Abstract
Dibenzo[a,l]pyrene (DB[a,l]P) (dibenzo[def,p]chrysene) is a highly carcinogenic polycyclic aromatic hydrocarbon (PAH) that has been identified in tobacco smoke and is found in our environment due to incomplete combustion of organic matter. Its metabolites are known to form stable DNA adducts in bacteria and mammalian cells, and can lead to tumors in animal models. Glucuronidation of major metabolites of DB[a,l]P by the uridine-5’-diphosphate glucuronosyltransferase (UGT) family of enzymes is an important route of detoxification of this pro-carcinogen. The focus of the current study was to characterize the glucuronidation of the pro-carcinogenic enantiomers DB[a,l]P-(+)-trans-11S,12S–diol and DB[a,l]P-(−)-trans-11R,12R–diol. Glucuronidation assays with HEK293 cell lines over-expressing individual human UGT enzymes demonstrated that UGTs 1A1, 1A4, 1A7, 1A8, 1A9, 1A10, and 2B7 glucuronidated one or both DB[a,l]P-trans-11,12-diol enantiomers. Three glucuronide conjugates were observed in activity assays with UGTs 1A1 and 1A10, while two glucuronides were formed by UGTs 1A7, 1A8, and 1A9, and one glucuronide was made by UGT1A4 and UGT2B7. Enzyme kinetic analysis indicated that UGT1A9 was the most efficient UGT at forming both the (+)-DB[a,l]P-11-Gluc and (−)-DB[a,l]P-11-Gluc products while UGTs 1A1 and 1A10 were the most efficient at forming the (+)-DB[a,l]P-12-Gluc product (as determined by the kcat/KM). Incubations with human liver microsomes showed formation of three diastereomeric glucuronide products: (+)-DB[a,l]P-11-Gluc, (+)-DB[a,l]P-12-Gluc, and (−)-DB[a,l]P-11-Gluc, with an average overall ratio of 31 : 32 : 37 in four liver specimens. Human bronchus and trachea tissue homogenates demonstrated glucuronidation activity against both DB[a,l]P-trans-11,12-diol enantiomers, with both tissues producing the (+)-DB[a,l]P-11-Gluc and (+)-DB[a,l]P-12-Gluc with little or no formation of (−)-DB[a,l]P-11-Gluc. These results indicate that multiple UGTs are involved in the stereospecific glucuronidation of DB[a,l]P-trans-11,12-diol in a pattern consistent with their expression in respiratory tract tissues, and that glucuronidation may be an important first-line detoxification mechanism of DB[a,l]P metabolites.
INTRODUCTION
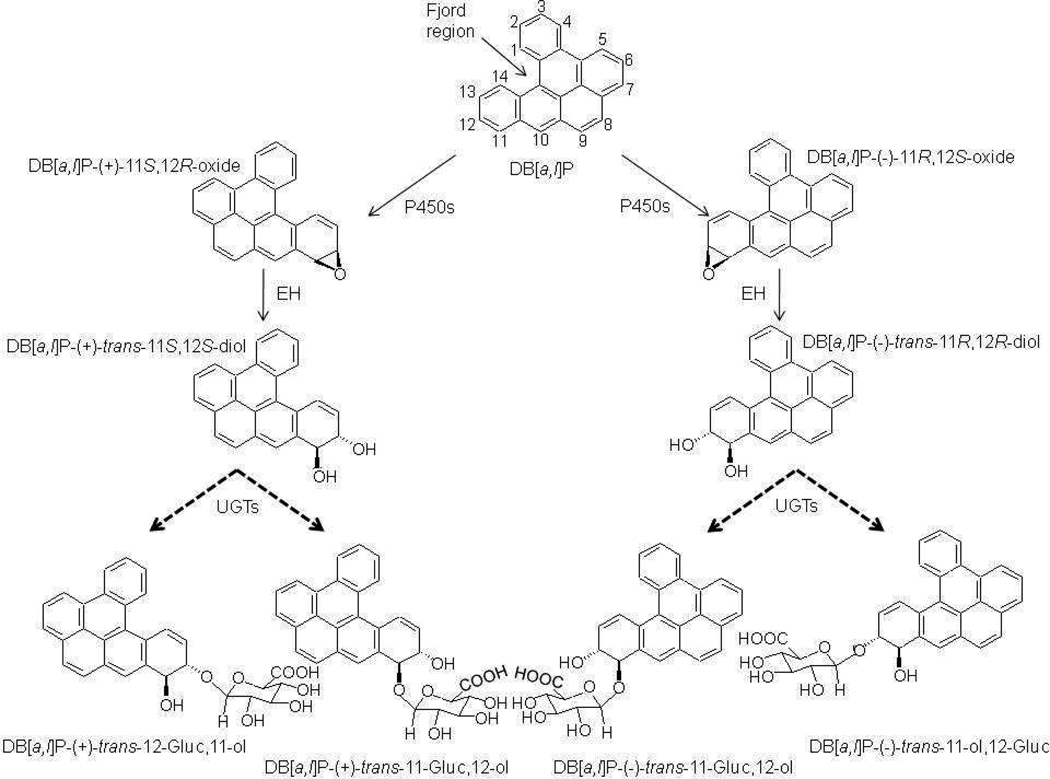
Dibenzo[a,l]pyrene (DB[a,l]P) (dibenzo[def,p]chrysene) is one of the most carcinogenic polycyclic aromatic hydrocarbons (PAH) yet identified. DB[a,l]P has been identified in tobacco smoke condensate 1, emissions from partially combusted organic matter such as smoky coal 2, and environmental soil and sediment samples 3. When DB[a,l]P enters mammalian cells, it can be metabolized to trans-11,12-diol and trans-11,12-diol-13,14-epoxide stereoisomers by cytochrome P450 (P450) and epoxide hydrolase (EH) enzymes, respectively (Scheme 1) 4,5. The P450-mediated metabolism of parent PAHs to the epoxide has been shown to favor the R,S-epoxide enantiomer up to 98%, which then leads to overwhelming conversion to the R,R-diol 6. This has been confirmed for several PAHs, which identify the R,S-diol-S,R-epoxide (product of the R,R-diol) as the main metabolite of the parent PAH 6. Furthermore, studies in mammalian cells have demonstrated that the majority of DNA adducts are formed by (−)-anti-(R,S,S,R)-DB[a,l]PDE, which suggests that DB[a,l]P is mainly converted to the (−)-trans-11R,12R–diol precursor 7–9 (Scheme 1).
Scheme 1. Simplified metabolism of DB[a,l]P.
The activation of DB[a,l]P involves formation of an epoxide by P450 enzymes, and then reduction to a diol by EH. Detoxification of each diol enantiomer by UGTs to make four potential diastereomeric monoglucuronides is shown. In mammalian cells, the majority of the DB[a,l]P compound is converted to DB[a,l]P-(−)-11R,12S–oxide. The dashed arrows indicate the potential glucuronides that would be formed by UGT enzymes. Cytochrome P450 (P450), epoxide hydrolase (EH), UDP-glucuronosyltransferase (UGT), glucuronide (Gluc).
DB[a,l]P belongs to a subclass of PAHs containing a structural feature composed of five carbon atoms called a fjord region (Scheme 1). Its high carcinogenicity is a result of its fjord region chemical structure, which causes it to adopt a non-planar conformation, allowing the electrophilic diol epoxide to react with and bind to purine nucleotides without distorting the DNA structure; this results in formation of adducts that are not well-recognized by DNA repair enzymes and are more persistent than the adducts derived from bay-region PAHs like benzo[a]pyrene (B[a]P) 10. DB[a,l]P has been shown to cause a high rate of mutagenesis in bacteria 11 and mammalian cells 12, and to initiate tumors in rodent models to a greater degree than other PAH diol epoxides tested 13–15, reacting with the exocyclic amine group of dAde over dGuo in a 4:1 ratio in the human A549 lung epithelial carcinoma cell line 16.
Glucuronidation is an important mechanism of detoxification for a variety of endogenous compounds including various steroids and bilirubin as well as a variety of exogenous compounds including many drugs and environmental carcinogens such as PAHS 17–21. Mediated by the uridine-5′-diphosphate glucuronosyltransferase (UGT) family of enzymes, the glucuronidation reaction involves the transfer of the glucuronic acid group from uridine-5′-diphosphate glucuronic acid (UDPGA) to a target substrate, increasing its water solubility and allowing elimination of the compound from the body through the bile and/or urine 17. UGTs are membrane-bound endoplasmic reticulum proteins 17 and can be classified into three major families based on sequence homology: UGT1A, UGT2A, and UGT2B 22. The UGT1A family is derived from a single gene locus in chromosome 2, coding for nine known functional proteins that differ only in their amino-terminus as a result of alternate splicing of independent exon 1 regions to a shared carboxy-terminus encoded by exons 2–5 23,24. Conversely, the UGT2B family members are derived from independent genes located in chromosome 4, encoding for six known functional proteins 17,23,24. The UGT2A family of enzymes are also located on chromosome 4, but UGTs 2A1 and 2A2 share a similar structural arrangement as the UGT1As with a unique exon 1 and shared exons 2–6; UGT2A3 is an independent gene locus comprised of six unique exons 22. Exon 1 exhibits less homology between UGTs and imparts the wide range of substrate specificities among UGTs 25 while the C-terminal region imparts the UDPGA co-factor-binding domain of the protein and is highly homologous between UGTs 17.
Prior to this study, the glucuronidation of metabolites of the bay-region PAH, B[a]P, has been investigated 19,20,26, including a report on the stereospecific glucuronidation of B[a]P-trans-7,8-diol enantiomers 21. An in vivo study of rats with a UGT1A1-null genotype demonstrated a great amount of covalent binding of B[a]P metabolites to DNA and liver microsomal protein than UGT1A–wild-type rats, indicating the importance of glucuronidation as a detoxification pathway for B[a]P 27. As DB[a,l]P metabolites are known to be among the most potent of carcinogens, the goal of the present study was to characterize the glucuronidation pathway of DB[a,l]P, to investigate whether diol precursors of the highly carcinogenic DB[a,l]PDE could also be glucuronidated by human UGT enzymes in a stereospecific manner, and to identify the individual UGT enzymes responsible for this activity.
MATERIALS AND METHODS
Chemicals and materials
DB[a,l]P-trans-11,12-diol was synthesized following our previously reported methods 28,29. Caution: Diol epoxides from polycyclic aromatic hydrocarbons are carcinogens, and thus experimental handling must be carried out under special safety conditions according to the National Cancer Institute guidelines. Alamethicin, UDPGA, and β-glucuronidase were purchased from Sigma (St. Louis, MO) and Dulbecco’s Modified Eagle’s Medium, fetal bovine serum and Geneticin (G418) were purchased from Invitrogen (Carlsbad, CA).
Human Tissues
Normal human liver tissue procured from the H. Lee Moffitt Cancer Center (when P Lazarus was faculty there) was used to make microsomes in a procedure that has been described elsewhere 30. Briefly, normal adjacent liver tissue samples were homogenized in Tris-buffered saline and the homogenate was centrifuged at 10,000g for 20 minutes at 4°C followed by ultracentrifugation of the supernatant at 100,000g for 1 hour at 4°C to pellet the microsomal fraction. The pellet was then resuspended in Tris-buffered saline and stored in aliquots at −70°C. Human tissue homogenates from the respiratory tract (bronchus and trachea) were purchased from Analytical Biological Services, Inc. (Wilmington, DE). Both tissues were normal tissues lying adjacent to tumors within their respective organ sites that were procured during cancer resection surgery.
Cell Lines and Cell Homogenate Preparation
HEK293 cells stably transfected with and over-expressing individual UGTs have been previously described 19,31–34. Briefly, the individual UGTs were transfected into HEK293 cells (purchased from ATCC; Rockville, MD) by electroporation. Stable transfectants that over-expressed the individual UGTs were selected by treatment with G418 (Invitrogen; Carlsbad, CA).
Cell homogenates were prepared by resuspending pelleted cells in Tris-buffered saline (25 mM Tris-HCl (pH 7.4), 138 mM NaCl, 2.7 mM KCl) and subjecting them to three rounds of freeze-thaw prior to gentle homogenization with a dounce homogenizer. The homogenates were aliquotted and stored in 500 µL aliquots at −70°C. Total cellular homogenate protein concentrations were determined using the BCA assay from Pierce Biotechnology (Rockford, IL) after protein extraction using standard protocols. The relative levels of UGT over-expression in each HEK293 cell line are continuously monitored and have been expressed at levels identical to those described previously in our laboratory by Western blot analysis 31. All of the UGT-over-expressing cell lines and homogenates used in these studies exhibited glucuronidation activity against known test substrates as previously described 19,31–34.
Separation of racemic DB[a,l]P-trans-11,12-diol
Separation of the DB[a,l]P-trans-11,12-diol enantiomers was performed using a high performance liquid chromatography (HPLC) instrument (Beckman-Coulter; Brea, CA) with a UV detector operated at 305 nm. The enantiomers were separated with a Pirkle Covalent (S,S) 25 cm × 4.6 mm Whelk-O 1 Reversible chiral column (Regis Technologies; Morton Grove, IL) and eluted with 92.5% hexane:5% ethanol:2.5% acetonitrile as shown in Figure S1 (panels A and B). Enantiomer verification was confirmed by circular dichroism analysis (see below) and compared to data from a previous publication 35.
Collection of glucuronide products
Homogenates of HEK293 cell lines over-expressing either UGT1A1 or 1A9 were pre-incubated with alamethicin (50 µg/mg protein) on ice for 15 minutes, followed by an incubation at 37°C overnight in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 4 mM UDPGA, and 200 µM DB[a,l]P-trans-11,12-diol (final volume = 0.5 mL). Assays were terminated by the addition of an equal volume of ice cold acetonitrile and reactions were centrifuged at 16,000 g for 10 min at 4°C. Supernatants were removed and evaporated in a Speedvac for 1 h and then resuspended in 50 µL of a water:acetonitrile (1:1) solution. Resuspensions (50 µL) were injected in sequential loadings onto a Luna 250 × 4.60 mm 5 µm C18 column (Phenomenex; Torrance, CA) using an HPLC instrument (Beckman-Coulter; Brea, CA) with a UV detector operated at 305 nm. Glucuronide products were separated as follows: 70% of buffer A (5 mM ammonium acetate pH 5.0/10% acetonitrile) for 10 min, then a linear gradient up to 75% buffer B (100% acetonitrile) over 10 min, followed by a gradient back to 25% buffer B over 20 min, all at a 1 mL/min flow rate. Eluted glucuronide products were collected and evaporated in a Speedvac and, after collection of >100 µg, glucuronide products were analyzed by NMR.
Glucuronidation assays
Glucuronidation rates were determined essentially as previously described, with up to 500 µg of total UGT-over-expressing cell homogenate or human liver microsomal protein, or 1 mg of homogenate protein from respiratory tract tissues, used in assays that contained a concentration range of DB[a,l]P-trans-11,12-diol enantiomers that encompassed their respective KM’s 19,21,30,36. Incubations were performed at 37°C for up to 4 h for human liver microsomes, 4 h for an initial activity screening of individual UGT-over-expressing cell lines, and 8 h for assessment of respiratory tract tissue homogenate glucuronidation activity. For the kinetic analysis of UGT-over-expressing cell homogenates that were active against DB[a,l]P-trans-11,12-diol, glucuronidation rate determinations, substrate concentrations, homogenate protein levels, and incubation times for individual assays were chosen to maximize levels of detection within a linear range of glucuronidation activity; a 2 h incubation was performed using 500 µg of total UGT-over-expressing cell homogenate or human liver microsomal protein except for UGT1A9-over-expressing cell homogenates, where a 25 min incubation was performed using 1 µg of UGT1A9-over-expressing cell homogenate protein. Glucuronidation reactions were analyzed by a Waters ACQUITY UPLC system (Milford, MA) equipped with an UV detector operated at 305 nm. Samples were loaded onto an ACQUITY UPLC BEH C18 column and eluted as follows: 75% of buffer A (5 mM ammonium acetate pH 5.0/10% acetonitrile) for 2 min with a linear gradient up to 75% buffer B (100% acetonitrile) over 2 min followed by a linear gradient back to 75% buffer A over 3 min, all at a flow rate of 0.5 mL/min. Glucuronide products were quantified using MassLynx software (Milford, MA). The kcat for all kinetic analysis was normalized to UGT levels for the respective cell line based upon Western blot analysis of protein expression for that cell line as described above. Reactions were treated with β-glucuronidase (37°C overnight incubation) to confirm the glucuronide peak. Experiments were always performed in triplicate as independent assays.
LC-MS analysis of DB[a,l]P-trans-11,12-diol glucuronides
Mass spectrometry analysis was performed on an ACQUITY SQD (Waters; Milford, MA) with electrospray ionization interface and an UPLC system. Samples were loaded onto an Acquity UPLC BEH C18 2.1 ×100 mm 1.7 µm column and eluted as follows: 75% of buffer A (5 mM ammonium acetate pH 5.0/10% acetonitrile) for 2 min with a linear gradient up to 75% buffer B (100% acetonitrile) over 2 min followed by a linear gradient back to 75% buffer A over 3 min at a flow rate of 0.5 mL/min. The UV detector was operated at 305 nm. The mass spectrometer operated in negative mode was set up to scan the daughter ion of m/z 511.5 in the range of m/z 200 to 600. The optimized mass spectrometry parameters used were as follows: capillary voltage, 3.14 kV; cone voltage, 70 V; collision energy, 30 V; source temperature, 140°C; and desolvation temperature, 450°C. Nitrogen was used as both the desolvation gas (a flow rate of 1001 L/h) and cone gas (20 L/h). Argon was used as the collision gas at a flow rate of 0.1 mL/min. Data acquisition and analysis were performed using the MassLynx NT 4.1 software with QuanLynx program (Waters; Milford, MA).
Circular Dichroism Analysis
Circular dichroism data were collected using a Jasco J-710 spectropolarimeter with a 1mm pathlength cell. Each of the DB[a,l]P-trans-11,12-diol enantiomers were dissolved in DMSO for analysis. The three glucuronide products collected from the large-scale glucuronidation reactions were dissolved in 100% ethanol for analysis.
NMR Analysis
NMR spectra for the DB[a,l]P-trans-11,12-diol racemic mixture and the three glucuronide products collected from the large-scale glucuronidation reactions were recorded in a Bruker Avance II 500 MHz spectrometer. The spectra were obtained in DMSO-d6 and the chemical shifts were indicated as δ values with reference to tetramethylsilane as internal standard. The standard Bruker cosygpprqf and selno pulse sequences were used with modification for COSY and selective NOE experiments, respectively.
Data Analysis
GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, CA) was employed to calculate kinetic values. Kinetic constants for glucuronidation of all the substrates were calculated using the Michaelis-Menten equation in equation 1:
| (1) |
where v0 is the initial rate of reaction, Vmax is the maximum velocity, KM is the Michaelis constant, and [S]0 is the initial substrate concentration. The Michaelis-Menten equation was fitted using non-linear regression, least squares (ordinary) fit. kcat was calculated from the Vmax value according to Western blot analysis of UGT expression from previous reports in our laboratory as cited and described above.
RESULTS
Characterization of glucuronide stereoisomers formed from the DB[a,l]P-trans-11,12-diol enantiomers
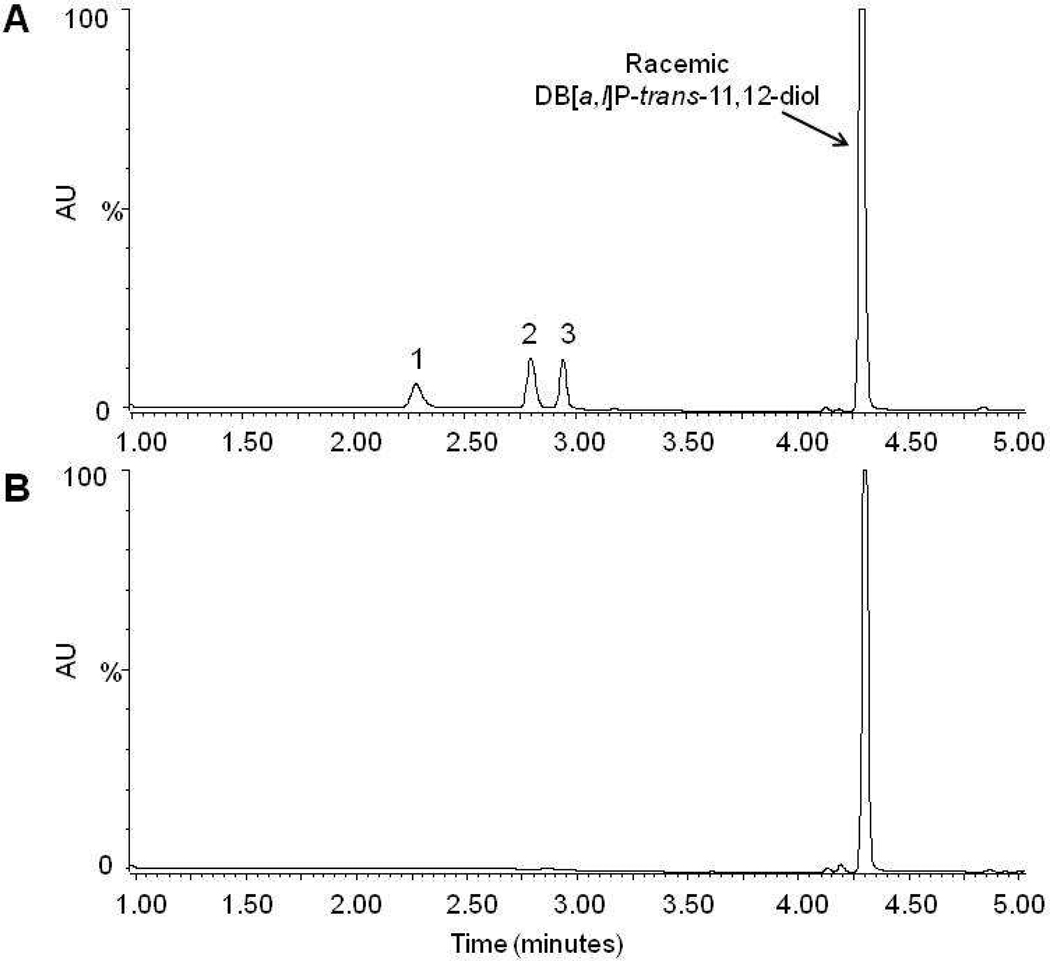
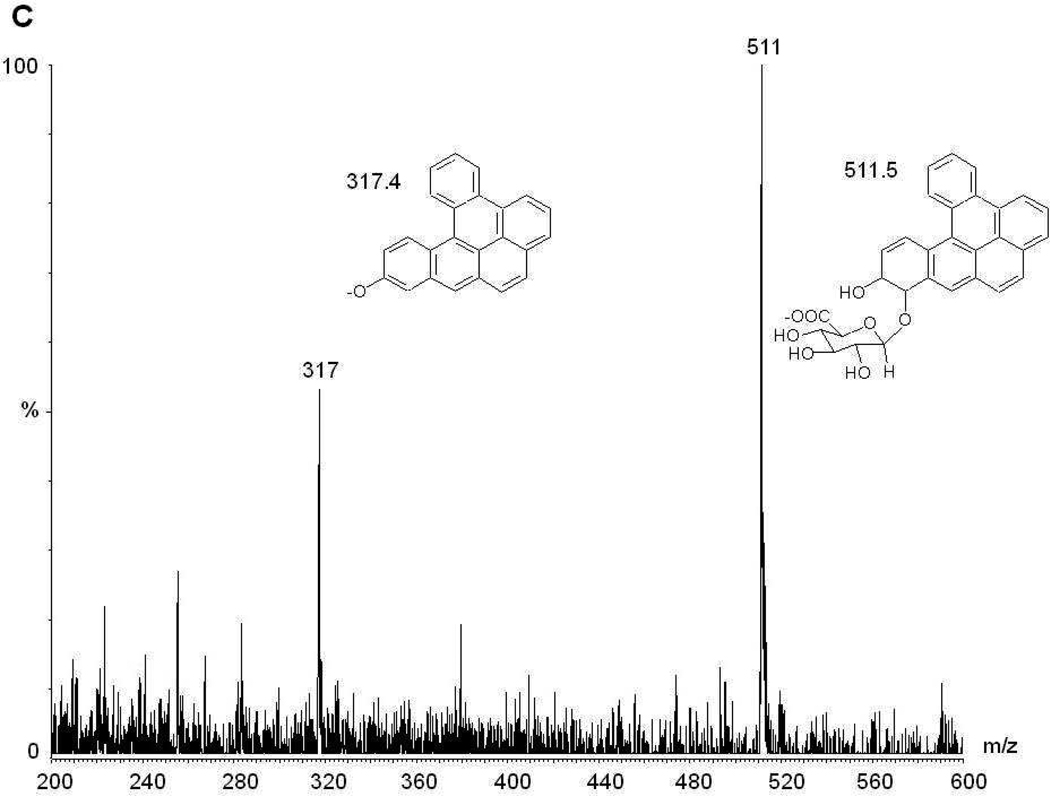
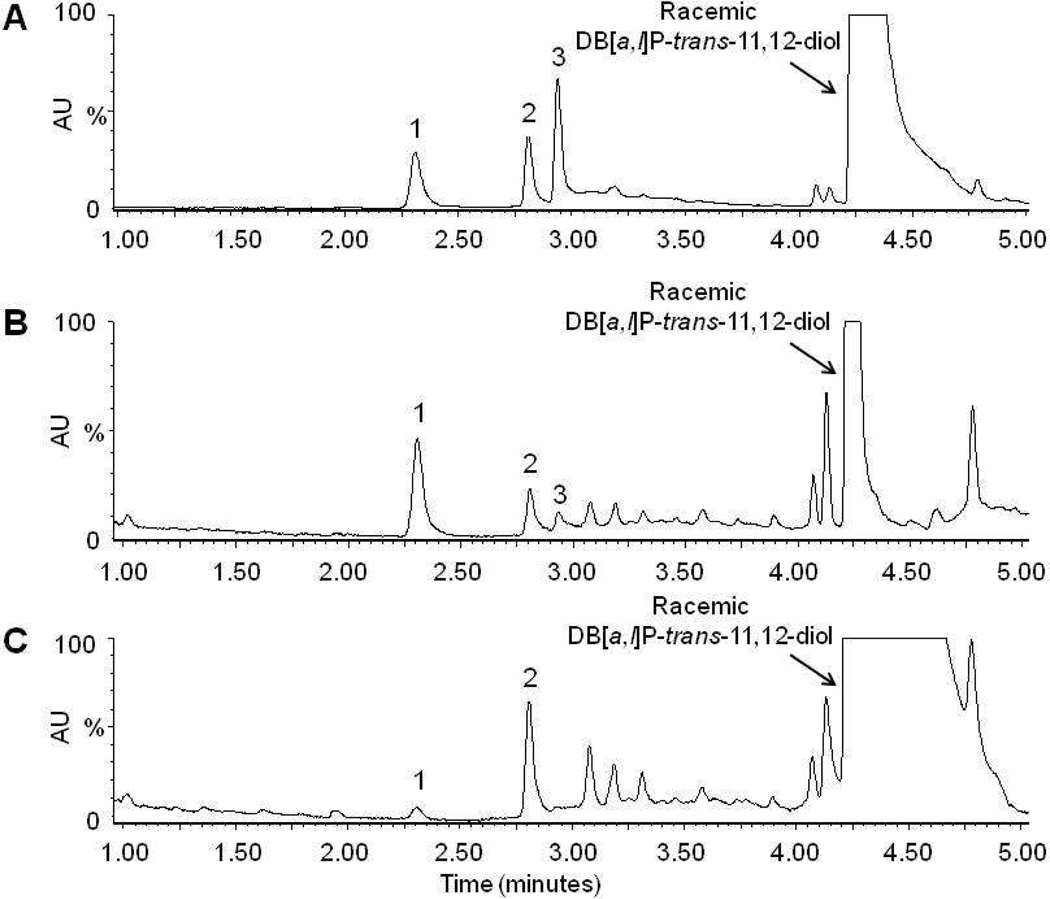
Four monoglucuronide conjugates could potentially be generated by UGT enzymes from the enantiomers of DB[a,l]P-trans-11,12-diol (Scheme 1): DB[a,l]P-(+)-trans-11-ol,12-Gluc [termed (+)-DB[a,l]P-12-Gluc], DB[a,l]P-(+)-trans-11-Gluc,12-ol [termed (+)-DB[a,l]P-11-Gluc], DB[a,l]P-(−)-trans-11-Gluc,12-ol [termed (−)-DB[a,l]P-11-Gluc], and DB[a,l]P-(−)-trans-11-ol,12-Gluc [termed (−)-DB[a,l]P-12-Gluc]. To initially characterize the glucuronidation of each DB[a,l]P-trans-11,12-diol enantiomer, glucuronidation assays with human liver microsomes and racemic DB[a,l]P-trans-11,12-diol were performed. Three peaks with retention times of 2.3, 2.7, and 2.9 min were observed by UPLC (Figure 1A). The three peaks were sensitive to treatment with β-glucuronidase, which cleaves the glucuronide from the parent compound, indicating that these peaks were glucuronides (Figure 1B). The products corresponding to the predicted monoglucuronides from isomers of DB[a,l]P-trans-11,12-diol were collected and analyzed by negative ion mass spectrometry. As shown in Figure 1C, the MS spectrum of the peak at 2.7 min (which was predicted to be a monoglucuronide, with a calculated molecular weight of 511.5 g/mol) showed a clear [M−] ion at m/z 511.3, and a fragment ion at m/z of 317.3, the latter corresponding to the parent compound –H with the loss of the glucuronide acid moiety (molecular weight =176 g/mol). Virtually identical patterns were observed for the other two glucuronide peaks (with retention times of 2.3 and 2.9 min respectively), exhibiting a clear [M−] ion at m/z 511.3 and fragment ion at m/z 317.3 (data not shown).
Figure 1. Characterization of glucuronide formation in Human Liver Microsomes with DB[a,l]P-trans-11,12-diol as substrate.
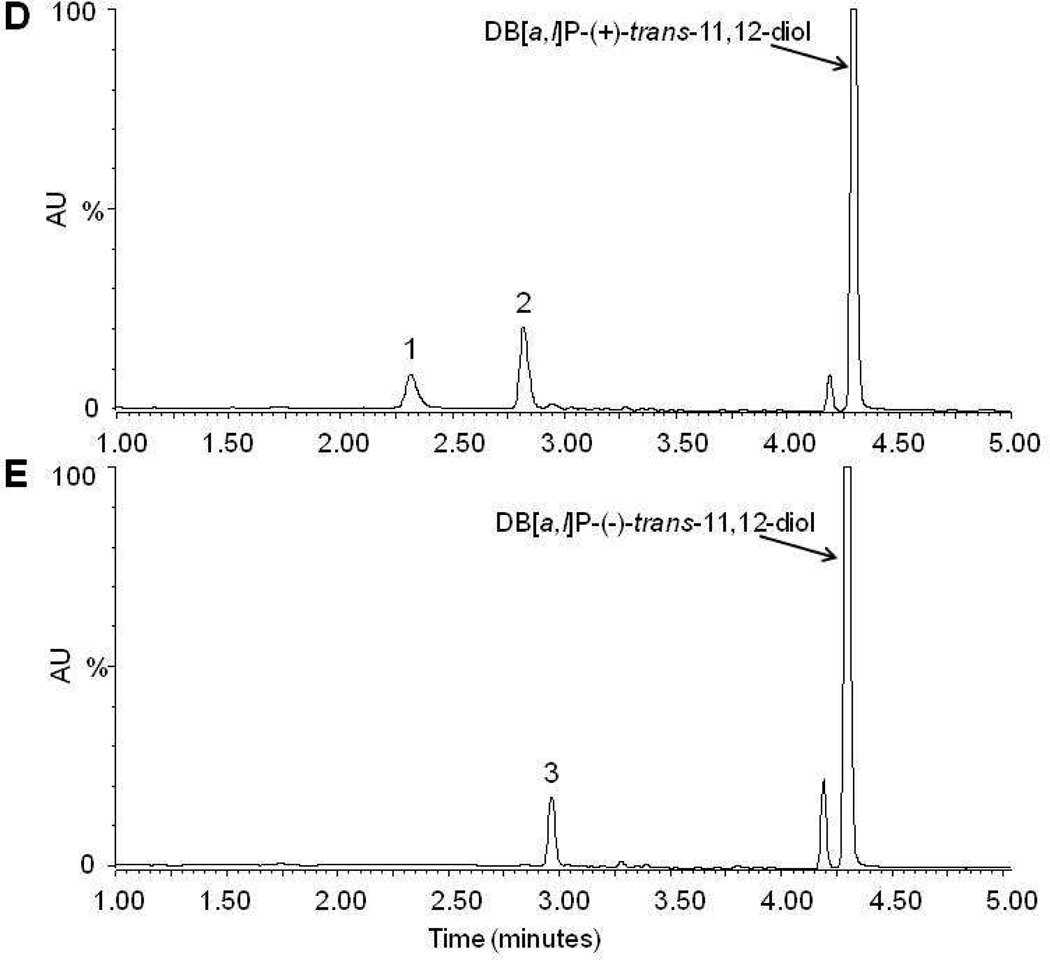
Panel A, representative UPLC chromatogram for glucuronidation assays of human liver microsomes incubated with racemic DB[a,l]P-trans-11,12-diol. Panel B, glucuronidation assay from panel A treated overnight with β-glucuronidase. Panel C, representative mass spectrometry trace of the putative glucuronide peak at 2.7 min from Figure 1A. The mass of 511.3 represents the parent ion while the peak at 317.4 is loss of the glucuronide. Panel D, glucuronidation assay of human liver microsomes and DB[a,l]P-(+)-trans-11,12-diol. Panel E, glucuronidation assay of human liver microsomes and DB[a,l]P-(−)-trans-11,12-diol. Glucuronide products are indicated by their respective numbers (peak 1 is (+)-DB[a,l]P-11-Gluc, peak 2 is (+)-DB[a,l]P-12-Gluc, and peak 3 is (−)-DB[a,l]P-11-Gluc).
In order to facilitate the stereochemical identification of the glucuronides, the racemic DB[a,l]P-trans-11,12-diol was separated into the individual enantiomers by HPLC using a chiral column, and the collected products were identified by circular dichroism analysis as described in the supplementary data (Figure S1A–C). After characterizing each enantiomer, a four hour incubation of the (+)-trans DB[a,l]P-diol with human liver microsomes yielded two glucuronide products with retention times of 2.3 (peak 1) and 2.7 (peak 2) min (Figure 1D) while the (−)-trans enantiomer yielded one peak with retention time of 2.9 min (peak 3) (Figure 1E). To better characterize each product, large-scale glucuronidation reactions were performed and individual peaks were collected by HPLC as described in the Materials and Methods. Circular dichroism analysis demonstrated that the glucuronides collected from reaction of either the (+)-trans or (−)-trans enantiomers had CD spectra similar to the parent compound (Figure S1D). This confirms that glucuronides were generated from their respective parent compounds with conservation of the stereochemistry on the PAH.
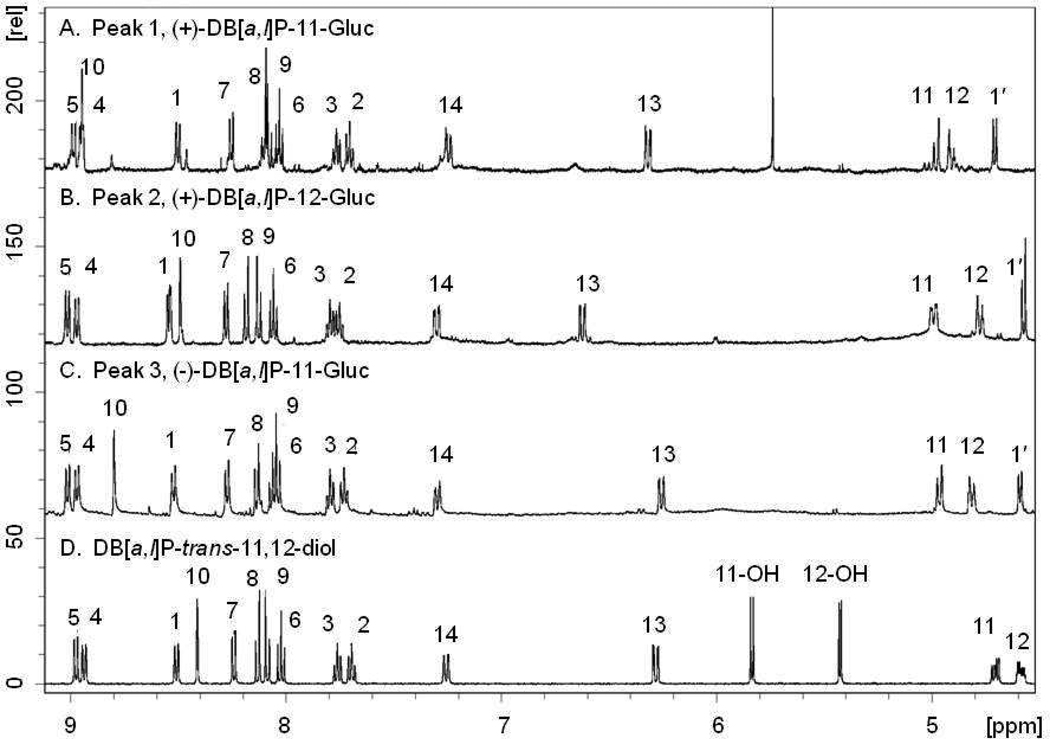
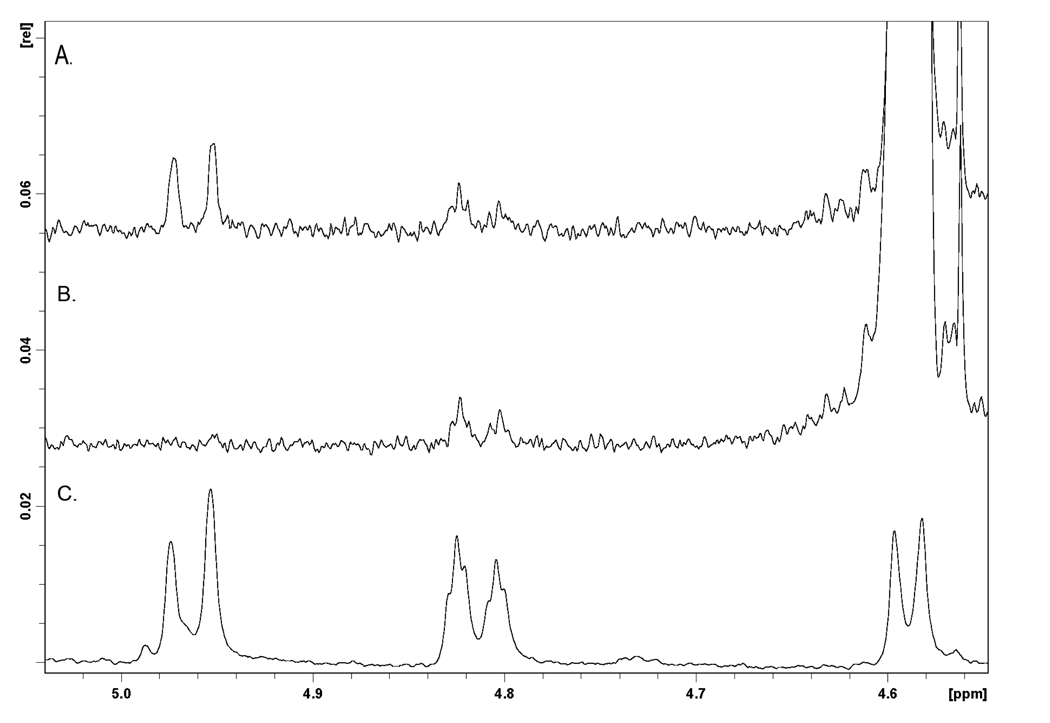
DB[a,l]P-trans-11,12-diol and the produced glucuronides were further characterized by NMR analysis. The downfield region of the 1H-NMR spectra of DB[a,l]P-trans-11,12-diol and the three peaks isolated by HPLC from incubations with human liver microsomes are shown in Figure 2 and summarized in Table 1. The proton assignments were made in association with COSY experiments. Comparison of the glucuronides (Figure 2A–C) with the parent diol (Figure 2D) shows the addition of the 1′-proton attached to the glucuronide at 5.6 to 5.7 ppm. Glucuronidation does not significantly alter the chemical shifts of most of the protons. For all three glucuronides, both protons at the 11 and 12 positions moved downfield ~0.2 ppm upon glucuronidation. For peak 1 (Figure 2A) and 3 (Figure 2C), the proton at the 10-position was shifted downfield from 8.41 ppm in the diol to 8.95 in peak 1 and 8.80 in peak 3. This shift would be most consistent with glucuronidation at the 11-position. For peak 2, the largest shift occurred at the proton at the 13-position, which moved from 6.28 ppm in the diol to 6.62 ppm (Figure 2B). This shift would be most consistent with glucuronidation at the 12-position. Selective NOE experiments confirmed these assignments. Figure 3 shows the selective NOE experiment with peak 3. Panel C is the 1H-NMR spectrum of peak 3, while panels A and B are the selective NOE experiments in which the NOE signal is generated by irradiating the 1′-position. Panel B shows a small signal of the proton at the 12-position (4.82 ppm) that is generated with 20 ms mixing time. As shown in panel A, this signal does not grow with increased mixing time and is due to the NOE pulse. Panel A shows the increase in the 11-position (4.96 ppm) upon 500 ms mixing time. This result indicates that the proton at the 11-position is closer to the proton at the 1′-position, consistent with the assignment of DB[a,l]P-11-Gluc to peak 3. Similarly, as shown in the supplemental data, peak 1 is a 12-glucuronide and peak 2 is an 11-glucuronide. These data confirm that each glucuronide product is a monoglucuronide, and that the (+)-trans DB[a,l]P enantiomer can be converted into both the 11-Gluc and 12-Gluc products (2.3 [peak 1] and 2.7 [peak 2] min retention time, respectively, on UPLC) while the (−)-trans enantiomer is only converted into the 11-Gluc product (2.9 [peak 3] minute retention time on UPLC).
Figure 2. 1H-NMR spectra in DMSO-d6.
Proton assignments were made in association with COSY experiments. Panels A–C show assignments for the glucuronides, while panel D shows the assignments for the parent diol.
Table 1.
1H-NMR data for the downfield region of DB[a,l]P-trans-11,12-diol and the isolated glucuronides.
| peak 1 | peak 2 | peak 3 | ||
|---|---|---|---|---|
| proton | DB[a,l]P-diol | (+)-DB [a,l]P-11 -Gluc | (+)-DB [a,l]P-12-Gluc | (−)-DB [a,l]P-11 -Gluc |
| 1 | 8.50 (d, J=8.4 Hz) | 8.50 (d, J=8.4 Hz) | 8.54 (d J=7.8 Hz) | 8.52 (d, J=8.1 Hz) |
| 2 | 7.70 (m) | 7.70 (m) | 7.76 (m) | 7.73 (m) |
| 3 | 7.76 (m) | 7.77 (m) | 7.80 (m) | 7.80 (m) |
| 4 | 8.94 (d, J=7.5 Hz) | 8.95(d) | 8.97 (d, J=7.9 | 8.97 d, J=8.1 Hz) |
| 5 | 8.98 (d, J=7.7Hz) | 8.99 (d, J= 8.4 Hz) | 9.01 (d, J=7.9 | 9.02 d 8.1 |
| 6 | 8.02 (t, J=7.7 Hz) | 8.03 (t, J=8.1 Hz) | 8.06 (t, J=8.0 Hz) | 8.05 (m) |
| 7 | 8.24 (d, J= 7.5 Hz) | 8.25 (d, J= 6.6 Hz) | 8.28 (d, J=7.60 Hz) | 8.27 (d, J= 9.0 Hz) |
| 8 | 8.13 (d, J= 9.3 Hz) | 8.05–8.12 (m) | 8.20 (d, J=8.80 Hz) | 8.14 (d, J=9.0 Hz) |
| 9 | 8.09 (d, J= 9.1 Hz) | 8.05–8.12 (m) | 8.12 (d, J=8.80 Hz) | 8.06 (m) |
| 10 | 8.41 (s) | 8.95 (s) | 8.49 (s) | 8.80 (s) |
| 11 | 4.71 (m) | 4.98 (m) | 4.99 (m) | 4.96 (m) |
| 12 | 4.59 (m) | 4.91 (m) | 4.78 (m) | 4.81 (m) |
| 13 | 6.28 (dd, J= 10.3, 2.1 Hz) | 6.32 (dd, J= 10.6, 2.2 Hz) | 6.62 (dd, J=10.4, 2.4 Hz) | 6.26 (dd, J=10.4, 2.4Hz) |
| 14 | 7.26 (dd, J= 10.6, 2.1 Hz) | 7.24 (dd, J= 10.6, 2.1 Hz) | 7.30 (dd, J=10.4, 2.4 Hz) | 7.30 (dd, J= 10.1, 2.4 Hz) |
| 11-OH | 5.84 (d, J= 5.6 Hz) | |||
| 12-OH | 5.43 (d, J= 4.8 Hz) | |||
| 1’ | 4.71 (d, J=6.8 Hz) | 4.58 (m) | 4.60 (d, J=7.1 Hz) | |
Figure 3. Selective NOE experiment for peak 3.
A. Irradiation of the 1’-proton at 4.59 ppm, with 500 ms mixing time. B. Irradiation of the 1′-proton with 20 ms mixing time. C. 1D experiment to show the chemical shifts of the 11, 12, and 1′ protons. The increase in intensity of the peaks at 4.97 ppm in panel A versus panel B indicates that the 1′ proton is closer to the proton at the 11-position than the proton at the 12-position.
As shown in Table 2, there was some variability in peak ratios in assays with different human liver microsomal specimens. In an analysis of four human liver microsomes incubated with racemic DB[a,l]P-trans-11,12-diol, the average ratio of (+)-DB[a,l]P-11-Gluc : (+)-DB[a,l]P-12-Gluc : (−)-DB[a,l]P-11-Gluc formation was 31 : 32 : 37. Kinetic analyses demonstrated that human liver microsomes formed (+)-DB[a,l]P-11-Gluc, (+)-DB[a,l]P-12-Gluc, and (−)-DB[a,l]P-11-Gluc products with observed KM’s, of 23 ± 5.4 µM, 48 ± 8.6 µM, and 9.3 ± 1.3 µM, respectively (results not shown).
Table 2.
DB[a,l]P-trans-11,12-diol glucuronide peak areas for individual human liver microsomal specimens.
| human liver microsomal specimen # | peak 1 | peak 2 | peak 3 |
|---|---|---|---|
| 2635 | 987 (27)a | 1090 (30) | 1523 (42) |
| 3333 | 1016 (31) | 1071 (32) | 1220 (37) |
| 3144 | 1153 (40) | 593 (21) | 1101 (39) |
| 972 | 1177 (25) | 2030 (44) | 1450 (31) |
| Mean percentages (± S.D.) | 31 ± 6.7 | 32 ± 9.3 | 37 ± 4.7 |
Shown are the areas under the curve for peaks 1 (DB[a,l]P-(+)-trans-11-Gluc), 2 (DB[a,l]P-(+)-trans-12-Gluc), and 3 (DB[a,l]P-(−)-trans-11-Gluc) for four individual human liver microsomal specimens. Shown in parentheses are percentages of the area under the curve for each peak versus total area under the curve for all peaks combined for each liver microsomal specimen.
DB[a,l]P-trans-11,12-diol glucuronidation in UGT-over-expressing cell homogenates
A comprehensive screening was performed to identify all UGT1A and 2B family member enzymes responsible for glucuronidating DB[a,l]P-trans-11,12-diol. Initially, each DB[a,l]P-trans-11,12-diol enantiomer was incubated separately with homogenate from cells expressing a single UGT. Of the fifteen individual UGTs examined for activity, UGTs 1A1, 1A4, 1A7, 1A8, 1A9, 1A10, and 2B7 were able to glucuronidate DB[a,l]P-trans-11,12-diol; UGTs 1A3, 1A5 1A6, 2B4, 2B10, 2B11, 2B15, and 2B17 did not produce detectable levels (≥ 0.8 pmol of glucuronidated substrate) of glucuronides, using up to 500 µg of UGT-over-expressing cell homogenate. The individual UGTs exhibited varied regio- and stereoselectivity. The (+)-DB[a,l]P-11-Gluc and (−)-DB[a,l]P-11-Gluc products were made by UGTs 1A1, 1A7, 1A8, 1A9, and 1A10 (Table 3). UGT2B7 only formed (+)-DB[a,l]P-11-Gluc while the (+)-DB[a,l]P-12-Gluc was only made by UGTs 1A1 and 1A10. In addition, UGT1A4 had very low glucuronidating activity against the (−)-trans enantiomer to form the (−)-DB[a,l]P-11-Gluc; no activity was observed against the (+)-trans enantiomer for this UGT (results not shown).
Table 3.
Kinetic analysis of glucuronide formation by UGT-over-expressing HEK293 cell lines.
| DB[a,l]P-(+)-trans-11-Gluca | DB[a,l]P-(+)-trans-12-Gluca | DB[a,l]P-(−)-trans-11-Glucb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Km (µM) |
kcat (min−1)c |
kcat/Km (mM−1.min−1) |
Km (µM) |
kcat (min−1)c |
kcat/Km (mM−1.min−1) |
Km (µM) |
kcat (min−1)c |
kcat/Km (mM−1.min−1) |
|
| UGT1A1 | 76 ± 21 | 0.44 ± 0.05 | 6.1 ± 1.5 | 78 ± 19 | 0.23 ± 0.04 | 3.1 ± 0.6 | 44 ± 8.0 | 0.22 ± 0.01 | 0.72 ± 0.07 |
| UGT1A7 | 43 ± 7.0 | 0.009 ± 0.003 | 0.23 ± 0.11 | nd d | nd | <0.001 | 28 ± 1.3 | 0.005 ± 0.001 | 0.18 ± 0.03 |
| UGT1A8 | 194 ± 39 | 0.15 ± 0.02 | 0.80 ± 0.12 | nd | nd | <0.001 | 243 ± 52 | 0.10 ± 0.02 | 0.46 ± 0.03 |
| UGT1A9 | 0.17 ± 0.01 | 0.48 ± 0.04 | 2980 ± 146 | nd | nd | <0.001 | 0.21 ± 0.05 | 0.70 ± 0.24 | 3290 ± 59 |
| UGT1A10 | 33 ± 9.8 | 0.30 ± 0.03 | 9.5 ± 2.0 | 66 ± 19 | 0.16 ± 0.01 | 2.6 ± 0.5 | 13 ± 1.7 | 0.17 ± 0.004 | 13 ± 1.3 |
| UGT2B7 | 175 ± 25 | 0.066 ± 0.007 | 0.38 ± 0.01 | nd | nd | <0.001 | nd | nd | <0.001 |
Glucuronides produced from DB[a,l]P-(+)-trans-11,12-diol enantiomer incubated with individual UGTs.
Glucuronide produced from DB[a,l]P-(−)-trans-11,12-diol enantiomer incubated with individual UGTs.
kcat was adjusted for UGT expression according to Western blot analysis.
nd indicates that glucuronide peaks were not detected, with a sensitivity of 0.8 pmol.
To better evaluate the stereoselectivity of individual UGTs against DB[a,l]P-trans-11,12-diol, UGT-over-expressing cell homogenate incubations were also performed with the racemic mixture. The ratio of (+)-DB[a,l]P-11-Gluc : (+)-DB[a,l]P-12-Gluc : (−)-DB[a,l]P-11-Gluc formation was 52 : 37 : 11 for UGT1A1 and 19 : 12 : 69 for UGT1A10. The ratio of (+)-DB[a,l]P-11-Gluc : (−)-DB[a,l]P-11-Gluc formation was 67 : 33 for UGT1A7, 48 : 52 for UGT1A8, and 30 : 70 for UGT1A9; as described above, no (+)-DB[a,l]P-12-Gluc formation was observed for these UGTs (results not shown).
Kinetic analysis was subsequently performed using a concentration range of DB[a,l]P-trans-11,12-diol enantiomers that encompassed the KM for a given UGT (Table 3; Michaelis-Menten curves shown in Figure S2) with incubation times that were within the linear range of catalysis for each UGT. Overall, UGT1A9 was the most efficient enzyme at forming both (+)-DB[a,l]P-11-Gluc and (−)-DB[a,l]P-11-Gluc as determined by the kcat/KM. The overall efficiencies of the UGT enzymes in forming both (+)-DB[a,l]P-11-Gluc and (−)-DB[a,l]P-11-Gluc were UGT1A9 >> UGT1A10 > UGT1A1 > UGT1A8 > UGT1A7. UGT2B7 had approximately the same glucuronidating efficiency as that of UGT1A8 for the formation of (+)-DB[a,l]P-11-Gluc while UGTs 1A1 and 1A10 both formed (+)-DB[a,l]P-12-Gluc with approximately the same efficiency. Kinetic analysis could not be performed for UGT1A4 due to relatively low glucuronidation activity against DB[a,l]P-trans-11,12-diol.
DB[a,l]P-trans-11,12-diol glucuronidation in respiratory tract tissue homogenates
To assess the potential physiological relevance of the glucuronidation pathway in the local elimination of DB[a,l]P-trans-11,12-diol in respiratory tract tissues, glucuronidation activity assays were performed using homogenates prepared from bronchus and trachea. In assays with racemic DB[a,l]P-trans-11,12-diol, glucuronidation activity was observed with homogenates from both bronchus and trachea tissue (Figure 4). Bronchus homogenates generated three glucuronides from racemic DB[a,l]P-trans-11,12-diol (Figure 4B) that were identical based on retention time to the glucuronides formed with human liver microsomes (Figure 4A; (+)-DB[a,l]P-11-Gluc, (+)-DB[a,l]P-12-Gluc, and (−)-DB[a,l]P-11-Gluc). The relative ratio of the three glucuronides was 74:21:5, respectively, for bronchus homogenates. Trachea homogenates generated only (+)-DB[a,l]P-11-Gluc and (+)-DB[a,l]P-12-Gluc, at a ratio of 8:92, respectively (Figure 4C). Treatment of bronchus and trachea homogenate glucuronidation assays with β-glucuronidase eliminated all glucuronide peaks (results not shown).
Figure 4. Representative chromatograms of glucuronidation activity by respiratory tract tissues.
Representative UPLC chromatograms are shown for human liver microsomes (panel A), bronchus (panel B), and trachea (panel C) incubated with racemic DB[a,l]P-trans-11,12-diol (indicated by an arrow). Glucuronide products are indicated by their respective numbers (peak 1 is (+)-DB[a,l]P-11-Gluc, peak 2 is (+)-DB[a,l]P-12-Gluc, and peak 3 is (−)-DB[a,l]P-11-Gluc).
DISCUSSION
Although phase I metabolism of DB[a,l]P by P450 enzymes has been studied previously, the phase II metabolism by glucuronidation of DB[a,l]P metabolites had not been characterized. In the present study, seven UGTs were found to have detectable levels of glucuronidation activity against DB[a,l]P-trans-11,12-diol enantiomers, with several of the UGTs displaying different regio- and stereospecific glucuronidation efficiencies. UGT1A9 was the most efficient UGT in generating both the (+)-DB[a,l]P-11-Gluc and (−)-DB[a,l]P-11-Gluc, with approximately 489- and 4570-fold higher activity, respectively (when comparing kcat/KM values), than UGT1A1, which was the next most efficient enzyme for both DB[a,l]P stereoisomers. However, UGT1A9 did not form detectable levels of (+)-DB[a,l]P-12-Gluc. This contrasts with UGT1A1 which was the only hepatic enzyme to form this DB[a,l]P glucuronide diastereomer. Two additional hepatic enzymes glucuronidated the DB[a,l]P-trans-11,12-diol enantiomers: UGT2B7 selectively formed (+)-DB[a,l]P-11-Gluc while UGT1A4 formed (−)-DB[a,l]P-11-Gluc.
Human liver microsomes from four individuals formed all three DB[a,l]P glucuronides [both the (+)- and (−)-DB[a,l]P-11-glucuronides and the (+)-DB[a,l]P-12-glucuronide]. While some variability was observed between individual liver microsomal specimens, the three glucuronides were formed in roughly equal amounts. While it is likely that UGTs 1A1 and 1A9 are the major contributors to the hepatic glucuronidation of the DB[a,l]P-trans-11,12-diol enantiomers, it is presently unclear why the (+)-DB[a,l]P-12-Gluc is formed in amounts similar to those of the (+)- and (−)-DB[a,l]P-11-glucuronides given that UGT1A9 does not form the (+)-DB[a,l]P-12-Gluc and that the overall activity of UGT1A1 for the formation of this glucuronide is similar to that observed for (+)-DB[a,l]P-11-Gluc formation. While both UGTs are well-expressed in human liver 37,38, a recent study by Bellemare et al. has shown that an alternate splicing of the UGT1A genes forms inactive protein products which dimerize with the active UGT1A protein and inhibit activity 39. Therefore, it is possible that the activity of specific UGTs could vary significantly regardless of wild-type expression of a given enzyme.
Three extra-hepatic UGTs (UGT1A7, UGT1A8, and UGT1A10) glucuronidated the DB[a,l]P-trans-11,12-diol enantiomers in our studies. These UGTs have been found to be expressed in tissues of the aerodigestive and respiratory tracts 17,20 including lung 19,38, trachea 37, and bronchial epithelial cells 40. Of these, only UGT1A10 formed all three DB[a,l]P-trans-11,12-diol glucuronides [the (+) and (−)-DB[a,l]P-11-glucuronides and the (+)-DB[a,l]P-12-glucuronide]. This is between 12–72-fold higher levels of activity as compared to the kcat/KM of UGTs 1A7 and 1A8 for the (+) and (−)-DB[a,l]P-11-glucuronides; neither UGTs 1A7 or 1A8 formed the (+)-DB[a,l]P-12-Gluc. In contrast to what was observed for human liver microsomes, the (+)-DB[a,l]P-12-Gluc was the predominant isomer formed in bronchus homogenate while the (+)-DB[a,l]P-11-Gluc was the primary isomer formed in trachea homogenate. This pattern is consistent with the fact that UGT1A10, which forms all three glucuronides including (+)-DB[a,l]P-12-Gluc, is expressed in tissues of the respiratory and aerodigestive tracts 20,37 and the detection of UGT1A enzymes in bronchial epithelial cells 40. It is also consistent with the fact that UGT1A9, the most active UGT in the formation of (+) and (−)-DB[a,l]P-11-glucuronides, is not expressed in trachea 37. However, given the peak formation pattern for UGT1A10 (forming all three DB[a,l]P glucuronides with high efficiency), it is possible that UGT1A1 may be playing a more important role in human bronchus given the similar peak pattern observed for UGT1A1 as compared to that observed for bronchus homogenates. This extrahepatic tissue pattern of glucuronidation was reported previously for B[a]P-trans-7,8,-diol in other extrahepatic tissues including larynx and esophagus, and is also consistent with the lack of UGT1A9 expression and the high expression of UGT1A10 in these extra-hepatic tissues 20,21. Further studies analyzing the expression of individual wild-type versus variant UGT1A enzymes are necessary to better evaluate these data. In addition, recent studies demonstrated that UGT2A1 was expressed in trachea and other respiratory tract tissues and that glucuronidating activity toward DB[a,l]P-trans-11,12-diol was detected for this enzyme 41. Although previous studies did not identify the individual DB[a,l]P glucuronides formed by this enzyme, this suggests tissues that UGT2A1 might also play an important role in detoxifying DB[a,l]P metabolites in these extra-hepatic tissues.
Studies have shown that B[a]P is present at about 9 ng/cigarette in mainstream smoke, and while DB[a,l]P has been identified in mainstream smoke, definitive quantifications have not yet been reported 42. Several studies have demonstrated that low doses of DB[a,l]P induced significantly more malignant tumors in mouse models when compared to the same treatment with B[a]P 13,14. DB[a,l]PDE stably bound to DNA has been found to cause significantly more mutational events in DNA compared to the bay region-PAH, benzo[a]pyrene-7,8-diol-9,10-epoxide (B[a]PDE) 43, further highlighting the potency of this DB[a,l]P metabolite. Interestingly, human cell culture studies in which MCF-7 cells were treated with each enantiomer as well as a racemic mixture of DB[a,l]P-trans-11,12-diol demonstrated that the diol-epoxide derived from the (+)-trans enantiomer does not bind to DNA in MCF-7 cells, suggesting that perhaps only the (−)-trans enantiomer, which is the major diol metabolite, is mutagenic 7,8.
Results of previous studies of B[a]P-trans-7,8-diol glucuronidation are consistent with the present finding for DB[a,l]P-trans-11,12-diol glucuronidation. B[a]P-trans-7,8,-diol formed diastereomeric glucuronide products by six of the UGTs that were identified in this study (the low-activity UGT1A4 being the exception) 21. Similar to that described in the present study for the 11-hydroxy group of DB[a,l]P-trans-11,12-diol, the 7-hydroxy group of B[a]P-trans-7,8-diol, which is analogous to the 11-hydroxy group of DB[a,l]P-trans-11,12-diol, was also preferentially glucuronidated 21.
In summary, this work demonstrates that the DB[a,l]P-trans-11,12-diol enantiomers are glucuronidated in a stereo-specific manner by several UGT enzymes. In addition to liver, bronchus and trachea tissue homogenates glucuronidated these enantiomers, further illustrating the important role UGT enzymes play in the local detoxification of tobacco carcinogens. Further studies examining functional polymorphisms in the relevant DB[a,l]P-trans-11,12-diol-metabolizing UGTs identified in the present study may therefore lead to a better understanding of susceptibility factors involved in exposure to combustion products and tobacco-related cancer risk.
Supplementary Material
Acknowledgements
We thank Nino Giambrone for his outstanding help with the mass spectrometry data. We also thank the NMR and Organic Synthesis Core Facilities at the Penn State College of Medicine.
Funding Support
This work was supported in part by the National Institutes of Health, National Institute of Dental and Craniofacial Research [Grant R01-DE13158, to P Lazarus]; the Pennsylvania Department of Health’s Health Research Formula Funding Programs [Grants 4100038714 to P. Lazarus and 4100038715 to P. Lazarus], and NCI Contract NO2-CB-81013-74 [to S. Amin].
ABBREVIATIONS
- (B[a]P)
Benzo[a]pyrene
- (B[a]PDE)
benzo[a]pyrene-7,8-diol-9,10-epoxide
- (P450)
cytochrome P450
- (DB[a,l]P)
dibenzo[a,l]pyrene
- (DB[a,l]P-11,12-diol)
dibenzo[a,l]pyrene-11,12-dihydrodiol
- (DB[a,l]PDE)
dibenzo[a,l]pyrene-11,12-diol-13,14-epoxide
- (EH)
epoxide hydrolase
- (GST)
glutathione-S-transferase
- (PAH)
polycyclic aromatic hydrocarbon
- (UPLC)
ultra performance liquid chromatography
- (UDPGA)
uridine 5’-diphosphate-glucuronic acid
- (UGT)
uridine 5’-diphosphate glucuronosyltransferase
REFERENCES
- 1.Rodgman A, Smith CJ, Perfetti TA. The composition of cigarette smoke: a retrospective, with emphasis on polycyclic components. Hum Exp Toxicol. 2000;19:573–595. doi: 10.1191/096032700701546514. [DOI] [PubMed] [Google Scholar]
- 2.Mumford JL, Li X, Hu F, Lu XB, Chuang JC. Human exposure and dosimetry of polycyclic aromatic hydrocarbons in urine from Xuan Wei, China with high lung cancer mortality associated with exposure to unvented coal smoke. Carcinogenesis. 1995;16:3031–3036. doi: 10.1093/carcin/16.12.3031. [DOI] [PubMed] [Google Scholar]
- 3.Kozin IS, Gooijer C, Velthorst NH. Direct determination of dibenzo[a,l]pyrene in crude extracts of environmental samples by laser-excited Shpol'skii spectroscopy. Analytical Chemistry. 1995;67:1623–1626. [Google Scholar]
- 4.Luch A, Seidel A, Glatt H, Platt KL. Metabolic activation of the (+)-S,S-and (−)-R,R-enantiomers of trans-11,12-dihydroxy-11,12-dihydrodibenzo[a,l]pyrene: stereoselectivity, DNA adduct formation, and mutagenicity in Chinese hamster V79 cells. Chem Res Toxicol. 1997;10:1161–1170. doi: 10.1021/tx970005i. [DOI] [PubMed] [Google Scholar]
- 5.Shou M, Krausz KW, Gonzalez FJ, Gelboin HV. Metabolic activation of the potent carcinogen dibenzo[a,l]pyrene by human recombinant cytochromes P450, lung and liver microsomes. Carcinogenesis. 1996;17:2429–2433. doi: 10.1093/carcin/17.11.2429. [DOI] [PubMed] [Google Scholar]
- 6.Luch A. On the impact of the molecule structure in chemical carcinogenesis. EXS. 2009;99:151–179. doi: 10.1007/978-3-7643-8336-7_6. [DOI] [PubMed] [Google Scholar]
- 7.Ralston SL, Coffing SL, Seidel A, Luch A, Platt KL, Baird WM. Stereoselective activation of dibenzo[a,l]pyrene and its trans-11,12-dihydrodiol to fjord region 11,12-diol 13,14-epoxides in a human mammary carcinoma MCF-7 cell-mediated V79 cell mutation assay. Chem Res Toxicol. 1997;10:687–693. doi: 10.1021/tx9700275. [DOI] [PubMed] [Google Scholar]
- 8.Luch A. The Carcinogenic Effects of Polycylic Aromatic Hydrocarbons. London: Imperial College Press; 2005. [Google Scholar]
- 9.Ralston SL, Seidel A, Luch A, Platt KL, Baird WM. Stereoselective activation of dibenzo[a,l]pyrene to (−)-anti (11R,12S,13S,14R)- and (+)-syn(11S,12R,13S,14R)-11,12-diol-13,14-epoxides which bind extensively to deoxyadenosine residues of DNA in the human mammary carcinoma cell line MCF-7. Carcinogenesis. 1995;16:2899–2907. doi: 10.1093/carcin/16.12.2899. [DOI] [PubMed] [Google Scholar]
- 10.Xue W, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol Appl Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Luch A, Glatt H, Platt KL, Oesch F, Seidel A. Synthesis and mutagenicity of the diastereomeric fjord-region 11,12-dihydrodiol 13,14-epoxides of dibenzo[a,l]pyrene. Carcinogenesis. 1994;15:2507–2516. doi: 10.1093/carcin/15.11.2507. [DOI] [PubMed] [Google Scholar]
- 12.Spencer WA, Singh J, Orren DK. Formation and differential repair of covalent DNA adducts generated by treatment of human cells with (+/−)-anti-dibenzo[a,l]pyrene-11,12-diol-13,14-epoxide. Chem Res Toxicol. 2009;22:81–89. doi: 10.1021/tx8001675. [DOI] [PubMed] [Google Scholar]
- 13.Higginbotham S, RamaKrishna NV, Johansson SL, Rogan EG, Cavalieri EL. Tumor-initiating activity and carcinogenicity of dibenzo[a,l]pyrene versus 7,12-dimethylbenz[a]anthracene and benzo[a]pyrene at low doses in mouse skin. Carcinogenesis. 1993;14:875–878. doi: 10.1093/carcin/14.5.875. [DOI] [PubMed] [Google Scholar]
- 14.Cavalieri EL, Higginbotham S, RamaKrishna NV, Devanesan PD, Todorovic R, Rogan EG, Salmasi S. Comparative dose-response tumorigenicity studies of dibenzo[alpha,l]pyrene versus 7,12-dimethylbenz[alpha]anthracene, benzo[alpha]pyrene and two dibenzo[alpha,l]pyrene dihydrodiols in mouse skin and rat mammary gland. Carcinogenesis. 1991;12:1939–1944. doi: 10.1093/carcin/12.10.1939. [DOI] [PubMed] [Google Scholar]
- 15.Amin S, Krzeminski J, Rivenson A, Kurtzke C, Hecht SS, el-Bayoumy K. Mammary carcinogenicity in female CD rats of fjord region diol epoxides of benzo[c]phenanthrene, benzo[g]chrysene and dibenzo[a,l]pyrene. Carcinogenesis. 1995;16:1971–1974. doi: 10.1093/carcin/16.8.1971. [DOI] [PubMed] [Google Scholar]
- 16.Dreij K, Seidel A, Jernstrom B. Differential removal of DNA adducts derived from anti-diol epoxides of dibenzo[a,l]pyrene and benzo[a]pyrene in human cells. Chem Res Toxicol. 2005;18:655–664. doi: 10.1021/tx0497090. [DOI] [PubMed] [Google Scholar]
- 17.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 18.Turgeon D, Carrier JS, Levesque E, Hum DW, Belanger A. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142:778–787. doi: 10.1210/endo.142.2.7958. [DOI] [PubMed] [Google Scholar]
- 19.Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab Dispos. 2006;34:943–949. doi: 10.1124/dmd.105.009100. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Z, Fang JL, Lazarus P. Glucuronidation: an important mechanism for detoxification of benzo[a]pyrene metabolites in aerodigestive tract tissues. Drug Metab Dispos. 2002;30:397–403. doi: 10.1124/dmd.30.4.397. [DOI] [PubMed] [Google Scholar]
- 21.Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, Marques MM, Lazarus P. Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res. 2002;62:1978–1986. [PubMed] [Google Scholar]
- 22.Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- 23.Owens IS, Ritter JK. Gene structure at the human UGT1 locus creates diversity in isozyme structure, substrate specificity, and regulation. Prog Nucleic Acid Res Mol Biol. 1995;51:305–338. doi: 10.1016/s0079-6603(08)60882-x. [DOI] [PubMed] [Google Scholar]
- 24.Nagar S, Remmel RP. Uridine diphosphoglucuronosyltransferase pharmacogenetics and cancer. Oncogene. 2006;25:1659–1672. doi: 10.1038/sj.onc.1209375. [DOI] [PubMed] [Google Scholar]
- 25.Testa B, Kramer SD. The biochemistry of drug metabolism--an introduction: part 4. reactions of conjugation and their enzymes. Chem Biodivers. 2008;5:2171–2336. doi: 10.1002/cbdv.200890199. [DOI] [PubMed] [Google Scholar]
- 26.Guillemette C, Ritter JK, Auyeung DJ, Kessler FK, Housman DE. Structural heterogeneity at the UDP-glucuronosyltransferase 1 locus: functional consequences of three novel missense mutations in the human UGT1A7 gene. Pharmacogenetics. 2000;10:629–644. doi: 10.1097/00008571-200010000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Hu Z, Wells PG. In vitro and in vivo biotransformation and covalent binding of benzo(a)pyrene in Gunn and RHA rats with a genetic deficiency in bilirubin uridine diphosphate-glucuronosyltransferase. J Pharmacol Exp Ther. 1992;263:334–342. [PubMed] [Google Scholar]
- 28.Krzeminski J, Lin JM, Amin S, Hecht SS. Synthesis of fjord region diol epoxides as potential ultimate carcinogens of dibenzo[a,l]pyrene. Chem Res Toxicol. 1994;7:125–129. doi: 10.1021/tx00038a002. [DOI] [PubMed] [Google Scholar]
- 29.Sharma AK, Kumar S, Amin S. A highly abbreviated synthesis of dibenzo[def,p]chrysene and its 12-methoxy derivative, a key precursor for the synthesis of the proximate and ultimate carcinogens of dibenzo[def,p]chrysene. J Org Chem. 2004;69:3979–3982. doi: 10.1021/jo0303822. [DOI] [PubMed] [Google Scholar]
- 30.Wiener D, Fang JL, Dossett N, Lazarus P. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 2004;64:1190–1196. doi: 10.1158/0008-5472.can-03-3219. [DOI] [PubMed] [Google Scholar]
- 31.Sun D, Sharma AK, Dellinger RW, Blevins-Primeau AS, Balliet RM, Chen G, Boyiri T, Amin S, Lazarus P. Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab Dispos. 2007;35:2006–2014. doi: 10.1124/dmd.107.017145. [DOI] [PubMed] [Google Scholar]
- 32.Ren Q, Murphy SE, Zheng Z, Lazarus P. O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos. 2000;28:1352–1360. [PubMed] [Google Scholar]
- 33.Balliet RM, Chen G, Gallagher CJ, Dellinger RW, Sun D, Lazarus P. Characterization of UGTs active against SAHA and association between SAHA glucuronidation activity phenotype with UGT genotype. Cancer Res. 2009;69:2981–2989. doi: 10.1158/0008-5472.CAN-08-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G, Dellinger RW, Sun D, Spratt TE, Lazarus P. Glucuronidation of tobacco-specific nitrosamines by UGT2B10. Drug Metab Dispos. 2008;36:824–830. doi: 10.1124/dmd.107.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y-S, Fang G-C, Moody J, Tungeln LSV, Fu PP, Hwang H-M, Yu H. In vitro metabolism of dibenzo[a,l]pyrene, 2-chlorodibenzo[a,l]pyrene and 10-chlorodibenzo[a,l]pyrene - effects of chloro substitution. Int. J. Mol. Sci. 2002;3:1008–1018. [Google Scholar]
- 36.Al-Zoughool M, Talaska G. High-performance liquid chromatography method for determination of N-glucuronidation of 4-aminobiphenyl by mouse, rat, and human liver microsomes. Anal Biochem. 2005;340:352–358. doi: 10.1016/j.ab.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 37.Ohno S, Nakajin S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2009;37:32–40. doi: 10.1124/dmd.108.023598. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T. Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos. 2008;36:1461–1464. doi: 10.1124/dmd.108.021428. [DOI] [PubMed] [Google Scholar]
- 39.Bellemare J, Rouleau M, Harvey M, Guillemette C. Modulation of the human glucuronosyltransferase UGT1A pathway by splice isoform polypeptides is mediated through protein-protein interactions. J Biol Chem. 285:3600–3607. doi: 10.1074/jbc.M109.083139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuzuya Y, Adachi T, Hara H, Anan A, Izuhara K, Nagai H. Induction of drug-metabolizing enzymes and transporters in human bronchial epithelial cells by beclomethasone dipropionate. IUBMB Life. 2004;56:355–359. doi: 10.1080/10258140412331286946. [DOI] [PubMed] [Google Scholar]
- 41.Bushey RT, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. Characterization of UDP-glucuronosyltransferase 2A1 (UGT2A1) variants and their potential role in tobacco carcinogenesis. Pharmacogenet Genomics. 21:55–65. doi: 10.1097/FPC.0b013e328341db05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hecht SS. Tobacco smoke carcinogens and breast cancer. Environ Mol Mutagen. 2002;39:119–126. doi: 10.1002/em.10071. [DOI] [PubMed] [Google Scholar]
- 43.Lagerqvist A, Hakansson D, Prochazka G, Lundin C, Dreij K, Segerback D, Jernstrom B, Tornqvist M, Seidel A, Erixon K, Jenssen D. Both replication bypass fidelity and repair efficiency influence the yield of mutations per target dose in intact mammalian cells induced by benzo[a]pyrene-diol-epoxide and dibenzo[a,l]pyrene-diol-epoxide. DNA Repair (Amst) 2008;7:1202–1212. doi: 10.1016/j.dnarep.2008.03.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.