Abstract
Phenobarbital (PB), a non-genotoxic carcinogen, activates the nuclear constitutive active/androstane receptor (CAR), resulting in the transcriptional induction or repression of various hepatic genes. We previously demonstrated that liver tumors developed following chronic PB treatment only when CAR is present. In order to understand the molecular mechanism of tumor promotion, cDNA microarray analysis was performed and we identified the Tubulin 8 (TUBA8) as the one of the candidate genes that may be involved in liver tumor promotion. TUBA8 mRNA was induced with PB treatment in mouse livers before tumor development as well as in tumor tissues. Since the functions of TUBA8 are unknown in liver, we investigated the effects of TUBA8 gene expression on cell growth, proliferation and cell migration. Sense- or anti-sense cDNA for mouse TUBA8 (mTUBA8) was stably transfected into Huh7 and HepG2 cells. Exogenous over-expression of mTUBA8 inhibited cell growth and proliferation in Huh7, but not in HepG2 cells, while cell migration was increased in HepG2 cells, but not Huh7 cells. These results indicate that TUBA8 can play a role in the regulation of cell growth, proliferation and cell migration in a cell specific manner in vitro, suggesting that TUBA8 may contribute to mouse liver tumorigenesis through these functions.
Keywords: CAR, Nuclear receptor, TUBA8, liver cancer, cell migration, cell growth
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common human cancers in the world, and the high rate of mortality is observed because there are no visible symptoms until the late stages of development. Various environmental risk factors, including hepatitis B virus or C virus infection, heavy alcohol consumption, dietary aflatoxin and exposure to oncogenic chemicals have been identified as causes of HCC [1, 2]. Such a wide range of causes makes it difficult to elucidate the molecular pathogenesis of HCC. The progression to HCC is usually promoted by a long-term period of virus infection or exposure to chemicals, which causes chronic inflammation. The resulting accumulation of genetic and epigenetic alterations in liver cells is thought to play an important role in liver tumorigenesis [3–5]; however the details of the molecular-based mechanism(s) remain unknown.
Rodent models have been used to clarify the underlying mechanism of HCC development, in particular the promotion of liver tumors that occurs without gene mutations. Liver tumors including HCC in rodents are usually produced by the chronic treatment of nongenotoxic agents such as phenobarbital (PB), dichlorodiphenyltrichloroethane (DTT) or ethinyl estradiol after initiation with genotoxic carcinogens (e.g., nitrosamines) [6–8]. Using this two-step mouse model of HCC development, we have reported that CAR, which is the nuclear receptor that is activated by PB in liver, was essential for liver tumor development. CAR is a member of the thyroid/steroid hormone nuclear receptor superfamily and plays an important role in regulating the expression of drug-metabolizing enzymes and transporters such as cytochrome P450s (CYPs) and multidrug resistance-associated proteins (MRPs) [9–11]. CAR is also an essential factor regulating the metabolism of bile acids and bilirubin, which cause liver injury [12, 13]. While CAR regulates these normal liver functions, the relationship between CAR and the development of liver diseases such as steatohepatitis, cholestasis and liver tumors has recently been shown [14]. These findings suggest that CAR is an important regulator of various liver functions. We have previously found that Car−/− mice did not develop liver tumors after chronic PB treatment for over 34 weeks compared to Car+/+ mice [15]. Based on this observation, we performed cDNA microarray analysis to identify the expression of candidate gene(s) correlating with liver tumor development. Over 90 genes were identified as being CAR-regulated, and TUBA8 was selected as a candidate that may be involved in CAR mediated liver tumor development.
TUBA8 was identified as a novel gene belonging to α-tubulin superfamily in 2000 [16], but its physiological function in vivo is still unknown. In general, the heterodimer complex of α- and β-tubulin assemble into microtubules (MTs) which constitute one of the major components of the cytoskeleton of eukaryotic cells and are involved in many essential processes including cell division, motility and intracellular transport [17]. Post-translational modification of α- or β-tubulin is important for controlling microtubule dynamics [18], and the mechanism of these modifications has been examined in detail in brain and neuronal cells [19]. However, TUBA8 is an atypical α-tubulin and does not have specific amino sequences involved in these modifications, suggesting that TUBA8 might have a unique function compared to other α-tubulins during the promotion of liver tumors by PB treatment.
Here we show the up-regulation of TUBA8 mRNA expression during the process of mouse liver tumor development in a CAR dependent manner. To examine the differences in malignant phenotypes including proliferation potency, colony-forming ability and cell migration, ectopic over-expression of TUBA8 in Huh7 and HepG2 cells (both human hepatocellular carcinoma cell lines) was tested. Interestingly, inhibition of cell growth and proliferation was observed in Huh7 cells, while up-regulation of cell migration was observed in HepG2 cells. Our results contribute to the understanding of TUBA8 function in the development of liver tumors.
2. Materials and methods
2.1. Animals
Car+/+ and Car−/− mice were previously generated in a C3H/HeNCrlBR (C3He) background [15]. C3H/HeNCrlBR (C3He) and C57BL/6 mice (5–7 weeks-old) were obtained from Charles River Laboratories International Inc. (MA, USA), housed in a specific pathogen-free rodent facility under a standard 12 h-light/12 h-dark cycle and were fed a standard rodent chow and water ad libitum. PB (100 mg/kg) was administered intraperitoneally. All animal procedures were approved by the Animal Ethics Committee of National Institute of Environmental Health Sciences.
2.2. Real-Time PCR
Total RNAs were extracted from mouse livers or culture cells using TRIzol reagent (Invitrogen, CA, USA), and used to synthesize cDNAs using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems Inc. CA, USA). Real-Time PCR was performed with the 7900HT Fast Real-Time PCR System (Applied Biosystems Inc. CA, USA). Forward and reverse primes for mouse TUBA8 cDNA were: 5’-CCGCCTTGACCATAAGTTTGA-3’ and 5’-TTCTTCCATACCCTCTCCAACATAC-3’, for human TUBA8 cDNA were: 5’-CATACGGAAGCTGACAGATGCT-3’ and CAGCAGAGAAGTGAAGCCG-3’, and for human TUBA1A cDNA were: 5’-GCCCGAGGGCACTACACCAT-3’ and 5’-CAGTTCCCCCGCCAAAGCT-3’. The TaqMan rodent GAPDH or human B-actin (Applied Biosystems Inc. CA, USA) was used as the reference housekeeping gene, and the expression level of each mRNA was normalized to that of mouse GAPDH or human B-actin mRNA.
2.3. cDNA cloning and construction of expression plasmids
A full-length cDNA of mouse TUBA8 (mTUBA8) was amplified using a set of primers, 5-ACCATGAGGGAATGCATATCG-3’ and 5’-TTAAAATTCCTCCCCCTCATTC-3’, and was subsequently cloned into pcDNA3.1-His-V5-TOPO plasmid (Invitrogen, CA, USA). Sense-mTUBA8 and anti-sense-mTUBA8 expression plasmids were selected after sequencing of their insert DNA. The expression plasmid of pEYFP-mTUBA8 fusion protein was generated using mTUBA8 cDNA, which was cloned into pcDNA3.1-His-V5-TOPO plasmid. The mTUBA8 cDNA was digested by using EcoRI and BamHI, and then cloned into the pEYFP-C1 vector (Clontech, CA, USA). The pECFP-hTUBA1 plasmid was purchased from Clontech (CA, USA).
2.4. Cell culture
HepG2 and Huh7 cells were cultured in 10% MEM (minimal essential medium supplement with 10% (v/v) fetal bovine serum, antibiotics (100 units/ml penicillin and 100 µg/ml streptomycin) and 2mM glutamine) in a 5% CO2 atmosphere at 37°C.
2.5. Colony formation assay
Huh7 cells were seeded in 10-cm culture dishes and 24 h later expression vector (sense- or anti-sense-mTUBA8 expression plasmid) was transfected using FuGENE 6 transfection reagent (Roche Diagnostics, IN, USA). 24 h after transfection, the harvested cells were diluted and reseeded on 10-cm culture dishes in triplicate. The transfected cells were grown in the presence of 0.8 mg/ml Geneticin (Invitrogen, CA, USA) for 2 weeks, after which the colonies formed from each cell were fixed with 10% formaldehyde, stained with 0.125% crystal violet and counted.
2.6. Selection of cell strains with stable expression of mTUBA8
HepG2 or Huh7 cells were pre-seeded on 10-cm culture dishes at a density of 1.0×106 cells/dish. 5 µg of either sense-mTUBA8, anti-sense-mTUBA8 or empty expression plasmid was transfected into the cells using FuGENE 6 transfection reagent (Roche Diagnostics, IN, USA); 24 h after transfection, cells were diluted and reseeded on 10-cm culture dishes in triplicate. Cells were grown in the presence of 0.8 mg/ml Geneticin (Invitrogen, CA, USA) for 2~3 weeks for stable cell selection. The expression of mTUBA8 mRNA or the presence of integrated plasmid DNA in each stably-transfected cells was confirmed by RT-PCR. Total RNA was extracted from culture cells using TRIzol reagent, and total genomic DNA was extracted from culture cells using DNeasy Blood & Tissue kit (QIAGEN Inc., CA, USA) as described in the manufacturer’s protocol. Integrated sense- or anti-sense mTUBA8 plasmid DNA was amplified using these primers, T7 primer: 5-TAATACGACTCACTATAGGG-3’, BGH reverse primer: 5’-TAGAAGGCACAGTCGAGG-3’ and R1 primer: 5’-ACTCTGTGAGGTCCACATTGA-3’.
2.7. MTT assay
Cell proliferation was determined by the MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Stably transfected Huh7 or HepG2 cells were seeded on 96 well plates at densities of 2.0×103 cells/well (Huh7 stable cells) or 4.0×103 cells/well (HepG2 stable cells) in 10% MEM medium. At selected time intervals, 10 µl of MTT working solution (5 mg/ml in ddH2O) was added and incubated at 37°C for 4 h. At the end of the incubation, the media was discarded and 100 µl of DMSO was added to each well for dissolving the dye. After incubation at room temperature for 1 h with gently shaking, the absorbance of the converted dye was measured using a microplate spectrophotometer (SpectraMax Plus 384; Molecular Device Inc., CA, USA) at 495 nm.
2.8. Analysis of cell cycle by flow cytometry
Stably transfected Huh7 cells were seeded on 6 cm dishes at a density of 5.0×105 cells/dish in 10% MEM medium. After 48 h, the cultured cells were trypsinized and harvested in PBS followed by two washes with PBS. Then the cells were resuspended in 70% ethanol overnight at 4°C.The fixed cells were centrifuged, resuspended in RNaseA solution (0.2 mg/ml RNaseA in PBS) and incubated at 37°C for 30 min. The cells were pelleted, incubated in Propidium Iodide (PI) staining solution (20µg/ml PI in PBS) for overnight at 4°C. The cells were analyzed by flow cytometer (BD FACSCalibur), and Cell Quest software (BD Biosciences) was used for data acquisition and analysis.
2.9. Recombinant adenoviruses
Based on the mTUBA8 expression plasmid, an Ad-mTUBA8 expression adenovirus vector was constructed with the AdEasy Vector System kit (Q-BIO gene, Montreal Canada) as described in the manufacturer’s protocol. Adenoviruses were propagated in HEK293 cells. Ad-β-gal encoding β-galactosidase previously constructed in our laboratory was used as control. Virus titers were measured and calculated using the 50% tissue culture infectious dose assay (TCID50 method) as described in the manufacturer’s application manual.
2.10. Cell migration assay
Transwell Chambers (8-µm pore size; Corning, MA, USA) were used for the in vitro cell migration assay. Stably transfected HepG2 cells were seeded into the upper part of chamber at a density of 5.0 × 104 cells in 300 µl of 10% MEM medium, and the lower compartment was filled with 600 µl of 10% MEM medium. In parallel experiments, HepG2 cells were infected with Ad-β-gal or Ad-mTUBA8 at 10 MOI (Multiplicity Of Infection) 24 h post-seeding, and cells were seeded into the chamber with the same method described above. After incubation for 72 h, cells were fixed with 10% formaldehyde. Non-migrating cells were gently removed from the upper chamber with cotton, and cells on the underside of the membrane were stained with 0.1% crystal violet. Migrating cells were counted in four microscopic fields at ×100 magnification.
2.11. Confocal microscopy
C3He male mice were euthanized at 8 h after tail-vein injection of pEYFP-mTUBA8 plasmid or pECFP-hTUBA1 plasmid using TransIT In Vivo Gene Delivery System (Mirus, WI, USA) with described protocol. The liver was then removed, embedded in Tissue-Tek OCT embedding compound (SAKURA Finetechnical Co., Ltd., Tokyo, Japan) and frozen on dry ice. Frozen liver sections of 30µm thickness were made using a cryostat by a routine procedure.
HepG2 or Huh7 cells were seeded on a 2-well chamber slide (Nalge Nunc International Corp., IL, USA) at a density of 1.2×105 cells or 0.8×105 cells per well in 10% MEM medium. The pEGFP-mTUBA8 plasmid was transfected into cells on the chamber slide using FuGENE 6 transfection reagent. After 48 hrs of transfection, cells were fixed with 100% cold methanol and permeabilized with 0.1% Triton-X100. The slides of liver sections or transfected cells were embedded with Hoechst 33258 solution (0.5 µg/ml in 80% glycerol), followed by confocal microscopic observation.
2.12. Statistical analysis
The statistical significance between two groups of data sets was determined by using the Student’s t test (two-tailed) with P<0.05 regarded as statistically significant.
Results
3.1. CAR-regulated induction of the mTUBA8 gene by PB in mouse liver
Our previous study demonstrated that CAR is an important factor in promoting the development of liver tumors following chronic PB treatment [15]. Car+/+ C3He mice treated with DEN followed by PB treatment for 23–32 weeks developed lesions and tumors, while DEN/32 weeks PB-treated Car−/− C3He mice didn’t show any liver tumors. To screen for genes that are induced during the development of mouse liver tumors in a CAR dependent manner, cDNA microarray analysis was first performed (unpublished data) and a number of candidate genes were detected. We focused on the mTUBA8 gene for further investigation, because there have been no previous reports on mTUBA8 in liver. We also found other tubulin family genes in the microarray analysis data, but most of them did not show any change in mRNA expression in PB treated liver. A slight induction of mTUBB2 mRNA was observed in tumor samples, although not as high compared with mTUBA8 (Table 1).
Table 1.
The list of fold change about tubulin family genes that were found out by microarray analysis
| CAR-KO (PB−) 32W V.S. CAR-KO (PB+) 32W |
CAR-KO (PB+) 32W V.S. CAR-WT (PB+) 32W |
CAR-KO (PB+) 32W V.S. CAR-WT Tumor 32W |
||
|---|---|---|---|---|
| Gene Name | Gene Description | Fold Change | Fold Change | Fold Change |
| TUBB3 | Tubulin, beta 3 | −1.01 | 1.04 | 1.09 |
| TUBG2 | Tubulin, gamma2 | 1.2 | −1.08 | −1.05 |
| TUBA4 | Tubulin, alpha 3 | 1.43 | 1.13 | 1.41 |
| TUBB4 | Tubulin, beta 4 | 1.26 | 1.13 | 1.42 |
| TUBA3 | Tubulin, alpha 3 | −1.07 | 1.37 | 1.44 |
| TUBA1 | Tubulin, alpha 1 | 1.11 | 1.14 | 1.23 |
| TUBD1 | Tubulin, delta 1 | −1.01 | 1.67 | −1.05 |
| TUBB5 | Tubulin, beta 5 | 1.26 | 1.24 | 2.05 |
| TUBB2 | Tubulin, beta 2 | 1.02 | 2.04 | 4.44 |
| TUBA8 | Tubulin, alpha 8 | −1.01 | 8.01 | 12.36 |
Microarray data are deposited to GEO: GSE29108.
The mRNA for mTUBA8 was already induced after 24 h of PB treatment in the livers of Car+/+ but not in Car−/− C3He mice, and continued to be induced in the DEN/32 weeks PB-treated Car+/+ C3He mice (Fig. 1A). Interestingly, the induction level of mTUBA8 mRNA was clearly higher in liver tumor tissues with more than 100-fold induction compared to control samples (Fig. 1A). These results indicated that the mTUBA8 gene undergoes CAR-mediated induction in the liver prior to tumor development and continues to be induced in tumor tissues.
Fig. 1.
Induction of mTUBA8 mRNA in the livers of PB treated mice. (A) The 24 h or 32 weeks indicate the period that the Car+/+ and Car−/− C3He mice were treated with PB. Open and closed bars show the levels of mTUBA8 mRNA in the non-tumor tissues from the non-PB treated and PB treated mice, respectively. The hatched bar shows the level of mTUBA8 mRNA in tumor tissues. The relative levels of mTUBA8 mRNA were calculated by setting the levels in the livers of non-PB treated Car+/+ C3He mice as one. The data are reported as mean ± SD, n=3 (24 h) or n=6 (32 weeks). (B) The 6 weeks indicate the period that the C3He or C57BL6 male mice were treated by PB. Open and closed bars show the levels of mTUBA8 mRNA in the liver tissues from the non-PB treated and PB treated mice, respectively. The relative levels of mTUBA8 mRNA were calculated by setting the levels in the livers of PBS-treated C3He male mice as one. The data are reported as mean ± SD, n=3.
3.2. Strain-dependent expressions
The C3He mouse is known to be more susceptible to developing liver tumors than the C57BL6 mouse [20, 21]. To examine the relationship between the induction level of mTUBA8 mRNA and liver tumor susceptibility, male mice were treated with PB for 6 weeks and liver RNA was prepared for real-time PCR to measure mTUBA8 mRNA. Compared with an over 6-fold increase of mTUBA8 mRNA in the PB-treated C3He males, the corresponding PB-treated C57BL6 mice showed a lower but still significant increase (2-fold) of mTUBA8 mRNA (Fig. 1B). This difference in induction rate between the two mouse strains suggests that mTUBA8 may correlate with the susceptibility of developing liver tumors.
3.3. TUBA8 Expression in human hepatocellular carcinoma cell lines
For further in vitro experiments using human hepatocellular carcinoma cell lines HepG2 or Huh7, we examined the basal expression level of human TUBA8 mRNA by real-time PCR. In addition to TUBA8, TUBA1A mRNA was also expressed in these cells (Fig. 2). Therefore, these cell lines can be useful for ectopic over-expression experiments to examine the function of mTUBA8.
Fig. 2.
Expression of TUBA8 in human hepatocellular carcinoma cells. Basal expression levels of human TUBA8 and TUBA1A mRNA in Huh7 or HepG2 cells were evaluated by real-time PCR, and normalized to that of human β-actin mRNA. Open and closed bars show the levels of human TUBA8 and TUBA1A mRNA in the cells, respectively. The data are reported as mean ± SD, n=3.
3.4. Suppression of colony formation ability by over-expression of mTUBA8
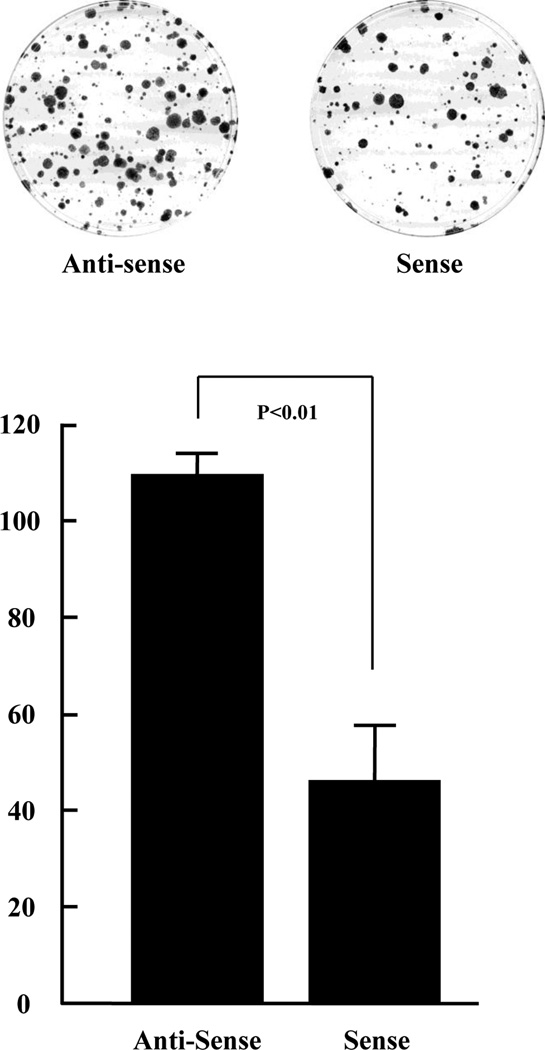
In order to explore the function of mTUBA8 in cell growth, we carried out colony formation assays after transfecting the sense-mTUBA8 and anti-sense-mTUBA8 plasmid vectors into Huh7 cells. There were 109.3 ± 4.5 (mean ± SD) colonies formed with the anti-sense-mTUBA8 plasmid transfected Huh7 cells and 46.0 ± 11.4 colonies formed with the sense-mTUBA8 expression plasmid transfected Huh7 cells (Fig. 3). This indicates that mTUBA8 over-expression in Huh7 cells inhibits cell growth.
Fig. 3.
Inhibition of Huh7 colony formation by ectopic expression of mTUBA8. Huh7 cells were transfected with anti-sense-mTUBA8 or sense-mTUBA8 expression plasmid, and grown on triplicate culture dishes in the presence of 0.8 mg/ml Geneticin for 2 weeks. Geneticin-resistant colonies were fixed, stained and counted. The data are reported as mean ± SD of three independent experiments.
3.5. Expression of mTUBA8 mRNA in transfected stable cells
We next established Huh7 and HepG2 cells that were stably expressing mTUBA8. We transfected empty, anti-sense-mTUBA8, and sense-mTUBA8 expression plasmids into Huh7 or HepG2 cells, and after G418 selection culture the expression of mTUBA8 mRNA was examined in three independent experiments by RT-PCR. A strong expression of mTUBA8 mRNA was detected in the sense-mTUBA8 plasmid stably transfected Huh7 cells, but not in empty-plasmid or anti-sense-mTUBA8 plasmid stably transfected Huh7 cells (Fig. 4A). Similar results were observed in stably transfected HepG2 cells, and a weak signal was detected in anti-sense-mTUBA8 plasmid transfected HepG2 cells. To confirm that the sense-mTUBA8 or anti-sense-mTUBA8 expression plasmids were integrated into genomic DNA of the stably transfected HepG2 cells, genomic DNA was isolated and PCR was carried out. As a result, we confirmed that the slight expression of mTUBA8 was derived from the anti-sense mTUBA8 expression plasmid (Fig. 4B). Finally, we compared the sense-mTUBA8 plasmid transfected Huh7 and HepG2 cells to empty or anti-sense-mTUBA8 plasmid transfectants selected at random as experimental controls. We could not see any morphological changes in stably transfected Huh7 or HepG2 cells (data not shown).
Fig. 4.
Confirmation of exogenous mTUBA8 expression in transfected Huh7 and HepG2 cells. (A) Empty-, Anti-sense, or Sense-mTUBA8 expression plasmid was transfected into Huh7 and HepG2 cells, followed by culture in G418 containing medium to establish stable transfectant cells. Expression of mTUBA8 in transfected cells was examined by RT-PCR. RT denotes Reverse Transcriptase, and M denotes size marker. (B) Genomic DNA was isolated from each set of transfected HepG2 cells, and the integration of plasmid DNA was examined by PCR. The T7/R1 primer set detects the sense-mTUBA8 plasmid, and the BGH rev/R1 primer set detects the anti-sense-mTUBA8 plasmid in transfected HepG2 cells.
3.6. Influence of mTUBA8 gene on the growth of Huh7 cells
In order to confirm the results of the colony formation assay which suggested that mTUBA8 plays a role in growth of Huh7 cells, the proliferation rate of transfected Huh7 cells was determined by MTT assay. A slight inhibition of cell proliferation was observed in sense-mTUBA8 transfected Huh7 cells after 72 hrs of culture compared with empty or anti-sense-mTUBA8 transfected Huh7 cells, and this became statistically significant after 96 hrs (P<0.01; Fig. 5A). We also examined the cell cycle distribution of mTUBA8-transfected cells by flow cytometry after 48 hrs culture. Although not statistically significant, an increase in the percentage of the cell population at the G0/G1 phase and decrease in the percentage of the cell population at the S phase and the G2/M phase was observed (Fig. 5B). These results suggest that mTUBA8 over-expression in Huh7 cells leads to an inhibition of cell growth.
Fig. 5.
Cell proliferation and cell cycle analysis in transfected Huh7 cells. (A) MTT assay was carried out 24, 48, 72 and 96 h after cell seeding. The results are reported as mean ± SD of six independent experiments. (B) Each set of stably transfected Huh7 cells was collected after 48 h of cell seeding, and the DNA content was analyzed by flow cytometry to determine the cell cycle distribution. Experiments were repeated three times independently and the average percentages are indicated in the bar graph.
3.7. Enhancement of cell migration in HepG2 cells by mTUBA8 over-expression
We also examined the effect of mTUBA8 over-expression in HepG2 cells. Stably transfected HepG2 cells were first examined for their rate of cell proliferation by MTT assay; however there was no important difference (Fig. 6A). This result suggested that mTUBA8 over-expression did not affect the proliferation of HepG2 cells. We next examined the effects of mTUBA8 expression on cell migration, since individual cell migration is an important characteristic of invasive tumor cells. Interestingly, sense-mTUBA8 transfected HepG2 cells had enhanced migration ability compared with other transfected cells (P<0.01; Fig. 6B), with the migration rates of the empty, anti-sense-mTUBA8, and sense-mTUBA8 transfected HepG2 cells at 181.3 ± 26.8 (mean ± SD), 180.3 ± 43.7, and 434.3 ± 39.0 cells, respectively (Fig. 6C).
Fig. 6.
Induction of cell migration ability in transfected HepG2 cells. (A) MTT assay was carried out 24, 48, 72 and 96 h after cell seeding. The results are reported as mean ± SD of six independent experiments. (B) Transfected cells were seeded on transwell chambers. After 72 h incubation, the migrated cells were fixed, stained with 0.1% crystal violet and photographed. (C) The number of migrated cells was counted in four fields for each group. The results are reported as mean ± SD of three independent experiments. (D) Parental HepG2 cells were infected with Ad-β-gal or Ad-mTUBA8 for 24 h at 10 MOI and then seeded on transwell chambers. After 72 h incubation, the migrated cells were fixed, stained and photographed (upper panel). The number of migrated cells was counted in four fields for each group (lower panel). The results are reported as mean ± SD of three independent experiments.
A similar induction of cell migration by ectopic mTUBA8 over-expression was observed in adenovirus-based infection experiments. The migration rates of Ad-β-gal and Ad-mTUBA8 infected HepG2 cells were statistically significant (P<0.01) at 117.5 ± 28.6 (mean ± SD) and 307.3 ± 68.6 respectively (Fig. 6D). This indicates that expression of mTUBA8 induced cell migration of HepG2 cells. We also examined cell migration rates in adenovirus based mTUBA8 over-expressed Huh7 cells, but there was no difference between Ad-mTUBA8 and Ad-β-gal infected (Supplemental Fig. 1). In addition to Huh7 and HepG2 cells, the neuroblastoma U373MG cells were utilized in the similar experiments, from which any changes in cell migration and cell proliferation were observed (data not shown).
3.8. Localization of mTUBA8 protein in hepatocytes and culture cells
We constructed a pEYFP-mTUBA8 expression plasmid to examine the localization of the fusion protein in liver cells since there is no commercially available mTUBA8 specific antibody. We co-injected the pEYFP-mTUBA8 and pECFP-hTUBA1 expression plasmids via the tail vein, and examined the localization of the fusion proteins in mouse liver. Most of the expressed pEYFP-mTUBA8 fusion protein was observed in the cytoplasm with several expression patterns described as fiber, smear or dotted (Fig. 7A). Moreover, the pEYFP-mTUBA8 and pECFP-hTUBA1 fusion proteins were co-localized in mouse liver (Fig. 7A). We also examined the localization of pEYFP-mTUBA8 fusion protein in Huh7 and HepG2 cells. Expressed pEYFP-mTUBA8 fusion protein were mainly observed in cytoplasm with a fiber-like structure (Fig. 7B), indicating that mTUBA8 may contribute to the construction or the maintenance of the cytoskeleton.
Fig. 7.
Subcellular localization of mTUBA8 in mouse liver and cultured cell lines. (A) pEYFP-mTUBA8 and pECFP-hTUBA1 expression plasmids were injected into mice via tail vein, and liver sections were prepared for examination using confocal microscopy. pEYFP-mTUBA8 expression is shown as a red signal, and pECFP-hTUBA1 expression is shown as a green signal. (B) pEYFP-mTUBA8 expression plasmids were transfected into Huh7 cells and HepG2 cells, and its expression was analyzed by confocal microscopy. pEYFP-mTUBA8 expression is shown as a yellow signal.
Discussion
Here, we demonstrate that mTUBA8 mRNA is induced in mouse liver by PB treatment in a CAR dependent manner. This gene, which is barely expressed in normal mouse livers, showed the highest expression in mouse liver tumors, suggesting that mTUBA8 has an important role in PB promoted liver tumorigenesis. CAR is a known transcriptional factor that regulates target genes, but we were unable to find candidate binding site(s) for CAR on the mTUBA8 promoter. Therefore CAR may not regulate mTUBA8 directly and how CAR regulates mTUBA8 mRNA expression in liver is an important theme of our future work.
In mammals, six α-tubulin genes have been cloned and sequenced, and human and mouse TUBA8 was identified and characterized as the 7th member of α-tubulin family [16]. Although higher TUBA8 expression was demonstrated in human heart, skeletal muscle and testis compared to other tissues including liver, its function remains unknown [16]. In general, α-tubulin undergoes posttranslational modifications in microtubules including: 1) acetylation of 40th lysine, 2) detyrosination or removal of the C-terminal tyrosine residue exposing glutamic acid as the new C-terminal residue, 3) phosphorylation, 4) polyglutamylation and 5) polyglycylation [18, 19, 22]. Acetylation and detyrosination of α-tubulins have been especially examined in brain tissue or neuronal cells. For example, acetylation of α-tubulins at the 40th lysine by the elongator complex is an important step to control the migration and differentiation of cortical neurons in the intermediate zone [23]. Detyrosination of C-terminal residues or polyglutamynation of α-tubulins around the C-terminal region seem to be important for the stabilization of microtubules, because C-terminal residues are involved in the interaction with many proteins such as microtubule associated proteins (MAPs) and motor proteins [19, 24]. However TUBA8 seems to be an atypical α-tubulin because TUBA8 doesn’t have the 40th lysine or the tyrosine residue located at the C-terminal. This suggests that TUBA8 may not be regulated by posttranslational modification of acetylation and of detyrosination, and allows for TUBA8 to regulate MAPs interactions and microtubule dynamics in a different manner from other α-tubulin isotypes.
As a potential function of mTUBA8, we demonstrated that ectopic over-expression of mTUBA8 decreased the rate of cell proliferation of Huh7 cells and accelerated cell migration in HepG2 cells. We hypothesized that these characteristic functions may be due to the lack of acetylation and/or detyrosination sites, so we constructed mutants of mTUBA8 that have a 40th lysine instead of the 40th alanine (A40K mutant), a 449th tyrosine instead of a 449th phenylalanine (F449Y mutant) or both double mutations (A40K/F449Y mutant). We performed colony formation assays in Huh7 cells and cell migration assays in HepG2 cells using these materials; however we could not detect any important difference between the mutants and wild type of mTUBA8 (data not shown). As a possible explanation for these results, the regulation system of acetylation and/or detyrosination of alpha-tubulin in these cell lines may not work well; we could not detect the acetylated or detyrosinated (Glu-terminaled) alpha-tubulin in parental Huh7 and parental HepG2 by western blotting analysis using specific antibodies (data not shown). Although we could not examine the effect of acetylation / detyrosination on cell growth or cell migration using these experimental conditions, our findings support the role of mTUBA8 in the inhibition of cell growth in Huh7 cells and the induction of cell migration in HepG2 cells.
How does mTUBA8 inhibit cell growth or accelerate migration of cells? As for the inhibition of cell growth, we demonstrated a slight increase in the cell population at the G0/G1 phase and decrease in the cell population at the S phase in mTUBA8 stably transfected Huh7 cells (Fig. 5B). This result may suggest that ectopic mTUBA8 over-expression affects the process of DNA replication rather than the process of mitosis or cytokinesis. In addition, a relationship between the dynamics of microtubules (MTs) and cell migration has been suggested from several reports. For example, destruction of MTs by the treatment of colcemid inhibited cell migration via inhibition of lamellipodial protrusion on the active edge of migrating fibroblast cells [25]. Moreover, recent studies of Adenomatous Polyposis Coli (APC) or p120-catenin showed that MT dynamics is important for cell migration [26, 27]. Interestingly, over-expression of MAP2, one of the MAP families, enhanced cell motility in oral cancer cells [28], suggesting that MTs and their interaction with α/β-tubulin are involved in the regulation of cell migration. Over-expressed mTUBA8 may interact with the MTs instead of endogenous α-tubulin and enhance cell migration in liver cells. In any case, further study will be required to solve these questions.
Recently, a relationship between a splicing mutation of TUBA8 and the disease of polymicrogyria with optic nerve hypoplasia has reported [29], but this is the only report described the potential physiological function of TUBA8. The detailed function of TUBA8 or the connection to other diseases including cancer remains unknown. We found that mTUBA8 was over-expressed during the development of liver tumors using cDNA microarray analysis. Additionally, we also detected a slight induction of mTUBB2 in mouse liver tumor (Table 1). Interestingly, human TUBB2 was found to be over-expressed in various normal tissues particularly in fetal and neonatal tissues or neoplastic tissues [30] and in a variety of cancers [31], suggesting that TUBA8 may be over-expressed in tumors as well as TUBB2. In preliminary observations, we examined the expression of TUBA8 mRNA in human hepatocellular carcinoma tissues by real-time PCR, and 8 cases out of a total 31 cases showed a higher expression of TUBA8 in tumor tissues compared to normal tissues derived from same the liver (unpublished data). Although the levels were not associated with the sex, age, etiology, multiplicity or stage of tumors, these findings suggest that TUBA8 may be linked to the development or promotion of human liver cancer.
In conclusion, we have demonstrated that mTUBA8 is up-regulated during mouse liver tumorigenesis in a CAR dependent manner. This is the first report that mTUBA8 may be involved in mouse liver tumorigenesis.
The biological process and detailed mechanism by which TUBA8 is involved in liver tumor development remains to be determined; for example, how the repression of cell growth by TUBA8 play a role in the liver during HCC development, while TCPOBOP activation of CAR increases proliferation of normal hepatocytes. Since both cell growth and death are simultaneously involved in tumor development, our findings suggest CAR-regulated repression of cell growth by TUBA8 may also be critical for tumor development and that the cell migration may be a factor for an intra-hepatic metastasis and a poor prognosis of liver cancer patients.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health, and National Institute of Environmental Health Sciences, Z01ES1005-01. We would like to acknowledge the efforts of all of the staff of the NIEHS DNA Sequencing Core, Flow Cytometry Center and Fluorescence Microscopy and Imaging Center. We especially thank to all our lab members for valuable discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 2.Marrero JA. Hepatocellular carcinoma. Curr Opin Gastroenterol. 2005;21:308–312. doi: 10.1097/01.mog.0000159817.55661.ca. [DOI] [PubMed] [Google Scholar]
- 3.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 4.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 5.Katoh H, Shibata T, Kokubu A, Ojima H, Fukayama M, Kanai Y, Hirohashi S. Epigenetic instability and chromosomal instability in hepatocellular carcinoma. Am J Pathol. 2006;168:1375–1384. doi: 10.2353/ajpath.2006.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dragon YP, Pitot HC. The role of the stages of initiation and promotion in phenotypic diversity during hepatocarcinogenesis in the rat. Carcinogenesis. 1992;13:739–750. doi: 10.1093/carcin/13.5.739. [DOI] [PubMed] [Google Scholar]
- 7.Preat V, de Gerlache J, Lans M, Taper H, Roberfroid M. Comparative analysis of the effect of phenobarbital, dichlorodiphenyltrichloroethane, butylated hydroxytoluene and nafenopin on rat hepatocarcinogenesis. Carcinogenesis. 1986;7:1025–1028. doi: 10.1093/carcin/7.6.1025. [DOI] [PubMed] [Google Scholar]
- 8.Mayol X, Perez-Tomas R, Cullere X, Romero A, Estadella MD, Domingo J. Cell proliferation and tumour promotion by ethinyl estradiol in rat hepatocarcinogenesis. Carcinogenesis. 1991;12:1133–1136. doi: 10.1093/carcin/12.6.1133. [DOI] [PubMed] [Google Scholar]
- 9.Sueyoshi T, Negishi M. Phenobarbital response element of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol. 2001;41:123–143. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- 10.Kodama S, Negishi M. Phenobarbital confers its dicerse effects by activating the orphan nuclear receptor CAR. Drug Metab Rev. 2006;38:75–87. doi: 10.1080/03602530600569851. [DOI] [PubMed] [Google Scholar]
- 11.Urquhart BL, Tirona RG, Kim RB. Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for inter- individual variability in response to drugs. J Clin Pharmacol. 2007;47:566–578. doi: 10.1177/0091270007299930. [DOI] [PubMed] [Google Scholar]
- 12.Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, Xie W. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol. 2004;65:292–300. doi: 10.1124/mol.65.2.292. [DOI] [PubMed] [Google Scholar]
- 13.Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc Natl Acad Sci USA. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakizaki S, Yamazaki Y, Takizawa D, Negishi M. New insights on the xenobiotic-sensing nuclear receptors in liver diseases –CAR and RXR–. Curr Drug Metab. 2008;9:614–621. doi: 10.2174/138920008785821666. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- 16.Stanchi F, Corso V, Scannapieco P, Ievolella C, Negrisolo E, Tiso N, Lanfranchi G, Valle G. TUBA8: A new tissue-specific isoforms of alpha-tubulin that is highly conserved in human and mouse. Biochem Biophys Res Commun. 2000;270:1111–1118. doi: 10.1006/bbrc.2000.2571. [DOI] [PubMed] [Google Scholar]
- 17.Wade RH. On and around microtubules: An overview. Mol Biotechnol. 2009;43:177–191. doi: 10.1007/s12033-009-9193-5. [DOI] [PubMed] [Google Scholar]
- 18.Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushima N, Furuta D, Hidaka Y, Moriyama R, Tsujiuchi T. Post-translational modifications of tubulin in the nervous system. J Neurochem. 2009;109:683–693. doi: 10.1111/j.1471-4159.2009.06013.x. [DOI] [PubMed] [Google Scholar]
- 20.Poole TM, Drinkwater NR. Strain dependent effects of sex hormones on hepatocarcinogenesis in mice. Carcinogenesis. 1996;17:191–196. doi: 10.1093/carcin/17.2.191. [DOI] [PubMed] [Google Scholar]
- 21.Ito A, Takahashi T, Watanabe H, Ogundigie PO, Okamoto T. Significance of strain and sex differences in the development of 252Cf Neutron-induced liver tumors in mice. Jpn J Cancer Res. 1992;83:1052–1056. doi: 10.1111/j.1349-7006.1992.tb02721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eipper BA. Rat brain microtubule protein: purification and determination of covalently bound phosphate and carbohydrate. Proc Natl Acad Sci USA. 1972;69:2283–2287. doi: 10.1073/pnas.69.8.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, Belachew S, Malgrange B, Chapelle JP, Siebenlist U, Moonen G, Chariot A, Nguyen L. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 24.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 25.Bershadsky AD, Vaisberg EA, Vasiliev JM. Pseudopodial activity at the active edge of migrating fibroblast is decreased after drug-induced microtubule depolymerization. Cell Motil Cytoskeleton. 1991;19:152–158. doi: 10.1002/cm.970190303. [DOI] [PubMed] [Google Scholar]
- 26.Kroboth K, Newton IP, Kita K, Dikovskaya D, Zumbrunn J, Waterman-Storer CM, Nathke IS. Lack of Adenomatous Polyposis coli protein correlates with a decrease in cell migration and overall changes in microtubule stability. Mol Biol Cell. 2007;18:910–918. doi: 10.1091/mbc.E06-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichii T, Takeichi M. p120-catenin regulates microtubule dynamics and cell migration in a cadherin-independent manner. Genes Cells. 2007;12:827–839. doi: 10.1111/j.1365-2443.2007.01095.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu SY, Chen YT, Tseng MY, Hung CC, Chiang WF, Chen HR, Shieh TY, Chen CH, Jou YS, Chen JY. Involvement of microtubule-associated protein 2 (MAP2) in oral cancer cell motility: a novel biological function of MAP2 in non-neuronal cells. Biochem Biophys Res Commun. 2008;366:520–525. doi: 10.1016/j.bbrc.2007.11.179. [DOI] [PubMed] [Google Scholar]
- 29.Abdollahi MR, Morrison E, Sirey T, Molnar Z, Hayward BE, Carr IM, Springell K, Woods CG, Ahmed M, Hattingh L, Corry P, Pilz DT, Stoodley N, Crow Y, Taylor GR, Bonthron DT, Sheridan E. Mutation of the variant alpha-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. Am J Hum Genet. 2009;85:737–744. doi: 10.1016/j.ajhg.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oda E, Nakamura Y, Yamamoto M, Kojiro M. Immunohistochemical distribution of tubulin beta II in human normal and neoplastic tissues. Kurume Med J. 2005;52:117–125. doi: 10.2739/kurumemedj.52.117. [DOI] [PubMed] [Google Scholar]
- 31.Yeh IT, Luduena RF. The beta II isotype of tubulin is present in the cell nuclei of a variety of cancers. Cell Motil Cytoskeleton. 2004;57:96–106. doi: 10.1002/cm.10157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.