Abstract
The sustained overproduction of reactive oxygen and nitrogen species results in an imbalance of cellular prooxidant-antioxidant systems and is implicated in numerous disease states, including alcoholic liver disease, cancer, neurological disorders, inflammation and cardiovascular disease. The accumulation of reactive aldehydes resulting from sustained oxidative stress and lipid peroxidation is an underlying factor in the development of these pathologies. Determining the biochemical factors that elicit cellular responses resulting from protein carbonylation remains a key element to developing therapeutic approaches and ameliorating disease pathologies. This review details our current understanding of the generation of reactive aldehydes via lipid peroxidation resulting in protein carbonylation, focusing on pathophysiologic factors associated with 4-hydroxynonenal-protein modification. Additionally, an overview of in vitro and in vivo model systems used to study the physiologic impact of protein carbonylation is presented. Finally, an update of the methods commonly used in characterizing protein modification by reactive aldehydes provides an overview of isolation techniques, mass spectrometry and computational biology. It is apparent that research in this area employing state-of-the-art proteomics, mass spectrometry and computational biology is rapidly evolving and yielding foundational knowledge concerning the molecular mechanisms of protein carbonylation and its relation to a spectrum of diseases associated with oxidative stress.
1. Introduction
Protein modification through carbonylation reactions is a documented consequence of oxidative stress, first defined as “a disturbance in the prooxidant-antioxidant balance in favor of the former leading to potential damage”. 1 In the healthy aerobic organism, there is a steady-state level of reactive oxygen species (ROS) and reactive nitrogen species (RNS) production that is readily buffered by effective cellular defense systems including glutathione as well as the antioxidant enzymes copper-zinc superoxide dismutase and glutathione peroxidase. However, the sustained overproduction of ROS and RNS overwhelms these defense systems and is implicated in a number of clinical conditions involving essentially every organ system. The resulting clinical conditions are characterized by chronic inflammation and include cardiovascular disease, alcoholic liver disease, diabetes and a spectrum of other disorders.
Identifying the carbonylation of proteins critical for cellular homeostasis could potentially provide important information concerning molecular mechanisms underlying the development and progression of diseases linked to oxidative stress. Generally, protein carbonylation is an irreversible non-enzymatic process resulting from ROS and downstream products of oxidative processes, such as metal-catalyzed oxidation (MCO) and the peroxidation of polyunsaturated fatty acids (PUFAs). 2, 3 Protein carbonyl groups are derived through two major mechanisms; where amino acid side chains are oxidized directly (direct) or through conjugation by reactive species such as advanced lipoxidation end products and advanced glycation end products (indirect). 2–4 The respective contribution of protein carbonyls derived directly or indirectly to disease pathologies remains unknown. It is likely that quantities of carbonyl derivatives generated and the length of time the generation is sustained are major factors. One point is that very few if any studies have been performed to monitor the time course of protein carbonyls and progression/regression of disease. Significant progress has been made in identifying certain proteins that are targets of carbonylation in order to understand the properties of these proteins that predispose them to this modification. Nevertheless, the molecular mechanisms involved in protein carbonylation via reactive aldehydes have proven complex and the experimental approaches used to recover and identify these modified proteins remain challenging. The purpose of this review is to provide an overview focusing on protein modification by reactive aldehydes derived from lipoxidation and their potential link to the initiation and progression of diseases associated with sustained oxidative stress. In addition, this perspective presents an update of the recent advances in experimental approaches employed to recover and identify proteins modified by oxidative damage.
2. Reactive Aldehydes and Factors Predisposing Proteins to Modification
Detrimental molecular consequences of oxidative stress can occur via two mechanisms, macromolecular damage and disruption of the regulation of redox signaling, resulting from free radical and non-radical induced damage, respectively. 5 Under basal conditions, these molecules with oxidant potential are produced at moderate concentrations. As such, these free radicals and nonradicals are involved in signaling cascades which upregulate numerous cellular pathways, including those involved in the generation of antioxidant defenses which maintain redox homeostasis under short-lived incidences of stress. Cellular redox auto-regulation yields an abundant supply of free radical scavenging enzymes (i.e. superoxide dismutase) as well as endogenous factors (i.e. GSH), which prevent a majority of free radical chain reactions from occurring.
A steady flow of ROS is generated through metabolic processes, as a consequence of oxidase activity, endoplasmic reticulum and mitochondrial electron transport, giving rise to superoxide anion (O2·-) and hydrogen peroxide (H2O2). Likewise, pro-oxidant environments created as a result of situations such as chronic ethanol intoxication exacerbate free radical production through dysregulation of hepatic enzymes such as cytochrome P450 2E-1 (CYP2E1), xanthine oxidase, superoxide dismutase, and aldehyde oxidases increase O2·- and H2O2 levels. 6 In the presence of transition metals such as iron, both O2·- and H2O2 generate highly reactive hydroxyl radicals (HO·) through Fenton-type reactions. 7 Nitric oxide (NO), the major precursor for cellular RNS, is produced in biological systems by specific nitric oxide synthases involved in a number of signaling pathways, including neurotransmission, defense mechanisms, and immune regulation. 8 Whereas NO and O2− are short-lived species, under certain conditions they react to produce significant amounts of an even more oxidatively active molecule, peroxynitrite anion (ONOO−). 9 Through the generation of ONOO−, NO contributes to the majority of biological RNS toxicity. 10 Consequently, the overproduction of HO· and ONOO− during oxidative stress leads to free radical mediated damage and the generation of nonradical oxidants, both of which may modify DNA, carbohydrates, proteins, and lipids.
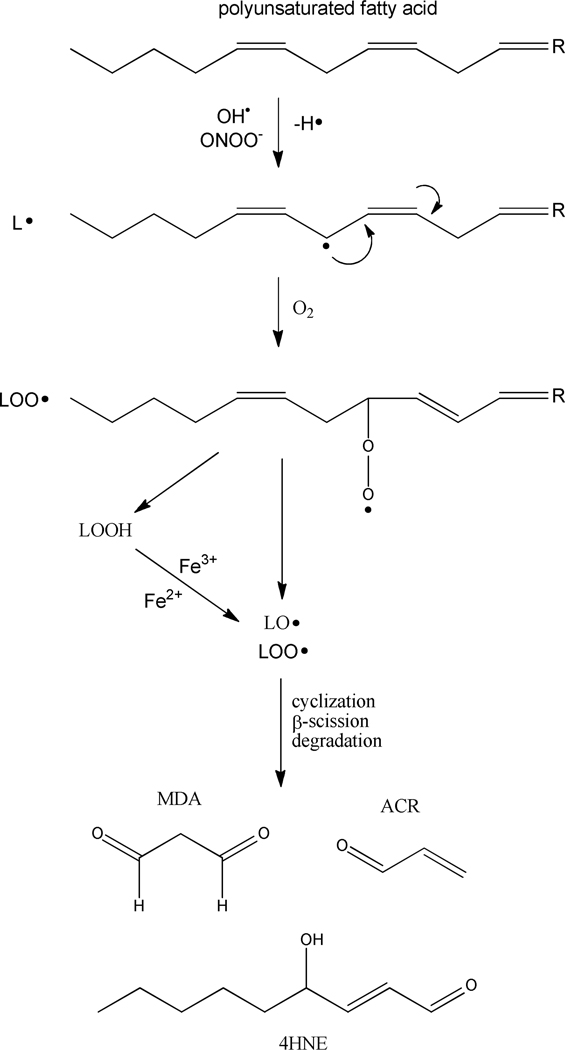
One mechanism of free radical mediated damage occurs through initiation events such as lipid peroxidation, and is of particular interest to researchers in applied toxicology, as protein carbonylation via toxic reactive aldehydes is known to result in altered protein function. 11–14 The generation of free radicals in close proximity to lipid rich cellular membranes containing PUFAs initiates lipid peroxidation, which occurs when HO· and ONOO− abstract an allylic hydrogen from PUFAs. The initial step in this process demonstrates the basic principle that reaction of a radical (HO· and ONOO−) with a non-radical species, typically results in the generation of a new, less reactive radical, which, in this case is a carbon-centered lipid radical (L·) (Figure 1). 15 This lipid radical, in turn, reacts with molecular oxygen to yield a lipid peroxyl radical (LOO·), resulting in the formation of lipid hydroperoxide (LOOH). These lipid hydroperoxides react with trace metals to form lipid alkoxyl radicals (LO·). 15 Both LO· and LOO· radicals generate reactive α,β-unsaturated aldehydes through cyclization and/or β-scission and degradation, most commonly generating highly electrophilic products such as 4-hydroxynonenal (4-HNE), malondialdehyde (MDA) and acrolein (ACR). As defined, these reactive aldehyde species result from free-radical initiated lipid peroxidation and are fairly long-lived within the cell, persisting for up to two minutes, allowing for intracellular diffusion and covalent modification of DNA, lipid, and protein throughout the cell. 16 The mobility and reactivity of these products of lipid peroxidation are central to their involvement in altered cellular signaling and diseases of oxidative stress.
Figure 1.
Lipid peroxidation of PUFAs results in the generation of biogenic aldehydes.
2.1. Pathophysiologic Factors Associated with 4-HNE-Protein Modification
Low to even moderate exposure of cells to pro-oxidative conditions is proposed to elicit a spectrum of cytoprotective responses. Under such circumstances, changes in steady state levels of 4-HNE are minimal and transient, resulting in controlled modification of cellular proteins. The modulation of transcriptional events through the Keap1-Nrf2 cascade and subsequent regulation of EpRE/ARE responses to 4-HNE is an example of such protective cellular networks. 17–19 It is not surprising that 4-HNE-modification of proteins is implicated in a spectrum of tissues obtained from humans and laboratory animals with pathophysiologic conditions characterized by sustained oxidative stress such as various neurodegenerative disorders including Alzheimer’s Disease. 20–26 Likewise, 4-HNE-modfied proteins have been detected in patients or animal models of diabetes, cardiovascular disease and a spectrum liver disorders including dysregulation of iron storage, nonalcoholic fatty liver disease or alcoholic liver disease. 27 These associations do not imply that 4-HNE production and subsequent protein modification is a primary cause of these diseases. However, the modification of certain proteins by this reactive biogenic aldehyde could play important roles in progression of these disorders, all of which are characterized by inflammation and oxidative stress.
Fundamentally it is important to determine if these protein modifications are simply a biomarker for inflammation and oxidative stress or whether they alter protein function and therefore regulate or control the biochemical pathways related to these altered proteins. Clearly 4-HNE modification of reactive thiols is reported to alter the enzymatic activity of certain proteins 11, 28, 29; however, characterizing the cause-effect association of these global changes with imparted cellular processes in vivo remains limited given the sensitivity and specificity of technologies available today. Currently, it appears as though only a small fraction of the protein pool is modified, further complicating our interpretation of how these modifications are mechanistically linked to the respective diseases.
2.2. Electrophile-Nucleophile Interactions
4-Hydroxynonenal is an α,β-unsaturated carbonyl. This conjugated system is characterized by an electron-withdrawing carbonyl group creating an area of electron deficiency at the β-carbon. This electrophilic center is the inherent property of 4-HNE rendering it a relatively strong electrophile. It is not surprising that this electron deficient species readily reacts with electron rich functional groups of nucleophiles. Therefore, predictable nucleophilic targets in proteins for 4-HNE adduction are cysteine, histidine and lysine. One primary determinant for 4-HNE reaction with these nucleophiles is the pKa value at physiological pH of the corresponding sulfhydryl (8.3), imidazole (6.04) and ε-amino side chain (10.5), respectively. The physiochemical and quantum mechanical parameters of the interactions of these nucleophiles with the electrophilic type 2 alkenes, acrolein and 4-HNE, are described in a recent comprehensive review. 30 In brief, all of the quantitative parameters unequivocally demonstrate the marked reactivity of the Cys thiolate with type 2 alkenes. It is noteworthy that these physiochemical calculations are consistent with a number of proteomic studies which identify cysteine sulfhydryl groups as favored targets for modification by 4-HNE, acrolein and a spectrum of type 2 alkenes. 31–36 One factor predisposing Cys residues in proteins to adduction by type 2 alkenes is that Cys thiols are frequently found in catalytic triads or diads facilitating shuttling of protons between flanking His, Lys and Arg residues, as well as their acidic equivalents (Asp, Glu) assisting in deprotonation of the sulfhydryl to the reactive thiolate anion.
It is important to note that the nucleophilic targets of 4-HNE modification in proteins occupy microenvironments within the protein that significantly impact their reactivity. For instance, those nucleophilic residues that are solvent accessible are predictably more reactive to addition reactions by 4-HNE. Conversely, amino acid nucleophilic residues that are buried within secondary and tertiary structures of a particular protein are less likely to be targets for type 2 alkene modification. For instance, a recent report describes the selective modification of Cys residues in the antioxidant protein, peroxiredoxin 6. 37 These investigators reported that the solvent accessible Cys91 was readily modified by 4-HNE while the active site Cys47 was not, even when exposed to high concentrations of this biogenic aldehyde.
Several investigators have explored the possibility that modification of human serum albumin (HSA) by end products of lipid peroxidation might be a reliable biomarker of oxidative stress. 31, 36, 38 Two reports 31, 36 suggest that that Cys34 is a particularly reactive nucleophile in HSA towards 4-HNE whereas Lys242 is reported to be especially susceptible for modification by this biogenic aldehyde. 38 In the latter case, selective Michael adduction of His residues of HSA by 4-HNE is an example of how protein microenvironments influence electrophile interactions with nucleophilic target residues. When exposed to 4-HNE, this 66-kDa protein displayed 10 sites of modification, 4 of which were Lys- Michael addition products with the remaining 6 modifications detected at His residues. Interestingly, through isotopeassisted adduction kinetic analysis, His242 present in the hydrophobic binding region of the HSA was demonstrated to be the most reactive. Analysis of the HSA crystal structure revealed that the hydrogen bonding of the 4-hydroxyl group of 4-HNE with an adjacent Lys199 facilitates formation of the Michael adduct with His242. These investigators employed an algorithm, in conjunction with the crystal structure of HSA, to estimate pKa values for the His and Lys target residues. Remarkably, the estimated pKa of His242 was predicted to be 0.81, further supporting its reactivity towards 4-HNE.
2.3. Protein Abundance
Studies with HSA demonstrate that modification by 4-HNE follows pseudo-first order kinetics 38. Therefore, it is predictable that the concentration of a cellular protein is an important factor in determining its interaction with and modification by lipid-derived electrophiles, including 4-HNE. In this context, a number of investigators report 4-HNE modification of extra- and intracellular proteins documented to be very abundant. For instance intracellular proteins such as creatine kinase and glyceraldehyde 3-phosphate dehydrogenase which are involved in metabolic processes are present in high concentrations and have been identified as targets of carbonylation in a number of independent studies. 39–42 Likewise, circulating HSA is present in concentrations upwards to 50 mg/ml making it one of the more abundant proteins present in serum or plasma and a potential biomarker for modification by 4-HNE. 38, 43
The results of a recent comprehensive study clearly show, however, that modification of intracellular proteins by 4-HNE proceeds on the basis of target specificity rather than abundance. 44 The basis for this experimental design is that proteins that are true targets for 4-HNE modification will be consistently detected in a 4-HNE concentration-dependent manner because the reaction follows pseudo-first order kinetics. 38 These investigators employed an experimental design using RKO cells exposed to subtoxic (0–100 µM) concentrations of 4-HNE for 1 hour. At the conclusion of the incubation period, intracellular proteins modified through 4-HNE Michael adduct formation were biotynylated, captured by streptavidin, subjected to proteolysis and identified by LC-mass spectrometry. Parallel experiments were performed using identical protein extracts from RKO cells not exposed to 4-HNE. As expected, there was some overlap in the proteins identified in cells treated with 4-HNE and those cells not treated. Interestingly, however, the correlation coefficient calculated for spectral counts between these overlapping proteins was not significant (0.044) indicating disassociation of the concentration-dependent relationship. The results of this study formulate a reproducible experimental approach to discriminate between proteins that are true targets for 4-HNE modification, independent of their respective abundance.
3. Characterizing Protein Carbonylation
Diseases of chronic oxidative stress involve the accumulation of reactive aldehydes. As described previously, this increase in lipid peroxidation products includes reactive aldehydes such as acrolein, 4-HNE and MDA, among numerous other ROS and RNS derived electrophiles. Endogenous and exogenous processes leading to the generation of these reactive aldehydes are well documented; however, their impact on cellular processes remains unclear. As noted, these electrophiles are known to play a role in intracellular signaling and, under stressed situations these electrophiles have the ability to modify nucleophilic amino acid side chains, potentially altering protein structure and function. Determining how cellular pathways are affected by these protein modifications is central to elucidating the molecular mechanisms of oxidative stress-related disease states and may provide avenues for potential therapeutics. In recent years, advances in proteomic techniques and mass spectrometry (MS) instrumentation have aided researchers in identifying these targets of carbonyl modification. The capacity to selectively isolate and identify both in vitro and in vivo protein carbonylation products relies heavily on the specificity and sensitivity of these methods. Incremental developments in ionization techniques and MS instrumentation are enabling researchers to identify protein modifications on a large scale and at a rapid pace.
3.1. In Vitro and In Vivo Model Systems
Diseases involving chronic oxidative stress and protein carbonylation are commonly examined through two models; cell culture and vertebrate model systems. Cell culture models include the treatment of an appropriate cell line with correlative agents for a specific physiologic insult, such as cigarette smoke condensate or ethanol in an attempt to replicate cellular stresses involved in chronic obstructive pulmonary disease and alcoholic liver disease, respectively. 45–47 An alternative in vitro model involves cell treatments using metabolic downstream products of oxidative stress, such as acrolein or 4-HNE. 44, 48–51 Using these models, researchers are able to identify potential causative relationships resulting from these reactive aldehydes and subsequent protein carbonylation. This approach allows for the enhanced detection and identification of protein carbonylation targets; treatments typically involve pathophysiologically relevant levels of reactive aldehydes or chemically synthesized analogs, while cellular antioxidant defenses are limited. Furthermore, performing these experiments with media in the presence or absence of serum may result in dramatically varied outcomes as serum albumin and other serum proteins are known reactive aldehyde scavengers. 31 While these cell culture models provide benefits in detecting targets of protein carbonylation, their relevance to physiologic insults remains limited, as verification through in vivo model systems is crucial to understanding the physiological impact of these biogenic aldehydes.
A spectrum of vertebrate models has been used for the evaluation of oxidative stress-related diseases and many have identified cellular pathways affected via protein carbonylation. 24, 52–54 Utilizing antibody-based methodologies in probing protein carbonylation, it is possible to further enhance our understanding of disease processes and evaluate the physiological consequences of chronic oxidative stress. However, in vivo characterization of these modified proteins has proven difficult as these methods involve subcellular fractionation or serum isolates, both of which contain complex protein mixtures. This requires a high level of specificity and sensitivity in detecting proteins expressed at a low-level. Developing both gel-based and gel-free strategies for separating and concentrating carbonylated proteins from these complex solutions has been a focus of many researchers, resulting in the development of numerous methods for isolating and characterizing modified proteins. Exhaustive lists of carbonyl-modified proteins identified through in vitro, and to a lesser extent, in vivo model systems are detailed elsewhere. 27, 44
3.2. Isolation of Carbonylated Protein
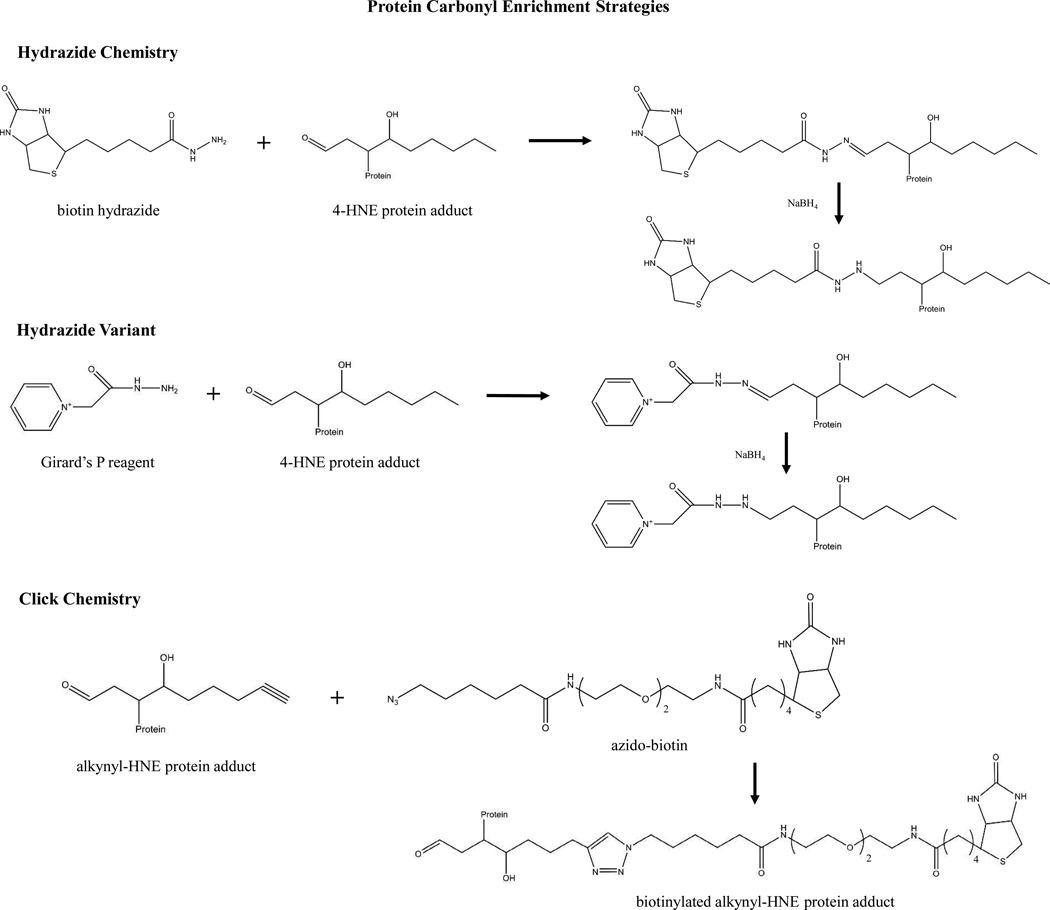
Understanding mechanisms of adduct formation and stability is central to developing strategies for the isolation of carbonylated protein from both cell lysates and tissue extracts. 4, 30, 55 As detailed above, these aldehydes react primarily through Michael-type additions with the sulfhydryl group of cysteine, the imidazole moiety of histidine and the ε-amino group of lysine in the general order of reactivity, Cys>>His>Lys. 56 Reactivity with arginine occurs at much lower levels than that of lysine and is rarely detected 57. Schiff base formation may also occur through lysine and arginine residues and is largely responsible for carbonyl-induced protein cross-linking; however both Michael-type and Schiff base adducts may initiate cross-linking. 22 Enzymatic processes capable of removing these adducts remain undiscovered, yet numerous studies have examined the reversibility and stability of Michael-type and Schiff base reactions in vivo and in vitro. 56, 58–61 A recent study by Xiaoxia Tang et al. examined the modification of cytochrome c by 4-HNE and reported that while Lys-4-HNE adducts were observed to be reversible, the 4-HNE-His33 adduct was quite stable. 61 Strategies for stabilizing and isolating carbonylated proteins are valuable tools aiding in the identification of cellular processes impacted under situations of chronic oxidative stress (Figure 2).
Figure 2. Strategies for isolating oxidative posttranslational protein modifications in vitro and in situ.
(A) Biotin hydrazide involves the formation of a reducible hydrazone bond with the carbonyl carbon of Michael-type protein adducts. This method provides specificity for protein carbonyls via avidin-biotin binding, eliminating non-specific binding which may occur with immunoprecipitation techniques. (B) Girard’s P reagent provides an example for modifying existing hydrazide chemistry to enhance specificity and selectivity. The presence of a positively charged quaternary amine in addition to the functional hydrazide allows for additional separation utilizing neutral pH strong cation exchange chromatography. (C) Click Chemistry is a recently developed strategy for isolating protein carbonyls. This method is applied to determine protein targets of reactive aldehyde species. Synthesized alkynyl-4-HNE is utilized to achieve protein carbonylation, which is then reacted with an azido-biotin reagent to form a triazole-linked product. Avidin isolation and subsequent proteomic analysis allows for identification of a broad spectrum of protein targets of 4-HNE.
The de facto method for isolating protein carbonyls is through chemical derivatization. The most common reagent employed is dinitrophenylhydrazine (DNPH), which forms a reducible hydrazone bond with carbonyl groups associated with aldehydes and ketones and is readily detectable via immunodetection. Protein carbonyl content is often determined in this manner with an “Oxyblot”, involving the treatment of cell lysates with DNPH followed by Western blotting using anti-DNPH antibody. A number of methodological variations exist that are based on these carbonyl-trapping hydrazides, including solid-phase hydrazides, biotin hydrazide and aldehyde reactive probes, which are often coupled with gel electrophoresis and MS analysis for protein identification (Figure 2A). 45, 53, 62–71 Once considered to be a highly specific agent, recent evidence questions DNPH specificity for protein carbonyls, as sulfenic acid derived thioaldehydes/aldehydes are also derivatized via DNPH treatment. 72
Another strategy for isolating protein carbonyls involves the application of Girard’s P reagent (GPR); developed initially to derivatize and solubilize insoluble steroids, this reagent also derivatizes oxidized proteins. 73–75 One noted advantage is the presence of both hydrazide and quaternary amine functional groups. While the hydrazide reacts as described above with protein carbonyls, the positively charged amine allows for derivatized protein enrichment via strong cation exchange chromatography (SCX) at neutral pH (Figure 2B). Recently, this technique provided a sensitive and selective method for protein carbonyl detection in yeast proteome stressed by hydrogen peroxide treatments, GPR was coupled to SCX and utilized in a stable-isotope dilution MS assay to identify 41 carbonylated peptides from 36 distinct proteins isolated from yeast proteome. 75
Recently, innovations in identifying protein targets has yielded the method of Click Chemistry, an azide-alkyne Huisgen cycloaddition that is a 1,3-dipolar cycloaddition between an azide and a terminal alkyne to yield a 1,2,3-triazole. 76 Demonstrated as a viable method for in vitro and ex vivo model systems, the addition of an alkynyl-aldehyde derivative (i.e., alkynyl-4-HNE) to cell culture or human plasma provides a method for identifying protein targets of 4-HNE modification. 77, 78 Once proteins are modified by the alkynyl-4-HNE, an azido-biotin agent is then applied to the sample, yielding a protein-4-HNE-alkynyl-azido-biotin derivative, which is readily characterized via avidin pulldown, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and LC-MS analysis (Figure 2C).
3.3. Applications of Gel Electrophoresis
Proteomic analysis requires the separation of complex protein mixtures resulting from cellular extracts and subcellular fractions. A recent report describing the chemical labeling and detection of proteins modified by lipid peroxidation products outlines a number of the more readily available approaches using gel electrophoresis and mass spectrometry. 79 Commonly applied in the detection of protein post-translational modifications (PTMs), one-dimensional and two-dimensional SDS-PAGE (1D-/2D-SDS-PAGE) provides efficient separation of complex protein mixtures for immunodetection, coomassie staining, proteolysis and MS characterization. Evaluating protein modifications via immunodetection and MS analysis is typically achieved through comparative Western blotting. Preferentially performed using 2D-SDS-PAGE immunodetection, an identical coomassie-stained gel is aligned to determine protein spots of interest, which are subsequently excised and digested for MS characterization. This technique enables the identification of covalently modified proteins through immunodetection while confirming protein identity via MS. One drawback of such methods is that the co-migration of immunoreactivity with a particular protein does not unambiguously demonstrate that the protein identified by MS is the same protein as that presenting the lipid-derived modification that is detected immunologically. Unfortunately, the adducted residue is rarely identified using this approach, as sensitivity and specificity may be rate-limiting. Although rare, identification of abundant and/or highly modified proteins does occur, providing in vivo adduct characterization by MS analysis. 11 Furthermore, although this technique is commonly adapted, it is generally misrepresented as evidence supporting the identification of modified proteins. However, it is only reliable for those very few instances in which the identification of the modified protein is established by sequencing that includes detection of the lipid-modified amino acid residue. Oftentimes a complementary in vitro analysis of recombinant protein treated with physiologically relevant concentrations of reactive electrophiles is helpful in confirming protein modification as well as identifying the exact residue of adduction.
3.4. Developments in Mass Spectrometry
Recent reviews have detailed the application of matrix assisted laser desorption time-of-flight (MALDI-TOF) and liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) in proteomic analysis and addressed standard MS techniques analyzing protein covalent modifications. 80, 81 Advances in protein separation and isolation with DNPH derivitization and Click Chemistry continue to enhance protein carbonyl detection. Technical developments in protein separation with two-dimensional liquid chromatography as well as increases in mass spectral resolving power and sensitivity have provided significant advancements in protein carbonyl mapping. Recently, high resolution Fourier Transform MS (LC-FTMS) using an LTQ-Orbitrap instrument was applied to analyze aldehyde modification of bovine serum albumin in vitro and human plasma proteins in vivo. 82 Applying a derivitization method using alkyl chloroformates, the investigators were able to qualitatively detect Michael-type and Schiff base adducts of individual modified amino acids after complete protein hydrolysis, which have the potential to be used as biomarkers for situations of chronic oxidative stress.
While progress in MS sensitivity and resolution has accelerated proteomics research in elucidating the physiological impact of protein carbonylation, the most important development has occurred in peptide fragmentation methodology. MS/MS fragmentation of adducted peptides allows for identifying the exact residue of carbonylation. In general, MS/MS analysis is performed via collision induced dissociation (CID). Unfortunately, CID introduces internal vibrational energy to the peptide to induce peptide backbone fragmentation and may result in a neutral loss of PTMs. CID fragmentation occurs at the least stable backbone amide or adduct bond, characteristically resulting in b- or y-type ions. Frequently, MS/MS fragmentation of peptides containing protein carbonyls results in the neutral loss of the modification, similar to other labile PTMs such as phosphorylation. PTM chemical stabilities and residue interactions vary widely, often rendering it quite difficult to confirm the location of the amino acid modification. In the more recently developed techniques of electron capture dissociation (ECD) and electron transfer dissociation (ETD), MS/MS peptide fragments retain PTMs more efficiently. ECD and ETD provide a more even distribution of fragmentation ions via backbone amine cleavage yielding c- and z-type ions. This presents the most comprehensive solution for complex PTM analysis because unstable modifications are usually retained during the fragmentation process. Recent publications describing ECD/ETD fragmentation for detection of 4-HNE adducts represent a potential paradigm shift in PTM characterization. 83–86
3.5. Computational Biology
Over the last few decades, advances in computational hardware and software have significantly advanced the predictive nature of computational biology. Highly specialized computational software packages such as Discovery Studio (Accelrys Inc., San Diego, CA) and Spartan (Wavefunction, Inc., Irvine, CA) are now widely available to aid in predicting and visualizing proteomic alterations due to reactive electrophiles. One of the many advantages in applying molecular modeling to examining chemical mechanisms of protein carbonylation is the ability to visualize adducted species in a 3-dimensional, physiologically-replicated microenvironment. Additionally, the accumulation of protein crystal structure data and homology modeling has contributed to a continually expanding PDB database (RCSB Protein Data Bank, http://www.rcsb.org) which is often used as a critical resource when modeling the predictive impact of protein carbonylation. Recent publications demonstrate the insights provided through computational modeling. For instance, mechanisms of 4-HNE- and 4-oxononenal (4-ONE)-induced protein crosslinking was elucidated in the study of peroxiredoxin 6. 37 Likewise, other investigators have used proteomics, in conjunction with molecular modeling, to elucidate inhibitory actions of 4-HNE-protein modification of SIRT3 28 and Akt2 29. In another study Wakita et al. employed computational modeling to provide structural insight into the stereoselective formation of 4-HNE-sulfhydryl modifications of a redox-regulated protein through visualizing electrostatic surface potential maps. 87 Furthermore, a recent review details the use of quantum mechanical analysis for the reactive nature of hard and soft electrophiles with potential target protein residues. 30 Importantly, the further integration of computational modeling with proteomics analysis of protein carbonylation will continue to contribute to our understanding of the molecular mechanisms of these reactive aldehydes and their role in altering physiological processes.
4. Challenges and Future Direction
A limited number of well-designed studies employing physiological and non-cytotoxic concentrations of reactive aldehydes have been performed with the goal of identifying specific proteins as targets of carbonyl modification. A modest group of carbonylated proteins are reported with respect to specific biochemical pathways and implications for human health and disease. This is not surprising given the small fraction of a given protein population that is directly modified by reactive biogenic aldehydes and the unstable or transient nature of the protein adducts. Thus, further advancements in isolation techniques and chemistries will be required to locate modified amino acid residues of protein targets in vivo. Finally, to clearly demonstrate the biological significance of such protein modification, it is also essential to develop quantitative analytical protocols that, at a minimum, provide a measure of the relative amounts of modified versus unmodified protein. A significant challenge going forward is the organization, integration and interpretation of information derived from protein carbonyl data sets. Importantly, a collective bioinformatics approach to systems biology will be needed to model and interpret such massive datasets in context of modulation of responsive genes, relevant transcription factors, changes in corresponding protein expression as well as impact on central metabolic pathways. An overview of an integrated approach to data analysis was recently presented 88 demonstrating how a bioinformatics analysis of data derived from cells exposed to 4-HNE revealed changes in gene expression functionally related to transcriptional changes which, in turn, were mechanistically linked with modulation of specific cellular signaling networks. The approach of identifying the impact of protein carbonylation on cell signaling is especially intriguing in that depending on the signaling pathway, small changes in a signaling cascade can markedly attenuate or amplify cellular responses to stimuli. A central goal of studies involving protein carbonylation is identification of protein targets that serve as biomarkers for disease states or adverse cellular responses to chemical toxins. Likewise, changes in protein carbonylation have the potential of providing information about beneficial responses to therapeutic agents. It is clear that cellular responses to protein carbonylation are multifactorial requiring refinement in the current techniques used to capture, identify and quantify carbonylated proteins. Similarly, application of the systems approach to facilitate integration and interpretation of cellular or organismic responses to carbonylated proteins will be essential.
Acknowledgments
Funding Support. This work was supported by the National Institutes of Health Grants 5R37AA009300 and 5R01DK074487 (D.R.P.).
Non-standard Abbreviations
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- MCO
metal catalyzed oxidation
- PUFA
polyunsaturated fatty acid
- GSH
glutathione
- O2·-
superoxide anion
- H2O2
hydrogen peroxide
- CYP2E1
cytochrome P450 2E-1
- HO·
hydroxyl radical
- NO
Nitric oxide
- ONOO−
peroxynitrite anion
- L·
lipid radical
- LOO·
lipid peroxyl radical
- LOOH
lipid hydroperoxide
- LO·
lipid alkoxyl radicals
- 4-HNE
4-hydroxynonenal
- MDA
malondialdehyde
- ACR
acrolein
- DNA
deoxyribonucleic acid
- Keap1
Kelch-like ECH-associated protein 1
- Nrf2
Nuclear factor (erythroid-derived 2)-like 2
- EpRE/ARE
electrophile/antioxidant response element
- HSA
human serum albumin
- MS
mass spectrometry
- DNPH
dinitrophenylhydrazine
- GPR
Girard’s P reagent
- SCX
strong cation exchange
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PTM
post-translational modification
- MALDI-TOF
matrix assisted laser desorption time-of-flight
- LC-ESI-MS/MS
liquid-chromatography electrospray-ionization tandem mass spectrometry
- FTMS
fourier transform mass spectrometry
- CID
collision-induced dissociation
- ETD
electron-transfer dissociation
- ECD
electron-capture dissociation
- PDB
protein databank
- 4-ONE
4-oxononenal.
References
- 1.Sies H. Oxidative Stress II. Oxidants and Antioxidants. London: Academic Press; 1991. [Google Scholar]
- 2.Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 2006;10:389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moller IM, Rogowska-Wrzesinska A, Rao RS. Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J Proteomics. 2011 doi: 10.1016/j.jprot.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Madian AG, Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J. Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albano E. Free radicals and alcohol-induced liver injury. In: Sherman VPaRW., editor. In Ethanol and the Liver. London: Taylor and Francis; 2002. pp. 153–190. [Google Scholar]
- 7.Liochev SI, Fridovich I. The Haber-Weiss cycle -- 70 years later: an alternative view. Redox Rep. 2002;7:55–57. doi: 10.1179/135100002125000190. author reply 59-60. [DOI] [PubMed] [Google Scholar]
- 8.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Pryor WA, Squadrito GL. The chemistry of peroxynitrite: a product from the reaction of nitric oxide with superoxide. Am. J. Physiol. 1995;268:L699–L722. doi: 10.1152/ajplung.1995.268.5.L699. [DOI] [PubMed] [Google Scholar]
- 10.Carr AC, McCall MR, Frei B. Oxidation of LDL by myeloperoxidase and reactive nitrogen species: reaction pathways and antioxidant protection. Arterioscler. Thromb. Vasc. Biol. 2000;20:1716–1723. doi: 10.1161/01.atv.20.7.1716. [DOI] [PubMed] [Google Scholar]
- 11.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 12.Carbone DL, Doorn JA, Kiebler Z, Sampey BP, Petersen DR. Inhibition of Hsp72-mediated protein refolding by 4-hydroxy-2-nonenal. Chem Res Toxicol. 2004;17:1459–1467. doi: 10.1021/tx049838g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lashin OM, Szweda PA, Szweda LI, Romani AM. Decreased complex II respiration and HNE-modified SDH subunit in diabetic heart. Free Radic Biol Med. 2006;40:886–896. doi: 10.1016/j.freeradbiomed.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 14.Luo J, Hill BG, Gu Y, Cai J, Srivastava S, Bhatnagar A, Prabhu SD. Mechanisms of acrolein-induced myocardial dysfunction: implications for environmental and endogenous aldehyde exposure. Am J Physiol Heart Circ Physiol. 2007;293:H3673–H3684. doi: 10.1152/ajpheart.00284.2007. [DOI] [PubMed] [Google Scholar]
- 15.Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res. 1998;39:1529–1542. [PubMed] [Google Scholar]
- 16.Petersen DR, Doorn JA. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radic Biol Med. 2004;37:937–945. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Tanito M, Agbaga MP, Anderson RE. Upregulation of thioredoxin system via Nrf2-antioxidant responsive element pathway in adaptive-retinal neuroprotection in vivo and in vitro. Free Radic. Biol. Med. 2007;42:1838–1850. doi: 10.1016/j.freeradbiomed.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, Yamamoto M, Mann GE. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ. Res. 2004;94:609–616. doi: 10.1161/01.RES.0000119171.44657.45. [DOI] [PubMed] [Google Scholar]
- 19.Iles KE, Dickinson DA, Wigley AF, Welty NE, Blank V, Forman HJ. HNE increases HO-1 through activation of the ERK pathway in pulmonary epithelial cells. Free Radic. Biol. Med. 2005;39:355–364. doi: 10.1016/j.freeradbiomed.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleuranceau-Morel P, Barrier L, Fauconneau B, Piriou A, Huguet F. Origin of 4-hydroxynonenal incubation-induced inhibition of dopamine transporter and Na+/K+ adenosine triphosphate in rat striatal synaptosomes. Neurosci. Lett. 1999;277:91–94. doi: 10.1016/s0304-3940(99)00652-7. [DOI] [PubMed] [Google Scholar]
- 21.Butterfield DA, Lauderback CM. Lipid peroxidation and protein oxidation in Alzheimer's disease brain: potential causes and consequences involving amyloid betapeptide-associated free radical oxidative stress. Free Radic. Biol. Med. 2002;32:1050–1060. doi: 10.1016/s0891-5849(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 22.Siegel SJ, Bieschke J, Powers ET, Kelly JW. The oxidative stress metabolite 4-hydroxynonenal promotes Alzheimer protofibril formation. Biochemistry (Mosc.) 2007;46:1503–1510. doi: 10.1021/bi061853s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sultana R, Perluigi M, Butterfield DA. Protein oxidation and lipid peroxidation in brain of subjects with Alzheimer's disease: insights into mechanism of neurodegeneration from redox proteomics. Antioxid. Redox. Signal. 2006;8:2021–2037. doi: 10.1089/ars.2006.8.2021. [DOI] [PubMed] [Google Scholar]
- 24.Reed TT, Pierce WM, Markesbery WR, Butterfield DA. Proteomic identification of HNE-bound proteins in early Alzheimer disease: Insights into the role of lipid peroxidation in the progression of AD. Brain Res. 2009;1274:66–76. doi: 10.1016/j.brainres.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Fahn S, Cohen G. The oxidant stress hypothesis in Parkinson's disease: evidence supporting it. Ann. Neurol. 1992;32:804–812. doi: 10.1002/ana.410320616. [DOI] [PubMed] [Google Scholar]
- 26.Zarkovic K. 4-hydroxynonenal and neurodegenerative diseases. Mol. Aspects Med. 2003;24:293–303. doi: 10.1016/s0098-2997(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 27.Poli G, Schaur RJ, Siems WG, Leonarduzzi G. 4-hydroxynonenal: a membrane lipid oxidation product of medicinal interest. Med Res Rev. 2008;28:569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 28.Fritz KS, Galligan JJ, Smathers RL, Roede JR, Shearn CT, Reigan P, Petersen DR. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem. Res. Toxicol. 2011;24:651–662. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shearn CT, Fritz KS, Reigan P, Petersen DR. Modification of Akt2 by 4-hydroxynonenal inhibits insulin-dependent Akt signaling in HepG2 cells. Biochemistry (Mosc.) 2011;50:3984–3996. doi: 10.1021/bi200029w. [DOI] [PubMed] [Google Scholar]
- 30.LoPachin RM, Gavin T, Petersen DR, Barber DS. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem Res Toxicol. 2009;22:1499–1508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aldini G, Vistoli G, Regazzoni L, Gamberoni L, Facino RM, Yamaguchi S, Uchida K, Carini M. Albumin is the main nucleophilic target of human plasma: a protective role against pro-atherogenic electrophilic reactive carbonyl species? Chem. Res. Toxicol. 2008;21:824–835. doi: 10.1021/tx700349r. [DOI] [PubMed] [Google Scholar]
- 32.Ishii T, Tatsuda E, Kumazawa S, Nakayama T, Uchida K. Molecular basis of enzyme inactivation by an endogenous electrophile 4-hydroxy-2-nonenal: identification of modification sites in glyceraldehyde-3-phosphate dehydrogenase. Biochemistry (Mosc.) 2003;42:3474–3480. doi: 10.1021/bi027172o. [DOI] [PubMed] [Google Scholar]
- 33.Cai J, Bhatnagar A, Pierce WM., Jr Protein modification by acrolein: formation and stability of cysteine adducts. Chem. Res. Toxicol. 2009;22:708–716. doi: 10.1021/tx800465m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopachin RM, Barber DS, Geohagen BC, Gavin T, He D, Das S. Structure-toxicity analysis of type-2 alkenes: in vitro neurotoxicity. Toxicol. Sci. 2007;95:136–146. doi: 10.1093/toxsci/kfl127. [DOI] [PubMed] [Google Scholar]
- 35.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem. Res. Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 36.Aldini G, Dalle-Donne I, Vistoli G, Maffei Facino R, Carini M. Covalent modification of actin by 4-hydroxy-trans-2-nonenal (HNE): LC-ESI-MS/MS evidence for Cys374 Michael adduction. J. Mass Spectrom. 2005;40:946–954. doi: 10.1002/jms.872. [DOI] [PubMed] [Google Scholar]
- 37.Roede JR, Carbone DL, Doorn JA, Kirichenko OV, Reigan P, Petersen DR. In Vitro and in Silico Characterization of Peroxiredoxin 6 Modified by 4-Hydroxynonenal and 4-Oxononenal. Chem. Res. Toxicol. 2008 doi: 10.1021/tx800244u. [DOI] [PubMed] [Google Scholar]
- 38.Szapacs ME, Riggins JN, Zimmerman LJ, Liebler DC. Covalent adduction of human serum albumin by 4-hydroxy-2-nonenal: kinetic analysis of competing alkylation reactions. Biochemistry (Mosc.) 2006;45:10521–10528. doi: 10.1021/bi060535q. [DOI] [PubMed] [Google Scholar]
- 39.Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer's disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 40.Boyd-Kimball D, Sultana R, Abdul HM, Butterfield DA. Gamma-glutamylcysteine ethyl ester-induced up-regulation of glutathione protects neurons against Abeta(1-42)-mediated oxidative stress and neurotoxicity: implications for Alzheimer's disease. J. Neurosci. Res. 2005;79:700–706. doi: 10.1002/jnr.20394. [DOI] [PubMed] [Google Scholar]
- 41.Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic. Biol. Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 42.Barreiro E, Gea J, Di Falco M, Kriazhev L, James S, Hussain SN. Protein carbonyl formation in the diaphragm. Am. J. Respir. Cell Mol. Biol. 2005;32:9–17. doi: 10.1165/rcmb.2004-0021OC. [DOI] [PubMed] [Google Scholar]
- 43.Aldini G, Gamberoni L, Orioli M, Beretta G, Regazzoni L, Maffei Facino R, Carini M. Mass spectrometric characterization of covalent modification of human serum albumin by 4-hydroxy-trans-2-nonenal. J. Mass Spectrom. 2006;41:1149–1161. doi: 10.1002/jms.1067. [DOI] [PubMed] [Google Scholar]
- 44.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol. Cell. Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin CC, Su TH, Wang TS. Protein carbonylation in THP-1 cells induced by cigarette smoke extract via a copper-catalyzed pathway. Chem. Res. Toxicol. 2009;22:1232–1238. doi: 10.1021/tx900008h. [DOI] [PubMed] [Google Scholar]
- 46.Osna NA, White RL, Todero S, McVicker BL, Thiele GM, Clemens DL, Tuma DJ, Donohue TM., Jr Ethanol-induced oxidative stress suppresses generation of peptides for antigen presentation by hepatoma cells. Hepatology. 2007;45:53–61. doi: 10.1002/hep.21442. [DOI] [PubMed] [Google Scholar]
- 47.Burcham PC, Raso A, Thompson CA. Toxicity of smoke extracts towards A549 lung cells: role of acrolein and suppression by carbonyl scavengers. Chem. Biol. Interact. 2010;183:416–424. doi: 10.1016/j.cbi.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Sarkar P, Hayes BE. Induction of COX-2 by acrolein in rat lung epithelial cells. Mol. Cell. Biochem. 2007;301:191–199. doi: 10.1007/s11010-007-9411-z. [DOI] [PubMed] [Google Scholar]
- 49.Stewart BJ, Roede JR, Doorn JA, Petersen DR. Lipid aldehydemediated cross-linking of apolipoprotein B-100 inhibits secretion from HepG2 cells. Biochim Biophys Acta. 2009;1791:772–780. doi: 10.1016/j.bbalip.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallagher EP, Huisden CM, Gardner JL. Transfection of HepG2 cells with hGSTA4 provides protection against 4-hydroxynonenal-mediated oxidative injury. Toxicol. In Vitro. 2007;21:1365–1372. doi: 10.1016/j.tiv.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarkar P, Hayes BE. Proteomic profiling of rat lung epithelial cells induced by acrolein. Life Sci. 2009;85:188–195. doi: 10.1016/j.lfs.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butterfield DA, Reed T, Sultana R. Roles of 3-nitrotyrosine- and 4-hydroxynonenal-modified brain proteins in the progression and pathogenesis of Alzheimer's disease. Free Radic. Res. 2011;45:59–72. doi: 10.3109/10715762.2010.520014. [DOI] [PubMed] [Google Scholar]
- 53.Chung WG, Miranda CL, Maier CS. Detection of carbonyl-modified proteins in interfibrillar rat mitochondria using N'-aminooxymethylcarbonylhydrazino-D-biotin as an aldehyde/keto-reactive probe in combination with Western blot analysis and tandem mass spectrometry. Electrophoresis. 2008;29:1317–1324. doi: 10.1002/elps.200700606. [DOI] [PubMed] [Google Scholar]
- 54.Sampey BP, Stewart BJ, Petersen DR. Ethanol-induced modulation of hepatocellular extracellular signal-regulated kinase-1/2 activity via 4-hydroxynonenal. J. Biol. Chem. 2007;282:1925–1937. doi: 10.1074/jbc.M610602200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maisonneuve E, Ducret A, Khoueiry P, Lignon S, Longhi S, Talla E, Dukan S. Rules governing selective protein carbonylation. PLoS ONE. 2009;4:e7269. doi: 10.1371/journal.pone.0007269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 57.Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. Oxidative stress and covalent modification of protein with bioactive aldehydes. J Biol Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab. Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 59.Uchida K, Stadtman ER. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra- and intermolecular cross-linking reaction. J. Biol. Chem. 1993;268:6388–6393. [PubMed] [Google Scholar]
- 60.Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol. Aspects Med. 2003;24:149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 61.Tang X, Sayre LM, Tochtrop GP. A mass spectrometric analysis of 4-hydroxy-2-(E)-nonenal modification of cytochrome c. J. Mass Spectrom. 2011;46:290–297. doi: 10.1002/jms.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan RN, Matharoo-Ball B, Shaw RW. Antioxidant enzyme expression, lipid peroxidation, and protein oxidation in human myometrium with parturition. Reprod Sci. 2010;17:78–84. doi: 10.1177/1933719109348027. [DOI] [PubMed] [Google Scholar]
- 63.Fenaille F, Tabet JC, Guy PA. Immunoaffinity purification and characterization of 4-hydroxy-2-nonenal- and malondialdehyde-modified peptides by electrospray ionization tandem mass spectrometry. Anal. Chem. 2002;74:6298–6304. doi: 10.1021/ac020443g. [DOI] [PubMed] [Google Scholar]
- 64.Roe MR, Xie H, Bandhakavi S, Griffin TJ. Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal. Chem. 2007;79:3747–3756. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 65.Soreghan BA, Yang F, Thomas SN, Hsu J, Yang AJ. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm. Res. 2003;20:1713–1720. doi: 10.1023/b:pham.0000003366.25263.78. [DOI] [PubMed] [Google Scholar]
- 66.Chavez J, Wu J, Han B, Chung WG, Maier CS. New role for an old probe: affinity labeling of oxylipid protein conjugates by N'-aminooxymethylcarbonylhydrazino d-biotin. Anal. Chem. 2006;78:6847–6854. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- 67.Wang Q, Zhao X, He S, Liu Y, An M, Ji J. Differential proteomics analysis of specific carbonylated proteins in the temporal cortex of aged rats: the deterioration of antioxidant system. Neurochem. Res. 2010;35:13–21. doi: 10.1007/s11064-009-0023-8. [DOI] [PubMed] [Google Scholar]
- 68.Breusing N, Arndt J, Voss P, Bresgen N, Wiswedel I, Gardemann A, Siems W, Grune T. Inverse correlation of protein oxidation and proteasome activity in liver and lung. Mech. Ageing Dev. 2009;130:748–753. doi: 10.1016/j.mad.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 69.Mirzaei H, Baena B, Barbas C, Regnier F. Identification of oxidized proteins in rat plasma using avidin chromatography and tandem mass spectrometry. Proteomics. 2008;8:1516–1527. doi: 10.1002/pmic.200700363. [DOI] [PubMed] [Google Scholar]
- 70.Han B, Stevens JF, Maier CS. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipidprotein conjugates. Anal. Chem. 2007;79:3342–3354. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 71.Yeh CC, Graham Barr R, Powell CA, Mesia-Vela S, Wang Y, Hamade NK, Austin JH, Santella RM. No effect of cigarette smoking dose on oxidized plasma proteins. Environ. Res. 2008;106:219–225. doi: 10.1016/j.envres.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dalle-Donne I, Carini M, Orioli M, Vistoli G, Regazzoni L, Colombo G, Rossi R, Milzani A, Aldini G. Protein carbonylation: 2,4-dinitrophenylhydrazine reacts with both aldehydes/ketones and sulfenic acids. Free Radic. Biol. Med. 2009;46:1411–1419. doi: 10.1016/j.freeradbiomed.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 73.Johnson DW. A modified Girard derivatizing reagent for universal profiling and trace analysis of aldehydes and ketones by electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:2926–2932. doi: 10.1002/rcm.3175. [DOI] [PubMed] [Google Scholar]
- 74.Mirzaei H, Regnier F. Enrichment of carbonylated peptides using Girard P reagent and strong cation exchange chromatography. Anal Chem. 2006;78:770–778. doi: 10.1021/ac0514220. [DOI] [PubMed] [Google Scholar]
- 75.Mirzaei H, Regnier F. Identification and quantification of protein carbonylation using light and heavy isotope labeled Girard's P reagent. J Chromatogr A. 2006;1134:122–133. doi: 10.1016/j.chroma.2006.08.096. [DOI] [PubMed] [Google Scholar]
- 76.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 77.Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem. Res. Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim HY, Tallman KA, Liebler DC, Porter NA. An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol. Cell. Proteomics. 2009;8:2080–2089. doi: 10.1074/mcp.M900121-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maier WCaC. Identification and Characterization of Oxylipid-Protein and Peptide Conjugates by Mass Spectrometry. Curr. Protoc. Toxicol. 2008;35:17.19.11–17.19.19. doi: 10.1002/0471140856.tx1709s35. [DOI] [PubMed] [Google Scholar]
- 80.Liebler DC. Proteomic approaches to characterize protein modifications: new tools to study the effects of environmental exposures. Environ. Health Perspect. 2002;110 Suppl 1:3–9. doi: 10.1289/ehp.02110s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem. Res. Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pournamdari M, Saadi A, Ellis E, Andrew R, Walker B, Watson DG. Development of a derivatisation method for the analysis of aldehyde modified amino acid residues in proteins by Fourier transform mass spectrometry. Anal. Chim. Acta. 2009;633:216–222. doi: 10.1016/j.aca.2008.11.070. [DOI] [PubMed] [Google Scholar]
- 83.Molina H, Matthiesen R, Kandasamy K, Pandey A. Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal Chem. 2008;80:4825–4835. doi: 10.1021/ac8007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rauniyar N, Stevens SM, Jr, Prokai L. Fourier transform ion cyclotron resonance mass spectrometry of covalent adducts of proteins and 4-hydroxy-2-nonenal, a reactive end-product of lipid peroxidation. Anal. Bioanal. Chem. 2007;389:1421–1428. doi: 10.1007/s00216-007-1534-2. [DOI] [PubMed] [Google Scholar]
- 85.Rauniyar N, Stevens SM, Prokai-Tatrai K, Prokai L. Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Anal. Chem. 2009;81:782–789. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wakita C, Maeshima T, Yamazaki A, Shibata T, Ito S, Akagawa M, Ojika M, Yodoi J, Uchida K. Stereochemical configuration of 4-hydroxy-2-nonenalcysteine adducts and their stereoselective formation in a redox-regulated protein. J. Biol. Chem. 2009;284:28810–28822. doi: 10.1074/jbc.M109.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jacobs AT, Marnett LJ. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res. 2010;43:673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]