Abstract
The ends of chromosomes are composed of a short repeat sequence and associated proteins that together form a cap, called a telomere, that keeps the ends from appearing as double-strand breaks (DSBs) and prevents chromosome fusion. The loss of telomeric repeat sequences or deficiencies in telomeric proteins can result in chromosome fusion and lead to chromosome instability. The similarity between chromosome rearrangements resulting from telomere loss and those found in cancer cells implicates telomere loss as an important mechanism for the chromosome instability contributing to human cancer. Telomere loss in cancer cells can occur through gradual shortening due to insufficient telomerase, the protein that maintains telomeres. However, cancer cells often have a high rate of spontaneous telomere loss despite the expression of telomerase, which has been proposed to result from a combination of oncogene-mediated replication stress and a deficiency in DSB repair in telomeric regions. Chromosome fusion in mammalian cells primarily involves nonhomologous end joining (NHEJ), which is the major form of DSB repair. Chromosome fusion initiates chromosome instability involving breakage-fusion-bridge (B/F/B) cycles, in which dicentric chromosomes form bridges and break as the cell attempts to divide, repeating the process in subsequent cell cycles. Fusion between sister chromatids results in large inverted repeats on the end of the chromosome, which amplify further following additional B/F/B cycles. B/F/B cycles continue until the chromosome acquires a new telomere, most often by translocation of the end of another chromosome. The instability is not confined to a chromosome that loses its telomere, the instability is transferred to the chromosome donating a translocation. Moreover, the amplified regions are unstable and form extrachromosomal DNA that can reintegrate at new locations. Knowledge concerning the factors promoting telomere loss and its consequences is therefore important for understanding chromosome instability in human cancer.
Keywords: Chromosome fusion, Chromosome healing, Chromosome instability, Double-strand break, Intrastrand annealing, Nonhomologous end joining, Telomere
1. Telomeres and the consequences of telomere loss
Seminal work by Hermann Muller in fruit flies [1] and Barbara McClintock in maize [2] demonstrated long ago that the ends of chromosomes did not fuse with chromosome breaks, and therefore must somehow be “capped.” These chromosome ends, which Muller termed telomeres, are now know to be composed of a short DNA repeat sequence [3] and a large number of proteins, named the shelterin complex [4], which together form the protective cap. Telomeres are maintained in germ line cells and embryonic stem (ES) cells by telomerase, but shorten during cell division in human somatic cells due to insufficient telomerase activity. Telomere shortening in human fibroblasts eventually results in cell senescence, as shown by the presence of DSB repair foci at telomeres in senescent cells [5, 6]. Fibroblasts that are unable to senesce continue to divide and undergo severe telomere shortening, which eventually results in “crisis,” involving massive chromosome fusion and cell death [7]. Some cells survive crisis by regaining the ability to maintain telomeres, either through expression of telomerase or through an alternative pathway involving recombination [8, 9]. Cancer cells typically demonstrate this ability to maintain telomeres [10], which is thought to be necessary for the extensive cell division required for the development of malignant cancer.
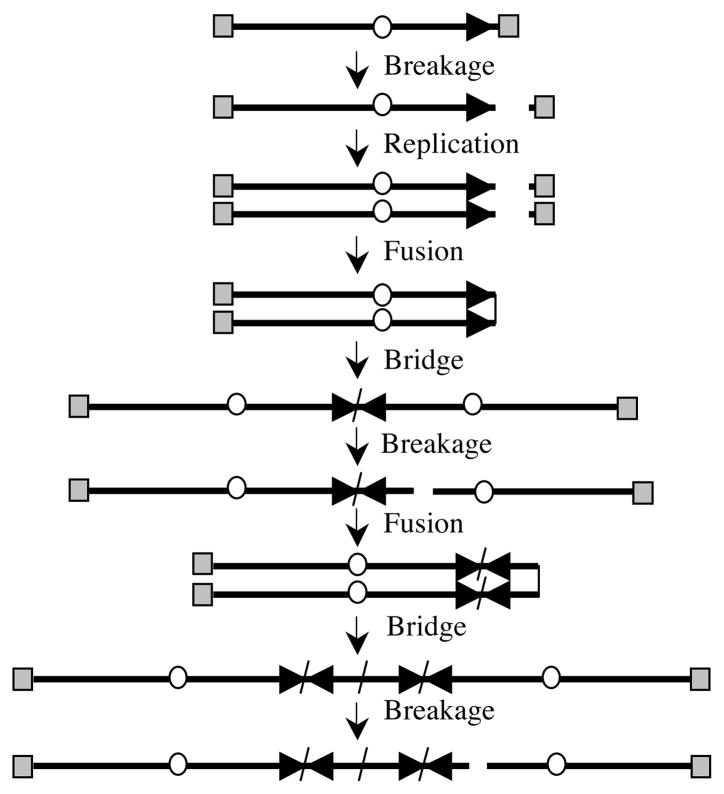
Barbara McClintock also demonstrated that fusions between two different chromosomes, or between sister chromatids following DNA replication, can result in chromosome instability involving breakage-fusion-bridge (B/F/B) cycles [2]. B/F/B cycles occur when fused chromosomes form a bridge during anaphase as their centromeres are pulled in opposite directions when the cell attempts to divide. The fused chromosomes eventually break at a location other than the site of fusion, resulting in the acquisition of DNA on the end of one chromosome and the loss of DNA on the end of the other chromosome. Because the broken chromosomes still do not have telomeres, they fuse again in the next cell cycle, continuing the B/F/B cycles. The consequences of B/F/B cycles depend on whether fusions occur between different chromosomes or between sister chromatids [11]. A high rate of telomere loss or telomere loss prior to DNA replication favors fusion between different chromosomes, while a low rate of telomere loss or telomere loss during DNA replication favors fusion between sister chromatids. Fusions between chromosomes can be detected by the presence of dicentric chromosomes at metaphase, and result in the transfer of DNA from one chromosome to another when the fused chromosomes break. On the other hand, sister chromatid fusions cannot be detected by conventional cytogenetic methods, because they appear normal at metaphase except for the lack of telomeres on one end of the sister chromatids. Unlike fusions between different chromosomes, sister chromatid fusion results in large inverted repeats on the end of the chromosome in one daughter cell and a large terminal deletion on the end of the chromosome in the other daughter cell (Fig. 1). In subsequent cell divisions, the chromosome containing the duplicated region can again undergo sister chromatid fusion, resulting in extensive DNA amplification consisting of large inverted repeats. Similarly, the chromosome containing a terminal deletion can also undergo sister chromatid fusions in subsequent cell cycles, leading to additional terminal deletions and amplification of sequences far from the original end of the chromosome. The extent of the amplification or terminal deletions can vary greatly depending on the number of B/F/B cycles that occur before the chromosome is stabilized by the acquisition of a new telomere. Chromosomes that go through a single B/F/B cycle will have a single large inverted repeat, while those that go through many cycles will experience extensive amplification, although the extent of amplification will also depend on the site of breakage and deletions within the amplified region [11].
Fig. 1. Chromosome instability involving B/F/B cycles.
B/F/B cycles are initiated when sister chromatids fuse following the replication of a chromosome that has lost a telomere. Due to the presence of two centromeres, the chromosome then forms a bridge during anaphase, which breaks when the centromeres are pulled in opposite directions during cell division. Breakage occurs at locations other than the site of fusion, resulting in large inverted repeats on the end of the chromosome in one daughter cell and a terminal deletion on the end of the chromosome in the other daughter cell. Because the chromosomes are missing a telomere in the next cell cycle, they will fuse again, resulting in further amplification of subtelomeric DNA on the chromosome containing the inverted repeat. Subsequent fusions on the chromosome containing the terminal deletion can also result in amplification of sequences far from original end of the chromosome. The B/F/B cycles will continue until the chromosome acquires a new telomere, most often by translocation. The telomeres (gray squares), centromeres (circles), and orientation of the subtelomeric sequences (horizontal arrows) are shown.
As predicted by the work of McClintock, chromosomes experiencing spontaneous telomere loss in human cancer cells also experience chromosome instability involving B/F/B cycles [12–14]. The analysis of individual marker chromosomes following telomere loss using chromosome-specific painting probes demonstrates that these B/F/B cycles primarily involve sister chromatid fusions, as shown by: 1) the absence of a telomere on the marker chromosome many cell generations after telomere loss, 2) anaphase bridges consisting of the fused sister chromatids of the marker chromosome, and 3) amplification of the subtelomeric DNA on the end of the marker chromosome. Importantly, the rate of this spontaneous telomere loss is low enough so that only a single chromosome is experiencing B/F/B cycles at any one time, and therefore is not lethal to these cells. In some cells in the population, the B/F/B cycles continue for many generations, resulting in extensive amplification of subtelomeric DNA, stopping only when the chromosome acquires a new telomere and once again becomes stable [14]. Importantly, these studies demonstrate that telomeres are often acquired by nonreciprocal translocations. Although these nonreciprocal translocations stabilize the chromosome that acquired the new telomere, they initiate chromosome instability in the chromosome donating the telomere [14]. Thus, the loss of a single telomere can result in chromosome instability in multiple chromosomes. Proof that chromosome instability involving B/F/B cycles is important in cancer is shown by the increase in carcinomas in telomerase deficient mice that are also deficient in p53, which show chromosome rearrangements containing amplified regions adjacent to translocations [15, 16]. Similar chromosome rearrangements are also found during amplification of c-Myc in pro-B lymphomas in mice [17]. Importantly, large inverted repeats have been found to be an early event in pancreatic cancer [18], and amplified regions in human cancer cells commonly contain inverted repeats [19, 20].
Although it is now clear that B/F/B cycles resulting from telomere loss can result in genomic instability, questions remain as to the role of B/F/B cycles in gene amplification in human cancer. B/F/B cycles were recognized long ago as a mechanism for gene amplification in rodent cells [21–23]; however, evidence was initially lacking that B/F/B cycles were associated with gene amplification in human cells. Gene amplification in rodent cells was obvious due to the breakage of the fused chromosomes far from the site of fusion, producing large “ladder-like” repeats visible by cytogenetic analysis. However, chromosome breaks during B/F/B cycles in human cells most often occur within 1 Mb of the site of fusion [13, 24], so that the size of the amplified regions are usually much smaller. In addition, the regions amplified during B/F/B cycles in human cells can be rapidly deleted out and form double minute chromosomes [25], which can also be involved in gene amplification and reintegrate back into chromosomes at other locations [26, 27]. The frequency of these large deletions demonstrates that the large inverted repeats formed during B/F/B cycles in human cells are especially unstable and susceptible to breakage, most likely due to their ability stimulate recombination and replication errors (see below). This instability can make the identification of rearrangements resulting from B/F/B cycles in human cells difficult to recognize in human cells.
Further confusion on the role of B/F/B cycles in gene amplification was generated by a study by Tanaka et al [28], which reported that only a small fraction of large inverted repeats in cancer cells are associated with gene amplification. This observation suggested that the formation of large inverted repeats is the rate-limiting step in gene amplification and occurs by a mechanism that is different than amplification itself. However, a more recent analysis by this group has now found that the number of large inverted repeats in cancer cells was much less than originally reported [29]. Thus, most large inverted repeats formed by sister chromatid fusion are likely to progress to more extensive DNA amplification.
2. Mechanisms of chromosome fusion following loss of telomere function
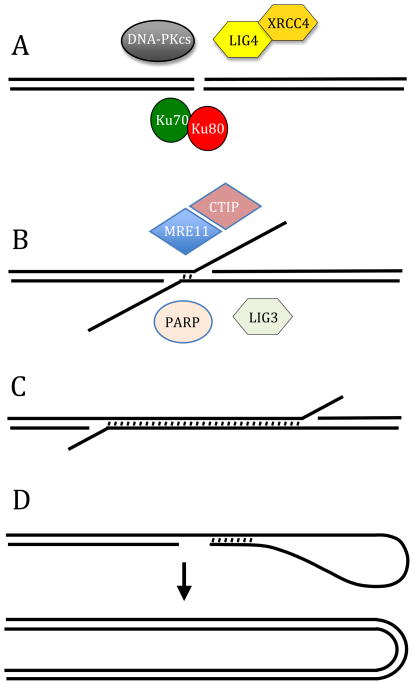
Chromosome fusions can occur through a variety of different mechanisms depending on the cell type and the mechanism of loss of telomere function. As will be discussed in detail below, most chromosome fusions in mammalian cells occur through double-strand break (DSB) repair involving nonhomologous end joining (NHEJ). This is not surprising, because NHEJ is the predominant form of repair of unprotected DNA ends in mammalian cells. There are at least two forms of NHEJ, classical (C-NHEJ), and alternative (A-NHEJ) (Fig. 2A and B). C-NHEJ (also called conservative or canonical NHEJ) is the major DSB repair pathway in mammalian cells, and involves proteins that are well characterized, including Ku70, Ku80, DNA-PKcs, LIG4, and XRCC4 [30]. A-NHEJ (also called deletional NHEJ) is characterized by the presence of microhomology at the recombination junctions, and is associated with large deletions and chromosome rearrangements [31–33]. A-NHEJ has been reported to utilize PARP-1 [34–36], LIG3 [35, 37, 38], MRE11 [39–41], and CtIP [42, 43]. A-NHEJ is increased in cells that are deficient in C-NHEJ due to the suppression of A-NHEJ by C-NHEJ [43, 44]. A-NHEJ has been proposed as a backup mechanism for C-NHEJ, although recent evidence demonstrating that A-NHEJ is much more efficient than originally thought suggests that it serves a more prominent role.
Fig. 2. Mechanisms of chromosome fusion.
(A) C-NHEJ is the most prominent mechanism of DSB repair in mammalian cells. C-NHEJ involves the direct rejoining of DSBs with minimal processing, utilizing a variety of well characterized proteins, including Ku70, Ku80, DNA-PKcs, LIG4, and XRCC4. C-NHEJ is involved in fusion of chromosomes as a result of a deficiency in TRF2. (B) A-NHEJ can also perform direct rejoining of DBS, although it commonly involves resection of 5′ ends and often utilizes 4 bps or less of microhomology to facilitate repair. A-NHEJ is associated with large deletions and chromosome rearrangements, and has been reported to utilize a variety of proteins, including PARP-1, MRE11, CtIP, and LIG3. A-NHEJ has been proposes as a major pathway of chromosome fusion following telomere loss in mammalian cells. (C) SSA is similar to A-NHEJ in requiring extensive resection of the 5′ stand to generate long single-stranded tails. However, SSA requires long complementary regions for end joining. (D) Intrastrand annealing involves resection of the end of an unprotected DNA end and the pairing of complementary sequences of short inverted repeats (4 to 12 bps). Long inverted repeats may also promote chromosome fusion by formation of hairpins or by promoting replication template switching. The role of short and long inverted repeats in chromosome fusion in mammalian cells is not known.
The fact that microhomology is commonly present at repair junctions generated by A-NHEJ suggests that it is related to the microhomology mediated end joining (MMEJ) mechanism that has been studied in yeast. However, the relationship between A-NHEj and MMEJ is controversial [45]. MMEJ in yeast utilizes 5 to 25 bps of microhomology for DSB repair. The recombination junctions generated during immunoglobulin class switching at the IgH locus in mouse B cells deficient in XRCC4 or LIG4 show increased microhomology compared to wild type cells [46]. However, the microhomology at most of these junctions is shorter (1 to 4 bps) than that observed with MMEJ in yeast, although a significant fraction have microhomology of 5 bps or longer. Similarly, the increased microhomology at recombination junctions resulting from A-NHEJ of DSBs induced with the I-SceI endonuclease in mouse and human cells often show no microhomology or microhomologies that are much shorter (1 to 4 bps) than that observed with MMEJ in yeast. Therefore, whether MMEJ and A-NHEJ are the same pathway, or whether there are multiple alternative NHEJ pathways for repair of DSBs, is unclear at this time.
Studies in yeast have demonstrated that chromosome fusion can occur through mechanisms other than NHEJ. Chromosome fusion due to telomere loss in yeast commonly involves single-strand annealing (SSA), in which resection of the 5′ strand on the end of one chromosome results in single-stranded DNA that pairs with a homologous single-stranded DNA on the end of another chromosome (Fig. 2C) [47]. In some respects, SSA is similar to A-NHEJ, in that both mechanisms involve extensive resection of the 5′ strand. However, while A-NHEJ typically involves only a few bps of microhomology for end joining, SSA requires large regions of homology. Chromosome fusion in yeast can also occur through the presence of inverted repeat sequences [48, 49]. DSBs occurring near 5.9 kb Ty1 inverted repeats in yeast result in sister chromatid fusions involving SSA [50]. In addition, inverted repeat sequences as short as 4 to12 bps can promote sister chromatid fusions involving intrastrand annealing in telomerase-deficient yeast [51]. Intrastrand annealing occurs following the resection of the 5′ strand, which allows the 3′ single-stranded DNA to loop back and pair with a complementary sequence located at a proximal site in the single-stranded DNA (Fig. 2D). Break-induced replication and DNA ligation is then proposed to complete the process, resulting in a chromosome with a hairpin on its end. Replication of the chromosome then results in sister chromatid fusion, converting the short inverted repeat into a very large inverted repeat (also called a palindrome). DSBs occurring near short inverted repeats as small as 4 to 6 bps have also been reported to result in chromosome fusions involving intrastrand annealing [52]. Other studies in yeast have found that longer inverted repeats consisting of 320 bp Alu sequences can cause chromosome fusion by promoting the formation of DSBs due to their ability to form cruciform structures that are recognized as recombination intermediates [53, 54]. Similar to intrastrand annealing, these cruciform structures on the ends of chromosomes then result in chromosome fusion when the chromosomes are replicated, resulting in sister chromatid fusion and large inverted repeats. Finally, inverted repeats in yeast can result in the formation of chromosome fusions in the absence of DSBs by promoting template-switching mechanisms during DNA replication. Chromosome fusions resulting from template switching involving very long inverted repeats of 5.3 kb are dependent on homologous recombination [55], while inverted repeats as short as 60 bps are capable of promoting template switching that is independent of homologous recombination [56, 57].
Although it is clear that SSA and inverted repeats can be involved in chromosome fusion in yeast, their role in chromosome fusion in mammalian cells has yet to be established. The presence of homologous sequences consistent with SSA annealing is not observed at the junctions of chromosome rearrangements in mammalian cells [58], leading to the hypothesis that it is prevented by sequence divergence and active suppression [59]. It has been proposed that intrastrand annealing involving short inverted repeats is likely to be responsible for most chromosome fusions in mammalian cells based on the fact that chromosome fusions occur in cells deficient in Ku70 or LIG4 [47, 53, 60]. However, as discussed above, it is now clear that A-NHEJ is a robust mechanism for repair of DSBs in mammalian cells that does not require Ku70 or LIG4. The study by Tanaka et al [60] also concluded that intrastrand annealing involving short inverted repeats is involved in sister chromatid fusions in mammalian cells based on their observation that one of the inverted repeats involved in sister chromatid fusions was often longer than the other, and by the presence of microhomology (0 to 3 bps) at the junctions. However, this microhomology profile is more typical of A-NHEJ [39, 40, 43, 61] than intrastrand annealing [51, 52]. Moreover, some sister chromatid fusions in the study by Tanaka et al occurred at the same location in both inverted repeats so that intrastrand annealing could not have been involved. Other studies have also found very short or no microhomology during sister chromatid fusion in mammalian cells (0 to 4 bps), as well as insertions from other chromosomes [62–64]. Therefore, sister chromatid fusions in these studies are likely to involve A-NHEJ, as has been proposed for chromosome fusions in other studies [65–67]. Mutations introduced artificially into the RNA template of mammalian telomerase that result in telomeres in which the two strands are complementary result in sister chromatid fusions [68], suggesting that as in yeast, long inverted repeats can promote sister chromatid fusion either through intrastrand annealing [51–54] or replication template switching [55–57]. Therefore, although there is no evidence that large inverted repeats initiate B/F/B cycles in mammalian cells, the large inverted repeats created by the breakage of the sister chromatid fusions are likely to contribute to chromosome instability in subsequent cell cycles.
3. Chromosome fusion due to a deficiency in telomeric proteins
Studies of cells deficient in shelterin proteins that are essential for the caps on the ends of chromosomes have been an important source of information on the mechanisms of chromosome fusion. Cells deficient in shelterin proteins undergo chromosome fusion despite the presence of telomeric repeat sequences due to a failure to protect the end of the chromosome. A deficiency in TRF2, which binds to double-stranded telomeric repeat sequences, results in the appearance of DSB repair foci at telomeres [69] and extensive chromosome fusion [70]. TRF2 works in combination with RAP1 [71] to regulate the nuclease activity of MRE11 [66, 72, 73] and Apollo [74, 75] to generate the 3′ single-stranded overhang required to form a T-loop on the end of the chromosome that prevents chromosome fusion. TRF2 also binds and inhibits ATM [76], a key protein in the mammalian cellular response to DSBs. Chromosome fusion resulting from a deficiency in TRF2 occurs by C-NHEJ, as shown by its dependence on LIG4 [66, 70, 73]. Chromosome fusion in TRF2-deficient cells is dependent on ATM [73, 77], despite the fact that only 10% of the DSBs repaired by NHEJ require ATM following ionizing radiation [78–80]. Chromosome fusions in TRF2-deficient cells may therefore require ATM-dependent modifications in chromatin structure to allow access for DSB repair proteins, because the DSBs that require ATM for NHEJ are those that occur within heterochromatin [78].
A deficiency in POT1, which binds to the single-stranded telomeric repeat sequences in the 3′ overhang, and/or TPP1, which associates with POT1, also promote chromosome fusion, although not to the same extent as a deficiency in TRF2 [66, 73, 77]. POT1 and TPP1 are essential in limiting the 5′-3′ resection of the telomere [81–84], and as a result, a deficiency in POT1 and/or TPP1 causes excessive resection and long single-stranded DNA on the ends of chromosomes. Unlike chromosome fusions resulting from a deficiency in TRF2, chromosome fusions resulting from a deficiency in POT1 and/or TPP1 are independent of LIG4, and are dependent on ATR, not ATM [66, 73, 77]. Based on these observations, chromosome fusion in cells deficient in POT1 and/or TPP1 was proposed to occur through A-NHEJ as a result of an inhibition of C-NHEJ by TRF2, either by the inhibition of ATM or by the regulation of resection by MRE11 [66]. Chromosome fusions resulting from gradual telomere shortening in wild-type cells were also proposed to occur by A-NHEJ, due to the absence of telomeric repeat sequences in the fused chromosomes, indicating that large deletions had occurred.
4. Monitoring telomere loss and chromosome fusion by PCR
Telomere shortening and its effect on chromosome fusion have been analyzed in human fibroblasts by single telomere length analysis (STELA). STELA uses polymerase chain reaction (PCR) to amplify individual telomeres utilizing one primer that is specific for the end of a chromosome, and a second primer that binds to the single-stranded 3′ overhang at the end of the telomere [85]. The results with STELA demonstrate that individual telomeres in human fibroblasts shorten gradually during cell division, and that telomeres are very short when cells reach senescence. STELA also demonstrates very clearly that normal cells experience rare rapid deletion events that result in dramatically shortened telomeres [65, 85]. A second PCR-based assay, called Fusion PCR, was also established to monitor the chromosome fusions resulting from the loss of individual telomeres [65, 86]. Fusion PCR uses primers specific for the ends of two different chromosomes. To detect chromosome fusions, the fibroblasts were infected with retroviral vectors containing human papilloma virus E6E7 oncoproteins to prevent the fibroblasts from entering senescence following telomere loss. The results demonstrate that chromosome fusions appear at the same time that STELA detects very short telomeres [65]. In addition, the analysis of the fusions show that the telomeric repeat sequences are invariably missing on one or both of the chromosomes. The longest telomeric repeat sequence found at the site of a fusion contained 12 5/6 repeats, indicating that 13 telomeric repeats are required to form a cap that is capable of protecting the end of a chromosome [65]. Large deletions of subtelomeric regions were common on one or both chromosomes involved in the fusions, showing that shortened telomeres are often associated with rapid degradation of the end of the chromosome. The results also suggest that these subtelomeric deletions are likely to be much larger, although they would not be detectable by this approach due to the loss of the DNA complementary to the primer used for Fusion PCR. The vast majority of chromosome fusions detected by Fusion PCR involved fusions between different chromosomes, and rarely if at all from fusions between sister chromatids. However, it was found that artificial fusions generated from the same chromosome could not be amplified by Fusion PCR, and therefore sister chromatid fusions are not detected by this method. Importantly, DNA sequence analysis demonstrated that microhomology was commonly present at the recombination junctions, which in combination with the presence of large deletions led the authors to propose that chromosome fusion occurs through A-NHEJ.
Fusion PCR also shows that rare chromosome fusions occur in replicating primary human fibroblasts and in senescent human fibroblasts [65, 86]. The fusions in replicating fibroblasts occur as a result of rapid deletion events within individual telomeres, and demonstrate that normal human cells can continue to divide despite the presence of a single chromosome fusion. Fusions also occur in some epithelial cancer cells and transformed epithelial cell lines that express telomerase [65]. However, these fusions only occur in cells with very short telomeres, suggesting this is a result of gradual telomere shortening due to a failure of telomerase to maintain sufficient telomere length. The results with fibroblasts and epithelial cancer cells demonstrate that chromosome fusions resulting from telomere loss are not limited to cells in crisis, which occurs when cells lose their ability to initiate senescence or apoptosis and continue to divide despite the presence of severely shortened telomeres. However, the high frequency of chromosome fusion detected by Fusion PCR in chronic lymphocytic leukemia, suggests that a crisis-like state caused by telomere shortening occurs during the progression of this disease [87].
5. Chromosome fusion due to rapid deletion events
Rapid deletion events similar to those detected by Fusion PCR were first reported in mammalian cells in an immortal human cell line that maintains its telomeres through a telomerase-independent mechanism [8]. These rapid deletion events were proposed to result from recombination, consistent with the demonstration that cell lines utilizing this “ALT” pathway maintain telomeres through recombination [9]. The rapid deletion events in ALT cells can result in complete loss of the telomeric repeat sequences, as shown by the fact that the frequency of rapid deletion events corresponds with the frequency of chromosome fusion [8]. Rapid deletion events resulting from recombination have subsequently been shown to result in the formation of DNA circles composed of telomeric repeat sequences [88]. Despite the expression of telomerase, human cancer cell lines can also demonstrate a high rate of spontaneous telomere loss that can initiate chromosome instability [12, 89]. This increased rate of telomere loss and chromosome fusion is found in a large number of early passage tumor cells in culture [90]. Telomere instability is also nearly universal in epithelial tumors [91], and tumor sections show a correlation between anaphase bridges and abnormal karyotypes in sarcomas, pancreatic carcinomas, and colon carcinomas [92].
The mechanism and consequences of spontaneous telomere loss in a cancer cell line expressing telomerase has been investigated using a plasmid containing a herpes simplex virus thymidine kinase (HSV-tk) gene integrated immediately adjacent to a telomere. The growth of cells in medium containing ganciclovir kills cells that express the HSV-tk gene, providing a means of selecting for cells that have lost the adjacent telomere. These studies demonstrate a 100-fold higher rate of loss of the HSV-tk gene near telomeres than at interstitial locations in the EJ-30 human tumor cell line [12]. The analysis of the types of rearrangements in the HSV-tk-deficient cells demonstrates that most (95%) involve the complete loss of the plasmid sequences. The remaining events involve either inverted repeats that result from sister chromatid fusion, or terminal deletions that result from the addition of a telomere at a new location. Importantly, Fusion PCR does not detect these types of events because they all commonly involve large deletions on the end of the chromosome and sister chromatid fusions. These sister chromatid fusions occur through NHEJ, not intrastrand annealing, as demonstrated by: 1) the fact that some fusions occurred at nearly the same location on both sister chromatids, 2) the insertion of fragments from another chromosome at the site of the break in some cells, and 3) the limited microhomology (0 to 4 bps) at the recombination junctions [62, 64].
6. The role of DSBs in telomere loss
The role of DSBs in telomere loss and chromosome fusion in mouse ES cells and the EJ-30 human tumor cell line has been investigated using DSBs introduced within subtelomeric regions with the I-SceI endonuclease. I-SceI endonuclease recognizes an 18 bp sequence that is not found in the mammalian genome, and has been used extensively to study DSB repair mammalian cells [93–96]. As with the analysis of spontaneous telomere loss, the consequences of I-SceI-induced DSBs near telomeres were investigated using plasmid sequences containing an HSV-tk gene. Interestingly, mouse ES cells selected with ganciclovir for the loss of the HSV-tk gene demonstrate the same three types of events observed in human tumor cells experiencing spontaneous telomere loss, i.e., complete loss of the plasmid sequences, sister chromatid fusions, and terminal deletions involving the addition of telomeric repeat sequences at the site of the break [62, 97, 98]. As with spontaneous telomere loss in the EJ-30 human tumor cell line, the DSB-induced sister chromatid fusion in mouse ES cells results in the amplification of subtelomeric sequences and translocations from other chromosomes, typical of B/F/B cycles. The sister chromatid fusions are associated with large deletions of up to 25 kb, which differ in length on the two sister chromatids, with 0 to 4 bps of microhomology at the recombination junctions. The association of microhomology and large deletions with these sister chromatid fusions suggests a mechanism involving A-NHEJ [67], consistent with other studies proposing a role for A-NHEJ in chromosome fusions following telomere loss [17, 65].
7. The sensitivity of subtelomeric regions to DSBs
The prevalence of large deletions, sister chromatid fusions, and the addition of telomeric repeat sequences at the site of the DSBs near telomeres in mouse ES cells suggests that DSBs in subtelomeric regions are processed differently from DSBs at interstitial sites. None of these events are common to DSBs generated by I-SceI at interstitial sites [93–96]. Consistent with this conclusion, a study in S. cerevisiae demonstrated that I-SceI-induced DSBs in subtelomeric regions were much more likely to result in gross chromosome rearrangements (GCRs) than DSBs at other locations, and that this was due to differences in DSB repair near telomeres [99]. A similar comparison of the types of events resulting from I-SceI-induced DSBs at interstitial and subtelomeric sites in the EJ-30 human tumor cell line demonstrated that subtelomeric regions in mammalian cells are also sensitive to DSBs, [64]. These studies utilized two different assay systems, both with and without selection, with nearly identical results. The frequency of small deletions of a few bps at the I-SceI site, which are the most common type of mutation generated by I-SceI at interstitial sites [93–96], are similar at subtelomeric and interstitial sites. However, the frequency of large deletions and GCRs are much more frequent at subtelomeric DSBs (40–50%) than at interstitial DSBs (5%). Moreover, Southern blot analysis demonstrated a qualitative difference in the type of rearrangements at subtlomeric and interstitial DSBs, in that degradation is much more extensive at DSBs near telomeres. A subsequent study found that the subtelomeric region that is sensitive to DSBs extends at least 100 kb from a telomere [100]. In view of the fact that there are 96 telomrees in a human cell, this represents a significant portion of the human genome that is sensitive to DSBs. Subtelomeric regions may therefore represent an important target for chromosome instability resulting from exposure to ionizing radiation or other agents that produce DSBs.
Using a reporter gene for green fluorescent protein that is activated following repair of I-SceI-induced DSBs, we have now demonstrated that the sensitivity of subtelomeric regions to DSBs is a result of a deficiency in NHEJ [101]. A similar deficiency in NHEJ was also observed at interstitial sites containing telomeric repeat sequences. A deficiency in NHEJ at DSBs occurring near telomeric repeat sequences suggests that shelterin proteins may somehow suppress NHEJ, possibly as part of their role in preventing chromosome fusion. There are multiple mechanisms by which telomeric proteins may suppress C-NHEJ. One mechanism is through the suppression of ATM by TRF2 [76]. Alternatively, the suppression of C-NHEJ could result from the fact that DSBs near telomeres are processed as though they are the DNA at the end of a telomere [101]. TRF2 regulates MRE11 [66, 72, 73] and Apollo [74, 75] to generate a single-stranded overhang on the leading strand on the ends of the newly replicated telomeres. POT1/TPP1 then binds to the single-stranded overhangs, preventing further resection and T-loop formation, and as a result, cells deficient in TPP1 and/or POT1 experience large single-stranded regions [81–84] and fusion by A-NHEJ [66]. A similar processing of subtelomeric DSBs as a result of the proximity of TRF2 might therefore also result in the formation of single-stranded overhangs. However, because the subtelomeric DNA is not composed of telomeric repeat sequences, POT1/TPP1 would not bind and limit resection. As a result, resection would continue, producing large deletions, telomere loss, and chromosome fusion by A-NHEJ, similar to that seen in cells deficient in TPP1 or POT1. A further characterization of DNA repair near telomeres is therefore important for understanding the mechanism of telomere loss and chromosome instability in cancer.
8. Chromosome healing
An important observation arising from our studies on the types of events resulting from spontaneous telomere loss or DSBs occurring near telomeres is that mammalian cells are capable of adding telomeric repeat sequences on to the ends of broken chromosomes, a process called chromosome healing. Chromosome healing has been extensively studied in yeast, where it has been demonstrated to involve telomerase [102]. Chromosome healing is known to occur in germ line cells in humans, since it is responsible for terminal deletions resulting in human genetic disease [103–105]. However, chromosome healing has not been observed as a result of either ionizing radiation or I-SceI-induced DSBs at interstitial sites, even in cells that express telomerase [93–96, 106]. It was somewhat surprising therefore, that chromosome healing is detectable as a result of spontaneous telomere loss or DSBs near telomeres in the EJ-30 human cancer cell line and mouse ES cells. In the EJ-30 human tumor cell line chromosome healing accounts for only about one percent of the total events [12, 64]; however, in mouse ES cells, chromosome healing is a frequent event at DSBs near telomeres, accounting for approximately one-third of the total events [97, 98]. As in yeast, chromosome healing in mouse ES cells involves telomerase, as demonstrated by the fact that it does not occur in ES cells from telomerase-knockout mice, although one ES cell clone that had acquired the ability to maintain telomeres through a telomerase-independent mechanism was also capable of performing chromosome healing. Unlike GCRs, which are usually associated with extensive degradation, chromosome healing nearly always occurs at the site of the DSB with minimal microhomology to telomeric repeat sequences [97, 98], consistent with in vitro studies demonstrating that mammalian telomerase is capable of de novo telomere addition with as little as a single bp of homology [107, 108]. Importantly, chromosome healing stabilizes the chromosome that has lost its telomere, as demonstrated by the fact that the marker chromosome is stable in subclones in which it has acquired a new telomere [14, 62]. Chromosome healing therefore prevents degradation, sister chromatid fusion, and B/F/B cycles, and as a result, has been hypothesized to be an important mechanism for dealing with DSBs within NHEJ-deficient subtelomeric regions to prevent chromosome instability [67, 97].
Chromosome healing in S. cerevisiae is inhibited by the DNA helicase PIF1 [109], which binds to TERT, the catalytic subunit of telomerase [110]. This inhibition of chromosome healing involves a DSB-induced phosphorylation of PIF1 by MEC1, the primary protein involved in the response to DSBs in S. cerevisiae [111]. This inhibition of chromosome healing by PIF1 has been proposed to be a mechanism for preventing chromosome healing from interfering with DSB repair [112]. Mammals also have PIF1, which also binds to TERT [113]. However, knockout of PIF1 has no effect on the sensitivity of mouse cells to ionizing radiation [113], and does not affect chromosome healing near telomeres (personal observation), suggesting that the regulation of chromosome healing is redundant in mammalian cells [113]. It therefore remains to be determined how chromosome healing is regulated in mammalian cells, including why chromosome healing occurs preferentially near telomeres, and why mouse ES cells are much more proficient in chromosome healing than the EJ-30 human tumor cell line. One possible explanation for the absence of chromosome healing at most interstitial DSBs is that the terminal deletions resulting from chromosome healing are lethal. However, this cannot be the only explanation for the prevalence of chromosome healing at DSBs near telomeres. Chromosome healing was not detectable at a DSB 100 kbps from a telomere despite the fact that Cre-mediated recombination showed that terminal deletions were not lethal at this location [64]. The prevalence of chromosome instability near telomeres could be a result of the inhibition of ATM by TRF2 [76], because ATM, like MEC1 in S. cerevisiae is a critical protein involved in the mammalian response to DSBs [114]. ATM would therefore not be activated near telomeres in response to DSBs, and therefore would not phosphorylate PIF1 and/or other proteins involved in preventing chromosome healing. On the other hand, the much lower frequency of chromosome healing in the EJ-30 human tumor cell line (1%) compared to mouse ES cells (30%) in response to DSBs near telomeres may be a result of the extensive degradation that occurs at DSBs near telomeres in EJ-30 [62, 64, 97], since resection of DSBs has been shown to also inhibit chromosome healing in S. cerevisiae [115].
9. Mechanism of spontaneous telomere loss
The types and proportions of DNA rearrangements that result from DSBs near telomeres in the EJ-30 human tumor cell line are very similar to what is observed as a result of spontaneous telomere loss, i.e., 95% of the events result in the loss of the plasmid, while approximately 4% result in GCRs, and 1% in chromosome healing [12, 64]. This similarity in spontaneous and I-SceI-induced events suggests that DSBs play a role in spontaneous telomere loss. However, while sister chromatid fusion was the predominant GCR observed with spontaneous telomere loss, DSBs near telomeres resulted in a variety of GCRs, including sister chromatid fusions, translocations, dicentrics, and ring chromosomes. A possible explanation for this difference is the timing of the spontaneous and I-SceI-induced DSBs. Whereas I-SceI-induced DSBs may occur anytime in the cell cycle, and therefore promote fusion between different chromosomes, DSBs leading to spontaneous telomere loss may occur during DNA replication, and therefore promote the fusion of sister chromatids. I have proposed that this increased rate of telomere loss in human cancer cells is a product of oncogene-mediated replication stress, which results in stalled replication forks at regions of DNA that are difficult to replicate [116]. Because stalled replication forks can collapse and form DSBs, these regions are unstable under replication stress, and are therefore referred to as fragile sites. Telomeres have been demonstrated to be fragile sites, both by the presence of stalled replication forks and by the presence of aberrant telomeres in response to aphidicolin, an inhibitor of DNA polymerase known to cause replication stress [117]. Although DSBs caused by replication stress would occur at many fragile sites, those occurring near telomeres would be more likely to result in chromosome instability due to the deficiency in DSB repair in subtelomeric regions. In addition, the low efficiency of chromosome healing near telomeres in human cancer cells could also contribute to this chromosome instability. Additional research on the factors promoting telomere loss, or factors or pathways involved in the regulation of chromosome healing, is therefore critical to our understanding and possibly intervention in this important pathway for chromosome instability in human cancer.
10. Future directions
The above studies leave little doubt that telomere loss, either during crisis or after the expression of telomerase, contributes to the chromosome instability leading to human cancer. However, much remains to be determined regarding the mechanisms involved in telomere loss and the chromosome instability that results. A primary focus of future studies will be to exploit this knowledge for anti-cancer therapy. Preventing chromosome instability resulting from telomere loss could serve to prevent genetic alterations leading to tumor cell progression and the development of resistance in cancer cells to anti-cancer therapies. One approach would be to somehow prevent or limit telomere loss a in cancer cells and thereby inhibit chromosome instability, although how this might be accomplished is not immediately clear. A second approach would be to inhibit the chromosome instability resulting from telomere loss. In this case, chromosome healing provides an excellent resource in that it can stabilize chromosomes that have lost a telomere. In view of the inefficient chromosome healing in the EJ-30 tumor cell line, promoting chromosome healing in human cancers could dramatically curtail chromosome instability resulting from telomere loss. This approach will require understanding how chromosome healing is regulated and why it is so inefficient in human cancer cells. Promoting chromosome healing could also lead to selective sensitization of human cancer cells to ionizing radiation by inhibiting DSB repair, since interfering with the regulation of chromosome healing would not affect DSB repair in normal somatic cells that are not be capable of chromosome healing due to insufficient telomerase activity.
Acknowledgments
This work was supported by National Institutes of Health grant CA120205.
Abbreviations
- A-NHEJ
alternative nonhomologous end joining
- B/F/B
breakage/fusion/bridge
- C-NHEJ
classical nonhomologous end joining
- DSBs
double-strand breaks
- GCRs
gross chromosome rearrangements
- HSV-tk
herpes simplex thymidine kinase
- NHEJ
nonhomologous end joining
- PCR
polymerase chain reaction
Footnotes
Disclosure of Potential Conflicts of Interest
No conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Muller HJ. The remaking of chromosomes. The collecting net-Woods Hole. 1938;13:181–198. [Google Scholar]
- 2.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;41:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 4.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 5.d’ Adda di Fagagna FdAd, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura AJ, Chiang YJ, Hathcock KS, Horikawa I, Sedelnikova OA, Hodes RJ, Bonner WM. Both telomeric and non-telomeric DNA damage are determinants of mammalian cellular senescence. Epigenetics Chromatin. 2008;1:6. doi: 10.1186/1756-8935-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murnane JP, Sabatier L, Marder BA, Morgan WF. Telomere dynamics in an immortal human cell line. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 10.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 11.Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst) 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 12.Fouladi B, Miller D, Sabatier L, Murnane JP. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia. 2000;2:540–554. doi: 10.1038/sj.neo.7900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo AWI, Sabatier L, Fouladi B, Pottier G, Ricoul M, Murnane JP. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia. 2002;6:531–538. doi: 10.1038/sj.neo.7900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in genomic instability involving multiple chromosomes in a human tumor cell line. Mol Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- 15.Artandi SE, Chang S, Lee S-L, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 16.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 17.Zhu C, Mills KD, Fergusion DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 18.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford M, Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986;45:425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 21.Ma C, Martin S, Trask B, Hamlin JL. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 22.Toledo F, Buttin G, Debatisse M. The origin of chromosome rearrangements at early stages of AMPD2 gene amplification in Chinese hamster cells. Curr Biol. 1993;3:255–264. doi: 10.1016/0960-9822(93)90175-n. [DOI] [PubMed] [Google Scholar]
- 23.Toledo F, Roscouet D, Buttin G, Debatisse M. Co-amplified markers alternate in megabase long chromosomal inverted repeats and cluster independently in interphase nuclei at early steps of mammalian gene amplification. EMBO J. 1992;11:2665–2673. doi: 10.1002/j.1460-2075.1992.tb05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp Cell Res. 2005;302:233–243. doi: 10.1016/j.yexcr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Singer MJ, Mesner LD, Friedman CL, Trask BJ, Hamlin JL. Amplification of the human dihydrofolate reductase gene via double minutes is initiated by chromosome breaks. Proc Natl Acad Sci USA. 2000;97:7921–7926. doi: 10.1073/pnas.130194897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol Cell. 1998;2:249–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- 27.Windle B, Draper BW, Yin Y, O’Gorman S, Wahl GM. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes and Development. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka H, Mendonca MS, Bradshaw PS, Hoelz DJ, Malkas LH, Meyn MS, Gilley D. DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proc Natl Acad Sci U S A. 2005;102:15539–15544. doi: 10.1073/pnas.0507915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diede SJ, Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Genome-wide analysis of palindrome formation. Nat Genet. 2010;42:279. doi: 10.1038/ng0410-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekiguchi JM, Ferguson DO. DNA double-strand break repair: a relentless hunt uncovers new prey. Cell. 2006;124:260–262. doi: 10.1016/j.cell.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Haber JE. Alternative endings. Proc Natl Acad Sci U S A. 2008;105:405–406. doi: 10.1073/pnas.0711334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Zha S, Boboila C, Alt FW. Mre11: roles in DNA repair beyond homologous recombination. Nat Struct Mol Biol. 2009;16:798–800. doi: 10.1038/nsmb0809-798. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 36.Audebert M, Salles B, Weinfeld M, Calsou P. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J Mol Biol. 2006;356:257–265. doi: 10.1016/j.jmb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 37.Liang L, Deng L, Nguyen SC, Zhao X, Maulion CD, Shao C, Tischfield JA. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 2008;36:3297–3310. doi: 10.1093/nar/gkn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 39.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 40.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhuang J, Jiang G, Willers H, Xia F. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J Biol Chem. 2009;284:30565–30573. doi: 10.1074/jbc.M109.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Baumann P. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol Cell. 2008;31:463–473. doi: 10.1016/j.molcel.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 48.Branzei D, Foiani M. Leaping forks at inverted repeats. Genes Dev. 2010;24:5–9. doi: 10.1101/gad.1884810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haber JE, Debatisse M. Gene amplification: yeast takes a turn. Cell. 2006;125:1237–1240. doi: 10.1016/j.cell.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 50.VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol. 2007;27:2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maringele L, Lydall D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004;18:2663–2675. doi: 10.1101/gad.316504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rattray AJ, Shafer BK, Neelam B, Strathern JN. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narayanan V, Lobachev KS. Intrachromosomal gene amplification triggered by hairpin-capped breaks requires homologous recombination and is independent of nonhomologous end-joining. Cell Cycle. 2007;6:1814–1818. doi: 10.4161/cc.6.15.4522. [DOI] [PubMed] [Google Scholar]
- 54.Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 55.Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 2009;23:2876–2886. doi: 10.1101/gad.1863009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paek AL, Kaochar S, Jones H, Elezaby A, Shanks L, Weinert T. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 2009;23:2861–2875. doi: 10.1101/gad.1862709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaochar S, Paek AL, Weinert T. Genetics. Replication error amplified. Science. 2010;329:911–913. doi: 10.1126/science.1194261. [DOI] [PubMed] [Google Scholar]
- 58.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinstock DM, Richardson CA, Elliott B, Jasin M. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst) 2006;5:1065–1074. doi: 10.1016/j.dnarep.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka H, Cao Y, Bergstrom DA, Kooperberg C, Tapscott SJ, Yao MC. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Mol Cell Biol. 2007;27:1993–2002. doi: 10.1128/MCB.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci U S A. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo AWI, Sprung CN, Fouladi B, Pedram M, Sabatier L, Ricoul M, Reynolds GE, Murnane JP. Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol Cell Biol. 2002;22:4836–4850. doi: 10.1128/MCB.22.13.4836-4850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okuno Y, Hahn PJ, Gilbert DM. Structure of a palindromic amplicon junction implicates microhomology-mediated end joining as a mechanism of sister chromatid fusion during gene amplification. Nucleic Acids Res. 2004;32:749–756. doi: 10.1093/nar/gkh244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zschenker O, Kulkarni A, Miller D, Reynolds GE, Granger-Locatelli M, Pottier G, Sabatier L, Murnane JP. Increased sensitivity of subtelomeric regions to DNA double-strand breaks in a human tumor cell line. DNA Repair. 2009;8:886–900. doi: 10.1016/j.dnarep.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rai R, Zheng H, He H, Luo Y, Multani A, Carpenter PB, Chang S. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. Embo J. 2010;29:2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murnane JP. Telomere loss as a mechanism for chromosomal instability in human cancer. Cancer Res. 2010;70:4255–4259. doi: 10.1158/0008-5472.CAN-09-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stohr BA, Xu L, Blackburn EH. The terminal telomeric DNA sequence determines the mechanism of dysfunctional telomere fusion. Mol Cell. 2010;39:307–314. doi: 10.1016/j.molcel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 70.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 71.Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. Embo J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu X-D, Kuster B, Mann M, Petrini JHJ, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 73.Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J. 2010;29:2230–2241. doi: 10.1038/emboj.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu P, van Overbeek M, Rooney S, de Lange T. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell. 2010;39:606–617. doi: 10.1016/j.molcel.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 78.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 79.Cornforth MN, Bedford JS. On the nature of a defect in cells from individuals with ataxia-telangiectasia. Science. 1985;227:1589–1591. doi: 10.1126/science.3975628. [DOI] [PubMed] [Google Scholar]
- 80.Kato TA, Nagasawa H, Weil MM, Little JB, Bedford JS. Levels of gamma-H2AX Foci after low-dose-rate irradiation reveal a DNA DSB rejoining defect in cells from human ATM heterozygotes in two at families and in another apparently normal individual. Radiat Res. 2006;166:443–453. doi: 10.1667/RR3604.1. [DOI] [PubMed] [Google Scholar]
- 81.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 82.Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 83.He H, Multani AS, Cosme-Blanco W, Tahara H, Ma J, Pathak S, Deng Y, Chang S. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. Embo J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 85.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 86.Letsolo BT, Rowson J, Baird DM. Fusion of short telomeres in human cells is characterized by extensive deletion and microhomology, and can result in complex rearrangements. Nucleic Acids Res. 2010;38:1841–1852. doi: 10.1093/nar/gkp1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, Fegan C, Pepper C, Baird DM. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 88.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 89.Nakamura AJ, Redon CE, Bonner WM, Sedelnikova OA. Telomere-dependent and telomere-independent origins of endogenous DNA damage in tumor cells. Aging (Albany NY) 2009;1:212–218. doi: 10.18632/aging.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meeker AK, Hicks JL, Iacoluzio-Donahue CA, Montgomery EA, Westra WH, Chan TY, Ronnett BM, De Marzo AM. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clin Can Res. 2004;10:3317–3326. doi: 10.1158/1078-0432.CCR-0984-03. [DOI] [PubMed] [Google Scholar]
- 92.Montgomery E, Wilentz RE, Argani P. Analysis of anaphase figures in routine histologic sections distinguishes chromosomally unstable from chromosomally stable malignancies. Cancer Biol Ther. 2003;2:248–252. doi: 10.4161/cbt.2.3.362. [DOI] [PubMed] [Google Scholar]
- 93.Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair (Amst) 2007;6:781–788. doi: 10.1016/j.dnarep.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 95.Rebuzzini P, Khoriauli L, Azzalin CM, Magnani E, Mondello C, Giulotto E. New mammalian cellular systems to study mutations introduced at the break site by non-homologous end-joining. DNA Repair (Amst) 2005;4:546–555. doi: 10.1016/j.dnarep.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 96.Varga T, Aplan PD. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair (Amst) 2005;4:1038–1046. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao Q, Reynolds GE, Wilcox A, Miller D, Cheung P, Artandi SE, Murnane JP. Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair. 2008;7:1233–1249. doi: 10.1016/j.dnarep.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sprung CN, Reynolds GE, Jasin M, Murnane JP. Chromosome healing in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1999;96:6781–6786. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ricchetti M, Dujon B, Fairhead C. Distance from the chromosome end determines the efficiency of double-strand break repair in subtelomeres of haploid yeast. J Mol Biol. 2003;328:847–862. doi: 10.1016/s0022-2836(03)00315-2. [DOI] [PubMed] [Google Scholar]
- 100.Kulkarni A, Zschenker O, Reynolds G, Miller D, Murnane JP. The effect of telomere proximity on telomere position effect, chromosome healing and sensitivity to DNA double-strand breaks in a human tumor cell line. Mol Cell Biol. 2010 doi: 10.1128/MCB.01137-09. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miller D, Reynolds GE, Mejia R, Stark JM, Murnane JP. Subtelomeric regions in mammalian cells are deficient in DNA double-strand break repair. DNA Repair. 2011 doi: 10.1016/j.dnarep.2011.03.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pennaneach V, Putnam CD, Kolodner RD. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol. 2006;59:1357–1368. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- 103.Flint J, Craddock CF, Villegas A, Bentley DP, Williams HJ, Galanello R, Cao A, Wood WG, Ayyub H, Higgs DR. Healing of broken human chromosomes by the addition of telomeric repeats. Am J Hum Genet. 1994;55:505–512. [PMC free article] [PubMed] [Google Scholar]
- 104.Varley H, Di S, Scherer SW, Royle NJ. Characterization of terminal deletions at 7q32 and 22q13.3 healed by de novo telomere addition. Am J Hum Genet. 2000;67:610–622. doi: 10.1086/303050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wong AC, Ning Y, Flint J, Clark K, Dumanski JP, Ledbetter DH, McDermid HE. Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am J Hum Genet. 1997;60:113–120. [PMC free article] [PubMed] [Google Scholar]
- 106.Latre L, Genesca A, Martin M, Ribas M, Egozcue J, Blasco MA, Tusell L. Repair of DNA broken ends is similar in embryonic fibroblasts with and without telomerase. Radiat Res. 2004;162:136–142. doi: 10.1667/rr3203. [DOI] [PubMed] [Google Scholar]
- 107.Morin GB. Recognition of a chromosome truncation site associated with α-thalassaimia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 108.Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 109.Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 110.Eugster A, Lanzuolo C, Bonneton M, Luciano P, Pollice A, Pulitzer JF, Stegberg E, Berthiau AS, Forstemann K, Corda Y, Lingner J, Geli V, Gilson E. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat Struct Mol Biol. 2006;13:734–739. doi: 10.1038/nsmb1126. [DOI] [PubMed] [Google Scholar]
- 111.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11:1383–1386. doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou J-Q, Monson EK, Teng S-C, Schultz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 113.Snow BE, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L. Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol. 2007;27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 115.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu XR, Papavassiliou AG, Gorgoulis VG. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 117.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]