Abstract
Ortholog identification is used in gene functional annotation, species phylogeny estimation, phylogenetic profile construction and many other analyses. Bioinformatics methods for ortholog identification are commonly based on pairwise protein sequence comparisons between whole genomes. Phylogenetic methods of ortholog identification have also been developed; these methods can be applied to protein data sets sharing a common domain architecture or which share a single functional domain but differ outside this region of homology. While promiscuous domains represent a challenge to all orthology prediction methods, overall structural similarity is highly correlated with proximity in a phylogenetic tree, conferring a degree of robustness to phylogenetic methods. In this article, we review the issues involved in orthology prediction when data sets include sequences with structurally heterogeneous domain architectures, with particular attention to automated methods designed for high-throughput application, and present a case study to illustrate the challenges in this area.
Keywords: phylogenomics, orthology, promiscuous domains, multi-domain architecture, function prediction, super-ortholog
INTRODUCTION
In recent years, DNA sequencing technologies have improved their throughput exponentially, leading to explosive growth in sequence databases. Unfortunately, experimental methods to elucidate gene function have not kept pace with the throughput of sequencing; analysis of Gene Ontology (GO) annotations [1] and evidence codes shows that <1% of genes have any experimental support for their annotations [2]. For these reasons, bioinformatics methods to predict gene function have played central roles in biological research. Problematically, the standard functional annotation protocol—transferring the annotation of the top BLAST [3] hit—has been shown to be fraught with systematic error [4]: as much as 25% of genes are estimated to be misannotated [5].
What accounts for such large error rates? The fundamental assumption underlying an annotation transfer protocol is that evolution conserves function, and that sequence similarity implies homology (i.e. a common ancestry) and can thus be used as a basis for inferring function. As in most of biology, the reality is a bit more complicated.
It is known that protein function is mediated by protein 3D structure, and that structural similarity is conserved over large evolutionary distances even when sequence similarity is undetectable. Protein structural domains—contiguous stretches of the polypeptide chain that fold independently into compact globular structures—comprise the building blocks of a protein’s overall structure. The ordered series of these domains is a protein’s ‘multi-domain architecture’ (reviewed in [6]). Changes in domain architecture, produced by gene fusion and fission events and other evolutionary processes, are a significant source of error in transitive annotation pipelines [4, 7, 8]. Of particular relevance to the task of protein function prediction is the presence of ‘promiscuous’ domains—domains found in many different combinations (kinase domains are a well-known example of this class) [9]. Individual domains in a multi-domain architecture can have very different evolutionary rates and functional roles, and taxonomic distributions can also vary widely, with some domains being conserved throughout the Tree of Life and others being restricted to particular lineages. See references [10–12] for reviews of structural domain distributions and characteristics and the CATH [13] and SCOP [14] databases for classifications of domains into structural hierarchies.
Proteins can also diverge functionally from a common ancestor through gene duplication events and mutations at key positions, producing protein superfamilies containing groups of orthologs and paralogs spanning many distinct functions; existing annotation errors also complicate any annotation-transfer protocol [4, 15–17].
Fortunately, a ‘structural phylogenomic’ analysis, combining evolutionary and structural analyses, provides an overarching framework to address the limitations of simple annotation transfer protocols [18]. For instance, knowledge of protein domain architecture can be used to restrict predicted orthologs to those that share the same series of structural or functional domains. Other applications of structural phylogenomics include using 3D structure and gene trees to predict enzyme active sites [19].
The term ‘phylogenomics’ was proposed initially by Eisen [20] to describe the use of phylogenetic analysis to improve the accuracy of gene functional annotation; it is also used to describe species phylogeny estimation using multiple genes (e.g. as in a concatenated gene matrix approach) [21]. A related approach was developed for the functional annotation of the human genome [22], using the SCI-PHY algorithm [23] to identify functional subfamilies, and subsequently extended into two phylogenomic databases of gene family phylogenies: the PANTHER tools [24] and the PhyloFacts resource [25, 26].
The term ‘ortholog’ was first proposed by Walter Fitch [27] to differentiate genes related by speciation from those related by duplication events: ‘Where the homology is the result of gene duplication so that both copies have descended side by side during the history of an organism…the genes should be called paralogous (para = in parallel). Where the homology is the result of speciation so that the history of the gene reflects the history of the species…the genes should be called orthologous (ortho = exact).’ Duplication events provide a release from evolutionary constraints, allowing genes to explore novel functions [28]. Note that orthology is a phylogenetic term, but is used in practice as a surrogate for functional equivalence; in fact, orthologs in distantly related species may have diverged functionally from their common ancestor.
These terms and concepts were quickly revealed to be insufficient to model the actual biological complexity of gene family evolution, and a host of new terms and concepts have since been developed. Ohno [28] proposed a model for functional diversification following gene duplication: in ‘neo-functionalization’, genes acquire novel functions (e.g. bind to new ligands), while in ‘sub-functionalization’, genes partition the ancestral function, potentially specializing for different tissues or developmental stages.
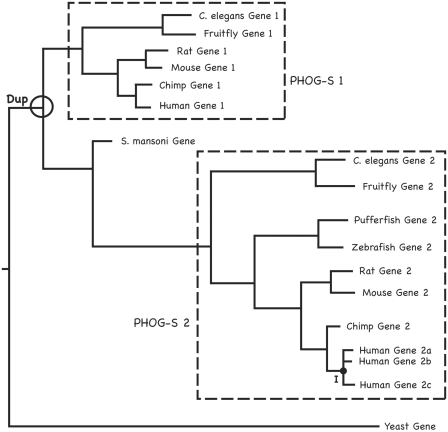
Since orthology is not transitive (i.e. if X and Y are orthologs, and Y and Z are orthologs, it does not necessarily follow that X and Z are orthologs) [29], Zmasek and Eddy [30] proposed a more restrictive definition of orthology that explicitly disallows any duplication events: two genes X and Y are ‘super-orthologs’ if and only if every node on the evolutionary tree relating them corresponds to a speciation event. The super-orthology relation has the advantage of being transitive as it partitions the gene family tree into super-orthologous subtrees. Sonnhammer and Koonin developed related terms to describe in-species duplication events (called ‘inparalogs’) and other types of paralogy [31]. Orthology, super-orthology and inparalog relationships are illustrated in Figure 1.
Figure 1:
Orthology and paralogy subtypes and the use of tree distances in PHOG. We present this toy example of gene family evolution to illustrate the main orthology subtypes and how the PHOG algorithm uses tree distances and topology jointly to infer orthologs. ‘Dup’ indicates a duplication event in the animal lineage, and ‘I’ represents a group of predicted inparalogs. Recall that super-orthology requires that all nodes on a path joining two sequences correspond to speciation events. The PHOG algorithm for super-orthology identification allows subtrees containing only members of a single species to be included in a PHOG super-orthology group; some of these will correspond to actual inparalogs while others will be multiple entries and/or isoforms of the same gene in protein sequence databases. The two boxed subtrees (PHOG-S 1 and PHOG-S 2) correspond to super-orthology groups by this definition, with PHOG-S 2 including a possible inparalogous subtree with human genes 2a, 2b and 2c. In contrast, the Schistosoma mansoni and yeast genes have no super-orthologs. Standard phylogenetic orthology prediction protocols consider only the tree topology, including the S. mansoni gene in an orthology group with the Gene 2 clade. However, PHOG uses both tree distance and topology to enhance orthology identification precision; since the tree distances between the S. mansoni gene and genes in PHOG-S 1 are smaller than those between it and genes in PHOG-S 2, it is excluded from PHOG-S 2. This toy example also illustrates the nontransitivity of the standard definition of orthology, which requires only that the most recent common ancestor of two genes correspond to a speciation event. By this definition, the yeast gene is orthologous to Mouse Gene 1 and Mouse Gene 2, and to Rat Gene 1 and Rat Gene 2 and to all of the other sequences in the tree. However, Mouse Gene 1 is clearly not orthologous to Rat Gene 2 (they are paralogs, since they are related by gene duplication).
Comparison of major orthology-prediction methods
Orthology-prediction methods fall into two main classes: ‘graph-based’ and ‘phylogenetic tree-based’ (or simply ‘phylogenetic’) [32, 33]. Graph-based methods perform pair-wise sequence comparisons between whole genomes, typically using all-versus-all BLAST, and then construct a graph with genes as nodes and edges weighted by pair-wise similarity scores. Each method uses its own technique to cluster this graph to identify orthologs. Reciprocal BLAST Hit (RBH), COGs [34], InParanoid [35], OrthoMCL [36], eggNOG [37], ClusTr [38], ProtoNet [39] and Systers [40] are examples of graph-based methods. Note that since graph-based orthology prediction methods are based on BLAST—a local alignment protocol—they are not designed to distinguish between sequences sharing a common domain architecture and those having only local matches, increasing the potential for annotation errors.
Phylogenetic methods of orthology prediction analyze gene trees (or, more precisely, multi-gene trees containing groups of paralogs and orthologs) to localize duplication events on the tree and separate orthologs from paralogs; phylogenetic methods also enable biologists to perform more fine-tuned analyses, e.g. to discriminate between orthologs and super-orthologs [30]. Gene trees are estimated from multiple sequence alignments (MSAs) of homologs, although co-estimation of a protein MSA and phylogeny is also possible, [41, 42]. As noted in numerous studies, phylogenetic methods have been shown to have greater precision than graph-based methods [43], but the combined dependency on human expertise and the computational cost of phylogenomic analyses has limited their large-scale application [44].
Most phylogenetic orthology prediction methods employ a process called ‘tree reconciliation’, overlaying the gene tree with a trusted species tree, to parsimoniously infer speciation, duplication and gene loss events; examples of these include EnsemblCompara [45], RIO [30], Orthostrapper [46] and NOTUNG [47]. Tree reconciliation may be complicated for any number of reasons. First, a reliable species tree may not be available; this is particularly true in microbes, due to rampant horizontal gene transfer. Second, incongruence between the gene tree and species tree [48] is a frequent problem. Incongruence may stem from ‘incomplete lineage sorting’ (see e.g. [49]), horizontal gene transfer, errors in the MSA, sequence fragments or insufficient information available to the phylogenetic reconstruction (e.g. a small number of sites [50, 51], as shown in Figure 2). Phylogenetic methods making use of a gene tree but not requiring reconciliation with a species tree include LOFT [52], PhylomeDB [53], COCO-CL [54] and PHOG [55].
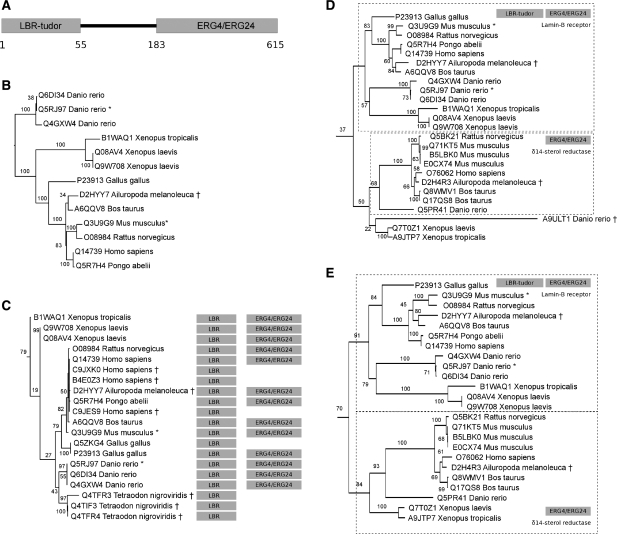
Figure 2:
Phylogenetic analysis of a human Lamin-B receptor (UniProt sequence Q14739). Orthologs selected by TreeFam in mouse and zebrafish (Danio rerio) are indicated with an asterisk. Sequence fragments are marked with a dagger. (A) Pfam domain architecture for Q14739. (B) Maximum likelihood (ML) tree of proteins sharing the same domain architecture identified using FlowerPower. (C) ML tree of proteins aligning to the N-terminal LBR_tudor domain; a subtree of the full tree is shown, restricted to the vertebrate lineage. Pfam domains found for the full-length amino acid sequences are displayed at right. (D) and (E) ML trees of sequences matching the C-terminal ERG4/ERG24 domain (restricted as in C to the vertebrate lineage) constructed using RAxML (D) and FastTree (E) respectively. Super-orthology groups are boxed with dashed lines; sequences within each super-orthology group have identical domain architectures and functions. In both D and E, the upper subtree contains the human Lamin-B receptor and orthologs; sequences in the lower subtree are missing the N-terminal LBR-tudor domain. Note that zebrafish protein A9ULT1 included by RAxML (albeit with low bootstrap support) was excluded by FastTree, allowing predicted super-orthologs in the lower subtree of E to expand to include the two Xenopus sequences. Homologs to Q14739 were retrieved using the PhyloBuilder webserver [25]; FlowerPower global–global homology clustering (i.e. requiring a common domain architecture) was used for the tree shown in B, and global–local mode was used for the domain phylogenies shown in C and D. Multiple sequence alignments for B–D were constructed with MAFFT [71], followed by masking columns with >70% gaps. Maximum likelihood trees were constructed using RAxML [64] with the JTT+Γ model and 20 discrete γ-rate categories, and for E using FastTree [72] with the same parameters. The statistical support of branches was evaluated by 100 bootstrap replicates. Trees were rooted using the mid-point method.
The Berkeley PhyloFacts Orthology Group (PHOG) [55] algorithm makes use of both tree topology and tree edge lengths to identify orthologs based on gene trees in the PhyloFacts Phylogenomic Encyclopedia [11]; a tree-distance threshold allows biologists control over the precision–recall trade-off and to target specific taxonomic distances. The PHOG webserver is available at http://phylofacts.berkeley.edu/orthologs/.
The KEGG resource [56] uses a novel approach to cluster proteins into orthologous groups that is distinct from both phylogenetic and graph-based approaches: new sequences are included based on local similarity to sequences already in a KEGG orthology group.
Orthology prediction based on phylogenies for individual domains
We present here a simple protocol that can be applied in high-throughput to identify orthologs for a protein sequence of interest.
Clustering protocols designed to retrieve proteins agreeing at the domain architecture level have been proposed. A popular solution to this problem uses a simple coverage criterion, e.g. requiring proteins to align over 70% of their lengths; this rule of thumb is reasonable for moderately sized proteins but may fail on longer proteins. FlowerPower [57] uses subfamily hidden Markov models [23] to iteratively retrieve and align homologous proteins followed by alignment analysis to provide high precision in selecting homologs with the same domain architecture.
However, there are circumstances under which a domain-based phylogeny may be preferable to one that is based on global similarity. First, requiring homologs to align well over their entire lengths—neither much longer nor shorter, and making very few insertions or gaps relative to other sequences in a cluster—can be overly restrictive, such that even orthologs from closely related species can sometimes be rejected. Disagreement with the consensus structure for the family most commonly arises from errors in the underlying gene model(s) but can also stem from natural structural variability, particularly at the N- and C-termini. In other cases, a reasonable number of homologs may be retrieved but none may be functionally informative. In such cases, it can be desirable to restrict the region used for phylogenetic analysis and orthology identification to one or more evolutionarily conserved subregions.
Given the ubiquity of domain architecture rearrangements and the problems associated with promiscuous domains, does it make sense to infer orthology on the basis of a single domain? In fact, orthology-prediction methods such as InParanoid and OrthoMCL are based on BLAST scores and thus, inherently local, and domain-based orthology prediction is the specific objective of some methods and resources (e.g. RIO [30], Orthostrapper [58], the HOPS resource [59] and PHOG [55]).
A domain-based phylogeny estimation protocol is relatively straightforward. In the first step, functional or structural domains are identified for a protein of interest; functional domains are typically identified using Pfam [60] while structural domains can be identified using protein structure prediction methods such as PHYRE [61]. Each functional or structural domain can then be used as a starting point to identify homologs in sequence databases such as UniProt [62] using BLAST, PSI-BLAST [63] or related tools. The homologous subregions of database hits (i.e. subsequences of the full-length proteins) are extracted, and a multiple sequence alignment is constructed. A gene tree can then be estimated from the MSA constructed for this domain; many methods are available for this step, including maximum likelihood (e.g. RAxML [64]), neighbor-joining [65] and Bayesian approaches (e.g. MrBayes [66]). Trees constructed using this protocol are likely to include sequences whose overall domain architectures differ, particularly if the selected domain is promiscuous. The tree is then used as input to a phylogenetic ortholog identification method.
The primary advantage of using a domain-based phylogenetic ortholog prediction protocol over a phylogenetic analysis based on whole proteins is the increased number of sequences that can be included in a phylogenetic reconstruction. A domain-based clustering protocol requires only that sequences agree along the selected domain; variability outside this region is tolerated. Due to this relaxed criterion for homolog selection, domain-based clustering protocols have increased robustness to both gene model errors and natural structural variation across a family, provided these occur outside the selected domain. Gene model errors are quite common in eukaryote genomes due to the presence of introns in many genes, but are also found in bacterial genomes (~10% of bacterial genes and a higher fraction of eukaryotic genes have gene model errors) [67]. As shown in Figure 2 and explained below, domain-based phylogenies can help biologists flag possible gene model errors for examination and revision.
Including additional homologs in a phylogenetic analysis is valuable for two reasons. First, thorough taxon sampling is known to be important for phylogenetic tree accuracy [68–70]. Second, because of the sparsity of experimental data [2], including additional homologs in a phylogenetic reconstruction increases the likelihood of a functionally informative ortholog being identified.
The main limitation of a domain-based phylogenetic analysis, whether for orthology identification or for other purposes, is the dependence of phylogenetic methods on sufficient site data (i.e. the length of the input multiple sequence alignment) as a source of phylogenetic signal [50, 51]. In phylogenomic methods of species phylogeny estimation, many orthologous genes can be concatenated into a supermatrix with thousands of sites providing ample phylogenetic signal [21], but in reconstructing phylogenies for protein superfamilies, we are limited to a far more finite quantity: protein structural domains range from a low of approximately 50 residues to a few hundred residues in length [12], and Pfam functional domains can be much smaller; some represent short repeat regions of only 20-odd amino acids. Phylogenies estimated from such short MSAs are rarely accurate, simply due to insufficient information. In some cases, the errors may be relatively minor, such that orthologous sequences cluster correctly into subtrees, but with errors in the branching order (i.e. the branching order relating these orthologs may not agree with the known species phylogeny). However, orthology prediction methods that require gene tree topologies for predicted orthologous groups to agree with trusted trees may fail on these data. This is illustrated in Figure 2.
Promiscuous domains present a significant challenge to orthology prediction: all methods of orthology prediction that are based either implicitly (as in graph-based methods) or explicitly (as in domain-based phylogenetic methods) on local alignment can incorrectly cluster proteins with different domain architectures into orthology groups. Since changes in domain architecture can dramatically change the function of a protein, and proteins with different multi-domain architectures will have nonhomologous regions, such predicted orthologs are clearly errors and should be rejected.
How robust are phylogenetic methods of orthology prediction to these data? In fact, domain-architecture intermingling is infrequent within subtrees corresponding to super-orthologs, due to the extreme stringency of this evolutionary relationship. We expect that this correspondence between domain architecture and proximity in the phylogenetic tree is due to the evolutionary pressures at the domain level to maintain a particular subfamily-specific function, i.e. a functional and/or structural variant that is tuned for that particular domain architecture.
A final complication in phylogenetic orthology prediction (whether based on a single domain or for full-length proteins) is the presence of sequence fragments and alternate isoforms or duplicate entries of the same gene. Each of these types of data complicates a phylogenetic analysis. For sequences in fully sequenced genomes, it can be possible to remove duplicate entries and to select one representative protein for each gene. But, when these duplicates are not culled at the outset, or when a whole genome is not available to enable this kind of redundancy filtering, duplicate entries can appear to be duplicated genes in the same genome (i.e. inparalogs) instead of the same gene in different forms. In fact, sequence fragments can cause actual errors in the phylogenetic tree topology; in some cases, these will result in discordance between the gene tree topology and a trusted species tree.
These issues are illustrated in Figure 2, in which we constructed phylogenies for homologs to human Lamin-B receptor, a 615-amino acid protein with an N-terminal LBR_tudor domain 55 amino acids in length and a C-terminal ERG4_ERG24 domain roughly 430 amino acids in length. Phylogenies were estimated based on sequences aligning globally to the human Lamin-B receptor and for the two domains separately. Comparing the three different estimated phylogenies and their impact on orthology prediction reveals the challenges of domain-based phylogenies versus those based on global similarity, and the advantages of using different domains for phylogenetic analysis.
As one would expect, all of the orthologs found in the common domain architecture phylogeny are also found in the Pfam domain trees. However, the N-terminal LBR_tudor domain phylogeny includes proteins from chicken (Gallus gallus), human and pufferfish (Tetraodon nigroviridis) not found in the two other phylogenies; all are fragments containing only the amino-terminal LBR_tudor domain. Genome locus analysis shows the novel human and chicken sequences to correspond to incorrect gene models for the same gene for which the correct (full length) protein was included in all three trees. In contrast, neither of the other phylogenies included any orthologs from pufferfish, demonstrating the utility of using domain-based phylogenies to increase ortholog-identification recall (of the three pufferfish proteins, two were removed from UniProt since the phylogeny was constructed; only Q4TIF3 remains, annotated by UniProt as a fragment).
Figure 2 also demonstrates the impact of limited site data on the accuracy of the phylogenetic tree topology. For instance, the phylogenetic placement of the mouse ortholog (Q3U9G9) is incorrect (albeit with high bootstrap support) in the tree estimated from the LBR_tudor domain, but is correct in the other two phylogenies, which were estimated using many more sites. Phylogenetic methods of ortholog identification that require subtree topologies to agree with trusted species phylogenies might reject these orthologs.
Finally, analysis of the Pfam domains of full-length proteins included in a phylogeny shows a close correspondence between proximity in the phylogenetic tree and agreement at domain architecture, allowing the inference of overall domain architecture for sequence fragments and increasing the reliability of domain-based orthology prediction.
DISCUSSION
In this article, we have presented some of the challenges involved in reconstructing phylogenies for protein functional domains and in inferring orthologs based on those phylogenies. We have focused on specific issues in automated methods of orthology identification for high-throughput application, e.g. for functional annotation of whole genomes.
There are two main advantages of domain-based orthology prediction. First, the relaxed criterion of local clustering protocols tends to result in many more sequences being included than when global similarity is required. This enhances taxon sampling, with resulting potential improvements to the phylogenetic tree topology accuracy and to ortholog prediction based on these trees. Second, if the aim of ortholog identification is functional annotation, including additional sequences in a phylogenetic reconstruction also increases the likelihood that functionally informative sequences will be retrieved.
Improvements to taxon sampling using domain-based phylogenies must be balanced against the dependency of phylogenetic reconstruction on sufficient site data. This is generally not a problem in reconstructing species phylogenies, where phylogenomic methods can incorporate data from many genes into a gene matrix with thousands of sites, but is a definite problem with protein sequences that are at most a few hundred residues in length. If the number of sites is restricted further to a single functional domain, which may be a few dozen amino acids in length, accuracy can degrade significantly. These short domains are also challenging to phylogenetic ortholog-identification methods that compare subtrees in gene trees against trusted species phylogenies. As we have shown, phylogenies based on short domains can have errors in branching order within orthologous subtrees due to limited phylogenetic signal; incongruities with trusted species phylogenies should be expected for these types of alignments.
A related issue is the presence of sequence fragments stemming from gene model errors; these can cause a subtree topology to disagree with a trusted species phylogeny with corresponding errors in phylogenetic orthology-prediction methods that require gene trees to agree with species phylogenies. However, as we have shown, domain-based phylogenies can help flag proteins with gene model errors so that these can be examined and potentially revised.
An additional challenge arises in the context of domain-based phylogenies: when sequences with different overall multi-domain architectures are included in a phylogenetic reconstruction, the potential for errors increases dramatically. This risk is mitigated by the strong tendency for sequences sharing a common domain architecture to cluster closely on a phylogenetic tree, provided that the domain selected as the basis for the phylogenetic tree topology is sufficiently long.
How long must a domain be for an accurate phylogeny? Our observations, admittedly based on a relatively small sample of phylogenetic trees estimated for Pfam domains of different lengths, support the findings reported in the literature that ‘size matters’. We cannot provide any general rules, but advise caution in using phylogenies based on domains of <70 amino acids in length. Additional studies are needed to quantify the correspondence between protein domain length and phylogenetic accuracy.
Interpreting domain-based phylogenies, when sequences included are drawn from different multi-domain architectures, requires particular attention: in a standard gene family phylogeny, internal nodes of a tree will correspond to either speciation or duplication events, but in domain-based phylogenies a third node label may be necessary to represent gene fusion and fission events.
In summary, domain-based phylogenetic ortholog identification can confer real advantages over phylogenetic methods based on whole proteins, but with some caveats: domains should be long enough to prevent problems with insufficient site data and care must be taken in interpreting phylogenies when data sets are drawn directly from standard sequence databases due to the high frequency of gene model errors, sequence fragments and multiple entries for the same gene. With these issues in mind, ortholog-prediction accuracy can be enhanced using a domain-based phylogenetic protocol.
Key Points.
Proteins are composed of structural domains that fold independently in solution; the ordered series of these structural domains is a protein’s ‘domain architecture’.
Domain architectures can be modified by evolutionary processes such as gene fusion and fission events; changes in domain architecture can be accompanied by dramatic shifts in protein function.
Most orthology-prediction methods are based on local alignment scores, and phylogenetic methods of orthology prediction can be derived from phylogenetic trees for individual functional domains; both graph-based and phylogenetic approaches can be prone to error when proteins included in the analysis have different domain architectures, with errors particularly likely to happen in the case of promiscuous domains.
Advantages of domain-based phylogenies include improved tree topology accuracy from increased taxon sampling, a greater degree of experimental data supporting functional annotations and the detection of gene model errors.
Phylogenetic ortholog identification methods that require gene tree topologies to agree with trusted species phylogenies may have limited accuracy when sequence fragments are included in an analysis or when the domain used as the basis of evolutionary tree construction is short, reducing the phylogenetic signal.
FUNDING
The National Science Foundation (grant 0732065 to K.S.); the Department of Energy (grant DE-SC0004916 to K.S.).
Acknowledgements
We are grateful to Tandy Warnow and to reviewers for helpful comments.
Biographies
Kimmen Sjölander is a computational evolutionary biologist specializing in algorithm development for various tasks associated with phylogenomic methods of protein structure and function prediction.
Ruchira Datta is a mathematician specializing in computational method development for orthology prediction.
Yaoqing Shen is a computational biologist working on protein family evolution.
Grant Shoffner is a bioinformaticist working on the PhyloFacts resource.
References
- 1.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.du Plessis L, Skunca N, Dessimoz C. The what, where, how and why of gene ontology–a primer for bioinformaticians. Brief Bioinform. 2011 doi: 10.1093/bib/bbr002. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sjölander K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20:170–9. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- 5.Schnoes AM, Brown SD, Dodevski I, et al. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Addou S, Rentzsch R, Lee D, et al. Domain-based and family-specific sequence identity thresholds increase the levels of reliable protein function transfer. J Mol Biol. 2009;387:416–30. doi: 10.1016/j.jmb.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 7.Galperin MY, Koonin EV. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1:55–67. [PubMed] [Google Scholar]
- 8.Abascal F, Valencia A. Automatic annotation of protein function based on family identification. Proteins. 2003;53:683–92. doi: 10.1002/prot.10449. [DOI] [PubMed] [Google Scholar]
- 9.Basu MK, Carmel L, Rogozin IB, et al. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18:449–61. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 11.Servant F, Bru C, Carrere S, et al. ProDom: automated clustering of homologous domains. Brief Bioinform. 2002;3:246–51. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- 12.Corpet F, Servant F, Gouzy J, et al. ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res. 2000;28:267–69. doi: 10.1093/nar/28.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuff AL, Sillitoe I, Lewis T, et al. The CATH classification revisited–architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–4. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreeva A, Howorth D, Chandonia JM, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–25. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilks WR, Audit B, De Angelis D, et al. Modeling the percolation of annotation errors in a database of protein sequences. Bioinformatics. 2002;18:1641–9. doi: 10.1093/bioinformatics/18.12.1641. [DOI] [PubMed] [Google Scholar]
- 16.Jones CE, Brown AL, Baumann U. Estimating the annotation error rate of curated GO database sequence annotations. BMC Bioinformatics. 2007;8:170. doi: 10.1186/1471-2105-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown D, Sjölander K. Functional classification using phylogenomic inference. PLoS Comput Biol. 2006;2:e77. doi: 10.1371/journal.pcbi.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sjölander K. Getting started in structural phylogenomics. PLoS Comput Biol. 2010;6:e1000621. doi: 10.1371/journal.pcbi.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sankararaman S, Sha F, Kirsch JF, et al. Active site prediction using evolutionary and structural information. Bioinformatics. 2010;26:617–24. doi: 10.1093/bioinformatics/btq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–7. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 21.Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–75. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 22.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 23.Brown DP, Krishnamurthy N, Sjölander K. Automated protein subfamily identification and classification. PLoS Comput Biol. 2007;3:e160. doi: 10.1371/journal.pcbi.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mi H, Guo N, Kejariwal A, et al. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–52. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glanville JG, Kirshner D, Krishnamurthy N, et al. Berkeley Phylogenomics Group web servers: resources for structural phylogenomic analysis. Nucleic Acids Res. 2007;35:W27–32. doi: 10.1093/nar/gkm325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnamurthy N, Brown DP, Kirshner D, et al. PhyloFacts: an online structural phylogenomic encyclopedia for protein functional and structural classification. Genome Biol. 2006;7:R83. doi: 10.1186/gb-2006-7-9-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 28.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 29.Dessimoz C, Boeckmann B, Roth AC, et al. Detecting non-orthology in the COGs database and other approaches grouping orthologs using genome-specific best hits. Nucleic Acids Res. 2006;34:3309–16. doi: 10.1093/nar/gkl433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zmasek CM, Eddy SR. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonnhammer EL, Koonin EV. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–20. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 32.Gabaldon T, Dessimoz C, Huxley-Jones J, et al. Joining forces in the quest for orthologs. Genome Biol. 2009;10:403. doi: 10.1186/gb-2009-10-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuzniar A, van Ham RC, Pongor S, et al. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–51. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–80. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen LJ, Julien P, Kuhn M, et al. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–4. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petryszak R, Kretschmann E, Wieser D, et al. The predictive power of the CluSTr database. Bioinformatics. 2005;21:3604–9. doi: 10.1093/bioinformatics/bti542. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan N, Sasson O, Inbar U, et al. ProtoNet 4.0: a hierarchical classification of one million protein sequences. Nucleic Acids Res. 2005;33:D216–8. doi: 10.1093/nar/gki007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meinel T, Krause A, Luz H, et al. The SYSTERS Protein Family Database in 2005. Nucleic Acids Res. 2005;33:D226–9. doi: 10.1093/nar/gki030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC, Sjölander K. SATCHMO: sequence alignment and tree construction using hidden Markov models. Bioinformatics. 2003;19:1404–11. doi: 10.1093/bioinformatics/btg158. [DOI] [PubMed] [Google Scholar]
- 42.Hagopian R, Davidson JR, Datta RS, et al. SATCHMO-JS: a webserver for simultaneous protein multiple sequence alignment and phylogenetic tree construction. Nucleic Acids Res. 2010;38:W29–34. doi: 10.1093/nar/gkq298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F, Mackey AJ, Vermunt JK, et al. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sjölander K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20:170–9. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- 45.Vilella AJ, Severin J, Ureta-Vidal A, et al. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–35. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storm CE, Sonnhammer EL. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics. 2002;18:92–9. doi: 10.1093/bioinformatics/18.1.92. [DOI] [PubMed] [Google Scholar]
- 47.Chen K, Durand D, Farach-Colton M. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol. 2000;7:429–47. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- 48.Hahn MW. Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution. Genome Biol. 2007;8:R141. doi: 10.1186/gb-2007-8-7-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollard DA, Iyer VN, Moses AM, et al. Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2006;2:e173. doi: 10.1371/journal.pgen.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. B.M.E. Moret UR, Warnow T. Sequence length requirements for phylogenetic methods. In: Guigo R, Gusfield D (eds). Proceedings of 2nd International Workshop on Algorithms in Bioinformatics (WABI'02), Lecture Notes in Computer Science 2452, Berlin: Springer, 2002;343–56.
- 51.Saitou N, Nei M. The number of nucleotides required to determine the branching order of three species, with special reference to the human-chimpanzee-gorilla divergence. J Mol Evol. 1986;24:189–204. doi: 10.1007/BF02099966. [DOI] [PubMed] [Google Scholar]
- 52.van der Heijden RT, Snel B, van Noort V, et al. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huerta-Cepas J, Bueno A, Dopazo J, et al. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008;36:D491–6. doi: 10.1093/nar/gkm899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jothi R, Zotenko E, Tasneem A, et al. COCO-CL: hierarchical clustering of homology relations based on evolutionary correlations. Bioinformatics. 2006;22:779–88. doi: 10.1093/bioinformatics/btl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta RS, Meacham C, Samad B, et al. Berkeley PHOG: PhyloFacts orthology group prediction web server. Nucleic Acids Res. 2009;37:W84–9. doi: 10.1093/nar/gkp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S, Hattori M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–7. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnamurthy N, Brown D, Sjölander K. FlowerPower: clustering proteins into domain architecture classes for phylogenomic inference of protein function. BMC Evol Biol. 2007;7:S12. doi: 10.1186/1471-2148-7-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hollich V, Storm CE, Sonnhammer EL. OrthoGUI: graphical presentation of Orthostrapper results. Bioinformatics. 2002;18:1272–3. doi: 10.1093/bioinformatics/18.9.1272. [DOI] [PubMed] [Google Scholar]
- 59.Storm CE, Sonnhammer EL. Comprehensive analysis of orthologous protein domains using the HOPS database. Genome Res. 2003;13:2353–62. doi: 10.1101/gr1305203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bateman A, Coin L, Durbin R, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, et al. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–25. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 62.Bairoch A, Apweiler R, Wu CH, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–9. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST–a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–7. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 64.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–63. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 65.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 66.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 67.Pati A, Ivanova NN, Mikhailova N, et al. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods. 2010;7:455–7. doi: 10.1038/nmeth.1457. [DOI] [PubMed] [Google Scholar]
- 68.Heath TA, Zwickl DJ, Kim J, et al. Taxon sampling affects inferences of macroevolutionary processes from phylogenetic trees. Syst Biol. 2008;57:160–6. doi: 10.1080/10635150701884640. [DOI] [PubMed] [Google Scholar]
- 69.Pollock DD, Zwickl DJ, McGuire JA, et al. Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol. 2002;51:664–71. doi: 10.1080/10635150290102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 2002;51:588–98. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
- 71.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–98. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 72.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]