Abstract
Characterization and functional manipulation of specific groups of neurons in the vertebrate central nervous system (CNS) remains a major hurdle for understanding complex circuitry and functions. In zebrafish, the Gal4/UAS system has permitted expression of transgenes and enhancer trap screens, but is often limited by broad expression domains. We have developed a method for cell-type specific expression using Gal80 inhibition of Gal4-dependent expression. We show that native Gal4 is able to drive strong expression, that Gal80 can inhibit this expression, and that overlapping Gal4 and Gal80 expression can achieve “intersectional” expression in spatially and genetically defined subsets of neurons. We also optimize Gal80 for expression in vertebrates, track Gal80 expression with a co-expressed fluorescent marker, and use a temperature-sensitive allele of Gal80 to temporally regulate its function. These data demonstrate that Gal80 is a powerful addition to the genetic techniques available to map and manipulate neural circuits in zebrafish.
Keywords: Gal80, zebrafish, Gal4, Gal4-VP16, intersectional expression, conditional expression
Introduction
Characterization and functional manipulation of subgroups of neurons in the vertebrate central nervous system (CNS) remain major hurdles for understanding complex nervous system function. Technology for targeted knock-ins, BAC transgenesis, transgenic enhancers, and enhancer traps in mouse and zebrafish has facilitated research on neuronal populations (Gong et al., 2003; Hatten and Heintz, 2005; Amsterdam and Becker, 2005; Asakawa et al., 2008). However, most of these techniques still rely upon expression dictated by a single gene or regulatory region, such that subgroups of neuronal types can not be differentiated. For example, methods to achieve neuronal sub-type specific expression have typically resorted to mosaic analyses that can be inefficient and difficult to genetically reproduce (Sato et al., 2007; Huberman et al., 2008).
In zebrafish, CNS transgene expression with the Gal4/UAS system is a popular means for expression of heterologous genes and for generating enhancer traps (Asakawa et al., 2008; Scheer and Campos-Ortega, 1999; Koester and Fraser, 2001; Scott 2009). However, we and others have noted that expression based on Gal4-VP16 enhancer traps or cloned enhancers can result in expression in non-targeted tissues, and that sub-groups of CNS neurons are often not distinguishable. Our goal was to improve the precision of genetically regulated expression in sub-types of neuronal cells, by developing a genetic method to “intersect” known expression patterns.
We considered two general strategies for genetically manipulating intersectional expression in subgroups of cells (Figure S1). The first, a positive intersectional method (Figure S1A), makes use of two enhancers with overlapping expression domains. Only cells with both components can express the target gene(s). Examples of this approach include the split Gal4, split GFP, and split Cre systems (Zhang et al., 2004; Luan et al., 2006; Hirrlinger et al., 2009). In the second, a negative intersectional method (Figure S1B), potential expression is established in a broad domain by the first enhancer, but is then restricted to cells in which a second enhancer is not expressed. The negative intersectional approach has been pioneered in Drosophila, using the Gal4/Gal80 system(Lee and Luo, 1999; Suster et al., 2004), but has not yet been used in a vertebrate system.
We sought a method to restrict Gal4-dependent expression, based on the use of Gal80-mediated inhibition of Gal4 (Lee and Luo, 1999; Lohr et al., 1995). We found that native (full-length) Gal4 was sufficient for strong expression in stable transgenic lines in zebrafish, and that Gal80 could be used to inhibit Gal4-dependent expression in the CNS. We demonstrated that Gal80 expression can be tracked with a co-expressed fluorescent marker, and optimized Gal80 for expression in vertebrates. Further, Gal4 and Gal80 could be driven by partially overlapping enhancers in order to achieve expression in spatially restricted, genetically defined subsets of neurons. Finally, using a temperature-sensitive allele of Gal80 (McGuire et al., 2003), we could temporally control the onset of Gal80-mediated Gal4 inhibition.
Results
We decided to investigate whether Gal80 could be used in zebrafish to inhibit Gal4-dependent expression. Gal80 is a yeast transcriptional repressor that binds to Gal4 and prevents it from activating transcription (Lohr et al., 1995; Ma and Ptashne, 1987). We chose a Gal80 negative intersectional method because of the widespread use of the Gal4/UAS system in zebrafish, and because we had enhancers with broader expression patterns in which we wished to analyze subgroups of neurons. For most experiments (except as otherwise described) we used stable transgenic lines that we generated.
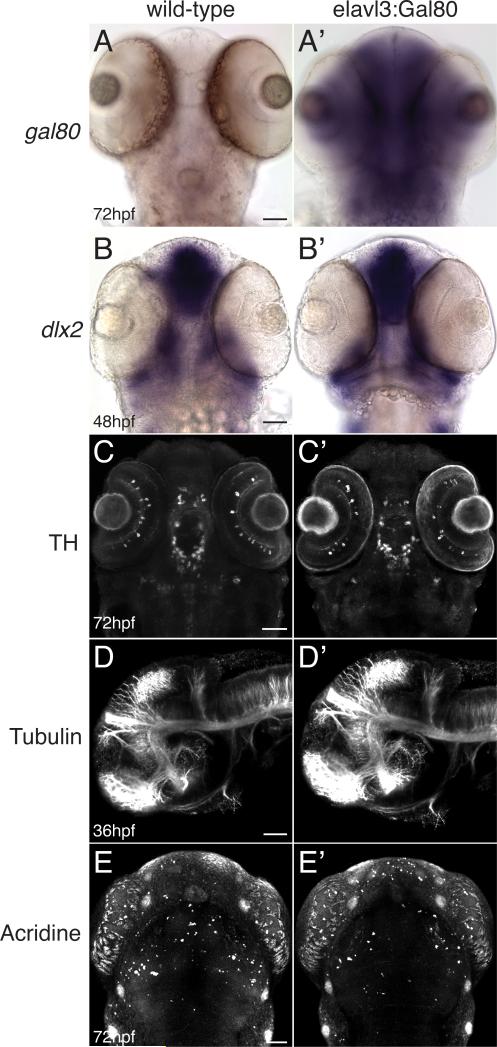
First, we examined whether broad expression of Gal80 would have any detrimental effects on CNS development. We expressed Gal80 using the pan-neuronal enhancer elavl3 (HuC) (Park et al., 2000) in stable transgenic lines (Figure 1A, A’); viability and fecundity were unaffected compared to non-transgenic siblings, and we did not note any gross morphological or developmental defects. Comparison of Tg(elavl3:Gal80) embryos with non-transgenic siblings (Figure 1 B-E’) showed no differences for markers of CNS fate specification (dlx2 in situ); for generation of neuronal transmitter identity (anti-tyrosine hydroxylase antibody); for axonal pathfinding (anti-acetylated tubulin antibody); or for apoptosis (acridine orange staining). Counts of apoptotic cells were not statistically different between wild-type and transgenic embryos (mean 6 and 6.7 respectively; SEM 0.33 and 0.26; n=12 embryos each; two-tailed t test p=0.12).
Figure 1.
Pan-neuronal expression of Gal80 does not affect CNS development. Whole-mount embryos (transgenic Tg(elavl3:Gal80)zc64 or non-transgenic sibling), ventral views (except D), anterior to the top, scale bar 50 μm. Images are confocal projections (except A and B). (A-A’) Brightfield images showing expression of gal80 by in situ in wild-type (A) and transgenic embryos (A’). (B-B’) dlx2 in situ expression in wild-type and transgenic embryos is similar. (C-C’) Pattern of tyrosine hydroxylase (TH) antibody expression is similar in wild-type and transgenic embryos. (D-D’) Axon tract architecture appears similar in wild-type and transgenic embryos (lateral views, anterior to the left). (E-E’) Acridine orange staining for apoptotic cells is similar in wild-type and transgenic embryos.
Native yeast Gal4 has been reported to be a poor activator of transcription in zebrafish, at least for transient transgenesis (Koester and Fraser, 2001; Ogura et al., 2009), although direct testing of stable transgenic lines carrying Gal4 has been limited (Scheer and Campos-Ortega, 1999; Scheer et al., 2002). To increase expression driven by Gal4, the transcriptional activation domain of herpes simplex virus VP16 is commonly used to replace Gal4's transcriptional activation domain (Gal4-VP16413-470, abbreviated hereafter as Gal4-VP16) (Koester and Fraser, 2001). To compare the activity of native Gal4 to Gal4-VP16, we injected Tg(UAS:GFP) embryos with a plasmid expressing either isl2b:Gal4 or isl2b:Gal4-VP16. At 72hpf we counted the total number of GFP-positive RGCs in the rostral-dorsal quadrant of the eye. Gal4 activated transcription in half as many RGCs as Gal4-VP16 (12 versus 24, p=0.011, two-tailed t test, n=13 embryos; see Methods) in these transient assays. When we compared expression on a cell-by-cell basis in Gal4 versus Gal4-VP16 expressing embryos, we observed no difference in GFP levels. In stable lines of Tg(otpb.A:Gal4) compared to Tg(otpb.A:Gal4-VP16), crossed to Tg(UAS:GFP), fluorescence (pixel intensity) was 143 vs 149 (p=0.6, SEM 9.2 and 7.2 respectively; >35 cells/genotype; 6 embryos/genotype).
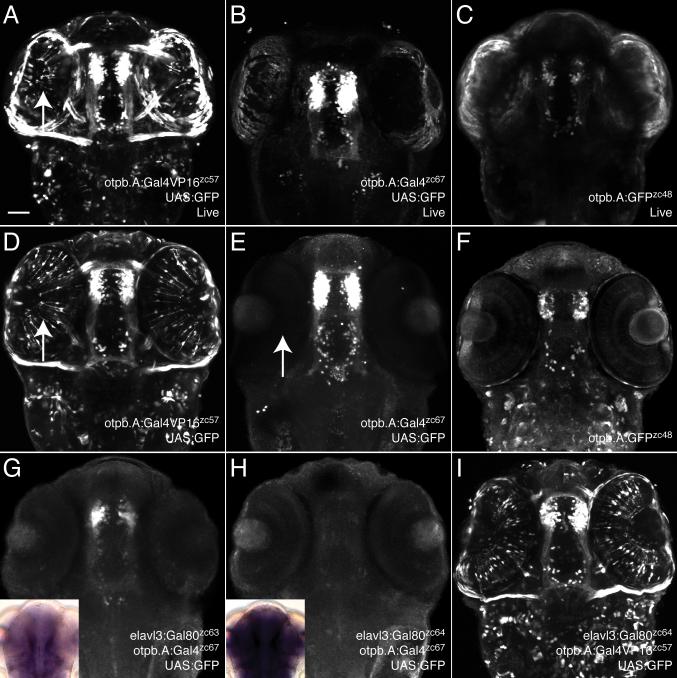
We next proceeded to compare levels of expression from stable UAS transgenic lines when driven by Gal4 versus Gal4-VP16, using an enhancer from the otpb gene, otpb.A (Fujimoto et al., 2011) to drive Gal4 or Gal4-VP16. We found that stable transgenic lines expressing either Gal4 or Gal4-VP16 drove strong CNS expression when crossed to a UAS:GFP transgenic line, visualized live or following immunohistochemistry (Figure 2A, B, D, E). Both Gal4 and Gal4-VP16 drove higher levels of expression from UAS:GFP than seen when driving GFP directly (otpb.A:GFP) (Figure 2C, F), as expected from the amplification of the Gal4/UAS system (Koester and Fraser, 2001). We determined that both Gal4 lines used in this paper (Tg(otpb.A:Gal4) and Tg(isl2b.3:Gal4) are single transgene insertions, so that Gal4 expression was not dependent on having multiple copies of Gal4. Thus, native Gal4 is a satisfactory alternative to Gal4-VP16.
Figure 2.
Gal4 drives strong expression in stable transgenic lines and can be inhibited by Gal80. Confocal maximum projections of 72hpf embryos; ventral views, anterior to the top. Embryos are heterozygous for the genotypes shown. Scale bar, 50μm. Arrow points to ectopic GFP expression in eyes driven by Gal4-VP16 (A, D) not noted using native Gal4 (E). (A-C) Live embryos imaged with identical laser power settings, showing that Gal4-driven expression (B) is stronger than a direct enhancer:GFP construct (C) and is comparable to Gal4-VP16-driven expression (A). (D-F) Immunohistochemistry for GFP in fixed embryos again shows that Gal4-dependent expression (E) is comparable to Gal4-VP16 (D), but has minimal expression in other tissues (such as eye muscles, arrow). (F) The original transgenic enhancer line Tg(otpb.A:egfp)zc48 shows dimmer expression compared to Gal4-driven expression in (E). (G) A weakly-expressing allele of elavl3:Gal80 only partially inhibits Gal4-dependent transgene expression. (H) A strongly-expressing elavl3:Gal80 allele completely inhibits Gal4-dependent expression. Insets for (G) and (H) show relative levels of Gal80 in situ expression. (I) Gal80 is unable to inhibit Gal4-VP16-driven expression.
Further, we found that the Tg(otpb.A:Gal4-VP16) transgenic line had expression in tissues not noted either in the original transgenic enhancer line Tg(otpb.A:GFP) or in the Tg(otpb.A:Gal4) line (Figure 2C, F). This expression included tissues in which the otpb gene is not detectably expressed, such as the eye and eye muscles (Del Giacco et al., 2006; Ryu et al., 2007). The transgenic line Tg(otpb.A:Gal4-VP16413-470)zc57 we used for experiments was the most specific found from screening of 3 stable transgenic lines. There are several possible explanations for this broadened expression in other tissues (Figure 2A, D). For example, the otpb.A:Gal4-VP16 transgene might be expressing ectopically due to a position effect. Or, Gal4-VP16 might be such a strong transcriptional activator that even low levels of expression in a tissue at any time could activate expression from UAS transgenes. We favor this latter explanation since we noted similar patterns in all three otpb.A:Gal4-VP16 transgenic lines, arguing against a position effect.
To test the ability of Gal80 to inhibit Gal4 in zebrafish, we used the pan-neuronal-expressing line Tg(elavl3:Gal80). Gal80 acts by binding Gal4's transcriptional activation domain, preventing recruitment of transcriptional machinery (Lohr et al., 1995; Lue et al.,1987). In Gal4-VP16, the most commonly used form of Gal4 in zebrafish (Asakawa et al., 2008; Kwan et al., 2007; Scott et al., 2007), the yeast Gal4 transcriptional activation domain has been replaced by that of VP16 (Koester and Fraser, 2001). Indeed, we found that Gal80 is unable to inhibit Gal4-VP16 driven expression (Figure 2I). However, Gal80 is able to partially or entirely inhibit native Gal4-dependent expression (Figure 2G, H). The degree of inhibition appeared to depend on the levels of Gal80 expression from different transgene alleles; the insets in Figure 2G, H show in situ expression of Gal80, with weaker Gal80 expression correlated to less inhibition.
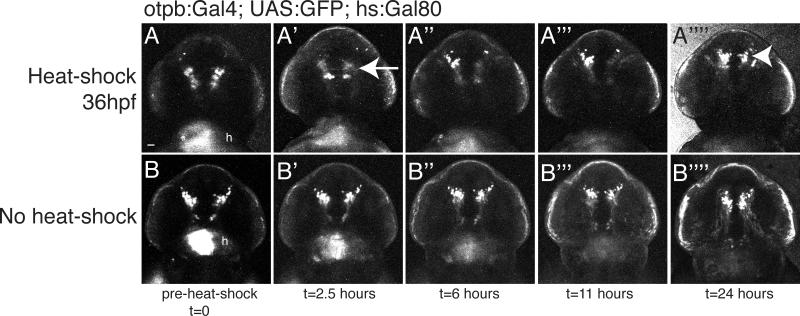
An important consideration for use of Gal80 is the dynamics of its inhibition of Gal4-dependent expression. To determine this, we used a stable transgenic line expressing Gal80 under the control of the inducible heat-shock promoter, Tg(hsp70l:Gal80)zc17, and crossed it to a line carrying Tg(otpb.A:Gal4)zc67; Tg(UAS:GFP). Following heat-shock at 48hpf, we found inhibition of Gal4-dependent expression within 2.5 hours, which lasted up to 24 hours before the return of Gal4-dependent GFP expression (Figure 3). Because we used only a short duration heat-shock, there was a mosaic pattern of repression. The intensity of GFP fluorescence diminished rapidly in the presence of Gal80 expression (Figure 3A-A””), possibly because of the 2-3-fold proliferation of these otpb-expressing diencephalic cells (Ryu et al., 2007; Russek-Blum et al., 2008), and subsequent dilution of GFP.
Figure 3.
Time course of Gal80-mediated inhibition of Gal4. Confocal images of live embryo GFP expression. Transgenic fish carrying Tg(hs:Gal80); Tg(otpb.A:Gal4); Tg(UAS:GFP) were imaged before a 45’ heat-shock, and then at defined time intervals following heat-shock. The same embryo is displayed in each respective set of panels (A1-A’’’’ or B-B’’’’). Arrow points to the inhibition of Gal4-dependent expression in (A’); arrowhead points to the return of GFP expression in (A’’’’) 24 hours following the end of heat-shock. 10 z-slices were used to create the image. Ventral views, rostral to the top, identical confocal settings for imaging both embryos. Scale bar, 50μm; “h”, GFP expression in the heart from the transgenesis marker.
Spatial and Temporal Regulation
Our results confirmed that Gal4 can drive transgene expression in zebrafish at sufficient levels to be experimentally useful, and that we could inhibit this transactivation using pan-neuronally expressed Gal80.
An important consideration was whether we could differentially label genetically similar subsets of neurons using Gal80. Further, we wanted to test if Gal80 expression could be visualized with a fluorescent tag, to show the cells in which expression was being inhibited. For these experiments we used a retinal ganglion cell (RGC) enhancer, isl2b.3, derived from the 17.6 kb isl2b enhancer (Pittman et al., 2008). Tg(isl2b.3:Gal4)zc65; Tg(UAS:GFP) embryos show strong expression in RGCs by 72hpf (Figure S2). We injected DNA for hsp70:Gal80, hsp70:Gal80-TagRFP, or hsp70:Gal80-2A-TagRFP Tol2 constructs along with Tol2 transposase mRNA into Tg(isl2b.3:Gal4)zc65; Tg(UAS:GFP) embryos at the 1-cell stage, heat-shocked them at 48hpf at 37°C for 1 hour, and analyzed them at 72hpf. Mosaic expression of a bicistronic Gal80-2A-TagRFP using the viral interrupting peptide 2A to express Gal80 and TagRFP from a single mRNA (Provost et al., 2007) (Figure S2B-B”), or of a direct Gal80-TagRFP fusion (Figure S2C-C”), led to inhibition of Gal4-driven expression and TagRFP fluorescence. Single confocal slice images showed that GFP (driven by Gal4) and TagRFP (marking Gal80) expression do not overlap (insets in Figure S2A”-C”, and magnified views in S2A””-4C”’). However, because of limitations with transient injections, we did perform experiments using stable lines as well (below, Figures 5 and 6).
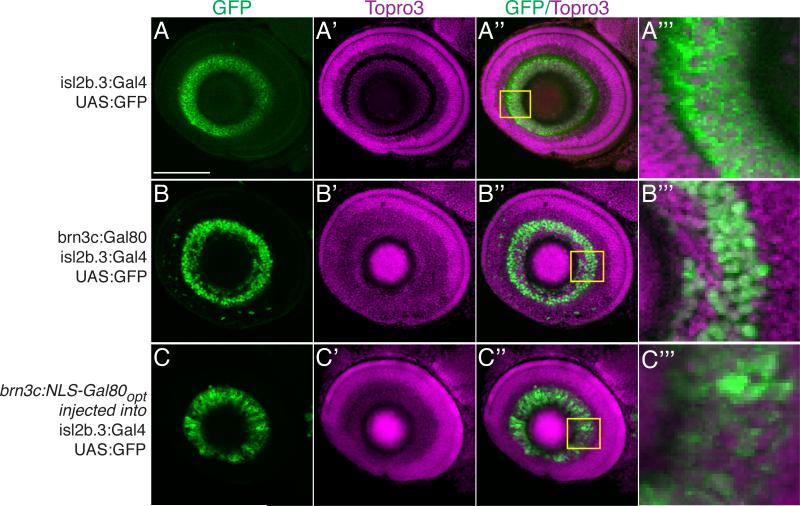
Figure 5.
Nuclear localization and codon optimization improves function of Gal80. Confocal maximum projections, lateral views, anterior to left, dorsal up, of eyes in Tg(isl2b.3:Gal4)zc65; Tg(UAS:GFP) 72hpf embryos. Scale bar, 50μm. Immunostaining for GFP, green; Topro3 nuclear stain, magenta. (A-A”’) Tg(isl2b.3:Gal4)zc65; Tg(UAS:GFP) transgenic embryos with no Gal80 show GFP expression in all RGCs. Inset and A”’ shows high power magnification of single confocal slice. (B-B”’) Triple transgenic Tg(isl2b.3:Gal4)zc65; Tg(UAS:GFP); Tg(brn3c:Gal80) shows inhibition of Gal4-dependent GFP expression in about 30% of RGCs. Inset and B”’ shows high power magnification of single confocal slice. (C-C”’) Transient injection with construct carrying “improved” Gal80 into Tg(isl2b.3:Gal4)zc65; Tg(UAS:GFP) embryos demonstrates inhibition similar to that of stable lines carrying native Gal80. Improved Gal80 has nuclear localization signal (NLS) and is codon optimized (“opt”).
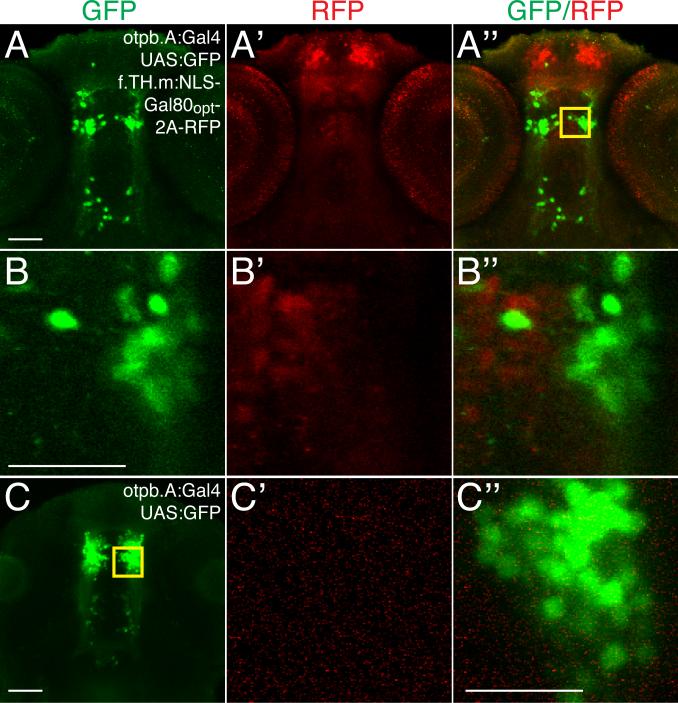
Figure 6.
Subgroups of neurons can be genetically defined by expressing Gal4 and Gal80 with partially overlapping enhancers. Confocal ventral views, anterior to top, of brain in 72hpf embryos. Scale bar, 50μm, 25μm in B and C. Immunostaining for GFP, green; TagRFP, red. (A-A”) Maximum intensity projections of Tg(otpb.A:Gal4)zc67; Tg(UAS:GFP); Tg(f.TH.m:NLS-Gal80opt-2A-TRFP)zc78 embryos shows non-overlap of Gal80 and GFP expression. (B-B”) Boxed inset area from A” shows single confocal slice of individual cells expressing either GFP or RFP but not both. (C) Single confocal slice of Tg(otpb.A:Gal4)zc67; Tg(UAS:GFP) embryo and inset (C’-C”) shows full complement of cells express GFP in absence of Gal80.
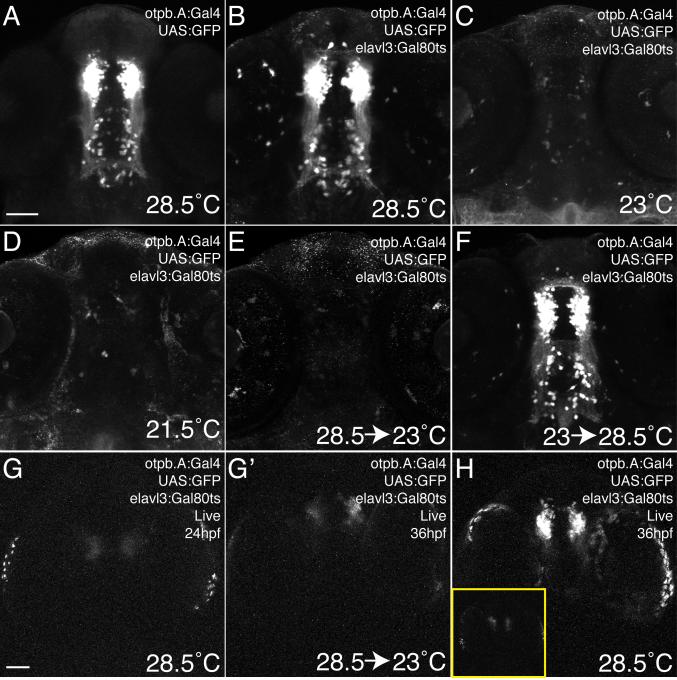
In order to achieve additional temporal control over Gal80 function, we explored the use of a temperature-sensitive Gal80 allele used in yeast and flies (McGuire et al., 2003). At higher temperatures in Drosophila (30°C), the Gal80ts protein is non-functional and Gal4-dependent expression occurs normally. At lower temperatures (19°C) in yeast and flies, however, Gal80ts binds Gal4 and prevents transcription. We generated a stable transgenic line to express Gal80ts pan-neuronally, Tg(elavl3:Gal80ts)zc68. When crossed to Tg(otpb.A:Gal4)zc67; Tg(UAS:GFP) fish and raised at 28.5°C, Gal80ts did not affect Gal4-dependent expression (Figure 4A, B). However, when raised at 23°C or 21.5°C from 24hpf to 72hpf, Gal80ts was able to effectively inhibit Gal4-dependent expression (Figure 4C, D). We were also able to perform temperature shift experiments by initially allowing Gal4-dependent expression to take place at 28.5°C through 48hpf and then shifting to 23°C to allow Gal80 to inhibit Gal4 (Figure 4E). Similarly, we could initially inhibit Gal4 at 23°C, and then relieve the inhibition by shifting to the non-permissive temperature (28.5°C) at 48hpf to permit Gal4-dependent expression (Figure 4F). We did note some variability in inhibition ability in these experiments, which depended at least in part on the strength of the Gal80ts transgene allele used (JLB, data not shown). Gal80ts was able to inhibit Gal4-dependent expression within 12 hours of shifting to the permissive temperature, even once Gal4-driven expression had started (Figure 4G-H).
Figure 4.
Temporal control of Gal4-dependent expression using a temperature sensitive version of Gal80. Confocal maximum intensity projections, ventral views, anterior to top, 72hpf Tg(otpb.A:Gal4)zc67; Tg(UAS:GFP) embryos stained with anti-GFP. Scale bar, 50μm. (A, B) Expression is indistinguishable between embryos without (A) or with (B) the Tg(elavl3:Gal80ts)zc68 transgene when raised at the restrictive temperature (28.5°C) (C, D) When raised at permissive temperatures (21.5°C or 23°C) starting at 24hpf, Gal80ts is able to effectively inhibit Gal4-dependent expression. (E) When Gal4-dependent expression is initially allowed to occur from 24 to 48hpf, shifting to the permissive temperature from 48-72hpf allows Gal80ts to inhibit Gal4-dependent expression. (F) Raising at a permissive temperature until 48hpf, then relieving Gal80-mediated inhibition at 48hpf permits Gal4-dependent expression to resume by 72hpf. (G, G’) Live confocal images of the same embryo before (at 24hpf) and after (at 36hpf) shifting to the permissive temperature, shows inhibition of Gal4-dependent expression. (H) In comparison, a control embryo raised at the restrictive temperature only shows robust expression. Inset shows this same embryo at 24hpf.
A central goal of the use of Gal80 was to genetically delineate subgroups of neurons. To do this we examined whether we could inhibit Gal4-depended expression in subgroups of RGCs. We used a stable Tg(isl2b.3:Gal4); Tg(UAS:GFP) line in which GFP is expressed in >95% of RGCs (Pittman et al., 2008) (Figure 5A-A”’), and crossed it to the stable transgenic line Tg(brn3c:Gal80). The brn3c enhancer expresses in only roughly 50% of RGCs (Xiang et al., 1995; Xiao et al., 2005). We found that GFP expression in triple transgenic embryos Tg(isl2b.3:Gal4); Tg(UAS:GFP); Tg(brn3c:Gal80) was found in about 70% of RGCs (Figure 5B-B”’). These results confirmed that Gal80 could be used with Gal4 to restrict expression to defined subsets of neurons when expressed with partially overlapping enhancers.
We did find, however, that Gal80 expression and ability to inhibit Gal4-dependent expression was variable with different enhancers. Strong enhancers, such as elavl3 or hsp70l, were able to drive robust levels of Gal80 and efficiently inhibit expression. In contrast, weaker enhancers such as brn3c or f.TH.m (Fujimoto et al., 2011) driving Gal80 did not inhibit at all using transient injections, and only strongly-expressing stable transgenic lines were able to inhibit Gal4-dependent expression. We tried several optimization strategies to improve the inhibition ability of Gal80. We found improved Gal80 inhibition by addition of a nuclear localizing signal (NLS) and by codon optimization of the Gal80 sequence for zebrafish codon usage (“Gal80opt”). Transient injection with the improved construct brn3c:NLS-Gal80opt resulted in inhibition of Gal4-dependent expression comparable to that of a stable transgenic line Tg(brn3c:Gal80) (Figure 5C-C’’’). To directly compare the efficacy of Gal80opt to Gal80, we performed transient injections of a plasmid carrying either brn3c:Gal80 or brn3c: Gal80opt into Tg(isl2b.3:Gal4); Tg(UAS:GFP) embryos. We then counted the number of GFP-positive RGCs at 72hpf in the rostral-dorsal quadrant of the eye. We found that the number of GFP-positive cells was significantly lower in embryos injected with brn3c: Gal80opt (mean number of cells 37 versus 24, p=0.03, two-tailed t test, n=11 embryos).
We tested whether Gal80opt could be used with a “weaker” enhancer, f.TH.m (Fujimoto et al., 2011), to limit Gal4-dependent expression in the CNS. We generated transgenic fish expressing NLS-Gal80opt-2A-TRFP in a subset of telencephalic and diencephalic neurons under the control of the f.TH.m enhancer (Figure 6). When crossed to animals expressing UAS:GFP driven by otpb.A:Gal4, we were able to restrict expression to a subset of genetically-defined neurons. Thus, using different enhancers to express Gal4 and Gal80, we can differentiate subsets of an otherwise homogeneous group of cells.
Discussion
We have demonstrated that Gal80 can be used in a vertebrate system to inhibit and refine Gal4-dependent expression. Native Gal4 is sufficient to drive UAS-dependent transgene expression at high levels in stable transgenic lines. One concern for the use of Gal4 has been that in vitro it has 100-fold less activity that Gal4-VP16413-490 (Sadowski et al., 1988), and in transient injections has been reported to be practically unable to drive transgene expression (Koester and Fraser, 2001; Ogura et al., 2009). In our hands, however, we found that transient injections with Gal4 resulted in similar levels of expression to that of Gal4-VP16-dependent expression. We did note that expression driven by Gal4 versus Gal4-VP16 appear to have an “all-or-nothing” phenomenon, and that once GFP expression from UAS was transactivated, levels of expression were similar. However, in transient injections, Gal4 did lead to transactivation of a lower percentage of cells than Gal4-VP16. Further, we found that stable Gal4 transgenic lines are able to drive UAS-dependent expression at levels comparable to Gal4-VP16 and higher than direct enhancer:transgene constructs. In zebrafish, Gal80 expression can be regulated temporally and spatially by use of different enhancers to specifically turn off Gal4 activity. In addition, we also showed that a temperature-sensitive Gal80ts transgene can be manipulated to permit Gal4-dependent expression at restrictive (higher) temperatures, and turn off Gal4-dependent expression at permissive (lower) temperatures. Either a direct Gal80-TagRFP fusion or a bicistronic Gal80-2A-TagRFP construct permit visualization of Gal80 expression.
The addition of Gal80 to use in the zebrafish Gal4/UAS system offers a new tool for dissection of complex developmental and functional roles of different neuronal groups in vertebrates. Another recent approach to inhibit undesired Gal4 function in the early embryo is to inject a Gal4-targeted morpholino (Faucherre and Lopez-Schier, 2011), who also convincingly demonstrated the ability to use Gal80 to inhibit Gal4-dependent expression in zebrafish. Binary expression systems in which transgene expression is regulated independently through an exogenous transcription factor already offer the potential for higher expression levels, inducible regulation, and ease in performing large-scale enhancer screens. Current binary systems include the Gal4/UAS system in Drosophila and zebrafish (Scheer and Campos-Ortega, 1999; Fischer et al., 1988; Brand and Perrimon, 1993), the tetracycline-inducible tTA/TRE in mice (Gossen and Bujard, 1992), the lexA/lexAO system in flies and zebrafish (Lai and Lee, 2006; Emelyanov et al., 2008), and recently, the Q system in Drosophila (Potter et al., 2010).
We did find that the effectiveness of wild-type Gal80, Gal80ts, or fluorescently-tagged Gal80, all depended on expression levels. Some alleles of the same transgene were weaker or stronger inhibitors of Gal4-dependent expression, presumably because different insertions expressed different levels of Gal80 due to position effects. Also, different enhancers were more or less effective at expressing Gal80 (JLB, data not shown). Another issue is that inhibition of Gal4-dependent expression takes several hours, depending on relative levels of Gal80 and Gal4 expression, and the rate of mRNA or protein turnover for the UAS-dependent transgene. The Gal4 protein itself has been shown to be quite stable in zebrafish and lasts up to 13 hours following transient induction of expression (Scheer et al., 2002). Both the Tg(otpb.A:Gal4) and Tg(isl2b.3:Gal4) lines were insertions at a single locus, and were the first stable lines recovered from screening relatively few potential founders (<15) in both cases. This suggests that expression from stable Gal4-expressing transgenes is similar to that of Gal4-VP16. That is, screening high numbers of founders in order to identify a Gal4-line with sufficient expression appears unnecessary.
For temporally-regulated expression, a potential advantage of Gal80ts is that shifting to the restrictive temperature will allow long-lasting Gal4-driven expression, unlike the very transient expression provided by the current heat-shock promoter method. In addition, Gal80ts can be expressed in spatially-restricted patterns, which is difficult with heat-shock-driven expression of Gal80. However, the use of Gal80ts may be limited by the temperature-sensitive nature of zebrafish development, especially at earlier stages of embryogenesis.
The use of Gal4/UAS in zebrafish has been limited by the available enhancers or enhancer-trap lines, which often drive expression in multiple types of cells. Intersectional expression using Gal80 offers the potential to restrict Gal4-dependent expression and more precisely define sub-groups of cells. Other experimental avenues are also made possible by the Gal4/Gal80 system. For example, heat-shock inducible expression of Gal80 in different locations or time-points, lineage-specific mosaic analysis (Lee and Luo, 1999), or repression of gene function late in development, can be used to analyze different developmental events of interest.
Experimental Procedures
Fish stocks and embryo raising
Adult fish were bred according to standard methods. Embryos were raised at 28.5°C (unless otherwise stated) in E3 embryo medium and staged by time and morphology (Kimmel et al., 1995). For in situ or immunohistochemistry staining, embryos were fixed in 4% paraformaldehyde (PFA) (in PBS) for 3 h at room temperature (RT) or overnight (O/N) at 4°C, washed briefly in PBS, dehydrated, and stored in 100% MeOH at -20°C until use.
Transgenic fish lines and alleles used in this paper are listed in Table 1. Previously existing lines were: Tg(5xUAS:GFP)nkuasgfp1a, listed in this paper as Tg(UAS:GFP), kind gift of K. Kawakami (Asakawa et al., 2008); Tg(otpb.A:GFP)zc48; and Tg(otpb.A:Gal4-VP16413-470, myl7:EGFP)zc57 (Fujimoto et al., 2011). New lines generated for this work were: Tg(elavl3:Gal80, myl7:EGFP)zc63 , Tg(elavl3:Gal80, myl7:TagRFP)zc64 , Tg(isl2b.3:Gal4, myl7:TagRFP)zc65, Tg(otpb.A:Gal4, myl7:EGFP)zc67, Tg(elavl3:Gal80ts, myl7:TagRFP)zc68, Tg(brn3c:Gal80, myl7:EGFP)zc70, Tg(hsp70l:Gal80, myl7:EGFP)zc71, and Tg(f.TH.m:NLS-Gal80opt-2A-TagRFP)zc78 Injection of DNA constructs and raising of stable transgenic lines was performed essentially as described (Bonkowsky et al., 2008). Determination of transgene genomic insertion sites was determined by splinkerette PCR and sequencing (Potter and Luo, 2010). Lines are available upon request.
TABLE 1.
List of DNA constructs and transgenic lines used in this paper.
| Name | Description | Plasmid | Transgenic line?1 |
|---|---|---|---|
| hs:RFP | TagRFP under control of hsp70l | + | |
| Gal4 | |||
| pME-Gal4 | full-length native Gal4 with stop in Gateway middle entry clone (pME) | + | n/a |
| otpb.A:Gal4 | native Gal4 driven by otpb enhancer* in the diencephalon | + | + (zc67) |
| otpb.A:Gal4VP16 | Gal4-VP16 driven by otpb enhancer* in the diencephalon | + | + (zc57) |
| isl2b.3:Gal4 | native Gal4 driven by isl2b.3 enhancer in RGCs | + | + (zc65) |
| Gal80 | |||
| pME-Gal80 | full-length Gal80 with stop | + | n/a |
| pME-Gal80ts | full-length temperature-sensitive Gal80 with stop | + | n/a |
| pME-NLS-Gal80 | NLS signal at N-terminus of Gal80 with stop | + | n/a |
| pUC57-Gal80opt | codon-optimized Gal80 with stop in pUC57 vector | + | n/a |
| pME-NLS-Gal80opt | NLS signal at N-terminus of codon-optimized Gal80 no stop | + | n/a |
| elavl3:Gal80 | Gal80 driven by pan-neuronal enhancer | + | + (zc63,zc64)^ |
| brn3c:Gal80 | Gal80 driven by the brn3c enhancer in the eye | + | + (zc70) |
| hs:Gal80 | Gal80 under control of hsp70l | + | + (zc71) |
| hs:Gal80-2A-RFP | Gal80 (no stop) and 2A-peptide linked TagRFP under control of hsp70l | + | - |
| hs:Gal80-RFP | Gal80 (no stop) fused to TagRFP under control of hsp70l | + | - |
| elavl3:Gal80ts | temperature-sensitive Gal80 driven by pan-neuronal enhancer | + | + (zc68) |
| f.TH.m:NLS-Gal80opt | NLS-Gal80opt driven in subset of telencephalon and diencephalon* | + | + (zc78) |
Comments:
Transgenic lines with no visible fluorescent marker were made using a Tol2 destination vector carrying a heart cmlc2 promoter driving either GFP or RFP.
zc63 is a weak allele (lower levels of Gal80 expression).
In situ hybridization and Immunohistochemistry
Whole-mount in situ labeling and immunohistochemistry were performed as described (Fujimoto et al., 2011; Bonkowsky et al., 2005; Bonkowsky et al., 2008). Antibodies used were rabbit polyclonal anti-tyrosine hydroxylase 1:400 (Millipore); mouse anti-acetylated tubulin 1:250; goat anti-mouse Alexa 488 1:500 (Invitrogen); and Cy-3 anti-rabbit 1:400. Staining for apoptotic cells was performed by incubating live embryos in 5μg/mL of acridine orange with gentle rocking for 30 minutes at room temperature, followed by washes in E3 for at least 30 minutes. Embryos were then embedded in 1.5% low-melt agarose and imaged live.
DNA Constructs
A list of clones generated is in Table 1. Standard Gateway (Invitrogen) cloning with attB recombination sequences appended to PCR primers was used to generate entry clones. Expression clones and final destination vectors were built using the Tol2 kit (Kwan et al., 2007). For destination vectors lacking an expressed fluorescent marker, either a cmlc2:EGFP (official nomenclature myl7:EGFP) or cmlc2:TagRFP transgenesis marker was used in the final construct. The identity of constructs was confirmed by restriction enzyme digests, and by sequencing on both strands (for coding sequences) or by partial end-sequencing (for enhancers). pME-Gal4 was cloned by PCR from pKG4021 (Guillemin et al., 2001) (kind gift of M. Metzstein) using primers 5’-AGACCATGAAGCTACTGTCTTCTATCGA-3’ and 5’-CTTACTCTTTTTTTGGGTTTGGTGG-3’. pME-Gal80 was cloned by PCR from TubPGAL80 (Lee and Luo, 1999) (obtained from Addgene, plasmid #17748) using primers 5’-ATGGACTACAACAAGAGATCTTCG-3’ and 5’-TTATAAACTATAATGCGAGATATTGCT-3’. pME-Gal80ts was cloned from pBPHLWL-Gal80ts (McGuire et al., 2003) by PCR (kind gift of T. Clandinin). pME-NLS-Gal80opt was generated by PCR using primers 5’-ATGGCTCCAAAGAAGAAGCGTAAGGTAATGGACTACAACAAAAGGAGCAG-3’ for the nuclear localization signal (Kwan et al., 2007) and 5’-CAGTGAGTAGTGAGAGATATTTG-3’ for codon-optimized Gal80 (GenScript; GenBank accession #JN314417). The p5E-elavl3 enhancer clone was generated by inserting the XhoI/SalI fragment from pCS2-HuC:Kaede (Sato et al., 2006) into the XhoI site of p5E-MCS. p5E-brn3c was cloned by PCR from brn3C:GAL4VP16 (Campbell et al., 2007) using primers 5’-CCGGATGCACTGTATATTGC-3’ and 5’-AATTCGTTGCGCACCTTGCA-3’. p5E-isl2b.3 was generated by digestion of p5E-isl2b(Δ16-22) (Ben Fredj et al., 2010) with BstXI and XhoI and blunt-end religation to remove an internal 5.1 kb fragment to generate a 9.6kb enhancer-promoter fragment. pTol2-hsp70l:TagRFP, pTol2-hsp70l:Gal80-2A-TagRFP, and pTol2-hsp70l:Gal80-TagRFP were generated using either pME-TagRFP, or pME-Gal80 with no stop codon, p5E-hsp70l (Kwan et al., 2007), and either p3E-2A-TagRFP, which uses the viral 2A peptide to interrupt translation to generate a bicistronic message (Provost et al., 2007) or p3ETagRFP, to generate a direct Gal80-TagRFP fusion protein. Plasmids and specific PCR conditions are available upon request.
Microscopy and image analysis
Image acquisition and analysis were performed as described previously (Fujimoto et al., 2011; Suli et al., 2006). Embryos were processed and placed in a solution of 80% glycerol/20% PBST, and mounted on a glass slide with a #0 coverslip. NIH ImageJ software was used to merge slices to create maximal intensity projections.
Heat shock and temperature shift experiments
For induction of heat-shock constructs, embryos were heat-shocked for 1 hour at 37°C at 48hpf, and collected at 72hpf for analysis. For Gal80ts experiments, embryos were raised at 28.5°C until 24hpf, then raised at either 21.5°C, 23°C, or 28.5°C from 24-48hpf or from 24-72hpf. Timing was based on embryo morphology, so duration of incubation was adjusted to correct for different rates of development at different temperatures. At 48hpf embryos were either raised in their current condition (21.5°C, 23°C, or 28.5°C), or shifted to a new temperature from 48-72hpf.
Supplementary Material
Figure S1. Schematic of positive and negative intersectional methods for labeling sub-groups of cells. (A) Positive intersectional method: a group of cells is labeled by partially overlapping expression domains of two different enhancers. Expression occurs only in cells in which both components (for example, of split Gal4) are present. (B) Negative intersectional method: a group of cells is labeled by two different enhancers; expression occurs in the cells in which only the positive activator is present.
Figure S2. Subgroups of neurons can be distinguished by inhibiting Gal4-driven expression with a fluorescently-tagged Gal80. Single slice confocal images, lateral views, anterior to left, dorsal up, of 72hpf eyes in Tg(isl2b.3:Gal4)zc65; Tg(UAS:GFP) embryos. Scale bar, 50μm. Immunostaining for GFP, green; TagRFP, red; Topro3 nuclear stain, magenta. (A-A”’) Embryos injected with hsp70l:TagRFP (no Gal80 expression). (B-B”’) Embryos injected with hsp70l:Gal80-2A-TagRFP and heat-shocked at 48hpf show expression of TagRFP (RFP) and inhibition of Gal4-dependent expression. (C-C”’) Embryos injected with hsp70l:Gal80-TagRFP and heat-shocked at 48hpf show RFP expression and concomitant inhibition of Gal4-dependent expression. (A””-C””) shows magnified view of the insets in (A”-C”), demonstrating disjoint GFP and RFP expression.
Acknowledgements
We would like to thank members of the Chien lab for their assistance in preparing this work; K. Kwan for generation of p3E-2A-TagRFP; T. Clandinin, K. Kawakami, M. Metzstein, and H. Okamoto for sharing clones and fish lines; R. Dorsky for helpful discussions; and H. Lopez-Schier for communication of results prior to publication. This work was supported by a PCMC Foundation grant to JLB, NIH R01 MH092256 to CBC, and NIH K12 5HD001410 and K08 DA024753 to JLB.
References
- Amsterdam A, Becker TS. Transgenes as screening tools to probe and manipulate the zebrafish genome. Dev Dyn. 2005;234:255–68. doi: 10.1002/dvdy.20541. [DOI] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci USA. 2008;105:1255–60. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Fredj N, Hammond S, Otsuna H, Chien CB, Burrone J, Meyer MP. Synaptic activity and activity-dependent competition regulates axon arbor maturation, growth arrest, and territory in the retinotectal projection. J Neurosci. 2010;30:10939–51. doi: 10.1523/JNEUROSCI.1556-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowsky JL, Chien CB. Molecular cloning and developmental expression of foxP2 in zebrafish. Dev Dyn. 2005;234:740–6. doi: 10.1002/dvdy.20504. [DOI] [PubMed] [Google Scholar]
- Bonkowsky JL, Wang X, Fujimoto E, Lee JE, Chien CB, Dorsky RI. Domain-specific regulation of foxP2 CNS expression by lef1. BMC Dev Biol. 2008;8:103. doi: 10.1186/1471-213X-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Stringham SA, Timm A, Xiao T, Law MY, Baier H, Nonet ML, Chien CB. Slit1a inhibits retinal ganglion cell arborization and synaptogenesis via Robo2-dependent and - independent pathways. Neuron. 2007;55:231–45. doi: 10.1016/j.neuron.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Del Giacco L, Sordino P, Pistocchi A, Andreakis N, Tarallo R, Di Benedetto B, Cotelli F. Differential regulation of the zebrafish orthopedia 1 gene during fate determination of diencephalic neurons. BMC Dev Biol. 2006;6:50. doi: 10.1186/1471-213X-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A, Parinov S. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev Biol. 2008;320:113–21. doi: 10.1016/j.ydbio.2008.04.042. [DOI] [PubMed] [Google Scholar]
- Faucherre A, López-Schier H. Delaying Gal4-driven gene expression in the zebrafish with morpholinos and Gal80. PLoS One. 2011;6:e16587. doi: 10.1371/journal.pone.0016587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–6. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Fujimoto E, Stevenson T, Chien CB, Bonkowsky JL. Identification of a dopaminergic enhancer indicates complexity in vertebrate dopamine neuron phenotype specification. Dev Biol. 2011;352:393–404. doi: 10.1016/j.ydbio.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–25. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin K, Williams T, Krasnow MA. A nuclear lamin is required for cytoplasmic organization and egg polarity in Drosophila. Nat Cell Biol. 2001;3:848–51. doi: 10.1038/ncb0901-848. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Large-scale genomic approaches to brain development and circuitry. Annu Rev Neurosci. 2005;28:89–108. doi: 10.1146/annurev.neuro.26.041002.131436. [DOI] [PubMed] [Google Scholar]
- Hirrlinger J, Scheller A, Hirrlinger PG, Kellert B, Tang W, Wehr MC, Goebbels S, Reichenbach A, Sprengel R, Rossner MJ, Kirchhoff F. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS One. 2009;4:e4286. doi: 10.1371/journal.pone.0004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, Barres BA. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron. 2008;59:425–38. doi: 10.1016/j.neuron.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Köster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–46. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–9. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–61. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–87. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–36. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF, Chasman DI, Buchman AR, Kornberg RD. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol Cell Biol. 1987;7:3446–51. doi: 10.1128/mcb.7.10.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–42. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–8. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, Miki N, Chitnis AB, Huh TL. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol. 2000;227:279–93. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- Ogura E, Okuda Y, Kondoh H, Kamachi Y. Adaptation of GAL4 activators for GAL4 enhancer trapping in zebrafish. Dev Dyn. 2009;238:641–55. doi: 10.1002/dvdy.21863. [DOI] [PubMed] [Google Scholar]
- Pittman AJ, Law MY, Chien CB. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development. 2008;135:2865–71. doi: 10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Luo L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS One. 2010;5:e10168. doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–48. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–9. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- Russek-Blum N, Gutnick A, Nabel-Rosen H, Blechman J, Staudt N, Dorsky RI, Houart C, Levkowitz G. Dopaminergic neuronal cluster size is determined during early forebrain patterning. Development. 2008;135:3401–13. doi: 10.1242/dev.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, Mahler J, Acampora D, Holzschuh J, Erhardt S, Omodei D, Simeone A, Driever W. Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr Biol. 2007;17:873–880. doi: 10.1016/j.cub.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–4. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44:136–42. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- Sato T, Hamaoka T, Aizawa H, Hosoya T, Okamoto H. Genetic single-cell mosaic analysis implicates ephrinB2 reverse signaling in projections from the posterior tectum to the hindbrain in zebrafish. J Neurosci. 2007;27:5271–9. doi: 10.1523/JNEUROSCI.0883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–8. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Scheer N, Riedl I, Warren JT, Kuwada JY, Campos-Ortega JA. A quantitative analysis of the kinetics of Gal4 activator and effector gene expression in the zebrafish. Mech Dev. 2002;112:9–14. doi: 10.1016/s0925-4773(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–6. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Scott EK. The Gal4/UAS toolbox in zebrafish: new approaches for defining behavioral circuits. J Neurochem. 2009;110:441–56. doi: 10.1111/j.1471-4159.2009.06161.x. [DOI] [PubMed] [Google Scholar]
- Suli A, Mortimer N, Shepherd I, Chien CB. Netrin/DCC signaling controls contralateral dendrites of octavolateralis efferent neurons. J Neurosci. 2006;26:13328–13337. doi: 10.1523/JNEUROSCI.2858-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster ML, Seugnet L, Bate M, Sokolowski MB. Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis. 2004;39:240–5. doi: 10.1002/gene.20051. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15:4762–85. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Roeser T, Staub W, Baier H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 2005;132:2955–67. doi: 10.1242/dev.01861. [DOI] [PubMed] [Google Scholar]
- Zhang S, Ma C, Chalfie M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell. 2004;119:137–44. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Schematic of positive and negative intersectional methods for labeling sub-groups of cells. (A) Positive intersectional method: a group of cells is labeled by partially overlapping expression domains of two different enhancers. Expression occurs only in cells in which both components (for example, of split Gal4) are present. (B) Negative intersectional method: a group of cells is labeled by two different enhancers; expression occurs in the cells in which only the positive activator is present.
Figure S2. Subgroups of neurons can be distinguished by inhibiting Gal4-driven expression with a fluorescently-tagged Gal80. Single slice confocal images, lateral views, anterior to left, dorsal up, of 72hpf eyes in Tg(isl2b.3:Gal4)zc65; Tg(UAS:GFP) embryos. Scale bar, 50μm. Immunostaining for GFP, green; TagRFP, red; Topro3 nuclear stain, magenta. (A-A”’) Embryos injected with hsp70l:TagRFP (no Gal80 expression). (B-B”’) Embryos injected with hsp70l:Gal80-2A-TagRFP and heat-shocked at 48hpf show expression of TagRFP (RFP) and inhibition of Gal4-dependent expression. (C-C”’) Embryos injected with hsp70l:Gal80-TagRFP and heat-shocked at 48hpf show RFP expression and concomitant inhibition of Gal4-dependent expression. (A””-C””) shows magnified view of the insets in (A”-C”), demonstrating disjoint GFP and RFP expression.