Summary
Eukaryotic RNases H2 consist of one catalytic and two accessory subunits. Several single mutations in any one of these subunits of human RNase H2 cause Aicardi-Goutières syndrome. To examine whether these mutations affect complex stability and activity of RNase H2, three mutant proteins of His-tagged Saccharomyces cerevisiae RNase H2 (Sc-RNase H2*) were constructed. Sc-G42S*, Sc-L52R*, and Sc-K46W* contain single mutations in Sc-Rnh2Ap*, Sc-Rnh2Bp*, and Sc-Rnh2Cp*, respectively. The genes encoding three subunits were co-expressed in E. coli and Sc-RNase H2* and its derivatives were purified in a heterotrimeric form. All of these mutant proteins exhibited enzymatic activity. However, only the enzymatic activity of Sc-G42S* was greatly reduced as compared to that of the wild-type protein. Gly42 is conserved as Gly10 in Thermococcus kodakareansis RNase HII (Tk-RNase HII). To analyze the role of this residue, four mutant proteins Tk-G10S, Tk-G10A, Tk-G10L, and Tk-G10P were constructed. All mutant proteins were less stable than the wild-type protein by 2.9–7.6°C in Tm. Comparison of their enzymatic activities, substrate binding affinities, and CD spectra suggest that introduction of a bulky side chain into this position induces a local conformational change, which is unfavorable for both activity and substrate binding. These results indicate that Gly10 is required to make the protein fully active and stable. The findings that the mutations in the accessory subunits of Sc-RNase H2* do not seriously affect the enzymatic activity suggest that the mutant forms of the protein are relatively unstable or interactions with other proteins are perturbed in human cells.
Keywords: Type 2 RNase H, Thermococcus kodakaraensis, Saccharomyces cerevisiae, heterotrimer, site-directed mutagenesis
Introduction
Ribonuclease H (RNase H, E.C. 3.1.26.4) is an enzyme that specifically cleaves the RNA moieties of RNA/DNA hybrids [1]. RNase H is widely present in prokaryotes, eukaryotes, and retroviruses. These RNases H are involved in DNA replication, repair, and transcription [2–8]. Because RNase H activity is required for proliferation of retroviruses, this activity is regarded as one of the targets for AIDS chemotherapy [9]. RNases H have been classified into two major families, Type 1 and Type 2 RNases H, which are evolutionarily unrelated, based on the difference in their amino acid sequences [10–12]. However, according to the crystal structures of Type 1 [13–21] and Type 2 [22–25] RNases H, these RNases H share a common folding motif, termed RNase H-fold, and share a common two-metal-ion catalysis mechanism. According to this mechanism, metal ion A is required for substrate-assisted nucleophile formation and product release, and metal ion B is required to destabilize the enzyme-substrate complex and thereby promote the phosphoryl transfer reaction [18,26,27].
Eukaryotic Type 2 RNases H (RNases H2) are distinguished from prokaryotic ones (RNases HII and HIII) by the subunit structure. Prokaryotic Type 2 RNases H are functional in a monomeric form [25,28], like prokaryotic [13,18,20] and eukaryotic [21] Type 1 RNases H. In contrast, eukaryotic Type 2 RNases H are functional as a complex of three different proteins [29,30]. One of these proteins (catalytic subunit) is a homologue of prokaryotic Type 2 RNase H, in which all of the active-site residues are conserved. Nevertheless, this subunit is active only when it forms a complex with two other accessory proteins. It has been suggested that two accessory proteins are required for correct folding of the catalytic subunit of RNase H2 [29].
It has been shown that certain mutations in any subunit of human RNase H2 cause Aicardi-Goutières syndrome (AGS) [30,31]. AGS is an autosomal recessive genetic disorder, that is phenotypically similar to in utero viral infection, leading to severe neurological defects. RNase H2 deficiency may promote the accumulation of RNA/DNA hybrids in cells, which may induce the innate immunity. Of these mutations, the Gly37→Ser mutation in the catalytic subunit (RNASEH2A) has been shown to greatly reduce enzymatic activity without seriously affecting the stability of the complex [30]. However, it remains to be determined whether other mutations in the accessory proteins (RNASEH2B and RNASEH2C) also reduce enzymatic activity without seriously affecting complex stability. In addition, the reason why the Gly37→Ser mutation in RNASEH2A reduces the enzymatic activity remains to be understood.
Saccharomyces cerevisiae RNase H2 (Sc-RNase H2) consists of one catalytic subunit (Sc-Rnh2Ap) and two accessory subunits (Sc-Rnh2Bp and Sc-Rnh2Cp) like human RNase H2 [29]. It has been overproduced in E. coli in an active form upon co-expression of the genes encoding these subunits [29]. Likewise, Thermococcus kodakaraensis RNase HII (Tk-RNase HII), which represents prokaryotic Type 2 RNases H and shows 30.4% amino acid sequence identity to the catalytic subunit of human RNase H2, has been overproduced in E. coli in an amount sufficient for structural and functional studies [32]. Its crystal structure has been determined [23]. Its stability has been determined thermodynamically [33,34].
In this study, we used Sc-RNase H2 as a model protein to analyze the effect of a disease-causing mutation on the activity and complex stability of human RNase H2. Information on the properties of this S. cerevisiae protein, together with the power of yeast genetics, will aid in both biochemical and functional assays of type 2 RNases H. We also used Tk-RNase HII as a model protein to analyze the role of Gly37 in the catalytic subunit of human RNase H2, which is fully conserved in prokaryotic RNases HII and eukaryotic RNases H2. Because Tk-RNase HII is catalytically active as a single polypeptide, we are able to gain more insight into the effects of the glycine residue near the active site of the protein. We showed that the mutation of the conserved glycine residue to Ser in Sc-Rnh2Ap greatly reduces enzymatic activity without seriously affecting complex stability. In contrast, neither the mutation in Sc-Rnh2Bp nor that in Sc-Rnh2Cp seriously affects enzymatic activity. The role of the conserved glycine residue in the catalytic subunit was further analyzed by constructing a number of the mutant proteins of Tk-RNase HII. Based on these results, we discuss on the structural importance of this glycine residue.
Results and Discussion
Overproduction and purification of Sc-RNase H2
The genes encoding the three subunits of Sc-RNase H2 have previously been co-expressed in an E. coli strain transformed with two plasmids (one for overproduction of one subunit and the other for overproduction of other two subunits) [29]. The complex of these subunits have been partially purified and used to analyze substrate specificity and cleavage-site specificity using various oligomeric substrates. The possibility that host-derived RNases H were co-purified with Sc-RNase H2, has not been completely ruled out. To avoid of this possibility, we used a mutant E. coli strain, MIC2067(DE3), which lacks all functional RNases H for overproduction of Sc-RNase H2. However, because of the limitation of the selection markers, it is difficult to use this strain as a host strain in this system. Therefore, in this study, we constructed plasmid pET-ABC, in which the transcription of the genes encoding all three subunits in a His-tagged form are controlled by the single T7 promoter, to facilitate the preparation of Sc-RNase H2 in an amount sufficient for biochemical characterization. All His-tagged proteins will be marked by asterisks hereafter (for example, Sc-Rnh2Ap* for His-tagged Sc-Rnh2Ap and Sc-RNase H2* for His-tagged Sc-RNase H2).
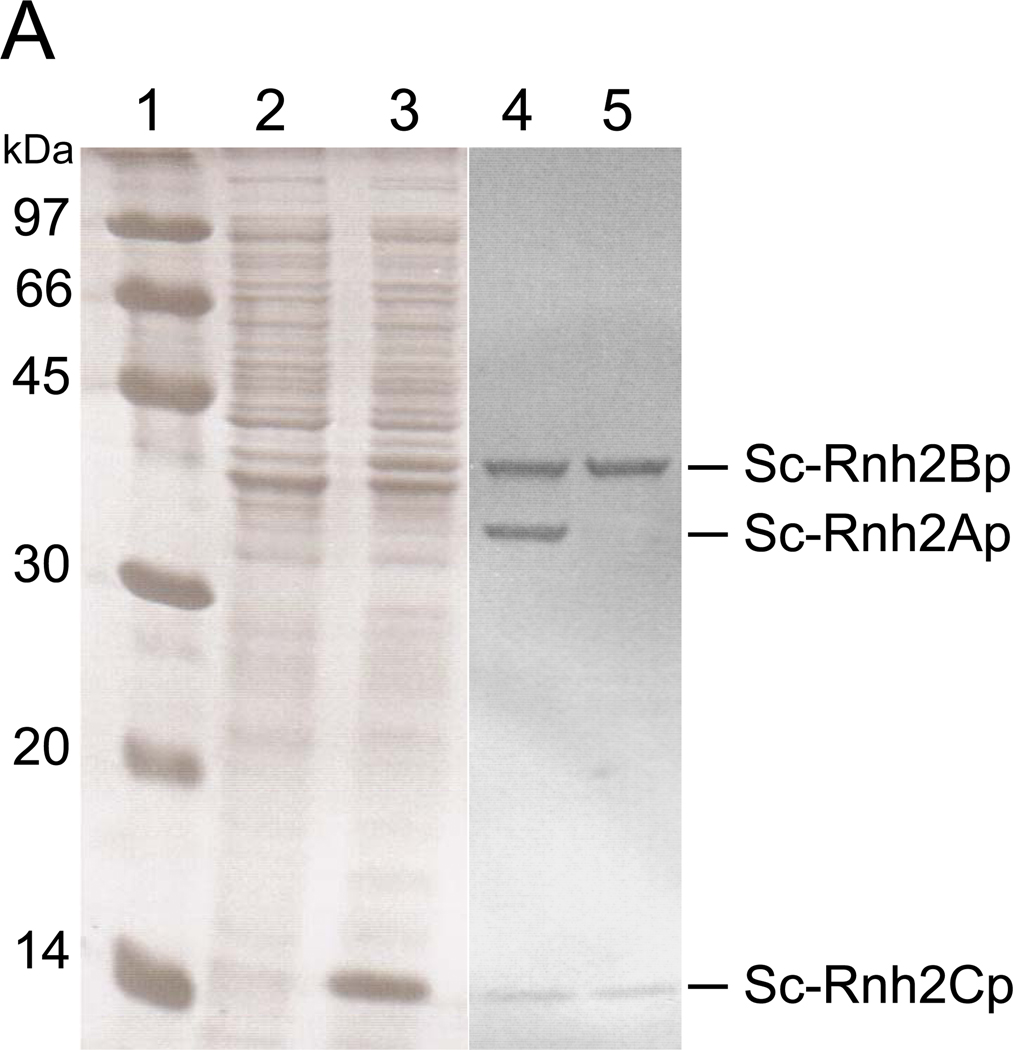
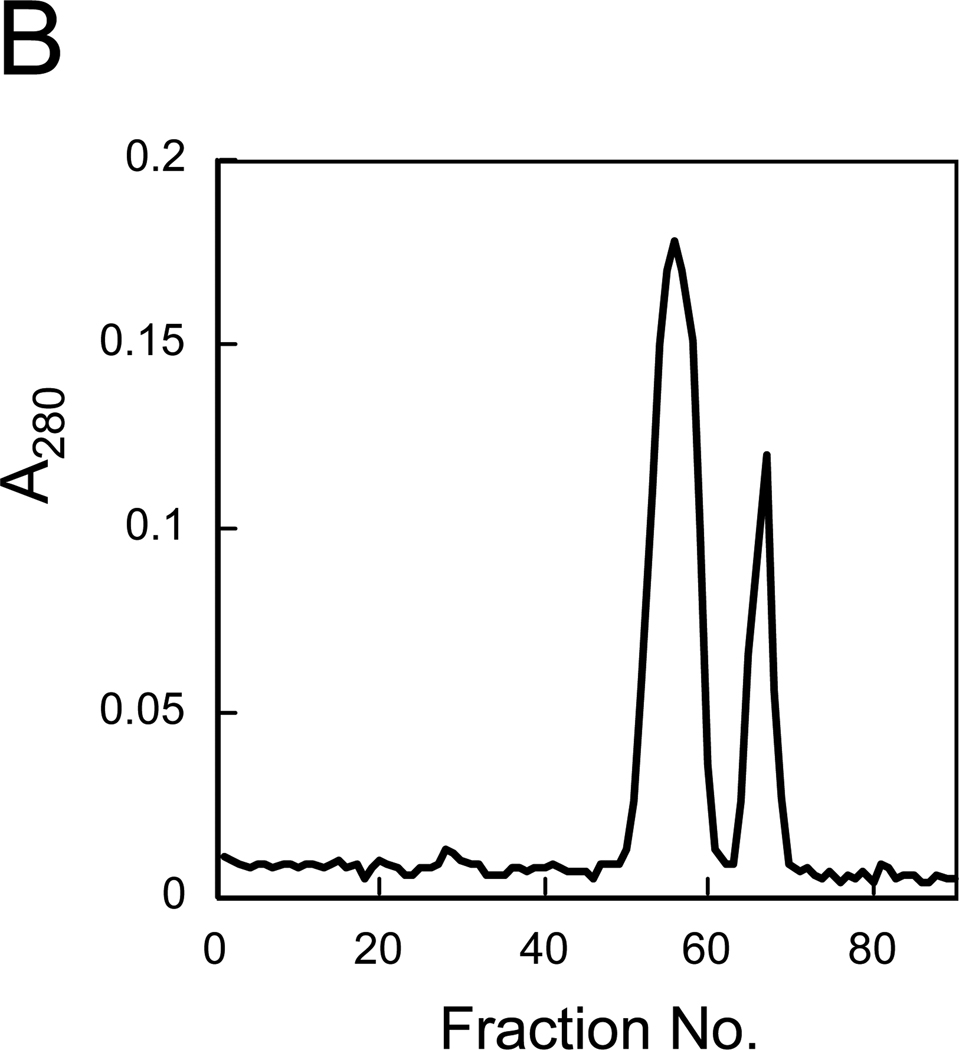
Upon overproduction, only Sc-Rnh2Cp* accumulated in the cells in abundance (Fig. 1A). The production levels of Sc-Rnh2Ap* and Sc-Rnh2Bp* were too low to clearly detect as a band on SDS-PAGE. Disruption of the cells, followed by centrifugation, indicated that Sc-Rnh2Cp* accumulated in the cells mostly in an insoluble form (data not shown). When all His-tagged proteins in a soluble form were purified by a Ni affinity column chromatography and subsequently applied to gel filtration column, two peaks were obtained (Fig. 1B). SDS-PAGE analyses indicated that the first peak consists of three subunits, while the second peak consists of Sc-Rnh2Bp* and Sc-Rnh2Cp* (Fig. 1A). No other peak was detected, suggesting that these proteins accumulate in the cells in a soluble form, only when they form a complex. The molecular masses of these peaks estimated from gel filtration column chromatography are 79 kDa for the first peak, which is slightly lower than but comparable to the sum of the molecular masses of three subunits in a His-tagged form (89,336), and 53 kDa for the second peak, which is comparable to the sum of the molecular masses of Sc-Rnh2Bp* and Sc-Rnh2Cp* (53,638). The molecular masses of three subunits estimated from SDS-PAGE are 36 kDa for Sc-Rnh2Ap*, 41 kDa for Sc-Rnh2Bp*, and 14 kD for Sc-Rnh2Cp*, which are comparable to the calculated ones (35,698 for Sc-Rnh2Ap*, 40,306 for Sc-Rnh2Bp*, and 13,332 for Sc-Rnh2Cp*). The intensities of the bands visualized by CBB staining also support the formation of a heterotrimer and heterodimer. Because only the first peak exhibited RNase H activity, the heterotrimeric complex of Sc-Rnh2Ap*, Sc-Rnh2Bp* and Sc-Rnh2Cp* will be simply designated as Sc-RNase H2*. The amount of Sc-RNase H2* purified from 1 liter culture was roughly 3 mg. The observation that Sc-Rnh2Bp* and Sc-Rnh2Cp* form a complex in the absence of Sc-Rnh2Ap* may suggest that formation of a heterotrimeric structure of Sc-RNase H2* is initiated by the formation of this complex.
Fig. 1.
Purification of Sc-RNase H2*. (A) SDS-PAGE of Sc-RNase H2* overproduced in E. coli cells. The genes encoding three subunits of Sc-RNase H2* were co-expressed using a polycistronic expression system. Samples were subjected to 15% SDS-PAGE and stained with Coomassie Brilliant Blue. Whole cell extracts before (lane 2) and after (lane 3) induction for overproduction, and purified complexes eluted from the gel filtration column as the first (lane 4) and second (lane 5) peaks were analyzed. Lane1, a low molecular weight marker kit (GE Healthcare). Numbers along the gel represent the molecular masses of individual marker proteins. (B) Gel filtration column chromatography of Sc-RNase H2*. The protein eluted from a HiTrap Chelating HP column was applied to a HiLoad 16/60 Superdex 200pg column equilibrated with 20 mM Tris-HCl (pH 8). The flow rate was 0.5 ml/min and fractions of 1 ml were collected.
Enzymatic activity of Sc-RNase H2*
The substrate and cleavage-site specificities of Sc-RNase H2 have previously been analyzed by using various oligomeric substrates, including RNA20/DNA20, DNA12-RNA4-DNA12/DNA28, RNA13-DNA27/DNA40, DNA12-RNA1-DNA27/DNA40, and RNA6-DNA38/DNA40 [29]. However, the metal ion preference, pH-dependence, and salt-dependence remain to be analyzed. In addition, the kinetic parameters for these substrates remain to be determined.
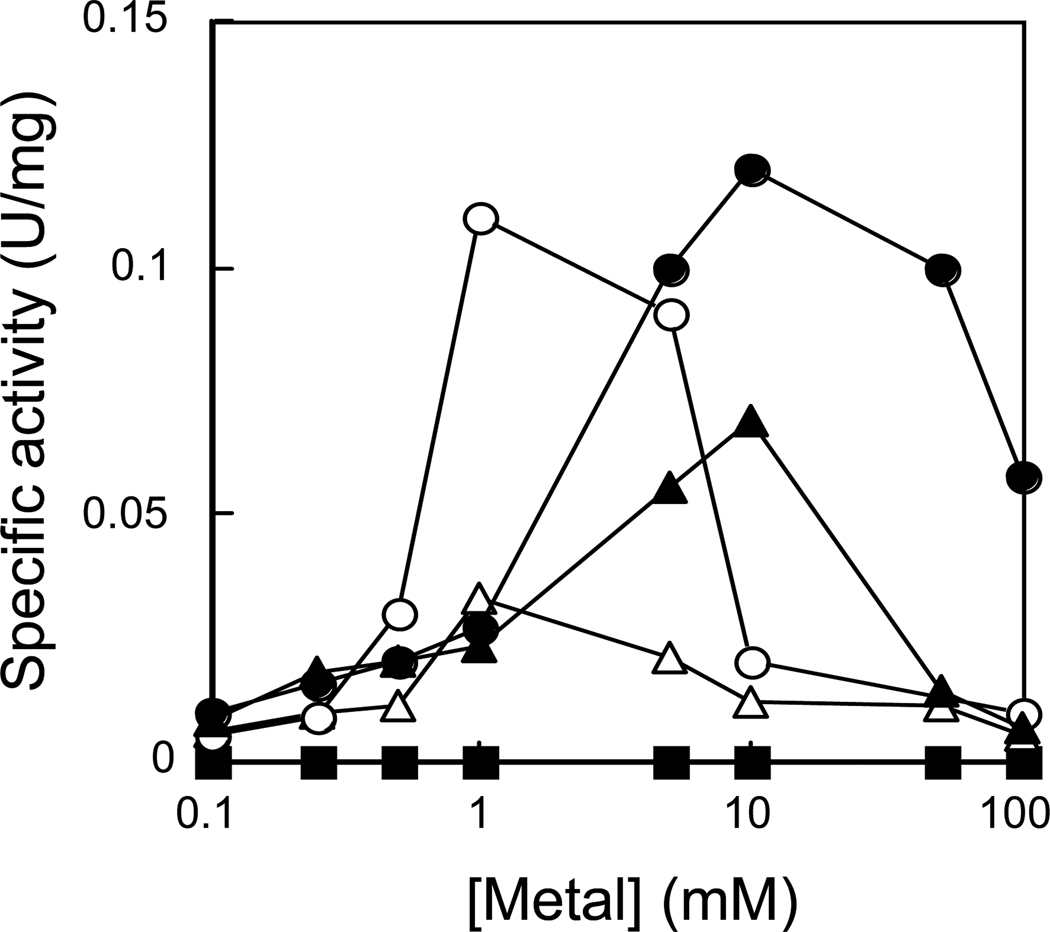
When the enzymatic activity of Sc-RNase H2* was determined in the presence of various concentrations of MgCl2, MnCl2, CoCl2, NiCl2, and CaCl2 at pH 8.0 by using DNA15-RNA1-DNA13/DNA29, which will be designated as [rA]1, as a substrate, Sc-RNase H2* exhibited maximum activity in the presence of 10 mM MgCl2 (Fig. 2A). It exhibited 92, 58, and 28% of the maximum activity in the presence of 1 mM CoCl2, 10 mM MnCl2, and 1 mM NiCl2, respectively, but was inactive in the presence of CaCl2. Enzymatic activity was always greatly reduced when the concentration of the metal ion exceeds the optimum, suggesting that metal ions are inhibitory at high concentrations. The pH- and salt-dependencies of the Sc-RNase H2* activity were analyzed in the presence of 10 mM MgCl2. Like other RNases H, Sc-RNase H2* exhibited enzymatic activity at alkaline pH with optimum pH of 8 (Fig. 2B). It exhibited maximum activity in the presence of 50 mM NaCl (Fig. 2C).
Fig. 2.
Metal ion preference, optimum pH, and optimum salt concentration of RNase H2*. (A) Dependence of Sc-RNase H2* activity on metal ion. The enzymatic activity of Sc-RNase H2* was determined at 30°C in 50 mM Tris-HCl (pH 8) containing 1 mM DTT, 0.01% BSA, and 50 mM NaCl, and various concentrations of MgCl2 (filled circle), CoCl2 (open circle), MnCl2 (filled triangle), NiCl2 (open triangle), and CaCl2 (filled square) using [rA]1 as a substrate. (B) pH-dependence of Sc-RNase H2* activity. The enzymatic activity of Sc-RNase H2* was determined in the presence of 10 mM MgCl2 as described above, except that the buffer was changed to MES (2-molpholinoethanesulfonic acid) (cross), PIPES [piperazine-1,4-bis(ethanesulfonic acid)] (open circle), and Tris-HCl (filled circle). (C) Dependence of Sc-RNase H2* activity on salt concentration. The enzymatic activity of Sc-RNase H2* was determined in the presence of 10 mM MgCl2 as described above, except that the NaCl concentration was changed to 10–200 mM.
Sc-RNase H2* cleaved [rA]1 and DNA13-RNA4-DNA12/DNA29 ([rA]4) most preferably at the RNA-DNA junction (a junction between the 3' side of RNA and 5' side of DNA) and at rA3–rA4 (phosphodiester bond between the third and fourth ribonucleotides), respectively (Fig. 3). These sites are identical with those reported for other similar substrates [29]. It also cleaved RNA29/DNA29 ([rA]29) at multiple sites, as reported for RNA20/DNA20 [29] (Fig. 3). Tk-RNase HII cleaved [rA]1 and [rA]4 at the same sites as Sc-RNase H2* (Fig. 3). It also cleaved [rA]29 at multiple sites, but with a slightly different cleavage-site preference (Fig. 3). The specific activities of Sc-RNase H2* determined at the substrate concentration of 1 µM and 30°C were 0.020 units/mg for [rA]1, 0.021 units/mg for [rA]4, and 0.031 units/mg for [rA]29, while those of Tk-RNase HII were 12 units/mg for [rA]1 and [rA]4, and 11 units/mg for [rA]29. These results indicate that Sc-RNase H2* exhibits very weak enzymatic activity as compared to Tk-RNase HII, but cleaves the substrate containing single ribobucleotide and RNA/DNA hybrid with comparable efficiencies like Tk-RNase HII.
Fig. 3.
Cleavage of 29 bp substrates with Sc-RNase H2* and Tk-RNase HII. The 5’-end labeled [rA]1, [rA]4, and [rA]29 were hydrolyzed by the enzyme at 30°C for 15 min and the hydrolysates were separated on a 20 % polyacrylamide gel containing 7 M urea as described in Experimental procedures. The reaction volume was 10 µl and the substrate concentration was 1.0 µM. Lane 1, no enzyme; lane 2, 10 ng of Sc-RNase H2*; lane 3, 100 ng of Sc-RNase H2*; lane 4, 18 pg of Tk-RNase HII; lane 5, 180 pg of Tk-RNase HII. The sequences of DNA15-RNA1-DNA13 of [rA]1, DNA13-RNA4-DNA12 of [rA]4, and RNA29 of [rA]29 around the cleavage sites are indicated along the gel. The major cleavage sites of [rA]1 and [rA]4 by both enzymes are shown by an arrow. The cleavage sites of [rA]29 are not shown, because this substrate is cleaved by these enzymes at all possible sites between g5 and a14.
Kinetic parameters of Sc-RNase H2* and Tk-RNase HII were determined by using [rA]1 and [rA]4 as a substrate. The cleavage of these substrates with Sc-RNase H2* followed Michaelis-Menten kinetics and the kinetic parameters were determined from a Lineweaver-Burk plot. The results are summarized in Table 1. The Km values of Sc-RNase H2* for both substrates, which were similar with each other, were comparable to those of Tk-RNase HII. In contrast, the kcat values of Sc-RNase H2* for both substrates, which were similar with each other, were lower than those of Tk-RNase HII by approximately 100 fold. These results indicate that the binding affinity of Sc-RNase H2* to substrate is comparable to that of Tk-RNase HII, while the turnover number of Sc-RNase H2* is much lower than that of Tk-RNase HII.
Table 1.
Kinetic parameters of Sc-RNase H2, Tk-RNase HII, and their derivatives. The enzymatic activity was determined at 30°C for 15 min in 50 mM Tris-HCl (pH 8.0) containing 10 mM MgCl2, 1 mM DTT, 50 mM NaCl, and 0.01% BSA using [rA]1, [rA]4, and [rA]29 as a substrate. The specific activities of the proteins for [rA]1 and [rA]4 are not shown, because the relative specific activities of the mutant proteins to that of the parent protein are nearly identical to their relative kcat values. The specific activities of Sc-RNase H2* determined at the substrate concentration of 1 µM are 0.020 units/mg for [rA]1 and 0.021 units/mg for [rA]4, and those of Tk-RNase HII are 12 units/mg for [rA]1 and [rA]4. Errors which represent 67% confidence limits are shown.
| [rA]1 | [rA]4 | [rA]29 | ||||||
|---|---|---|---|---|---|---|---|---|
| Protein |
Km (µM) |
kcat (min−1) |
Relative kcata (%) |
Km (µM) |
kcat (min−1) |
Relative kcata (%) |
Specific activityb (units/mg) |
Relative sp. act.c (%) |
| Sc-RNase H2* | 0.56±0.10 | 2.4±0.29 | 100 | 0.87±0.16 | 2.9±0.50 | 100 | 0.031±0.005 | 100 |
| Sc-G42S* | 0.34±0.06 | 0.003±0.001 | 0.1 | 0.72±0.10 | 0.13±0.002 | 4.5 | 0.004±0.001 | 13 |
| Sc-L52R* | 0.54±0.10 | 2.1±0.32 | 88 | 0.70±0.06 | 2.0±0.20 | 70 | 0.023±0.002 | 74 |
| Sc-K46W* | 0.52±0.05 | 2.1±0.23 | 88 | 0.71±0.09 | 2.0±0.20 | 70 | 0.022±0.002 | 71 |
| Tk-RNase HII | 0.82±0.07 | 270±3.7 | 100 | 0.73±0.06 | 280±3.8 | 100 | 11±0.80 | 100 |
| Tk-G10A | 0.81±0.09 | 270±4.6 | 100 | 0.81±0.09 | 230±8.9 | 87 | 11±0.75 | 100 |
| Tk-G10S | 0.93±0.15 | 25±2.9 | 9.3 | 0.95±0.06 | 110±4.4 | 40 | 2.8±0.53 | 25 |
| Tk-G10L | (<0.01)c | (<0.01)c | <0.001 | <0.01 | ||||
| Tk-G10P | (<0.01)c | (<0.01)c | <0.001 | <0.01 | ||||
The kcat values of the mutant proteins relative to that of the parent protein.
The specific activities were determined at the substrate concentration of 1 µM.
The specific activities of the mutant proteins relative to that of the parent protein.
Construction of mutant proteins of Sc-RNase H2*
The Gly37→Ser mutation is the only disease-causing mutation identified in the catalytic subunit of human RNase H2 (Hs-RNASEH2A) [30,31]. This residue, which is fully conserved in various Type 2 RNase H sequences [11], is conserved as Gly42 in Sc-Rnh2Ap (Fig. 4). To examine whether the mutation of this residue to Ser affects the activity and stability of Sc-RNase H2*, G42S-Rnh2Ap* was constructed. Likewise, L52R-Rnh2Bp* with the Leu52→Arg mutation in Sc-Rnh2Bp* and K46W-Rnh2Cp* with the Lys46→Trp mutation in Sc-Rnh2Cp* were constructed. The corresponding mutations (Leu60→Arg in Hs-RNASEH2B and Arg69→Trp in Hs-RNASEH2C) are not the only disease-causing mutations identified in these subunits. Thirteen single disease-causing mutations have so far been identified in total in Hs-RNASEH2B [30,31]. The parent residues at these mutation sites are well conserved among mammals. However, of these residues, only Leu60 is conserved as Leu52 in Sc-Rnh2Bp. Sc-Rnh2Bp shows a low amino acid sequence identity to Hs-RNASEH2B. However, a sequence motif around this conserved leucine residue is relatively well conserved in these sequences (Fig. 4). Likewise, of the six residues in Hs-RNASEH2C, only Arg69 and Pro76 are conserved as Lys46 and Pro53 in Sc-Rnh2Cp, respectively. Sc-Rnh2Cp also shows low amino acid sequence identity to Hs-RNASEH2C. However, a sequence motif around these conserved arginine and proline residues is relatively well conserved in these sequences (Fig. 4). P53L-Rnh2Cp* with the Pro53→Leu mutation in Sc-Rnh2Cp* was not constructed in this study, because this mutation has only recently been identified as a disease-causing mutation [31].
Fig. 4.
Alignment of the amino acid sequences around the mutation sites. The accession numbers for these sequences are P53942 for Sc-Rnh2Ap, Q05635 for Sc-Rnh2Bp, Q12338 for Sc-Rnh2Cp, O75792 for Hs-RNASEH2A, Q5TBB1 for Hs-RNASEH2B, Q8TDP1 for Hs-RNASEH2C, and AB012613 for Tk-RNase HII. The identical and similar amino acid residues among these sequences are highlighted by black and gray, respectively. The amino acid residues that are mutated in this study are denoted by open inverted triangles. Two of the four active site residues are denoted by filled inverted triangles. The numbers represent the positions of the amino acid residues relative to the initiator methionine for each protein.
The mutant proteins Sc-G42S*, Sc-L52R*, and Sc-K46W*, in which one of the subunits of Sc-RNase H2* is replaced by G42S-Rnh2Ap*, L52R-Rnh2Bp*, and K46W-Rnh2Cp*, respectively, were overproduced in E. coli MIC2067(DE3) using a polycistronic expression system. The production levels of these subunits in the cells and the amount of the mutant proteins of Sc-RNase H2* purified from 1 liter culture were not seriously changed regardless of the loci of the mutations (data not shown). These results indicate that a disease-causing mutation introduced into any subunit does not seriously affect the complex formation or stability. The far-UV CD spectra of these mutant proteins were almost identical to that of Sc-RNase H2* (data not shown), suggesting that these mutations do not seriously affect protein conformation.
Enzymatic activities of mutant proteins of Sc-RNase H2*
To examine whether the Gly37→Ser mutation in Sc-Rnh2Ap*, Leu52→Arg mutation in Sc-Rnh2Bp*, or Lys46→Trp mutation in Sc-Rnh2Cp* affects substrate binding and turnover number of Sc-RNase H2*, the kinetic parameters of Sc-G42S*, Sc-L52R*, and Sc-K46W* for [rA]1 and [rA]4 were determined. The results are summarized in Table 1. The Km values of all mutant proteins for both substrates were comparable to those of Sc-RNase H2*. The kcat values of Sc-L52R* and Sc-K46W* for both substrates were also comparable to those of Sc-RNase H2*, indicating that neither the Leu52→Arg mutation in Sc-Rnh2Bp* nor the Lys46→Trp mutation in Sc-Rnh2Cp* seriously affects substrate binding and turnover number of Sc-RNase H2*. In contrast, the kcat values of Sc-G42S* for both substrates were greatly reduced as compared to those of Sc-RNase H2*, suggesting that this mutation greatly reduces the turnover number of the protein without seriously affecting substrate binding. The specific activity of Sc-G42S* for [rA]29 was also greatly reduced as compared to that of the wild-type protein (Table 1). Nevertheless, Sc-G42S* could complement the RNase H-dependent temperature sensitive growth phenotype of MIC2067(DE3) like Sc-RNase H2* (data not shown), indicating that Sc-G42S* is still functional in vivo. These results are consistent with the finding that the corresponding mutation does not fully inactivate Hs-RNase H2, but greatly reduces its activity [30].
It is noted that Sc-Rnh2Bp* and Sc-Rnh2Cp* show very low amino acid sequence identities to the human counterparts. It may be that the lack of similarity in primary sequence will make studies on the yeast enzyme useful as a model for the human RNase H2 only when the structure of the ABC complex is known. However, it seems unlikely that the mutations corresponding to the Leu52→Arg and Lys46→Trp mutations seriously affect the enzymatic activity of human RNase H2, because the amino acid sequences around these mutation sites are relatively well conserved in both proteins (Fig. 4). The observation that Sc-L52R* and Sc-K46W* are as active as the wild-type protein suggests that great reduction of RNase H2 activity is not the only reason why mutations in the RNase H2 subunits cause AGS.
Construction of mutant proteins of Tk-RNase HII
Four mutant proteins Tk-G10S, Tk-G10A, Tk-G10L, and Tk-G10P were constructed to analyze the role of Gly10 of Tk-RNase HII, which is conserved as Gly37 in Hs-RNASEH2A and Gly42 in Sc-Rnh2Ap (Fig. 4). Tk-G10S was constructed because the corresponding mutation in Hs-RNASEH2A has been identified as one of the disease-causing mutations [30]. Tk-G10A was constructed because Ala has the smallest side chain among all amino acid residues, except Gly. Tk-G10L was constructed because Leu has a bulky hydrophobic side chain. Tk-G10P was constructed because Pro is expected to limit the flexibility of the loop containing Gly10. Upon overproduction, all mutant proteins accumulated in the E. coli cells in a soluble form. Their production levels were similar to that of the wild-type protein. They were purified to give a single band on SDS-PAGE (data not shown). The amount of the protein purified from 1 liter culture was approximately 10 mg for all mutant proteins.
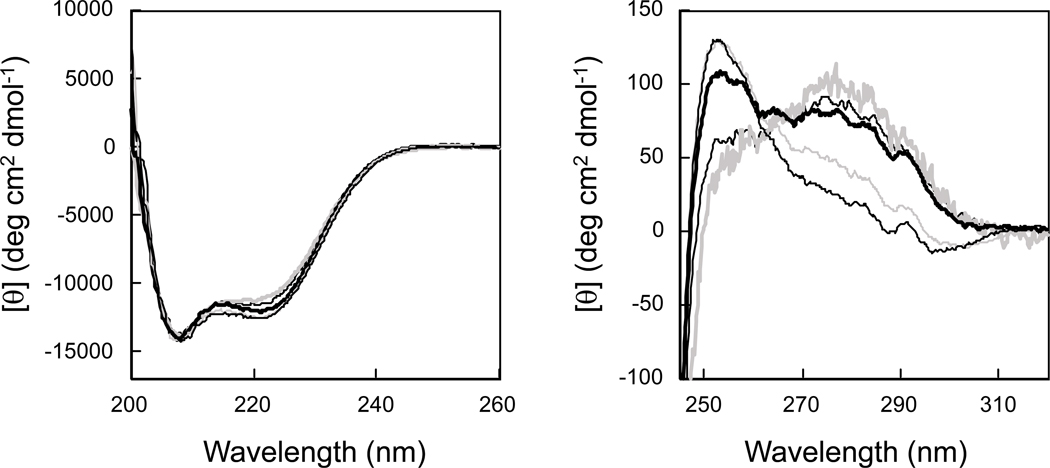
The CD spectra of all mutant proteins in the far-UV region (200–250 nm) were nearly identical to that of the wild-type protein (Fig. 5). On the other hand, the CD spectra in the near-UV region (250–300 nm) varied for different mutant proteins (Fig. 5). The near-UV CD spectrum of Tk-G10A is similar to that of the wild-type protein, which gives a positive peak at around 255 nm. The near-UV CD spectrum of Tk-G10S shows similarity to that of the wild-type protein at <260 nm but is different at >260 nm. The near-UV CD spectra of Tk-G10L and Tk-G10P are different from that of the wild-type protein in the entire region and gives a positive peak at around 275 nm. These spectra show a similarity to that of Tk-G10S at >260 nm. These results suggest that the mutation at Gly10 does not seriously affect the main chain fold of the protein, but affects a local conformation around the mutation site. The extent of this local conformational change seems to increase as the size of the side chain introduced into this position increases (Ala<Ser<pro<Leu).
Fig. 5.
CD spectra. The far-UV (left) and near-UV (right) CD spectra of Tk-RNase HII (thin solid dark line), Tk-G10S (thick solid dark line), Tk-G10A (thin solid gray line), Tk-G10L (thick solid gray line), and Tk-G10P (dashed dark line) are shown. These spectra were measured at pH 8.0 and 20°C as described in Experimental procedures.
Enzymatic activities of mutant proteins of Tk-RNase HII
Enzymatic activities of the mutant proteins were determined by using [rA]1, [rA]4, and [rA]29 as a substrate. Tk-G10A and Tk-G10S cleaved these substrates at the same sites as the wild-type protein (data not shown). In contrast, Tk-G10L and Tk-G10P did not cleave these substrates, suggesting that these mutant proteins are inactive. Tk-G10A and Tk-G10S complemented the RNase H-dependent temperature-sensitive growth phenotype of E. coli MIC2067(DE3), while Tk-G10L and Tk-G10P did not (data not shown). These results indicate that Tk-G10A and Tk-G10S are functional both in vivo and in vitro, while Tk-G10L and Tk-G10P are not functional either in vivo or in vitro.
The kinetic parameters of Tk-G10A and Tk-G10S were determined by using [rA]1 and [rA]4 as a substrate. The results are summarized in Table 1. The Km and kcat values of Tk-G10A were highly similar to those of the wild-type protein for both substrates, indicating that the mutation of Gly10 to Ala does not seriously affect the substrate binding affinity and turnover number of the protein for both substrates. The Km values of Tk-G10S for both substrates were also comparable to those of the wild-type protein. However, The kcat values of Tk-G10S for [rA]1 and [rA]4 were 40 and 10% of those of the wild-type protein. Similar results were obtained for [rA]29. The specific activities of Gly10A and Gly10S for this substrate were also 100 and 25% of that of the wild-type protein (Table 1). These results suggest that the mutation of Gly10 to Ser significantly reduces the turnover number of the protein without seriously affecting the substrate binding affinity.
Binding analyses using BIAcore system
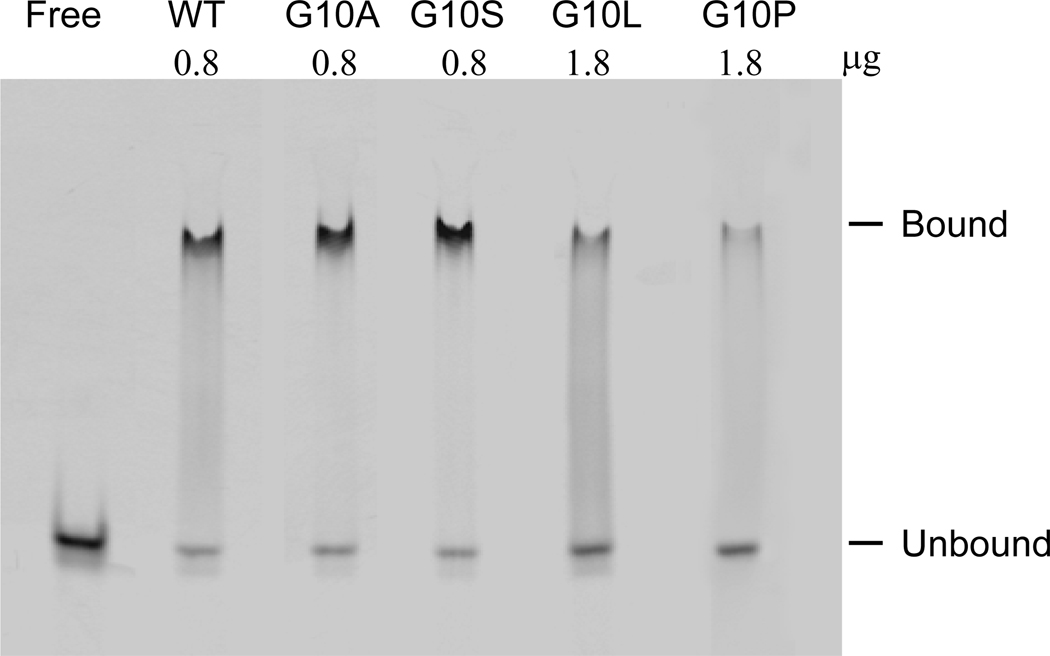
To examine whether the mutation of Gly10 to Leu or Pro affects the substrate binding affinity of the protein, the interaction between the protein and substrates ([rA]1 and [rA]4) were analyzed in the absence of the metal cofactor by BIAcore. The dissociation constants of the proteins estimated from the equilibrium binding level to the substrates are summarized in Table 2. The KD values of Tk-G10A for both substrates were comparable to those of the wild-type protein. The KD values of Tk-G10S were higher than those of the wild-type protein, but only by 5.6 fold for ([rA]1 and 2.5 fold for [rA]4. In contrast, the KD values of Tk-G10L and Tk-G10P were much higher than those of the wild-type protein. The KD values of Tk-G10L, which were slightly higher than those of Tk-G10P for both substrates, were increased by roughly 1000 fold for [rA]1 and 100 folds for [rA]4 as compared to those of the wild-type protein. These results indicate that the binding affinity of the protein to the substrate is not seriously affected or only slightly affected by the mutation of Gly10 to Ala or Ser, but is greatly decreased by that to Leu or Pro.
Table 2.
The KD values for binding of Tk-RNase HII and its derivatives to [rA]1 and [rA]4. Each experiment was carried out in duplicate, and the average value and the error from the average value are shown.
| Protein | KD (µM) | |
|---|---|---|
| [rA]1 | [rA]4 | |
| Tk-RNase HII | 0.055±0.0085 | 0.039±0.0037 |
| Tk-G10A | 0.079±0.0065 | 0.038±0.0053 |
| Tk-G10S | 0.31±0.014 | 0.10±0.007 |
| Tk-G10L | 55±1.4 | 3.7±0.37 |
| Tk-G10P | 41±7.2 | 1.4±0.18 |
It is noted that the Km values of Tk-G10S for these substrates are comparable to those of the wild-type protein, unlike its KD values (Table 1). This disagreement may be caused by the difference in the conditions, in which the interactions between the protein and substrate are analyzed. The Km values were determined in the presence of metal cofactor, while the KD values were determined in the absence of metal cofactor. However, the difference in the KD values between Tk-G10S and wild-type protein is negligible as compared to that between Tk-G10L or Tk-G10P and wild-type protein.
Stabilities of mutant proteins of Tk-RNase HII
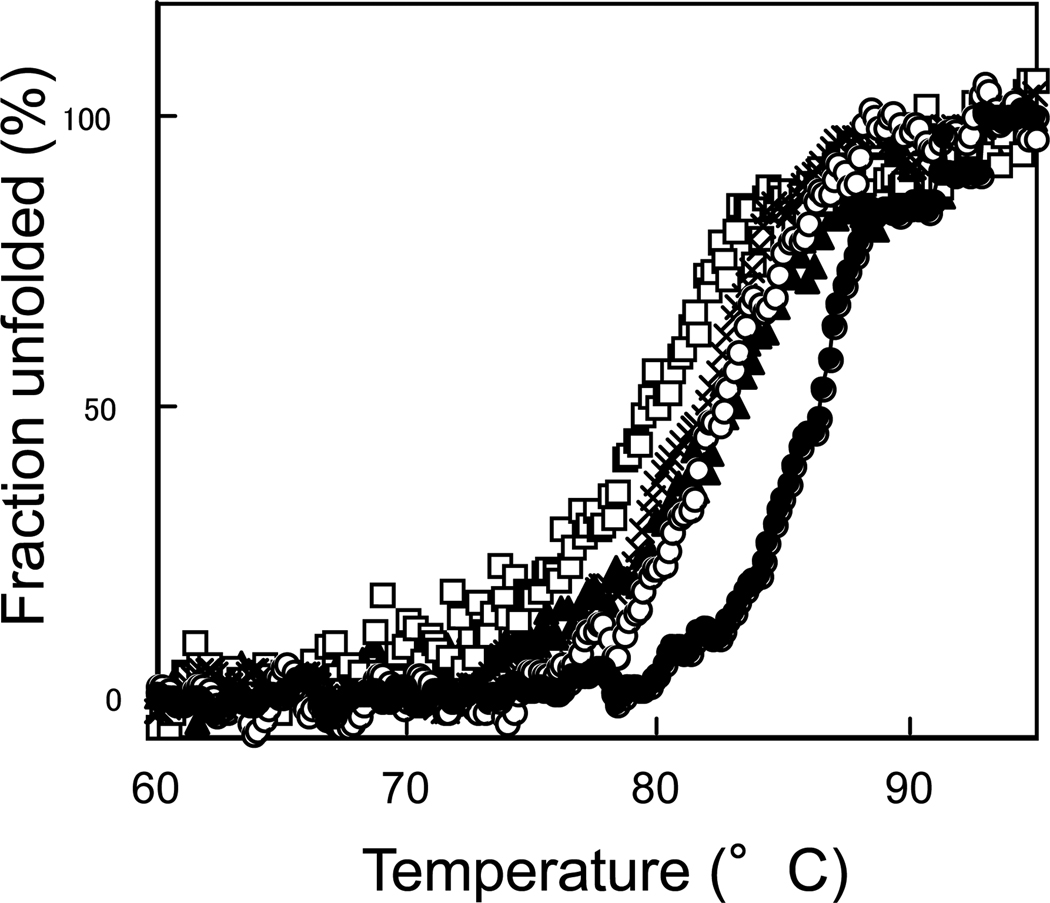
To examine whether the mutation at Gly10 affects the stability of Tk-RNase HII, thermal stabilities of the mutant proteins were determined by monitoring changes of the CD values at 220 nm. At pH 9, all mutant and wild-type proteins unfolded in a single cooperative fashion in a reversible manner. The thermal denaturation curves of the mutant proteins are compared with that of the wild-type protein in Fig. 6. The parameters characterizing the thermal denaturation of the wild-type and mutant proteins are summarized in Table 3. Comparison of these parameters indicate that all mutant proteins are less stable than the wild-type protein by 2.9–7.6°C in Tm and 1.0–2.6 kcal/mol in ΔG. No clear correlation is observed between the size or hydrophobicity of the residue at position 10 and stability, although Tk-G10L with the largest and most hydrophobic side chain at position 10 is most unstable among four mutant proteins.
Fig. 6.
Thermal denaturation curves. Thermal denaturation curves of Tk-RNase HII (filled circle), Tk-G10A (open circle), Tk-G10S (cross), Tk-G10L (open square), and Tk-G10P (filled triangle) are shown. These curves were obtained at pH 9.0 by monitoring the change in the CD value at 220 nm as described in Experimental procedures.
Table 3.
Parameters characterizing the thermal denaturation of Tk-RNase HII and its derivatives. The melting temperature (Tm) is temperature of the mid-point of the thermal denaturation transition. The difference in the melting temperature between the wild-type and mutant proteins (ΔTm) is calculated as Tm (mutant) – Tm (wild-type). ΔHm is the enthalpy change of unfolding at Tm calculated by van’t Hoff analysis. The difference between the free energy change of unfolding of the mutant protein and that of the wild-type protein at Tm of the wild-type protein (ΔΔGm) was estimated by the equation, ΔΔGm = ΔTm × ΔSm(wild-type), where ΔSm(wild-type) is the entropy change of the wild-type protein at Tm. Errors are within ±0.3°C for Tm, ±12 kcal/mol for ΔHm, ±0.04 kcal/mol/K for ΔSm, and ±0.1 kcal/mol for ΔΔGm.
| Protein |
Tm (°C) |
ΔTm (°C) |
ΔHm (kcal/mol) |
ΔSm (kcal/mol/K) |
ΔΔGm (kcal/mol) |
|---|---|---|---|---|---|
| Tk-RNase HII | 87.2 | – | 125.1 | 0.347 | – |
| Tk-G10A | 82.5 | −4.7 | 120.8 | 0.339 | −1.6 |
| Tk-G10S | 81.5 | −5.7 | 114.7 | 0.323 | −1.9 |
| Tk-G10L | 79.6 | −7.6 | 91.1 | 0.258 | −2.6 |
| Tk-G10P | 84.3 | −2.9 | 90.2 | 0.252 | −1.0 |
Role of Gly10 of Tk-RNase HII
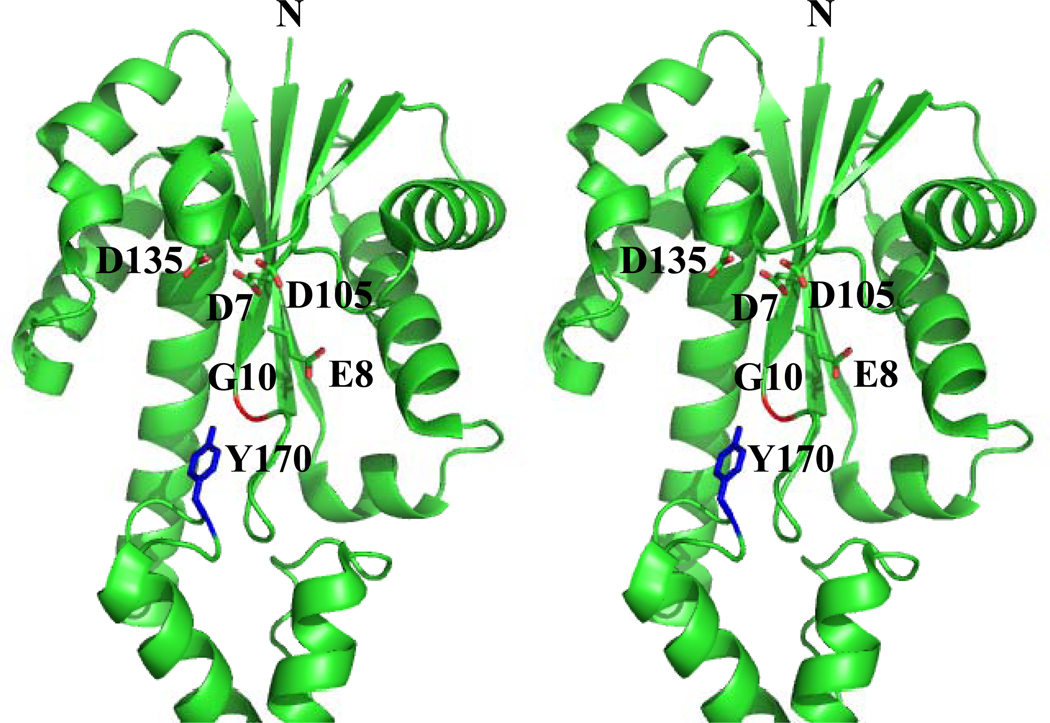
According to the crystal structure of Tk-RNase HII [23], Gly10 is located at the turn region just behind the β1-strand (Fig. 7). The (ϕ, ψ) values of this residue are (80.8°, 43.8°). According to the statistical analysis of the backbone conformational angles by Nicholson et al. [35] and designation by Efimov [36], the backbone conformation of Gly10 in Tk-RNase HII is defined as the left-handed αL conformation. It has been reported that non-glycine residues are energetically unfavorable for left-handed helical conformation because of the local steric interaction of the backbone atoms and the side-chain Cβ atom [35,37–40]. This may be the reason why the mutation at Gly10 destabilizes the protein. Thus, Gly10 contributes to the stabilization of Tk-RNase HII by assuming a left-handed helical structure.
Fig. 7.
Stereoview of the crystal structure of Tk-RNase HII. The side chains of the four acidic active site residues (Asp7, Glu8, Asp105, and Asp135) are shown as stick models, in which the oxygen atoms are shown in red. The side chain of Tyr170 is also shown by blue stick model. The main chain of Gly10 is shown in red. N represents the N terminus. The PDB code for this structure is 1IO2.
Gly10 is also important to make Tk-RNase HII fully active. Tyr170 is located in the vicinity of this residue (Fig. 7). Therefore, it is likely that introduction of a bulky residues into this position forcibly shifts the position of Tyr170 to overcome steric hindrance between these residues. Significant changes in the near-UV CD spectrum of Tk-RNase HII by the mutation of Gly10 to Ser, Leu, and pro supports this possibility. This conformational change may reduce both the substrate binding affinity and turnover number of the protein, because Gly10 is located near the active site and Tyr170 is located at the putative substrate binding site. In fact, Tyr164 of Archaeoglobus fulgidus RNase HII, which corresponds to Tyr170 of Tk-RNase HII, has been reported to be important for substrate binding [24]. Tk-G10A is almost fully active, but is less stable than the wild-type protein, probably because the mutation of Gly10 to Ala neither seriously affects the left-handed backbone structure of this residue nor the local conformation around this residue, as revealed by CD spectra.
Conclusion
In this report, we used Sc-RNase H2* and Tk-RNase HII as model proteins to analyze the effect of a disease-causing mutation on the activity, stability, and structure of human RNase H2 and the role of the glycine residue fully conserved in prokaryotic RNases HII and eukaryotic RNases H2. We showed that Gly10 is required to make Tk-RNase HII fully stable and active. Introduction of the bulky side chains of Pro and Leu in this position causes a significant conformational change around the substrate binding site and active site and thereby inactivates the protein. However, the side chain of Ser seems to be too small to induce a conformational change that is sufficient to inactivate the protein but does result in a great reduction in enzymatic activity. These results suggest that replacement of Gly37 with a bulky amino acid in Hs-RNASEH2A also inactivates human RNase H2. It has been proposed that an individual carrying an inactive mutant protein will exhibit more severe AGS phenotype or is lethal at the early stage of embryonic development [31]. Our results that all mutant proteins of Sc-RNase H2 exhibit at least partial enzymatic activity support the latter possibility. The reason the mutations in the accessory proteins do not seriously affect the activity of Sc-RNase H2 but cause AGS remains to be clarified. The mutant forms of the protein may be relatively unstable or interactions with other proteins are perturbed in human cells.
In this report, we also showed that Sc-RNase H2 is overproduced in E. coli in a heterotrimeric form upon co-expression of the genes encoding three subunit proteins and purified by two column chromatographic procedures. Availability of this overproduction system will facilitate not only the crystallographic studies of Sc-RNase H2 but also its physicochemical studies. These studies will facilitate understanding of the role of the accessory proteins for folding of the catalytic domain.
Experimental procedures
Cells and plasmids
E. coli MIC2067 [F−, λ−, IN(rrnD–rrnE)1, rnhA339::cat, rnhB716::kam] [4] was kindly donated by M. Itaya. E. coli MIC2067(DE3) was constructed by lysogenizing E. coli MIC2067 with λDE3 using a λDE3 Lysogenization Kit (Novagen, Madison, WI, USA). Plasmid pJAL700K containing the Tk-RNase HII gene was previously constructed [32]. Plasmid pVANPH2 containing the Sc-Rnh2Ap gene [5] and plasmids pET279 and pAC154-2 containing the Sc-Rnh2Bp and Sc-Rnh2Cp genes, respectively [29], were previously constructed. Plasmid pET25b was obtained from Novagen. E. coli MIC2067(DE3) transformants were grown in NZCYM medium (Novagen) containing 50 mg/l ampicillin and 0.1 % (w/v) glucose.
Plasmid construction
Plasmid pET-ABC for polycistronic expression of the genes encoding Sc-Rnh2Ap*, Sc-Rnh2Bp*, and Sc-Rnh2Cp* was constructed as described below. The DNA fragments containing the genes encoding Sc-Rnh2Ap*, Sc-Rnh2Bp*, and Sc-Rnh2Cp* were amplified by PCR using the combinations of the primers RNH2A-F and RNH2A-R, RNH2B-F and RNH2B-R, and RNH2C-F and RNH2C-R, respectively. Plasmids pVANPH2, pET279, and pAC154-2 were used as a template. The sequences of these primers are 5’-ATTATCATATGGGTACCCCCACGG-3’ f o r R N H 2 A-F, 5’-TGTGGAATTCAGTGGTGGTGGTGGTGGTGCCGGTACCAATTATCTAGGG-3’ f o r R N H 2 A-R, 5’-ATATGAATTCTCTCTAAGGAGATATACTTATGACCGTTTCCAACATTGGG-3’ f o r R N H 2 B-F, 5’-GGGGAAGCTTCTAGTGGTGGTGGTGGTGGTGCTTACGTTTAAAAAATCCATC-3’ f o r R N H 2 B-R, 5’-ATATAAGCTTCTCTCAAGGAGATATACTTATGACCAAAGATGCCGTG-3’ for RNH2C-F, and 5’-GGAGCTCGAGTTAGTGGTGGTGGTGGTGGTGCTGATTTATGACATCGATGAGG-3’ for RNH2C-R. In these sequences, underlined bases show the positions of the NdeI (for RNH2A-F), EcoRI (for RNH2A-R and RNH2B-F), HindIII (for RNH2B-R and RNH2C-F), and XhoI (for RNH2C-R) sites, double underlined bases show the region of the gene encoding a histidine tag, italic bases show the position of the ribosome binding site, and bold-faced bases show the initiation codon for translation. The resultant DNA fragments containing the Sc-Rnh2Ap*, Sc-Rnh2Bp*, and Sc-Rnh2Cp* genes were sequentially ligated into the NdeI-EcoRI, EcoRI-HindIII, and HindIII-XhoI sites of pET25b and its derivatives, respectively, one by one in this order, to generate pET-ABC. In pET-ABC, the Sc-Rnh2Ap*, Sc-Rnh2Bp*, and Sc-Rnh2Cp* genes are located between the NdeI and EcoRI sites, between the EcoRI and HindIII site, and between the HindIII and XhoI sites, respectively.
Plasmid pET700K for overproduction of Tk-RNase HII was constructed by ligating the 700 bp DNA fragment into the NdeI-EcoRI sites of pET25b. This DNA fragment was amplified by PCR using pJAL700K as a template. The sequences of the PCR primers are 5’-ATATTCATATGAAGATAGCGGGCATTGACGAGGC-3’for 5’ primer and 5’-ATTATAGAATTCTCACTTTTTCTCGCTCTCAACTTTTTC-3’ for 3’ primer, where underlined bases show the positions of the NdeI (5’-primer) and EcoRI (3’-primer) sites.
PCR was performed with a GeneAmp PCR system 2400 (PerkinElmer, Tokyo, Japan) using a KOD DNA polymerase (Toyobo, Kyoto, Japan) according to the producers recommended by the supplier. All DNA oligomers for PCR were synthesized by Hokkaido System Science (Sapporo, Japan). The DNA sequence was confirmed with a Prism 310 DNA sequencer (GE Healthcare, Tokyo, Japan).
Site-directed mutagenesis
Site-directed mutagenesis was carried out by the PCR overlap extension method [41] for the mutations of the Sc-RNase H2 genes or by PCR using a long 5'- mutagenic primer, instead of 5'-primer, for the mutations of the Tk-RNase HII gene. Plasmids pET-ABC and pET-700K were used as a template. The mutagenic primers were designed such that the GGC codon for Gly42 of Sc-Rnh2Ap is changed to AGT for Ser, CTT codon for Leu52 of Sc-Rnh2Bp is changed to CGC for Arg, AAG codon for Lys46 of Sc-Rnh2Cp is changed to TGG for Trp, and GGG codon for Gly10 of Tk-RNase HII is changed to GCG for Ala, TCG for Ser, CTC for Leu, and CCG for Pro. PCR was carried out as mentioned above and the nucleotide sequences were confirmed by a Prism 310 DNA sequencer (Perkin-Elmer). All oligonucleotides were synthesized by Hokkaido System Science.
Overproduction and purification
For overproduction of Sc-RNase H2* and its derivatives, E. coli MIC2067(DE3) was transformed with pET-ABC and its derivatives and grown at 32°C. When the absorbance at 600 nm reached around 0.5, 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) was added to the culture medium and cultivation was continued for an additional 3 hours. Cells were harvested by centrifugation at 8000 rpm for 10 min, suspended in 10 mM sodium phosphate (pH 7.4) containing 30 mM imidazol and 0.5 M NaCl, disrupted by French press, and centrifuged at 30,000 g for 30 min. The supernatant was collected and applied to a HiTrap Chelating HP column (5 ml, GE Healthcare) equilibrated with the same buffer. The protein was eluted from the column with a linear gradient from 30 to 500 mM imidazole in 10 mM sodium phosphate (pH 7.4) containing 0.5 M NaCl. The fractions containing the protein were collected, dialyzed against 20 mM Tris-HCl (pH 8), and applied to a HiLoad 16/60 Superdex 200pg column (GE Healthcare) equilibrated with the same buffer. The flow rate was 0.5 ml/min. The fractions containing the protein were collected and used for further analyses. The molecular mass of the protein was estimated by this gel filtration column chromatography using bovine serum albumin (BSA) (67 kDa), ovalbumin (44 kDa), chymotrypsinogen A (25 kDa), and RNase A (14 kDa) as standard proteins. All the purification procedures were carried out at 4°C.
For overproduction of Tk-RNase HII and its derivatives, E. coli MIC2067(DE3) was transformed with pET700K and its derivatives. Overproduction was carried out as described above for Sc-RNase H2* and its derivatives. Sonication lysis of the cells and purification of the protein were carried out as described previously [32].
The purity of the protein was analyzed by SDS-PAGE on a 15 % polyacrylamide gel [42], followed by staining with Coomassie Brilliant Blue. The protein concentration was determined from UV absorption using an A280 value of 0.1% solution of 0.56 cm−1 for Tk-RNase HII and its variants, 0.94 for Sc-RNase H2*, Sc-G42S*, and Sc-L52R*, and 1.01 for Sc-K46W*. These values were calculated by using ε of 1576 M−1cm−1 for Tyr and 5225 M−1cm−1 for Trp at 280 nm [43].
Enzymatic activity
The RNase H activity was determined by using 29 bp RNA29/DNA29 ([rA]29), DNA13-RNA4-DNA12/DNA29 ([rA]4), and DNA15-RNA1-DNA13/DNA29 ([rA]1) as a substrate. These oligomeric substrates were prepared by hybridizing 1 µM of the 5’-FAM-l abeled 29 base DNA13-RNA4-DNA12 (5’-AATAGAGAAAAAGaaaaAAGATGGCAAAG-3’) and DNA15-RNA1-DNA13 (5’-AATAGAGAAAAAGAAaAAAGATGGCAAAG-3’) with 1.5 molar equivalent of the complementary DNA, respectively, as described previously [10]. In these sequences, DNA and RNA are represented by uppercase and lowercase letters, respectively. FAM represents 6-carboxyfluorescein. All oligonucleotides were synthesized by Hokkaido System Science. Hydrolysis of the substrate at 30°C for 15 min and separation of the products on a 20% polyacrylamide gel containing 7 M urea were carried out as described previously [10]. The reaction buffer was 50 mM Tris-HCl (pH 8) containing 10 mM MgCl2, 1 mM DTT, 0.01% BSA, and 50 mM NaCl, and substrate concentration was 1 µM. The products were detected by Typhoon 9240 Imager (GE Healthcare) and identified by comparing their migration on the gel with those of the oligonucleotides generated by partial digestion of 5’-FAM-labeled 29 base D13-R4-D12 or D15-R1-D13 with Crotalus atrox phosphodiesterase (Sigma, Tokyo, Japan) [44]. One unit is defined as the amount of enzyme degrading 1 µmol of the substrate per min at 30°C. The specific activity was defined as the enzymatic activity per milligram of protein. For determination of the kinetic parameters, the substrate concentration was varied from 0.25 to 2.0 µM. The protein concentration was 6.1 nM for Sc-RNase H2*, Sc-L52R*, and Sc-K46W*, 50 nM for Sc-G42S*, 0.08 nM for Tk-RNase HII and Tk-G10A, and 0.8 nM for Tk-G10S.
Binding analysis of proteins to RNA/DNA hybrid
Binding of the proteins to the substrate was analyzed using the Biacore X instrument (Biacore, Uppsala, Sweden). Twenty-nine bp [rA]1 and [rA]4 were prepared so that the RNA strand was biotinylated at the 5'-end. These substrates were immobilized on the SA sensor chip (BIAcore), on which streptavidin is covalently linked, by injecting 20 µl of TBS buffer (10 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.005% Tween P20, pH 8.0) containing 100 nM of biotinylated [rA]1 and [rA]4. The proteins were dissolved in TBS buffer and injected at 25°C at a flow rate of 50 µl/min onto the sensor chip surface on which [rA]1 or [rA]4 has been immobilized. Binding surfaces were regenerated by washing with 2 M NaCl.
To determine the dissociation constant, KD, the concentration of the protein injected onto the sensor chip was varied from 20 to 100 nM for Tk-RNase HII and Tk-G10A, 30–150 nM for Tk-G10S, and 20–100 µM for Tk-G10L and Tk-G10P. From the plot of the equilibrium binding responses as a function of the concentrations of the proteins, the KD value was determined using steady state affinity program of BIAevaluation Software (Biacore).
Circular dichroism (CD) spectra
The CD spectra were measured on a J-725 spectropolarimeter (Japan Spectroscopic, Tokyo, Japan) at 20°C. The far-UV CD spectra were obtained using solutions containing protein at 0.1 mg/ml in 20 mM Tris-HCl (pH 8.0) in a cell with an optical path length of 2 mm. For near-UV CD spectra, the protein concentration and optical path length were increased to 0.5 mg/ml and 10 mm, respectively. The mean residue ellipticity, θ, which has the units of deg cm2 dmol−1, was calculated by using an average amino acid molecular weight of 110.
Thermal denaturation
Thermal denaturation curves Tk-RNases HII and its derivatives were obtained by monitoring the change in CD values at 220 nm as the temperature was increased. The proteins were dissolved in 20 mM Tris-HCl (pH 9.0). The protein concentration and optical path length were 0.1 mg/ml and 2 mm, respectively. The temperature of the protein solution was linearly increased by approximately 1.0°C/min. The thermal denaturation of these proteins was reversible under this condition. The temperature of the midpoint of the transition, Tm, was calculated from curve fitting of the resultant CD values versus temperature data on the basis of a least squares analysis. The enthalpy (ΔHm) and entropy (ΔSm) changes for thermal denaturation at Tm were calculated by van’t Hoff analysis. The difference in the free energy change of unfolding between the wild-type and mutant proteins at the Tm of the wild-type protein (ΔΔGm) was estimated by the relationship, ΔΔGm = ΔTm ΔSm [45], where ΔTm is the change in ΔTm of a mutant protein relative to that of the wild-type protein and ΔSm is the entropy change of the wild-type protein at the Tm.
Acknowledgments
We thank Drs. T. Tadokoro and A. Mukaiyama for helpful discussions. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas "Systems Genomics" from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by an Industrial Technology Research Grant Program from the New Energy and Industrial Technology Development Organization (NEDO) of Japan, and also by the Intramural Research Program of the Eunice Kennedy Shriver National Institutes of Child Health and Human Development, NIH.
Abbreviations
- RNase H
ribonuclease H
- Sc-RNase H2
RNase H2 from S. cerevisiae
- Tk-RNase HII
RNase HII from T. kodakareansis
- [rA]29
RNA29/DNA29
- [rA]4
DNA13-RNA4-DNA12/DNA29
- [rA]1
DNA15-RNA1-DNA13/DNA29
- CD
circular dichroism
- bp
base pair
proteins marked by asterisk (*), proteins with a His-tag
References
- 1.Crouch RJ, Dirksen ML. Ribonuclease H. In: Linn SM, Roberts RJ, editors. Nuclease. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1982. pp. 211–241. [Google Scholar]
- 2.Kogoma T, Foster PL. Physiological functions of E. coli RNase HI. In: Crouch RJ, Toulme JJ, editors. Ribonucleases H. Paris: INSERM; 1998. pp. 39–66. [Google Scholar]
- 3.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itaya M, Omori A, Kanaya S, Crouch RJ, Tanaka T, Kondo K. Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J. Bacteriol. 1999;181:2118–2123. doi: 10.1128/jb.181.7.2118-2123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5:789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- 6.Haruki M, Tsunaka Y, Morikawa M, Kanaya S. Cleavage of a DNA-RNA-DNA/DNA chimeric substrate containing single ribonucleotide at the DNA-RNA junction with prokaryotic RNases HII. FEBS Lett. 2002;531:204–208. doi: 10.1016/s0014-5793(02)03503-2. [DOI] [PubMed] [Google Scholar]
- 7.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci USA. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 9.Hughes SH, Arnold E, Hostomsky Z. RNase H of retroviral reverse transcriptases. In: Crouch RJ, Toulme JJ, editors. Ribonucleases H. Paris: INSERM; 1998. pp. 195–224. [Google Scholar]
- 10.Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: Classification of RNases H into three families. Biochemistry. 1999;38:605–618. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- 11.Ohtani N, Haruki M, Morikawa M, Kanaya S. Molecular diversity of RNases H. J Biosci Bioeng. 1999;88:12–19. doi: 10.1016/s1389-1723(99)80168-6. [DOI] [PubMed] [Google Scholar]
- 12.Ohtani N, Yanagawa H, Tomita M, Itaya M. Cleavage of double-stranded RNA by RNase HI from a thermoacidophilic archaeon, Sulfolobus tokodaii 7. Nucleic Acids Res. 2004;32:5809–5819. doi: 10.1093/nar/gkh917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M, Matsuzaki T, Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990;347:306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- 14.Yang W, Hendrickson WA, Crouch RJ, Satow Y. Structure of ribonuclease H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 15.Davies JF, Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 16.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa K, Okumura M, Katayanagi K, Kimura S, Kanaya S, Nakamura H, Morikawa K. Crystal structure of ribonuclease HI from Thermus thermophilus HB8 refined at 2.8 Å resolution. J Mol Biol. 1993;230:529–542. doi: 10.1006/jmbi.1993.1169. [DOI] [PubMed] [Google Scholar]
- 18.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 19.Lim D, Gregorio GG, Bingman C, Martinez-Hackert E, Hendrickson WA, Goff SP. Crystal structure of the moloney murine leukemia virus RNase H domain. J Virol. 2006;80:8379–8389. doi: 10.1128/JVI.00750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You D-J, Chon H, Koga Y, Takano K, Kanaya S. Crystal structure of type 1 RNase H from hyperthermophilic archaeon Sulfolobus tokodaii: role of Arg118 and C-terminal anchoring. Biochemistry. 2007;46:11494–11503. doi: 10.1021/bi700830f. [DOI] [PubMed] [Google Scholar]
- 21.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Lai L, Yokota H, Hung L-W, Kim R, Kim S-H. Crystal structure of archeal RNase HII: a homologue of human major RNase H. Structure. 2000;8:897–904. doi: 10.1016/s0969-2126(00)00179-9. [DOI] [PubMed] [Google Scholar]
- 23.Muroya A, Tsuchiya D, Ishikawa M, Haruki M, Morikawa M, Kanaya S, Morikawa K. Catalytic center of archaeal type 2 ribonuclease H as revealed by X-ray crystallographic and mutational analyses. Protein Sci. 2000;10:707–714. doi: 10.1110/ps.48001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapados BR, Chai Q, Hosfield DJ, Qiu J, Shen B, Tainer JA. Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J Mol Biol. 2001;307:541–556. doi: 10.1006/jmbi.2001.4494. [DOI] [PubMed] [Google Scholar]
- 25.Chon H, Matsumura H, Koga Y, Takano K, Kanaya S. Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N-terminus. J Mol Biol. 2005;356:165–178. doi: 10.1016/j.jmb.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 26.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Kanaya S. Prokaryotic type 2 RNases H. Methods Enzymol. 2001;341:377–394. doi: 10.1016/s0076-6879(01)41165-7. [DOI] [PubMed] [Google Scholar]
- 29.Jeong HS, Backlund PS, Chen H-C, Karavanov AA, Crouch RJ. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 2004;32(2):407–414. doi: 10.1093/nar/gkh209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Jackson AP, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 31.Rice G, Patrick T, Parmer R, Taylor CF, Aeby A, Aicardi J, Crow YJ, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am J Hum Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haruki M, Hayashi K, Kochi T, Muroya A, Koga Y, Morikawa M, Imanaka T, Kanaya S. Gene cloning and characterization of recombinant RNase HII from hyperthermophilic archaeon. J Bacteriol. 1998;180:6207–6214. doi: 10.1128/jb.180.23.6207-6214.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukaiyama A, Takano K, Haruki M, Morikawa M, Kanaya S. Kinetically robust monomeric protein from hyperthermophile. Biochemistry. 2004;43:13859–13866. doi: 10.1021/bi0487645. [DOI] [PubMed] [Google Scholar]
- 34.Mukaiyama A, Haruki M, Ota M, Koga M, Takano K, Kanaya S. A hyperthermophilic protein acquires function at the cost of stability. Biochemistry. 2006;45:12673–12679. doi: 10.1021/bi060907v. [DOI] [PubMed] [Google Scholar]
- 35.Nicholson H, Soderlind E, Tronrud DE, Matthews BW. Contributions of left-handed helical residues to the structure and stability of bacteriophage T4 lysozyme. J Mol Biol. 1989;210:181–193. doi: 10.1016/0022-2836(89)90299-4. [DOI] [PubMed] [Google Scholar]
- 36.Efimov AV. Standard conformations of a polypeptide chain in irregular protein regions. (Russian) Mol Biol (Moskow) 1986;20:250–260. [PubMed] [Google Scholar]
- 37.Kimura S, Kanaya S, Nakamura H. Thermostabilization of Escherichia coli ribonuclease HI by replacing left-handed helical Lys95 with Gly or Asn. J Biol Chem. 1992;267:22014–22017. [PubMed] [Google Scholar]
- 38.Ishikawa K, Kimura S, Kanaya S, Morikawa K, Nakamura H. Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability. Protein Eng. 1993;6:85–91. doi: 10.1093/protein/6.1.85. [DOI] [PubMed] [Google Scholar]
- 39.Takano K, Yamagata Y, Yutani K. Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein. Proteins. 2001;45:274–280. doi: 10.1002/prot.1147. [DOI] [PubMed] [Google Scholar]
- 40.Pulido M, Tanaka S, Sringiew C, You D-J, Matsumura H, Koga Y, Takano K, Kanaya S. Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses. J Mol Biol. 2007;374:1359–1373. doi: 10.1016/j.jmb.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 41.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 42.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Goodwin TW, Morton RA. The spectrophotometric determination of tyrosine and tryptophan in proteins. Biochem J. 1946;40:628–632. doi: 10.1042/bj0400628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jay E, Bambara R, Padmanabham P, Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974;1:331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]