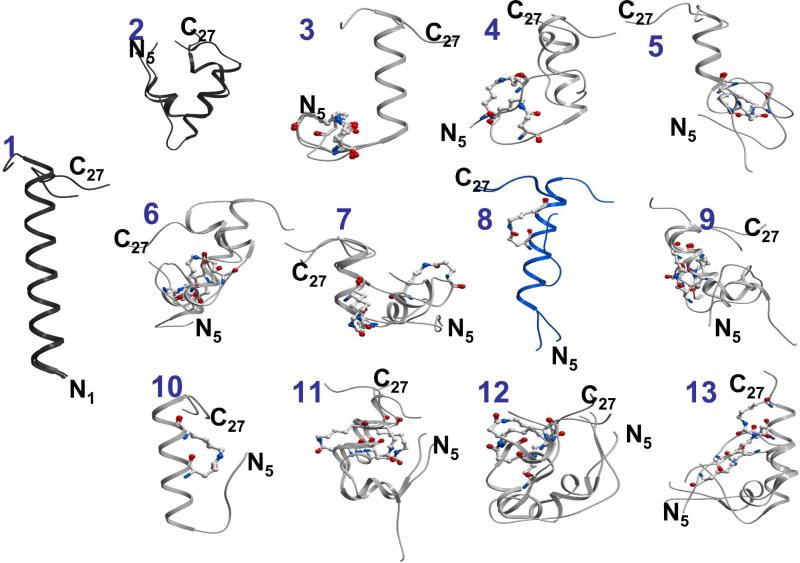
The isolated fully extended forms of the secretin peptides were allowed to fold using BPMC molecular mechanics simulations. The lowest energy conformations in three independent simulations for each of the peptides are shown in Figure 5, with their properties quantified in Table 2. Of note, the lowest energy conformations from three of the lactam-constrained peptides, c[K5,E9][Y10]sec(5-27) (3), c[E16,K20][Y10]sec(5-27) (8), and c[E17,K21][Y10]sec(5-27) (10), revealed that these peptides had a tendency to assume significantly more α-helical conformation than the unconstrained analogous parental peptide, [Y10]sec(5-27) (2), or than any of the other secretin analogues containing lactams. The numbers of residues involved in continuous α-helical structure were 12.7 ± 1.2, 14.7 ± 2.5 and 14 ± 0 for the three lactam-containing peptides noted above, respectively. These helices most consistently extended from peptide residue 11 to 22 for c[K5,E9][Y10]sec(5-27) (3), from residue 8 to 22 for c[E16,K20][Y10]sec(5-27) (8), and from residue 11 to 24 for c[E17,K21][Y10]sec(5-27) (10). When analyzed in the same way, the full length peptide, [Y10]sec(1-27) (1), had 21.7 ± 1.2 residues involved in continuous α-helical structure, extending from residue 2 to 22, although this represents a higher helical content than found for this peptide in a membrane-mimetic solvent using NMR approaches 11-14.
Figure 5.
Molecular mechanics simulations of the soluble peptides. Shown are the lowest energy conformations from three independent molecular mechanics simulations for each of the unbound lactam-constrained secretin analogues (3-13) shown in gray, with the c[E16,K20][Y10]sec(5-27) peptide (8) highlighted in blue, and the lactam bridges expanded and colored by atomic type. Also shown are the lowest energy conformations of the full length secretin peptide (1) and the truncated secretin(5-27) (2) in darker gray.
Table 2.
Properties of the unbound peptide from the lowest conformations of three independent runs per peptide as determined by molecular mechanics simulations in ICM.
| No. | Secretin analogues | Best energy (kcal/mol, ± S.D.) | Accessible surface area (Å2, ± S.D.) |
|---|---|---|---|
| 1 | [Y10]sec(1-27) | -408.5 ± 0.3 | 3116.3 ± 67.3* |
| 2 | [Y10]sec(5-27) | -356.1 ± 1.5 | 2608.8 ± 76.5 |
| 3 | c[K5,E9][Y10]sec(5-27) | -337.7 ± 0.4 | 2743.9 ± 108.1 |
| 4 | c[K6,E9][Y10]sec(5-27) | -336.8 ± 0.9 | 2694.8 ± 90.7 |
| 5 | c[E11,K14][Y10]sec(5-27) | -323.8 ± 0.5 | 2681.4 ± 100.1 |
| 6 | c[K11,E15][Y10]sec(5-27) | -336.7 ± 2.8 | 2669.1 ± 108.4 |
| 7 | c[E15,K18][Y10]sec(5-27) | -311.7 ± 1.4 | 2408.3 ± 142.3* |
| 8 | c[E16,K20][Y10]sec(5-27) | -347.8 ± 2.3 | 2745.6 ± 135.8 |
| 9 | c[E17,K20][Y10]sec(5-27) | -344.6 ± 0.3 | 2622.5 ± 55.6 |
| 10 | c[E17,K21][Y10]sec(5-27) | -331.5 ± 0.3 | 2489.6 ± 12.9* |
| 11 | c[E18,K21][Y10]sec(5-27) | -296.2 ± 0.1 | 2418.4 ± 151.3* |
| 12 | c[E21,K24][Y10]sec(5-27) | -312.6 ± 2.9 | 2476.6 ± 5.3* |
| 13 | c[E21,K25][Y10]sec(5-27) | -327.8 ± 2.8 | 2602.7 ± 175.9 |
significant differences from value for c[E16,K20][Y10]sec(5-27), p< 0.05
Molecular Dynamics of the Secretin Peptide-Receptor Complexes
Twenty ns MD simulations were performed for the six lactam-constrained secretin analogues in which the location of the lactam bridge is within the putative binding cleft of the receptor amino-terminal domain. The stability of the peptide-receptor complexes was determined through the root-mean-square fluctuation (RMSF) of the Cα in the residues for the last 10 ns of these simulations, a time period when the lactam analogues would have had enough time to diverge from their initial structures. The RMSF of the carboxyl-terminal region of these peptides were comparable, ranging from 0.6-3.8 Å, reflecting the stability of these complexes through the given simulation time. Similarly, the receptor amino terminus in these simulations also exhibited stability, with most residues having an RMSF in the same range. However, the amino-terminal region of the peptides, known to interact with the receptor transmembrane domain 4, 70, displayed substantially higher RMSF values in these simulations (up to 6 Å), because that portion of the receptor was not present to stabilize it in the structures used for analysis.
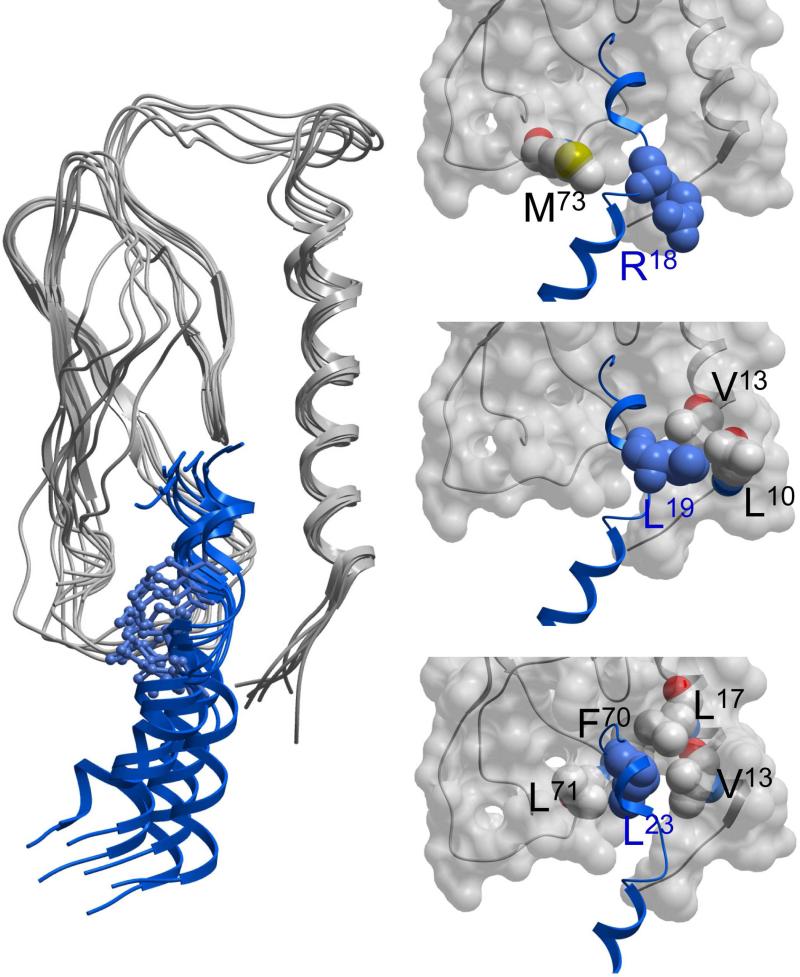
The distances from the receptor to specific residues within the secretin peptides (Arg18, Leu19, and Leu23) that were most closely approximated with residues within the homology model of the receptor amino-terminal domain in the complexes were also determined (shown in Table 3). Ligand residues 19 and 23 of the lactam-constrained secretin analogues seemed to be similarly oriented relative to the receptor amino-terminal residues as were [Y10]sec(5-27) (2) and [Y10]sec(1-27) (1). These secretin residues have been shown to represent critical binding determinants in alanine scanning studies of secretin in the peptide-receptor complex 10. The distances between these ligand residues and their closest receptor residues were not significantly different for any of the lactams than for the unconstrained peptide. Of note, Arg18 within the c[E16,K20][Y10]sec(5-27) peptide (8) was found to have a shorter distance to the proximate receptor residue, Met73, than any of the other lactam-constrained peptides, with this distance similar to that of the full length secretin peptide. Figure 6 illustrates the docking of this peptide with the amino-terminal domain of the secretin receptor. The relationships between the key residues in ligand and receptor are illustrated for a sample conformation in the MD simulation.
Table 3.
Minimum distances between particular residues within peptide-receptor pairs. The average distances (in Å units ± S.D.) were determined during the last 10 ns of the MD simulations.
| No. 1 | 2 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|
| [Y10]sec(1-27) | [Y10]sec(5-27) | c[E16,K20][Y10]sec(5-27) | c[E17,K20][Y10]sec(5-27) | c[E17,K21][Y10]sec(5-27) | c[E18,K21][Y10]sec(5-27) | c[E21,K24][Y10]sec(5-27) | c[E21,K25][Y10]sec(5-27) | |
| Arg18-Met73 | 9.55 ± 1.20 | 8.16 ± 0.87 | 9.82 ± 1.26 | 18.46 ± 1.60 | 16.08 ± 1.39 | 17.55 ± 1.27 | 11.79 ± 2.06 | 15.49 ± 0.69 |
| Leu19-Leu10 | 3.97 ± 0.28 | 3.53 ± 0.52 | 4.02 ± 0.35 | 4.13 ± 0.33 | 6.33 ± 0.79 | 4.28 ± 0.50 | 4.48 ± 0.95 | 9.37 ± 1.11 |
| Leu19-Val13 | 4.19 ± 0.56 | 4.63 ± 0.30 | 4.00 ± 0.32 | 4.10 ± 0.44 | 4.00 ± 0.33 | 3.88 ± 0.24 | 4.19 ± 0.43 | 7.40 ± 0.93 |
| Leu23-Val13 | 3.89 ± 0.29 | 8.07 ± 0.57 | 4.06 ± 0.35 | 4.06 ± 0.45 | 5.64 ± 0.73 | 4.28 ± 0.45 | 9.42 ± 0.50 | 4.21 ± 0.50 |
| Leu23-Leu17 | 4.05 ± 0.39 | 4.86 ± 0.65 | 4.87 ± 0.84 | 7.78 ± 1.17 | 4.37 ± 0.50 | 4.35 ± 0.56 | 7.62 ± 0.39 | 9.91 ± 0.59 |
| Leu23-Phe70 | 4.56 ± 0.78 | 5.37 ± 0.49 | 3.97 ± 0.38 | 3.96 ± 0.49 | 4.46 ± 0.69 | 3.87 ± 0.31 | 8.53 ± 0.66 | 4.17 ± 0.38 |
| Leu23-Leu71 | 8.95 ± 2.04 | 7.53 ± 0.85 | 4.25 ± 0.55 | 6.82 ± 1.00 | 4.49 ± 0.75 | 6.05 ± 0.84 | 4.00 ± 0.30 | 6.06 ± 0.35 |
Figure 6.
Molecular dynamics simulations of complexes including docked peptide and receptor amino terminus. Shown are snapshots of the MD simulations of the docked complexes of c[E16,K20][Y10]sec(5-27) (8) peptide (blue) and secretin receptor amino-terminal domain (gray) taken every 2 ns for t = 10 ns to 20 ns (left image), along with magnified views of the residues proposed as being involved in the interactions between this peptide and this region of the receptor (right three images). Residues of interest that are potentially involved in interactions are displayed as CPK representations, with receptor residues labeled in blue and peptide residues labeled in black.
The solvent-accessible surface areas (ASA) of the peptides during the last 10 ns of their MD simulations are shown in Table 4. Included are values for the intact complexes and for the peptide and receptor components of the complexes, as well as the change in accessible surface area (ΔASA = ASAcomplex - ASAreceptor - ASApeptide) that reflects the surface of the interface.
Table 4.
Features of the peptide-receptor complexes during the last 10 ns of the MD simulations. Shown are the solvent-accessible surface areas (ASA, in Å2 units ± S.D.), as well as the van der Waals, hydrogen bond, and electrostatic energies between residues 15 to 25 of the peptide and residues in the receptor with any atom within 5 Å of the peptide region and the change in surface energy between the complex and its components determined using ICM. All energies are in kcal/mol ± S.D.
| No. 1 | 2 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|
| Sec(1-27) | [Y10]sec(5-27) | c[E16,K20][Y10]sec(5-27) | c[E17,K20][Y10]sec(5-27) | c[E17,K21][Y10]sec(5-27) | c[E18,K21][Y10]sec(5-27) | c[E21,K24][Y10]sec(5-27) | c[E21,K25][Y10]sec(5-27) | |
| Hydrophobic ASA | ||||||||
| Complex | 3172 ± 80 | 2943 ± 109 | 3143 ± 84 | 3207 ± 85 | 2967 ± 80 | 3071 ± 84 | 2924 ± 111 | 3073 ± 91 |
| Receptor | 2650 ± 72 | 2459 ± 109 | 2675 ± 62 | 2546 ± 80 | 2504 ± 76 | 2576 ± 70 | 2543 ± 87 | 2451 ± 80 |
| Peptide | 915 ± 47 | 869 ± | 874 ± 32 | 853 ± 32 | 821 ± 30 | 777 ± 34 | 800 ± 32 | 865 ± 34 |
| ΔASA | -393 | -385 | -406 | -192 | -358 | -282 | -419 | -243 |
| Hydrophilic ASA | ||||||||
| Complex | 6171 ± 132 | 6129 ± 131 | 6053 ± 125 | 6161 ± 111 | 5880 ± 152 | 6144 ± 123 | 5934 ± 141 | 6128 ± 117 |
| Receptor | 5063 ± 102 | 5018 ± 116 | 5024 ± 94 | 5082 ± 115 | 4991 ± 116 | 5120 ± 104 | 5108 ± 115 | 5078 ± 99 |
| Peptide | 1874 ± 97 | 1711 ± 48 | 1686 ± 52 | 1583 ± 47 | 1507 ± 46 | 1545 ± 43 | 1577 ± 49 | 1641 ± 50 |
| ΔASA | -766 | -600 | -657 | -504 | -618 | -521 | -751 | -591 |
| Energy Analysis | ||||||||
| Van der Waals energy | -27.5 ± 3.1 | -20.9 ± 3.0 | -34.2 ± 3.1 | -22.4 ± 3.3 | -32.8 ± 3.4 | -24.4 ± 2.2 | -35.8 ± 3.6 | -20.8 ± 2.3 |
| Hydrogen bond energy | -0.9 ± 0.8 | -1.3 ± 1.3 | -3.7 ± 1.2 | -0.4 ± 0.8 | -3.1 ± 1.1 | -3.5 ± 1.6 | -0.9 ± 1.3 | -3.5 ± 2.5 |
| Electrostatic energy | 0.1 ± 0.4 | -0.3 ± 0.2 | -1.7 ± 0.4 | -0.2 ± 0.3 | -1.0 ± 0.4 | -1.4 ± 0.6 | -0.3 ± 0.3 | -1.0 ± 0.8 |
| ΔSurface energy | -28.6 ± 2.2 | -20.1 ± 1.4 | -27.7 ± 2.4 | -17.1 ± 1.6 | -24.8 ± 1.6 | -21.1 ± 2.2 | -28.8 ± 1.7 | -21.1 ± 1.4 |
In addition, the van der Waals, hydrogen bond, and electrostatic energies were calculated between peptide residues 15 to 25 and all neighboring receptor residues having atoms within 5.0 Å of this peptide region. Interestingly, c[E16,K20][Y10]sec(5-27) peptide (8) had more favorable hydrogen bond and electrostatic energies than the other lactam-constrained peptides (9-13). The van der Waals energy of peptide (8) was also more favorable than most lactam analogues, with the exception of peptide (12) (Table 4). Furthermore, the surface energy of the complex (ΔEsf = Esf,complex - Esf,receptor - ΔEsf,peptide) was determined in ICM as the product of the total solvent-accessible area and the surface tension parameter. The surface energy of the c[E16,K20][Y10]sec(5-27) peptide was more favorable than the other lactam analogues, with the exception of peptide (12). Since the tension parameter is constant, the favorable surface energy reflects the change in accessible surface area due to the formed interface.
DISCUSSION
Natural peptide ligands for receptors can provide leads for the development of drugs acting at their orthosteric binding sites, with primary structure-activity relationships helping to direct such efforts. For the natural ligands of family B GPCRs, it is recognized that the amino terminus is most responsible for biological activity and that truncations of that region result in competitive antagonists 39-41. However, due to the loss of important binding determinants that are also contributed by the ligand amino terminus, such truncated ligands often bind to their receptors with low affinity 39-44. Binding affinity of such truncated analogues may be further reduced by the potential loss of the helix N-capping motif, thought to contribute to the stabilization of the α-helical conformation of the portion of these peptides that docks within the peptide-binding cleft of the receptor amino-terminal domain.
In the current work, we have utilized an amino-terminally truncated form of secretin in which the first four residues were deleted, representing the shortest truncation that totally eliminated agonist activity 40, 42-44. However, that peptide had a very low binding affinity, representing a loss of greater than 200-fold. We designed and synthesized 11 analogues of this peptide that each incorporated a lactam bond constraint linking amino acids spaced three or four residues apart in an attempt to stabilize its α-helical structure. This had the additional advantage of providing insights into conformations compatible with binding of this peptide to its receptor.
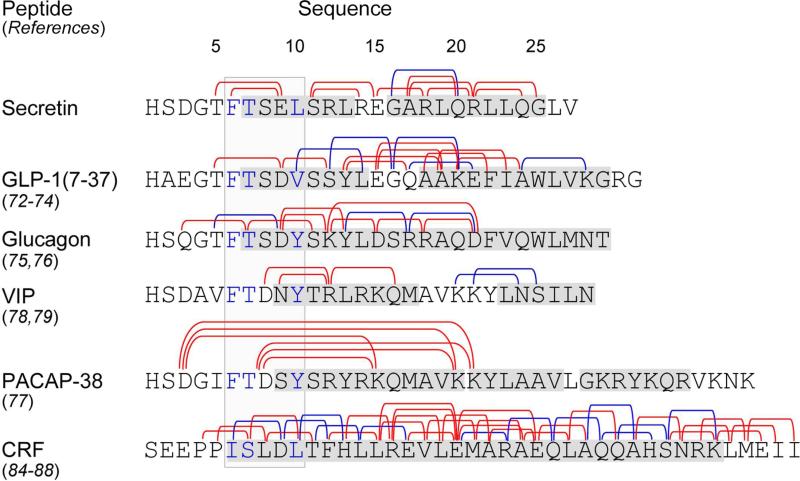
Such constraints have been incorporated into other members of this family, including receptors for GLP-1 71-73, glucagon 74, 75, PACAP 76, VIP 77, 78, calcitonin 79-82, CRF 83-87, PTH 88, 89, and PTH-related protein 90; however, these have typically been utilized in full length analogues that already bound to their receptors with high affinity 71-75, 79-82. Such series have been useful in determining which structural constraints might be compatible with receptor binding and biological activity. Figure 7 summarizes a large amount of data from the studies performed with the peptides that can be best aligned with secretin, illustrating that most such constraints have been found to interfere with binding and biological activity despite their possible stabilizing effect on helical structure. A large number of lactam-constrained PTH analogues have also been studied, but the structural differences between PTH and secretin make it difficult to directly compare those results.
Figure 7.
Summary of literature for family B GPCR lactam-constrained peptide ligands. Shown are the sequences of secretin and several closely-related family B GPCR ligands and the positions of lactam bridges incorporated in each peptide. Lactam constraints interfering with binding and biological activity are shown in red and those maintaining binding and biological activity are shown in blue. Shown in the shaded light gray box are the helix N-capping motifs, with key residues in positions 6, 7, and 10 highlighted in blue. Shaded in dark gray in each peptide sequence are residues shown to form α-helical structures in solution-phase NMR studies.
The peptides most closely related to secretin in which this experimental approach has been applied are GLP-1 and glucagon. It is encouraging that the single constrained secretin peptide that exhibited improved binding affinity, with its lactam bridge linking residues in positions 16 and 20, was analogous to one of the constrained GLP-1 peptides that was also effective in retaining the binding and biological activity of that ligand, although it did not improve those parameters in the full length GLP-1 analogue 71, 72. However, other lactams that were analogous to those that were also tolerated in GLP-1 and glucagon, such as those linking positions Gln23-Glu27 of GLP-1 72, 73 and positions Thr5-Asp9 and Arg17-Asp21 of glucagon 74, 75, did not improve secretin binding to its receptor in the current studies.
Review of the other data illustrated in Figure 7 reveals several successful applications of similar lactam constraints to other members of this family, with most of these stabilizing the mid-region and carboxyl terminus of those peptides. Indeed, this is the region of these ligands expected to be in α-helical conformation when docked within the peptide-binding cleft of these receptors. However, like the data described above, the absolute positions of the successful lactam constraints are not consistent across the family. While lactam bonds with this spacing likely stabilize α-helices in all of these peptides, this modification might disrupt binding if the bond were on the side of the helix that contacts the receptor within its peptide-binding cleft or if it involved a residue critical to the binding of that peptide. Apparently, the specific details of the binding of each of these peptides to their receptors vary in a receptor-specific manner.
In the current work, we have utilized the incorporation of lactam constraints into secretin analogues in a different way and for a different purpose than was attempted in many of these studies. While, like many of the other studies in family B GPCR ligands, it was critical for the lactams to be tolerated and to not eliminate critical binding determinants, for the current effort it was hoped that such a constraint would also enhance the stability and binding affinity of the peptide in which it was present. Of note, only one of these lactam-constrained [Y10]sec(5-27) (2) analogues achieved this goal, exhibiting significant improvement in binding affinity. A comprehensive computational strategy was therefore employed in an attempt to understand these results with this particular receptor. It was hoped that an analogous approach could ultimately be applied to other family members to gain insights into receptor-specific differences.
Natural peptide ligands bound to family B GPCRs are known to have amphipathic structures, whereby the hydrophobic residues face the receptor binding cleft, while the hydrophilic residues are more likely to be solvent-exposed. This suggests that hydrophobic interactions represent an important component of the binding mechanism. Indeed, in the computational simulations, all of the peptides were found to be capable of docking in such a manner. The extent of hydrophobic atoms being buried within this cleft in c[E16,K20][Y10]sec(5-27) was comparable to that in the docked truncated and full-length secretin peptides and, while greater than that in some of the lactam-constrained peptides, other lactam analogues also had substantial buried surface areas that were not significantly different from that of the 16-20 lactam analogue. The surface areas of hydrophilic atoms in the interface of the peptide-receptor complexes were not significantly different for any of the docked peptides. This suggests that most of the lactam-constrained peptides were theoretically capable of assuming an appropriate α-helical conformation and that their lactam bridges did not interfere with binding, however the experimental data clearly show that most of these ligands bound to the secretin receptor with very low affinities.
The effectiveness of the c[E16,K20][Y10]sec(5-27) peptide (8) in binding to the receptor might be due to a favorable helical conformation in the unbound form of the peptide or due to specific effective molecular interactions formed in the complex with the receptor or conceivably both of these factors. Molecular mechanics simulations of the isolated peptides in implicit solvent were performed to determine propensity of these peptides to form helical structures, while molecular dynamics simulations of the peptide-receptor complexes in explicit solvent were performed to gain insights into the energetic factors contributing to their receptor binding.
It should be noted that each molecular mechanics simulation of an isolated peptide produces only a single representative low energy structure. Furthermore, the unfavorable entropic contribution to the free energy for the coil to helix transition is not accounted for in this technique, resulting in conformations that may suggest the presence of excessive helical content. This is evident from the predicted helical conformation of the full-length secretin peptide, which has been experimentally found to be largely disordered in aqueous solution 13, 91, 92. However, the energetic preference for this peptide to be in the disordered state may be small, since changing the system slightly by the addition of trifluoroethanol or addition of its receptor can result in substantially more helical content 12, 13. Because the secretin analogues are all the same length and have a structurally similar single lactam bridge, the entropic contributions to folding that are not included in this calculation would be expected to be of similar magnitude, thus supporting the relative helical propensities predicted for the peptides in this series.
It is interesting that only three of the truncated secretin analogues (3, 8, and 10) were found to converge toward substantial α-helical structures, and the 16-20 lactam-containing peptide (8) was indeed one of these. This is expected to contribute to increased binding affinity because lactam-induced stabilization of the α-helical segment present in the unbound peptide conformation reduces the entropic penalty for forming the complex. It is possible that the other two peptide analogues had some steric clash or loss of a critical binding determinant when it came to docking at the receptor. Indeed, the lactam bridge of peptide (3) that extends from residues 5 to 9 may constrain the region involved in the putative helix N-capping motif (residues 6, 7, and 10) in that peptide, and thereby negatively affect its conformation. It is also noteworthy that the α-helix in the simulation of the 16-20 lactam-constrained peptide (8) extended further toward the amino terminus of this peptide than did the α-helix in the other two helical peptides.
The receptor interactions and proximity with peptide residues Arg18, Leu19, and Leu23 were all suggested to be effective in the c[E16,K20][Y10]sec(5-27) peptide (8) as sampled through MD simulations, with none of these distances significantly different from those in docked natural full-length secretin. When different energy terms were determined between residues 15 to 25 of the peptide and its neighboring receptor residues, c[E16,K20][Y10]sec(5-27) peptide (8) showed more favorable van der Waals, hydrogen bond, and electrostatic energies than most of the other lactam-constrained analogues. A comparison of the energy components of peptides (8) and (10), both of which were shown to be α-helical in the molecular mechanics simulations of the unbound peptide, showed that peptide (8) had slightly more favorable van der Waals, hydrogen bond, and electrostatic energies and change in surface energy than peptide (10), all of which might combine to favor the complex formation of (8) over (10). In addition, while peptide (12) was shown to have the most favorable van der Waals energy in the series, it had less favorable hydrogen bonding and electrostatic energies and significantly lower helicity than peptide (8).
The lactam bridge in the c[E16,K20][Y10]sec(5-27) peptide (8) should provide a useful constraint to incorporate in developing a more effective, high affinity receptor antagonist. The approaches to molecular modeling of this peptide and its complex with the amino terminus of the secretin receptor have provided important clues to the effectiveness of this peptide. It will be important to confirm this conformation experimentally using direct approaches, such as NMR analysis. These insights should ultimately contribute to the development of a more effective secretin receptor antagonist.
Supplementary Material
Supplementary Figure 1. HPLC profiles reflecting the purification of one of the lactam analogues of the secretin peptide, c[K6,E9][Y10]sec(5-27) (4). Shown are representative A280 absorbance profiles of the crude products of synthesis on a semi-preparative reversed-phase column and the final product on an analytical reversed-phase column (inset). The positions of migration of the compound of interest are marked with arrowheads in the profiles. This compound had the expected mass by mass spectrometry.
ACKNOWLEDGEMENTS
The authors thank Ms. Mary Lou Augustine for her technical assistance and Dr. Scott Struthers of Crinetics Pharmaceuticals for helpful discussions during the conception of this project.
This work was supported by a grant from the National Institutes of Health (DK46577) and by the Mayo Clinic. Salary support was provided by the Peking Union Medical College Hospital (XX and JW).
Abbreviations
- ASA
accessible surface area
- BMPC
biased-probability Monte Carlo
- CHO-SecR
Chinese hamster ovary cell line expressing the human secretin receptor
- CRF
corticotrophin releasing factor
- GLP-1
glucagon-like peptide-1
- GPCR
G protein-coupled receptor
- ICM
Internal Coordinate Mechanics
- KRH
Krebs-Ringers/HEPES
- MD
molecular dynamics
- PACAP
pituitary adenylate cyclase-activating peptide
- PTH
parathyroid hormone
- RMSF
root-mean-square fluctuation
- VIP
vasoactive intestinal polypeptide
Footnotes
Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
REFERENCES
- 1.Mayo KE, Miller LJ, Bataille D, Dalle S, Goke B, Thorens B, Drucker DJ. International Union of Pharmacology. XXXV. The glucagon receptor family. Pharmacol. Rev. 2003;55:167–194. doi: 10.1124/pr.55.1.6. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich CD, 2nd, Wood P, Hadac EM, Kopras E, Whitcomb DC, Miller LJ. Cellular distribution of secretin receptor expression in rat pancreas. Am. J. Physiol. 1998;275:G1437–1444. doi: 10.1152/ajpgi.1998.275.6.G1437. [DOI] [PubMed] [Google Scholar]
- 3.Bisello A, Adams AE, Mierke DF, Pellegrini M, Rosenblatt M, Suva LJ, Chorev M. Parathyroid hormone-receptor interactions identified directly by photocross-linking and molecular modeling studies. J. Biol. Chem. 1998;273:22498–22505. doi: 10.1074/jbc.273.35.22498. [DOI] [PubMed] [Google Scholar]
- 4.Dong M, Li Z, Pinon DI, Lybrand TP, Miller LJ. Spatial approximation between the amino terminus of a peptide agonist and the top of the sixth transmembrane segment of the secretin receptor. J. Biol. Chem. 2004;279:2894–2903. doi: 10.1074/jbc.M310407200. [DOI] [PubMed] [Google Scholar]
- 5.Dong M, Pinon DI, Cox RF, Miller LJ. Molecular approximation between a residue in the amino-terminal region of calcitonin and the third extracellular loop of the class B G protein-coupled calcitonin receptor. J. Biol. Chem. 2004;279:31177–31182. doi: 10.1074/jbc.M404113200. [DOI] [PubMed] [Google Scholar]
- 6.Segre GV, Goldring SR. Receptors for secretin, calcitonin, parathyroid hormone (PTH)/PTH-related peptide, vasoactive intestinal peptide, glucagonlike peptide 1, growth hormone-releasing hormone, and glucagon belong to a newly discovered G-protein-linked receptor family. Trends Endocrinol. Metab. 1993;4:309–314. doi: 10.1016/1043-2760(93)90071-l. [DOI] [PubMed] [Google Scholar]
- 7.Chey WY, Chang TM. Secretin, 100 years later. J. Gastroenterol. 2003;38:1025–1035. doi: 10.1007/s00535-003-1235-3. [DOI] [PubMed] [Google Scholar]
- 8.Hacki WH. Secretin. Clin. Gastroenterol. 1980;9:609–632. [PubMed] [Google Scholar]
- 9.Dong M, Miller LJ. Molecular pharmacology of the secretin receptor. Receptors Channels. 2002;8:189–200. [PubMed] [Google Scholar]
- 10.Dong M, Le A, Te JA, Pinon DI, Bordner AJ, Miller LJ. Importance of Each Residue within Secretin for Receptor Binding and Biological Activity. Biochemistry. 2011;50:2983–2993. doi: 10.1021/bi200133u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blankenfeldt W, Nokihara K, Naruse S, Lessel U, Schomburg D, Wray V. NMR spectroscopic evidence that helodermin, unlike other members of the secretin/VIP family of peptides, is substantially structured in water. Biochemistry. 1996;35:5955–5962. doi: 10.1021/bi9601520. [DOI] [PubMed] [Google Scholar]
- 12.Clore GM, Nilges M, Brunger A, Gronenborn AM. Determination of the backbone conformation of secretin by restrained molecular dynamics on the basis of interproton distance data. Eur. J. Biochem. 1988;171:479–484. doi: 10.1111/j.1432-1033.1988.tb13814.x. [DOI] [PubMed] [Google Scholar]
- 13.Gronenborn AM, Bovermann G, Clore GM. A 1H-NMR study of the solution conformation of secretin. Resonance assignment and secondary structure. FEBS Lett. 1987;215:88–94. doi: 10.1016/0014-5793(87)80119-9. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann M, Gondol D, Bovermann G, Nilges M. Conformation of secretin in dimethyl sulfoxide solution. NMR studies and restrained molecular dynamics. Eur. J. Biochem. 1989;186:95–103. doi: 10.1111/j.1432-1033.1989.tb15182.x. [DOI] [PubMed] [Google Scholar]
- 15.Grace CR, Perrin MH, DiGruccio MR, Miller CL, Rivier JE, Vale WW, Riek R. NMR structure and peptide hormone binding site of the first extracellular domain of a type B1 G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12836–12841. doi: 10.1073/pnas.0404702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grace CR, Perrin MH, Gulyas J, Digruccio MR, Cantle JP, Rivier JE, Vale WW, Riek R. Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4858–4863. doi: 10.1073/pnas.0700682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koth CM, Abdul-Manan N, Lepre CA, Connolly PJ, Yoo S, Mohanty AK, Lippke JA, Zwahlen J, Coll JT, Doran JD, Garcia-Guzman M, Moore JM. Refolding and characterization of a soluble ectodomain complex of the calcitonin gene-related peptide receptor. Biochemistry. 2010;49:1862–1872. doi: 10.1021/bi901848m. [DOI] [PubMed] [Google Scholar]
- 18.Parthier C, Kleinschmidt M, Neumann P, Rudolph R, Manhart S, Schlenzig D, Fanghanel J, Rahfeld JU, Demuth HU, Stubbs MT. Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13942–13947. doi: 10.1073/pnas.0706404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pioszak AA, Parker NR, Suino-Powell K, Xu HE. Molecular recognition of corticotropin-releasing factor by its G-protein-coupled receptor CRFR1. J. Biol. Chem. 2008;283:32900–32912. doi: 10.1074/jbc.M805749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pioszak AA, Xu HE. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5034–5039. doi: 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Runge S, Thogersen H, Madsen K, Lau J, Rudolph R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J. Biol. Chem. 2008;283:11340–11347. doi: 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- 22.Sun C, Song D, Davis-Taber RA, Barrett LW, Scott VE, Richardson PL, Pereda-Lopez A, Uchic ME, Solomon LR, Lake MR, Walter KA, Hajduk PJ, Olejniczak ET. Solution structure and mutational analysis of pituitary adenylate cyclase-activating polypeptide binding to the extracellular domain of PAC1-RS. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7875–7880. doi: 10.1073/pnas.0611397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ter Haar E, Koth CM, Abdul-Manan N, Swenson L, Coll JT, Lippke JA, Lepre CA, Garcia-Guzman M, Moore JM. Crystal structure of the ectodomain complex of the CGRP receptor, a class-B GPCR, reveals the site of drug antagonism. Structure. 2010;18:1083–1093. doi: 10.1016/j.str.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Underwood CR, Garibay P, Knudsen LB, Hastrup S, Peters GH, Rudolph R, Reedtz-Runge S. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 2010;285:723–730. doi: 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao YJ, Gimpl G, Fahrenholz F. The amino-terminal fragment of the adenylate cyclase activating polypeptide (PACAP) receptor functions as a high affinity PACAP binding domain. Biochem. Biophys. Res. Commun. 1995;212:673–680. doi: 10.1006/bbrc.1995.2021. [DOI] [PubMed] [Google Scholar]
- 26.Gourlet P, Vilardaga JP, De Neef P, Waelbroeck M, Vandermeers A, Robberecht P. The C-terminus ends of secretin and VIP interact with the N-terminal domains of their receptors. Peptides. 1996;17:825–829. doi: 10.1016/0196-9781(96)00107-6. [DOI] [PubMed] [Google Scholar]
- 27.Graziano MP, Hey PJ, Strader CD. The amino terminal domain of the glucagon-like peptide-1 receptor is a critical determinant of subtype specificity. Receptors Channels. 1996;4:9–17. [PubMed] [Google Scholar]
- 28.Holtmann MH, Hadac EM, Miller LJ. Critical contributions of amino-terminal extracellular domains in agonist binding and activation of secretin and vasoactive intestinal polypeptide receptors. Studies of chimeric receptors. J. Biol. Chem. 1995;270:14394–14398. doi: 10.1074/jbc.270.24.14394. [DOI] [PubMed] [Google Scholar]
- 29.Juppner H, Schipani E, Bringhurst FR, McClure I, Keutmann HT, Potts JT, Jr., Kronenberg HM, Abou-Samra AB, Segre GV, Gardella TJ. The extracellular amino-terminal region of the parathyroid hormone (PTH)/PTH-related peptide receptor determines the binding affinity for carboxyl-terminal fragments of PTH-(1-34) Endocrinology. 1994;134:879–884. doi: 10.1210/endo.134.2.8299582. [DOI] [PubMed] [Google Scholar]
- 30.Stroop SD, Nakamuta H, Kuestner RE, Moore EE, Epand RM. Determinants for calcitonin analog interaction with the calcitonin receptor N-terminus and transmembrane-loop regions. Endocrinology. 1996;137:4752–4756. doi: 10.1210/endo.137.11.8895343. [DOI] [PubMed] [Google Scholar]
- 31.Dong M, Lam PC, Gao F, Hosohata K, Pinon DI, Sexton PM, Abagyan R, Miller LJ. Molecular approximations between residues 21 and 23 of secretin and its receptor: Development of a model for peptide docking with the amino terminus of the secretin receptor. Mol. Pharmacol. 2007;72:280–290. doi: 10.1124/mol.107.035402. [DOI] [PubMed] [Google Scholar]
- 32.Dong M, Pinon DI, Cox RF, Miller LJ. Importance of the amino terminus in secretin Family G protein-coupled receptors: Intrinsic photoaffinity labeling establishes initial docking constraints for the calcitonin receptor. J. Biol. Chem. 2004;279:1167–1175. doi: 10.1074/jbc.M305719200. [DOI] [PubMed] [Google Scholar]
- 33.Tan YV, Couvineau A, Murail S, Ceraudo E, Neumann JM, Lacapere JJ, Laburthe M. Peptide agonist docking in the N-terminal ectodomain of a class II G protein-coupled receptor, the VPAC1 receptor. Photoaffinity, NMR, and molecular modeling. J. Biol. Chem. 2006;281:12792–12798. doi: 10.1074/jbc.M513305200. [DOI] [PubMed] [Google Scholar]
- 34.Chorev M. Parathyroid hormone 1 receptor: insights into structure and function. Receptors Channels. 2002;8:219–242. [PubMed] [Google Scholar]
- 35.Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, Spedding M, Harmar AJ. International Union of Pharmacology. XLVI. G protein-coupled receptor list. Pharmacol. Rev. 2005;57:279–288. doi: 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- 36.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 37.Frimurer TM, Bywater RP. Structure of the integral membrane domain of the GLP1 receptor. Proteins. 1999;35:375–386. [PubMed] [Google Scholar]
- 38.Dong M, Lam PC, Pinon DI, Orry A, Sexton PM, Abagyan R, Miller LJ. Secretin occupies a single protomer of the homodimeric secretin receptor complex: insights from photoaffinity labeling studies using dual sites of covalent attachment. J. Biol. Chem. 2010;285:9919–9931. doi: 10.1074/jbc.M109.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pozvek G, Hilton JM, Quiza M, Houssami S, Sexton PM. Structure/function relationships of calcitonin analogues as agonists, antagonists, or inverse agonists in a constitutively activated receptor cell system. Mol. Pharmacol. 1997;51:658–665. doi: 10.1124/mol.51.4.658. [DOI] [PubMed] [Google Scholar]
- 40.Robberecht P, Conlon TP, Gardner JD. Interaction of porcine vasoactive intestinal peptide with dispersed pancreatic acinar cells from the guinea pig. Structural requirements for effects of vasoactive intestinal peptide and secretin on cellular adenosine 3':5'-monophosphate. J. Biol. Chem. 1976;251:4635–4639. [PubMed] [Google Scholar]
- 41.Turner JT, Jones SB, Bylund DB. A fragment of vasoactive intestinal peptide, VIP(10-28), is an antagonist of VIP in the colon carcinoma cell line, HT29. Peptides. 1986;7:849–854. doi: 10.1016/0196-9781(86)90105-1. [DOI] [PubMed] [Google Scholar]
- 42.Bodansky M, Natarajan S, Gardner JD, Makhlouf GM, Said SI. Synthesis and some pharmacological properties of the 23-peptide 15-lysine-secretin-(5--27). Special role of the residue in position 15 in biological activity of the vasoactive intestinal polypeptide. J. Med. Chem. 1978;21:1171–1173. doi: 10.1021/jm00209a018. [DOI] [PubMed] [Google Scholar]
- 43.Gardner JD, Rottman AJ, Natarajan S, Bodanszky M. Interaction of secretin5-27 and its analogues with hormone receptors on pancreatic acini. Biochim. Biophys. Acta. 1979;583:491–503. doi: 10.1016/0304-4165(79)90066-7. [DOI] [PubMed] [Google Scholar]
- 44.Robberecht P, De Neef P, Waelbroeck M, Camus JC, Scemama JL, Fourmy D, Pradayrol L, Vaysse N, Christophe J. Secretin receptors in human pancreatic membranes. Pancreas. 1988;3:529–535. doi: 10.1097/00006676-198810000-00004. [DOI] [PubMed] [Google Scholar]
- 45.Neumann JM, Couvineau A, Murail S, Lacapere JJ, Jamin N, Laburthe M. Class-B GPCR activation: is ligand helix-capping the key? Trends Biochem. Sci. 2008;33:314–319. doi: 10.1016/j.tibs.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Ulrich CD, 2nd, Pinon DI, Hadac EM, Holicky EL, Chang-Miller A, Gates LK, Miller LJ. Intrinsic photoaffinity labeling of native and recombinant rat pancreatic secretin receptors. Gastroenterology. 1993;105:1534–1543. doi: 10.1016/0016-5085(93)90162-6. [DOI] [PubMed] [Google Scholar]
- 47.Gardner JD, Conlon TP, Beyerman HC, Van Zon A. Interaction of synthetic 10-tyrosyl analogues of secretin with hormone receptors on pancreatic acinar cells. Gastroenterology. 1977;73:52–56. [PubMed] [Google Scholar]
- 48.Kofod H. Synthesis of biologically active porcine secretin and [ITyr10] porcine secretin. Int. J. Pept. Protein Res. 1991;37:185–190. doi: 10.1111/j.1399-3011.1991.tb00269.x. [DOI] [PubMed] [Google Scholar]
- 49.Powers SP, Pinon DI, Miller LJ. Use of N,O-bis-Fmoc-D-Tyr-ONSu for introduction of an oxidative iodination site into cholecystokinin family peptides. Int. J. Pept. Protein Res. 1988;31:429–434. doi: 10.1111/j.1399-3011.1988.tb00899.x. [DOI] [PubMed] [Google Scholar]
- 50.Li D, Elbert DL. The kinetics of the removal of the N-methyltrityl (Mtt) group during the synthesis of branched peptides. J. Pept. Res. 2002;60:300–303. doi: 10.1034/j.1399-3011.2002.21018.x. [DOI] [PubMed] [Google Scholar]
- 51.Harikumar KG, Pinon DI, Miller LJ. Transmembrane segment IV contributes a functionally important interface for oligomerization of the Class II G protein-coupled secretin receptor. J. Biol. Chem. 2007;282:30363–30372. doi: 10.1074/jbc.M702325200. [DOI] [PubMed] [Google Scholar]
- 52.Hadac EM, Ghanekar DV, Holicky EL, Pinon DI, Dougherty RW, Miller LJ. Relationship between native and recombinant cholecystokinin receptors: role of differential glycosylation. Pancreas. 1996;13:130–139. doi: 10.1097/00006676-199608000-00003. [DOI] [PubMed] [Google Scholar]
- 53.Jensen RT, Lemp GF, Gardner JD. Interactions of COOH-terminal fragments of cholecystokinin with receptors on dispersed acini from guinea pig pancreas. J. Biol. Chem. 1982;257:5554–5559. [PubMed] [Google Scholar]
- 54.Abagyan R, Totrov M, Kuznetsov D. ICM-A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994;15:488–506. [Google Scholar]
- 55.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 56.Momany FA, McGuire RF, Burgess AW, Scheraga HA. Energy parameters in polypeptides. VII. Geometric parameters, partial atomic charges, nonbonded interactions, hydrogen bond interactions, and intrinsic torsional potentials for the naturally occurring amino acids. J. Phys. Chem. 1975;79:2361–2381. [Google Scholar]
- 57.Nemethy G, Pottle MS, Scheraga HA. Energy parameters in polypeptides. 9. Updating of geometrical parameters, nonbonded interactions, and hydrogen bond interactions for the naturally occurring amino acids. J. Phys. Chem. 1983;87:1883–1887. [Google Scholar]
- 58.Nemethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA. Energy Parameters in Polypeptides. 10. Improved Geometrical Parameters and Nonbonded Interactions for Use in the Ecepp/3 Algorithm, with Application to Proline-Containing Peptides. J. Phys. Chem. 1992;96:6472–6484. [Google Scholar]
- 59.Abagyan R. Protein structure prediction by global energy optimization. In: van Gunseteren WF, Weiner PK, Wilkinson AJ, editors. Computer Simulation of Biomolecular Systems: Theoretical and Experimental Applications. Kluwer Academic; 1997. pp. 363–394. [Google Scholar]
- 60.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 61.MacKerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 62.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 63.Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM force field in GROMACS: Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theor. Comput. 6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 64.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 65.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 66.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 2009;52:7182–7190. [Google Scholar]
- 67.Hess B, Bekker H, Berendsen HJC, Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 68.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 69.Munson PJ, Rodbard D. Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- 70.Dong M, Lam PC, Pinon DI, Sexton PM, Abagyan R, Miller LJ. Spatial approximation between secretin residue five and the third extracellular loop of its receptor provides new insight into the molecular basis of natural agonist binding. Mol. Pharmacol. 2008;74:413–422. doi: 10.1124/mol.108.047209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miranda LP, Winters KA, Gegg CV, Patel A, Aral J, Long J, Zhang J, Diamond S, Guido M, Stanislaus S, Ma M, Li H, Rose MJ, Poppe L, Veniant MM. Design and synthesis of conformationally constrained glucagon-like peptide-1 derivatives with increased plasma stability and prolonged in vivo activity. J. Med. Chem. 2008;51:2758–2765. doi: 10.1021/jm701522b. [DOI] [PubMed] [Google Scholar]
- 72.Murage EN, Gao G, Bisello A, Ahn JM. Development of potent glucagon-like peptide-1 agonists with high enzyme stability via introduction of multiple lactam bridges. J. Med. Chem. 2010;53:6412–6420. doi: 10.1021/jm100602m. [DOI] [PubMed] [Google Scholar]
- 73.Murage EN, Schroeder JC, Beinborn M, Ahn JM. Search for alpha-helical propensity in the receptor-bound conformation of glucagon-like peptide-1. Bioorg. Med. Chem. 2008;16:10106–10112. doi: 10.1016/j.bmc.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 74.Trivedi D, Lin Y, Ahn JM, Siegel M, Mollova NN, Schram KH, Hruby VJ. Design and synthesis of conformationally constrained glucagon analogues. J. Med. Chem. 2000;43:1714–1722. doi: 10.1021/jm990559d. [DOI] [PubMed] [Google Scholar]
- 75.Ahn JM, Gitu PM, Medeiros M, Swift JR, Trivedi D, Hruby VJ. A new approach to search for the bioactive conformation of glucagon: positional cyclization scanning. J. Med. Chem. 2001;44:3109–3116. doi: 10.1021/jm010091q. [DOI] [PubMed] [Google Scholar]
- 76.Bitar KG, Somogyvari-Vigh A, Coy DH. Cyclic lactam analogues of ovine pituitary adenylate cyclase activating polypeptide (PACAP): discovery of potent type II receptor antagonists. Peptides. 1994;15:461–466. doi: 10.1016/0196-9781(94)90206-2. [DOI] [PubMed] [Google Scholar]
- 77.Moreno D, Gourlet P, De Neef P, Cnudde J, Waelbroeck M, Robberecht P. Development of selective agonists and antagonists for the human vasoactive intestinal polypeptide VPAC(2) receptor. Peptides. 2000;21:1543–1549. doi: 10.1016/s0196-9781(00)00309-0. [DOI] [PubMed] [Google Scholar]
- 78.Bolin DR, Michalewsky J, Wasserman MA, O'Donnell M. Design and development of a vasoactive intestinal peptide analog as a novel therapeutic for bronchial asthma. Biopolymers. 1995;37:57–66. doi: 10.1002/bip.360370203. [DOI] [PubMed] [Google Scholar]
- 79.Taylor JW, Jin QK, Sbacchi M, Wang L, Belfiore P, Garnier M, Kazantzis A, Kapurniotu A, Zaratin PF, Scheideler MA. Side-chain lactam-bridge conformational constraints differentiate the activities of salmon and human calcitonins and reveal a new design concept for potent calcitonin analogues. J. Med. Chem. 2002;45:1108–1121. doi: 10.1021/jm010474o. [DOI] [PubMed] [Google Scholar]
- 80.Kazantzis A, Waldner M, Taylor JW, Kapurniotu A. Conformationally constrained human calcitonin (hCt) analogues reveal a critical role of sequence 17-21 for the oligomerization state and bioactivity of hCt. Eur. J. Biochem. / FEBS. 2002;269:780–791. doi: 10.1046/j.0014-2956.2001.02689.x. [DOI] [PubMed] [Google Scholar]
- 81.Kapurniotu A, Kayed R, Taylor JW, Voelter W. Rational design, conformational studies and bioactivity of highly potent conformationally constrained calcitonin analogues. Eur. J. Biochem. / FEBS. 1999;265:606–618. doi: 10.1046/j.1432-1327.1999.00708.x. [DOI] [PubMed] [Google Scholar]
- 82.Kapurniotu A, Taylor JW. Structural and conformational requirements for human calcitonin activity: design, synthesis, and study of lactam-bridged analogues. J. Med. Chem. 1995;38:836–847. doi: 10.1021/jm00005a011. [DOI] [PubMed] [Google Scholar]
- 83.Miranda A, Lahrichi SL, Gulyas J, Koerber SC, Craig AG, Corrigan A, Rivier C, Vale W, Rivier J. Constrained corticotropin-releasing factor antagonists with i-(i + 3) Glu-Lys bridges. J. Med. Chem. 1997;40:3651–3658. doi: 10.1021/jm970311t. [DOI] [PubMed] [Google Scholar]
- 84.Miranda A, Koerber SC, Gulyas J, Lahrichi SL, Craig AG, Corrigan A, Hagler A, Rivier C, Vale W, Rivier J. Conformationally restricted competitive antagonists of human/rat corticotropin-releasing factor. J. Med. Chem. 1994;37:1450–1459. doi: 10.1021/jm00036a010. [DOI] [PubMed] [Google Scholar]
- 85.Koerber SC, Gulyas J, Lahrichi SL, Corrigan A, Craig AG, Rivier C, Vale W, Rivier J. Constrained corticotropin-releasing factor (CRF) agonists and antagonists with i-(i+3) Glu-Xaa-DXbb-Lys bridges. J. Med. Chem. 1998;41:5002–5011. doi: 10.1021/jm980350k. [DOI] [PubMed] [Google Scholar]
- 86.Rivier J, Lahrichi SL, Gulyas J, Erchegyi J, Koerber SC, Craig AG, Corrigan A, Rivier C, Vale W. Minimal-size, constrained corticotropin-releasing factor agonists with i-(i+3) Glu-Lys and Lys-Glu bridges. J. Med. Chem. 1998;41:2614–2620. doi: 10.1021/jm980164e. [DOI] [PubMed] [Google Scholar]
- 87.Rivier J, Gulyas J, Kirby D, Low W, Perrin MH, Kunitake K, DiGruccio M, Vaughan J, Reubi JC, Waser B, Koerber SC, Martinez V, Wang L, Tache Y, Vale W. Potent and long-acting corticotropin releasing factor (CRF) receptor 2 selective peptide competitive antagonists. J. Med. Chem. 2002;45:4737–4747. doi: 10.1021/jm0202122. [DOI] [PubMed] [Google Scholar]
- 88.Barbier JR, Neugebauer W, Morley P, Ross V, Soska M, Whitfield JF, Willick G. Bioactivities and secondary structures of constrained analogues of human parathyroid hormone: cyclic lactams of the receptor binding region. J. Med. Chem. 1997;40:1373–1380. doi: 10.1021/jm960743o. [DOI] [PubMed] [Google Scholar]
- 89.Barbier JR, MacLean S, Morley P, Whitfield JF, Willick GE. Structure and activities of constrained analogues of human parathyroid hormone and parathyroid hormone-related peptide: implications for receptor-activating conformations of the hormones. Biochemistry. 2000;39:14522–14530. doi: 10.1021/bi001527r. [DOI] [PubMed] [Google Scholar]
- 90.Bisello A, Nakamoto C, Rosenblatt M, Chorev M. Mono- and bicyclic analogs of parathyroid hormone-related protein. 1. Synthesis and biological studies. Biochemistry. 1997;36:3293–3299. doi: 10.1021/bi9619029. [DOI] [PubMed] [Google Scholar]
- 91.Gandhi S, Rubinstein I, Tsueshita T, Onyuksel H. Secretin self-assembles and interacts spontaneously with phospholipids in vitro. Peptides. 2002;23:201–204. doi: 10.1016/s0196-9781(01)00596-4. [DOI] [PubMed] [Google Scholar]
- 92.Robinson RM, Blakeney EW, Jr., Mattice WL. Lipid-induced conformational changes in glucagon, secretin, and vasoactive intestinal peptide. Biopolymers. 1982;21:1271–1228. doi: 10.1002/bip.360210615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. HPLC profiles reflecting the purification of one of the lactam analogues of the secretin peptide, c[K6,E9][Y10]sec(5-27) (4). Shown are representative A280 absorbance profiles of the crude products of synthesis on a semi-preparative reversed-phase column and the final product on an analytical reversed-phase column (inset). The positions of migration of the compound of interest are marked with arrowheads in the profiles. This compound had the expected mass by mass spectrometry.