Abstract
Recent development of deep sequencing technologies has facilitated de novo genome sequencing projects, now conducted even by individual laboratories. However, this will yield more and more genome sequences that are not well assembled, and will hinder thorough annotation when no closely related reference genome is available. One of the challenging issues is the identification of protein-coding sequences split into multiple unassembled genomic segments, which can confound orthology assignment and various laboratory experiments requiring the identification of individual genes. In this study, using the genome of a cartilaginous fish, Callorhinchus milii, as test case, we performed gene prediction using a model specifically trained for this genome. We implemented an algorithm, designated ESPRIT, to identify possible linkages between multiple protein-coding portions derived from a single genomic locus split into multiple unassembled genomic segments. We developed a validation framework based on an artificially fragmented human genome, improvements between early and recent mouse genome assemblies, comparison with experimentally validated sequences from GenBank, and phylogenetic analyses. Our strategy provided insights into practical solutions for efficient annotation of only partially sequenced (low-coverage) genomes. To our knowledge, our study is the first formulation of a method to link unassembled genomic segments based on proteomes of relatively distantly related species as references.
Keywords: Chondrichthyes, trained gene prediction, next generation sequencing, genome assembly, orthology
INTRODUCTION
Recent innovations in massively parallel sequencing technologies have enabled individual laboratories to conduct de novo genome sequencing projects [1]. However, due to the shorter reads they produce compared with the traditional Sanger method, genome sequencing based on new technologies will yield more genome sequences that are not well assembled [2, 3], which in turn will hinder comprehensive annotation of the protein-coding landscape. In particular, a major challenge is the identification of protein-coding sequences split into multiple unassembled genomic segments. Currently, the main approach to overcoming this problem is to assemble the low-coverage genome using a high-coverage reference genome as template (see [4] for a review). Several methods based on this approach have been proposed [5–8], but due to their strong reliance on the template genome, they do not cope well with duplication, loss and translocation of genomic segments, and require the reference genome to be evolutionarily close to the low-coverage genome. Ensembl pipeline [9] also detects split genes (http://www.ensembl.org/info/docs/compara/homology_method.html), but this approach is not readily applicable to newly sequenced genomes by individual researchers, because it is not yet well documented or available outside the Ensembl pipeline.
In this study, we focused on the genome of a species in the order of chimaeras (Chimaeriformes, Holocephali, Chondrichthyes), Callorhinchus milii (also called elephant shark or ghost shark; Figure 1), previously sequenced as genome survey sequence (GSS) only with 1.4× coverage [10]. This species was initially selected as a target of shot-gun genome sequencing solely because of its small genome size [11]. The available assembly for the C. milii genome includes 647 131 contigs that amount to 0.77 Gb in total, for its estimated haploid genome size of 0.91 Gb [10]. Although sequenced with the Sanger method, the N50 length of the contigs is 912 bp, suggesting that many protein-coding genes are split into different contigs that have remained unassembled. Chondrichthyans (cartilaginous fishes, namely chimaeras, sharks, rays and skates) have been studied in diverse biological fields including immunology [12], developmental biology [13, 14] and endocrinology [15]. A reliable annotation of the C. milii genome would answer the growing demand for molecular sequence resources for this animal group.
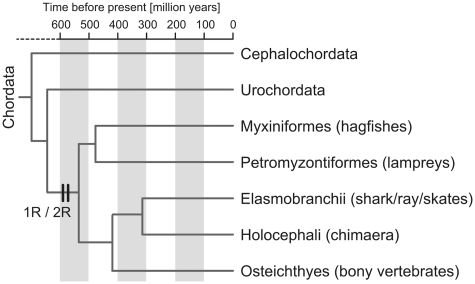
Figure 1:
Phylogenetic position of the C. milii. Overview of phylogenetic relationships between major vertebrate lineages are depicted with their estimated divergence times [16, 17]. The target of this study, C milii, is a species in the subclass Holocephali. The timing of the first-round (1R) and second-round (2R) of whole genome duplications is based on [18].
Using the C. milii genome as a test case, we constructed a repeat library for this species and trained gene prediction program AUGUSTUS [19]. To better annotate this fragmentary assembly, we implemented a novel algorithm ESPRIT (Establishing Split Protein-coding Regions In Tentative genomes) to identify possible linkages between protein-coding portions derived from a single genomic locus (mostly corresponding to exons) split into unassembled contigs. We validated the approach and fitted parameters using an artificially fragmented human genome and an early mouse genome assembly (NCBI m34 assembly). We then applied ESPRIT to the C. milii genome and evaluated its performance based on full-length C. milii sequences from GenBank and phylogenetic analyses. As we elaborate below, the strategy applied to a genome only partially sequenced provides insights into practical solutions for efficient genome annotation.
METHODS
Preparation of repeat-masked genome assembly
The 1.4× coverage C. milii genomic assembly was downloaded from the Elephant Shark Genome Project website (http://esharkgenome.imcb.a-star.edu.sg/resources.html). To construct a repeat library customized for this genome, we ran RepeatModeler with default parameters (http://www.repeatmasker.org/RepeatModeler.html), which detected 501 types of repeat elements that are ≥15-nt long and present as >15 copies in the C. milii genome. Based on that, RepeatMasker (http://www.repeatmasker.org/) masked 42.2% of the C. milii genome, consisting of 38.7% of repeat elements from the library and 3.5% of simple repeats, low complexity stretches, small RNAs and satellites.
Training gene set
To prepare a gene set used for training by the gene prediction software AUGUSTUS [19], we employed three different approaches described below. First, we surveyed the database NCBI GenBank [20] (as of 7 June 2010) and identified 149 entries of C. milii protein-coding genes. Those lacking the N-terminus were excluded. To remove redundant sequences, we used BLASTP [21] in order to cluster those proteins into similarity groups (sequences that had a hit with bits score >70 in BLASTP constituted a similarity group). From each of these similarity groups, only one representative protein was retained. Finally, we selected 22 C. milii proteins from GenBank. Second, we referred to the C. milii genome assembly itself to identify full-length protein-coding genes using ab initio gene prediction tools, GENSCAN [22] and MAKER [23]. Inside MAKER, we implemented SNAP [24] and est2genome [25] components with default parameters. To run est2genome, we input the C. milii cDNA data set available at the Elephant Shark Genome Project website (http://esharkgenome.imcb.a-star.edu.sg). All C. milii peptides predicted by GENSCAN and SNAP were subjected to BLASTP searches against human Ensembl peptides (version 57; http://www.ensembl.org/). C. milii query peptides that were aligned with a human peptide with identical lengths with no gaps and had a ≥70% similarity were retained. As a result, we identified 22, 4 and 17 genes that satisfied these criteria, based on SNAP, est2genome, and GENSCAN, respectively. The last approach employed was the CEGMA pipeline which searches for 458 core eukaryotic genes (CEGs) in a given genome based on hidden Markov models [26]. After applying this to the C. milii genomic sequences, we retrieved 37 genes.
After merging genes identified in the different approaches described above into one data set, we filtered it with a BLASTP-based search against itself to remove homogeneity within it. As a result, 90 sequences that do not have a similarity of >70% to any other sequence in the data set were retained. Because seven genes identified in GenBank were not included in the C. milii genome assembly, the final training gene set contained 83 genes (Supplementary Table S1). With this training gene set, the training module of AUGUSTUS was run to produce ‘.prob’ files customized for the C. milii genome.
Trained gene prediction
Gene prediction was performed with AUGUSTUS, using the C. milii genome assembly in which repetitive sequences are masked. Our prediction setting trained for this genome is available in the AUGUSTUS installation package as well as its web interface (http://bioinf.uni-greifswald.de/augustus/submission).
Detection of split protein-coding regions
Split protein-coding regions were inferred by comparative genomics, as a new step in the OMA orthology detection pipeline [27, 28]. The protein-coding landscape of the genome survey sequence (GSS) of C. milii was compared with those of nine chordate species (human, mouse, anole lizard, chicken, African clawed frog, zebrafish, medaka, Ciona intestinalis and Branchiostoma floridae). After identifying all pairs of putative homologs (the all-against-all step of the OMA algorithm [28]), we perform an exhaustive search of triplets of proteins such that two proteins in the partial genome map to a common gene in one of the reference genomes. The two putative fragments are not allowed to overlap for more than 5 amino acids (we assume that overlapping contigs have already been merged as part of the assembly process). If the distances between the two putative fragments and the reference gene vary more than a given threshold, the triad is discarded. Indeed, given the hypothesis that both the two or more protein-coding regions in the genome to be annotated emerge from one split gene and assuming a similar average rate of evolution for the two parts, their distance to homologs should not be much different.
Formally, we search for all triads (x1, x2, y) such that
x1, x2 are proteins in the partial genome encoded by different genes, y is a protein from a reference genome;
the pairs (x1, y) and (x2, y) have significant Smith-Waterman pairwise alignments over at least 60% of the length of x1, x2 respectively (Gonnet-matrix score ≥250, which approximately corresponds to an E-value of 1e–18 [29]).
the Smith-Waterman pairwise alignments (x1, y) and (x2, y) have at most five overlapping residues in terms of sequence y;
the evolutionary distances (x1, y) and (x2, y) are not significantly different. This is implemented using a Z-test:
where d(x,y) is the maximum-likelihood estimator (Gonnet matrices over amino acids; [29]) of the evolutionary distance between sequence x and y based on the Smith-Waterman pairwise alignments, σ2(x,y) its variance, and tol a tolerance parameter (in units of normal standard deviation).
Once all triads are identified, ESPRIT identifies candidate pairs of split proteins (x1, x2) under the additional conditions that x1 and x2 be part of a common triad in at least MinRefGen reference genomes. The final requirement for candidate pairs to be predicted by ESPRIT is that neither x1 nor x2 is part of a candidate pair involving a third protein x3. This is to ensure that the predictions are consistent, at the expense of a potential decrease in recall.
Genomic PCR
Genomic DNA of C. milii was provided by Byrappa Venkatesh. Based on C. milii genomic contigs AAVX01108858 and AAVX01626565, we designed two gene-specific primers, 5′-TCAAGTTCCAGGAGGTCA-3′ and 5′-CCACGAGGAAGATGATGAT-3′, respectively. PCR was performed using 100 ng of the genomic DNA with GC-rich PCR System (Roche) following the manufacturer’s instruction. The amplified DNA fragment was purified with MinElute PCR purification kit (Qiagen) and sequenced on Genetic Analyzer 3130 (Applied Biosystems). The obtained sequence was deposited in the EMBL sequence database under the accession ID FR872381.
RESULTS
Assessing coverage of the protein-coding landscape in the C. milii genome
The coverage of the C. milii genome assembly was previously estimated to be ~75% [11]. We used the CEGMA pipeline [26] to estimate the percentage of genes covered by the current assembly [32]. CEGMA searches for 248 core eukaryotic genes (CEGs) in a given genome and can report which of them are completely or partially covered. In the C. milii genome, the software identified only 49 genes (including 35 partial genes) out of the 248 CEGs (19.8%). To assess whether this low coverage might be due to fast evolving proteins that have diverged beyond recognition, we restricted the analysis to the subgroup of 65 CEGs with high conservation (‘Group 4’ as defined in [26]). Of those, the software identified 14 (22%) in the C. milii genome. These observations suggest that the coverage of protein-coding regions is considerably lower than the coverage estimated for the entire genome (~75%).
Gene prediction
To train the gene prediction program AUGUSTUS [19], we prepared a set of 83 non-redundant genes from C. milii entries in GenBank, from high-confidence de novo gene predictions, and from the core eukaryotic genes identified by the CEGMA pipeline [26] (Supplementary Table S1). This was followed by the execution of the AUGUSTUS gene prediction module on the C. milii genome with repetitive sequences masked (see Methods section). This gene prediction produced 22 079 gene models (Supplementary Table S2). The total coding sequence of the predicted genes was ~8-Mb long (~1% of the assembly), and the median length of individual coding sequences (CDSs) was 267 bp.
Detecting split protein-coding regions: ESPRIT
The main challenge with low-coverage genomes that we address here is the issue of split protein-coding regions, i.e. individual genes present on several unassembled contigs. As a result, fragments of the same gene are wrongly annotated as being distinct genes. This can confound orthology identification because these fragments erroneously appear as duplicates, and duplicates within the same genome are typically used to identify paralogous relations across species (e.g. [33, 34]). To identify split protein-coding regions, we took a comparative genomics approach and searched full-length homologous counterparts in other genomes. If any two non-overlapping fragments of the low-coverage genome consistently map to a single gene in other reference genomes, it suggests that the two fragments might be parts of the same gene (Figure 2). To minimize spurious predictions, we further require that the two candidate fragments be at approximately the same evolutionary distance to the reference gene, that such appropriate reference genes be found in several reference genomes, and that the two candidate fragments be not involved in another, potentially conflicting, prediction (see Methods section).
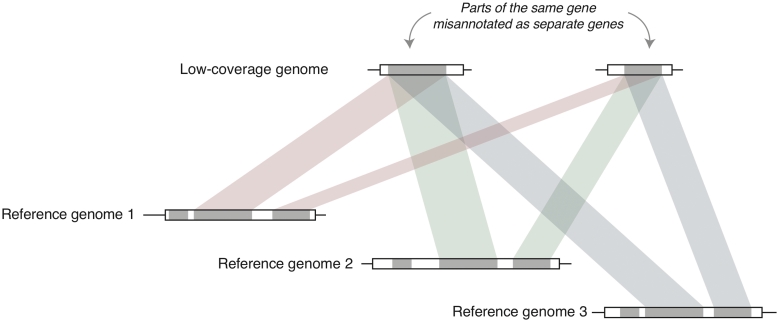
Figure 2:
Method overview to identify split protein-coding regions. Genes are depicted as boxes, protein-coding regions are indicated with gray areas. White areas indicate introns or untranslated regions (UTRs). If two CDSs annotated as part of different genes in the partial genome consistently map to non-overlapping parts of a common gene in several reference genomes, this suggests that the two CDSs are part of a split protein-coding region and should be merged (refer to Methods section for details).
Validation and parameter sensitivity analysis
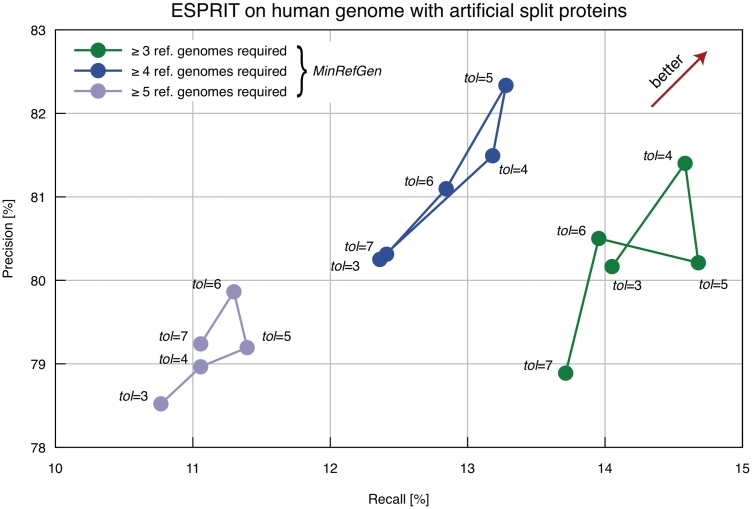
To evaluate ESPRIT and estimate suitable parameters, we used two complementary approaches. First, we introduced artificial splits into 8% of all human CDS (Ensembl v55; [35]) and sought to recover these splits using ESPRIT. As reference genomes, we used 6 other vertebrates (mouse, anole lizard, chicken, African clawed frog, zebrafish, medaka) and two invertebrates (Ciona intestinalis and Branchiostoma floridae). We evaluated precision and recall of ESPRIT for various combinations of the two main parameters. Overall, precision was ~80%, while recall was ~10–15% (Figure 3). Manual inspection of some of the false positives revealed that practically all mistakes were due to the confounding effect of naturally occurring CDSs belonging to paralogous genes. As for the false negative predictions, most of them were due to the short size of one or both artificial fragments: in 64% of all artificially-introduced splits, at least one of the resulting CDS fragments was <80 amino acid long. The short length of these fragments makes them more prone to spurious alignment and precludes accurate estimation of evolutionary distances. Increasing the parameter MinRefGen (minimal number of required matching reference genomes) reduced recall but did not always result in higher precision. Likewise, we observed local optima for the parameter tol (tolerance in the difference in evolutionary distance between two fragments and their corresponding full-length homolog in the reference genome, expressed in normal standard deviations) both in terms of precision and recall. In general, note that relaxing parameters does not necessarily increase coverage because this can lead to an increase in the number of conflicting candidate pairs as well, and those are excluded from predictions by ESPRIT (see Methods section).
Figure 3:
Evaluation of ESPRIT based on human genome with artificial split introduced in 8% of all CDSs. The y-axis depicts the percentage of accurate predictions [(true positive)/(true positive + false positive)]. The x-axis depicts the percentage of all artificial splits (2071 cases) that were covered by the predictions [(true positive)/(true positive + false negative)]. The figure shows performance for various combinations of the two main parameters: the minimum number of reference genome with full-length homolog (parameter MinRefGen) and tolerance value (parameter tol) for the difference in distance between two fragments and their full-length homolog.
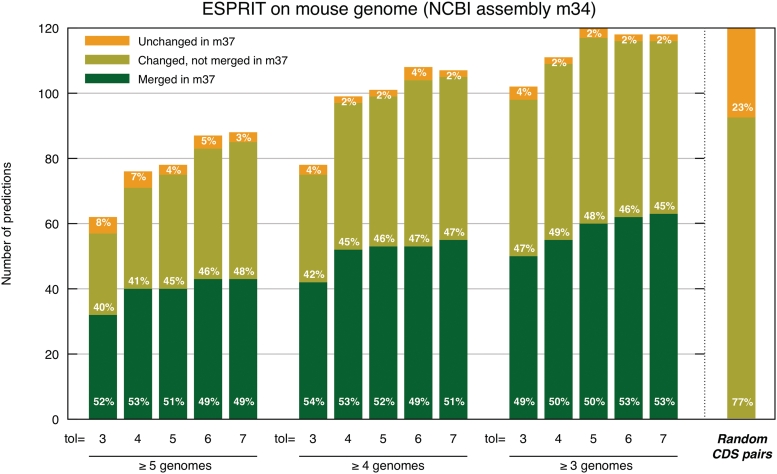
Second, we exploited improvements in the NCBI assembly of the mouse genome between 2005 (m34 release) and 2007 (m37 release, the most recent one to date). We ran ESPRIT on the m34 assembly using as reference genomes the same set as above, but with human and without mouse. Predicted split proteins were divided into three categories depending on their fate in the m37 assembly: pairs of predicted split proteins that were merged in the m37 assembly (confirmed cases), pairs of which one or both proteins changed in m37 (i.e. deleted, altered, split, or merged with another fragment not predicted by ESPRIT), and pairs unchanged in m37. Overall, about half the predictions on m34 were merged outright in m37, with most of the other half having changed in one or both sequences in m37 (Figure 4). This is in stark contrast to the distribution of randomly selected CDS pairs from m34, of which <0.1% were merged, 77% changed in one or both sequences, and 23% stayed unchanged. For example, illustrated in Figure 5a, ESPRIT correctly predicted that Ensembl genes ENSMUSG00000057751 and ENSMUSG00000029031 in Ensembl mouse v35 (m34 assembly) are merged into the gene ENSMUSG00000057751 (multiple EGF-like-domains 6 gene) in Ensembl mouse v48 (m37 assembly). Note however that in terms of coverage, ESPRIT is only able to predict a small fraction of all gene pairs merged between m34 and m37 (between 32 and 63 pairs out of a total of 1917).
Figure 4:
Evaluation of ESPRIT based on comparison between NCBI m34 and m37 mouse assemblies (CDS from Ensembl v35 and v48 respectively). The y-axis measures predictions of split genes made by ESPRIT on the m34 assembly according to their fate in the m37 assembly. Contrary to random pairs (far right), about half the pairs identified by ESPRIT were merged in the m37 assembly. In addition, the fraction of pairs that did not undergo any change between m34 and m37 is considerably lower than for random pairs.
Figure 5:
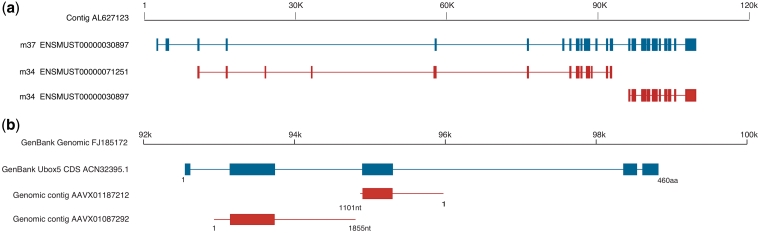
Confirmation of predicted split genes (in red) from full-length reference counterparts (in blue). (a) The fragments from m34 assembly (Ensembl v.35) are correctly predicted as being part of the same gene, as they have been merged in the m37 assembly (in blue, Ensembl v48). (b) The two C. milii genomic contigs (in red) contain fragments of Ubox5 gene. The scale depicts a 8-kb stretch of the GenBank genomic DNA entry FJ185172. Blue boxes show protein-coding sequences encoding the GenBank peptide entry ACN32395.1. Red lines show the two C. milii genomic contigs, each of which contains a single exon (red boxes) of the Ubox5 gene.
Application of ESPRIT to the C. milii genome
Based on what emerged as the best parameters from the analyses on human and mouse genomes (MinRefGen = 4, tol = 5), we applied ESPRIT to the C. milii genome with the same set of reference genomes as above (mouse, anole lizard, chicken, African clawed frog, zebrafish, medaka, Ciona intestinalis and Branchiostoma floridae). ESPRIT predicted 642 pairs of split protein coding regions (Supplementary Data S3).
To evaluate these predictions with independent information, we referred to GenBank which contained 172 entries for C. milii protein-coding genes on the nuclear genome (as of 11 March 2011). While most GenBank entries had zero or one identical match among peptide sequence data set predicted on the partial C. milii genome assembly, we identified nine C. milii protein-coding gene entries that were represented by more than one C. milii contigs and used them to evaluate the predictions. Of the nine, ESPRIT identified only one of the GenBank entries, namely the genomic sequence FJ185172 harboring the U box containing 5 (Ubox5) gene encoding a ubiquitin ligase [36]. For this protein, ESPRIT identified two peptides predicted on two different C. milii genomic contigs, AAVX01087292 and AAVX01187212 as a continuous pair (Figure 5b). This prediction was confirmed by the fact that the two genomic contigs show 100% identity to the single GenBank entry (Figure 5b).
To detect the eight other cases, we relaxed the distance tolerance parameter tol from 5 to 2. This increased the number of predictions from 642 to 666 (Supplementary Data S4), and allowed identification of three additional fragments part of the same GenBank entry, namely tubulin tyrosine ligase-like family member 6 (ttll6; FJ824599, genomic), rhodopsin (EF565167, mRNA), and long wavelength sensitive opsin 1 (LWS1; EF565165, mRNA) genes. As with the Ubox5 gene fragments, these cases were all supported by the continuity of their respective genomic contigs. More generally, this analysis indicates that parameters optimized for a given set of genomes do not necessarily perform well in other settings (see Discussion section below).
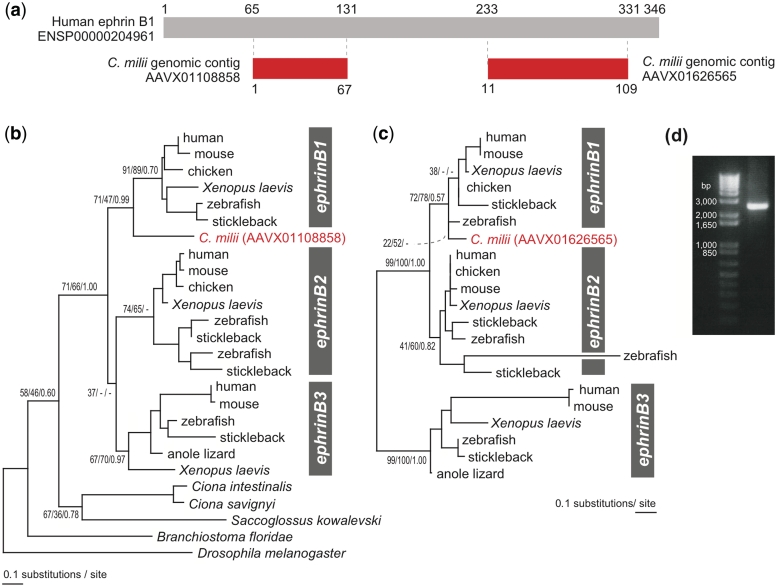
To evaluate predictions without corresponding sequences in GenBank, we performed molecular phylogenetic analyses. In principle, multiple fragments derived from a single locus are expected to show the same phylogenetic relationship with homologs although confidence in phylogenetic inference tends to be low when the sequences are short. Figure 6 shows an example of C. milii ephrin B1 gene covered by two distinct, non-overlapping genomic contigs. Molecular phylogenetic reconstruction based on the two predicted peptide sequences resulted in closer relationship of the C. milii sequence with the ephrin B1 group than with ephrin B2 or B3 groups (Figure 6b and c). Our genomic PCR using primers designed on the two contigs successfully amplified a DNA stretch bridging them (Figure 6d).
Figure 6:
Callorhinchus milii genomic contigs containing fragments of ephrin B1 gene. (a) Portions of the human ephrin B1 peptide (gray box) homologous to C. milii peptides (red boxes) predicted on two genomic contigs. Numbers below and above the boxes indicate amino acid positions. (b) Molecular phylogenetic tree of ephrin B group members based on the portion (56 amino acid residues) covered by the C. milii contig AAVX01108858. (c) Molecular phylogenetic tree of ephrin B group members based on the portion (75 amino acid residues) covered by the C. milii contig AAVX01626565. In (b) and (c), support values at nodes are bootstrap probabilities in the ML method, bootstrap probabilities in the NJ method, and posterior probabilities in the Bayesian inference. Maximum-likelihood and neighbor joining trees were inferred using PhyML [30] with the WAG+Γ4 model. We employed MrBayes for Bayesian inference [31]. In (c), invertebrate sequences are excluded because amino acid sequences in the range of the peptides on which the tree is based are not conserved in invertebrates. (d) A gel image of amplification of a DNA stretch bridging the two C. milii fragments in (a).
Finally, we assessed the effect of ESPRIT on the coverage of the C. milii proteome as estimated by the CEGMA pipeline. We bridged the 666 pairs of contigs identified by ESPRIT (MinRefGen = 4, tol = 2) with an intervening stretch of 100 unknown bases (‘N’) in the orientation predicted by ESPRIT. On this prediction-based version of the genome assembly, the number of CEGs identified by CEGMA increased from 49 genes (including 35 partial genes) to 54 genes (including 38 partial genes), which constitutes an improvement of 9%. Given that CEGMA only considers 248 genes in the C. mili proteome, the magnitude of this increase is in line with our previous observations that the predictions by ESPRIT are mostly correct.
DISCUSSION
Challenges to annotating de novo non-model genome sequences
As previous studies demonstrate, the basic features of gene models differ largely among animals [37], and gene prediction programs such as AUGUSTUS can be trained effectively to adapt to these species-specific features [38]. In a previous effort, protein-coding regions had been identified with BLASTX, and repetitive sequences had been detected with RepeatMasker as well as with intra-genomic BLASTN searches for high copy number (>500) repeats [10]. To annotate the protein-coding and repetitive landscape of the genome in a more specific manner, we constructed a species-specific repeat library including low-copy de novo repeats and ran the trained gene prediction program AUGUSTUS on the masked genomic sequences.
Challenges to annotating low-coverage genomes
The biggest difficulty ESPRIT faces in detecting split genes is posed by the confounding effect of fragments that belong to phylogenetically closely related but distinct paralogous genes. They can be indistinguishable from bona fide split genes. Nevertheless, to minimize this problem, ESPRIT implements two key ideas. First, it requires absence of substantial overlap (>5 amino acids) between potential split protein-coding sequences. Indeed, it can be expected that any regions with significant overlap have already been merged during assembly of raw genomic reads, perhaps with the exception of highly polymorphic regions. Thus, most annotated peptide sequences with substantial overlap are likely to come from distinct genes. Still, this does not exclude all confounding cases because fragments from distinct paralogs might happen to be non-overlapping. Second, ESPRIT requires similar evolutionary distances between two potential fragments and their full-length counterpart in reference genomes (modulated with the tolerance parameter tol). However, this idea is also not infallible as the rate of sequence evolution can vary considerably among different parts of the same gene, and paralogs resulting from gene duplication specific to the partial genome lineage can be similarly distant to all their homologs in reference genomes.
At a more practical level, a considerable challenge lies in identifying optimal parameters. Our analyses on the manipulated human genome show that the two parameters of ESPRIT cannot be optimized in isolation of one another: there is a compensatory effect between MinRefGen and tol. Thus, tightening one parameter may require relaxing the other. More importantly, the parameters fitted in the analyses on human and mouse genomes did not translate well for the partial C. milii genome in which we obtained superior results by considerably lowering tol. This could be partly due to much shorter lengths of predicted C. milii CDSs. Furthermore, C. milii represents a unique evolutionary lineage, namely Chondrichthyes, that has no large-scale genomic sequence resource for any other species. As a result, all reference genomes used in this study were relatively distant from C. milii, with time of divergence over 400 million years ago (Figure 1) [16]. In contrast, the human and mouse lineages separated <100 million years ago. Another confounding factor specific to early vertebrates could be the timing of whole genome duplications at the base of the vertebrate radiation, which exacerbates the challenges with paralogs discussed above [18]. Together, these observations suggest that optimal parameters vary depending on the coverage of the partial genome of interest, the availability of closely related reference genomes, and the peculiarities of the underlying phylogeny.
Because of these challenges, the predictions of ESPRIT cannot be expected to be error-free. This is especially true if the genome of a species in question is susceptible to lineage-specific genome duplications, such as teleost fish genomes with numerous duplicates with respect to tetrapods [39]. In such cases additional caution is advised. Nonetheless, under the notion that computational analyses cannot fully compensate for low-quality or incomplete data, ESPRIT can be effective at identifying plausible targets to be experimentally verified, for instance by RT–PCR or genomic PCR to fill the gap between the split parts as demonstrated in Figure 6d. Previously, scaffolding a genome with RNA-seq was attempted [40], and scaffolding transcript sequences with the proteome of closely related species was also shown to be effective [41]. To our knowledge, this study formulates for the first time an algorithm to link unassembled genomic segments based on proteomes of relatively distantly related species as references.
FUTURE PERSPECTIVES
The present study lays the groundwork for developments in several directions. In particular, ESPRIT could be improved by exploiting information beyond the amino acid sequence of annotated CDSs. A straightforward development would be to use the nucleotide sequences, which might be beneficial when analyzing relatively close sets of species (e.g. primates, flies). The location of potential fragments on their respective contigs might also be exploited to improve split gene detection as genes close to the ends might be more likely to be fragmented. The use of features pertaining to typical gene structure such as absence of start/stop codons may also enable improvements. More generally, the gene confidence scores determined by gene predictors such as AUGUSTUS might already consider some of these aspects and integrate them in a way that could be readily used in this context as well.
In terms of algorithmic development, ESPRIT currently rejects conflicting candidate predictions. A more efficient approach would be to resolve these conflicts and report them along with confidence scores. This could be implemented by refining the current threshold-based approach into a probabilistic model, and using the relative probabilities of conflicting scenarios to decide which to report, if any.
Key Points.
High-throughput sequencing is enabling de novo genome sequencing projects, but due to technical constraints and diminishing returns, many projects are limited to poorly assembled, low-coverage genome surveys. As a result, fragments of the same gene can be present on multiple, unassembled contigs, and are wrongly annotated as being distinct genes.
We present ESPRIT, a novel method that identifies such split genes on the basis of full-length counterparts in reference genomes.
We validate ESPRIT based on artificially fragmented human genome, on improvements between early and recent mouse genome assemblies.
We applied ESPRIT to an only partially sequenced genome of Callorhinchus milii, and validated its prediction on comparison with experimentally validated sequences from GenBank and on phylogenetic analyses.
SUPPLEMENTARY DATA
Supplementary data are available online at http://bib.oxfordjournals.org/. The code for ESPRIT is available as part of the OMA stand-alone program (http://omabrowser.org/standalone), and is free for non-commercial use.
FUNDING
This study was supported by grants from German Research Foundation (DFG) (program ID: KU2669/1-1) and Young Scholar Fund from University of Konstanz (to S.K.) and also supported by Konstanz Research School Chemical Biology (KoRS-CB) (to T.M.) and an ETH Independent Investigators' Research Award (to C.D.).
Supplementary Material
Acknowledgements
The authors thank Mario Stanke for suggestions in using the gene prediction program AUGUSTUS and Byrappa Venkatesh for gifting genomic DNA of C. milii. The authors gratefully acknowledge infrastructure support by the ETH Zurich cluster Brutus, on which part of the computations were performed. Finally, the authors thank Julia Jones, Naoki Irie, Adrian Altenhoff, Maria Anisimova, Manuel Gil, Jean Muller, Olivier Poch, Nives Škunca and Julie Thompson for helpful discussions and/or comments on the manuscript.
Biographies
Christophe Dessimoz is a senior postdoc and lecturer in the CBRG group at ETH Zurich, Switzerland. He strives to understand the forces that shape genes, genomes and species, using computational and statistical methods.
Stefan Zoller is a PhD student in the CBRG group at ETH Zurich, Switzerland. He is interested in combining computer science and genomics to learn about evolution.
Tereza Manousaki is a PhD student in Konstanz Graduate School Chemical Biology (KoRS-CB) and the Chair of Zoology and Evolutionary Biology, Department of Biology, University of Konstanz, Germany. She is interested in exploring various aspects of vertebrate genomes.
Huan Qiu was a postdoc in the Chair of Zoology and Evolutionary Biology, Department of Biology, University of Konstanz, Germany and is now a postdoc in Bigelow Laboratory for Ocean Sciences, West Boothbay Harbor, Maine, USA.
Axel Meyer is a principal investigator in KoRS-CB and a professor in the Chair of Zoology and Evolutionary Biology, Department of Biology, University of Konstanz, Germany. He is interested in phenotypic and molecular evolution.
Shigehiro Kuraku is a principal investigator in KoRS-CB and an assistant professor in the Chair of Zoology and Evolutionary Biology, Department of Biology, University of Konstanz, Germany.
References
- 1.Kircher M, Kelso J. High-throughput DNA sequencing—concepts and limitations. Bioessays. 2010;32:524–36. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 2.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–5. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birney E. Assemblies: the good, the bad, the ugly. Nat Methods. 2011;8:59–60. doi: 10.1038/nmeth0111-59. [DOI] [PubMed] [Google Scholar]
- 4.Pop M. Genome assembly reborn: recent computational challenges. Brief Bioinform. 2009;10:354–66. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bainbridge MN, Warren RL, He A, et al. THOR: targeted high-throughput ortholog reconstructor. Bioinformatics. 2007;23:2622–4. doi: 10.1093/bioinformatics/btl513. [DOI] [PubMed] [Google Scholar]
- 6.Husemann P, Stoye J. Phylogenetic comparative assembly. Algorithms Mol Biol. 2010;5:3. doi: 10.1186/1748-7188-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter DC, Schuster SC, Huson DH. OSLay: optimal syntenic layout of unfinished assemblies. Bioinformatics. 2007;23:1573–9. doi: 10.1093/bioinformatics/btm153. [DOI] [PubMed] [Google Scholar]
- 8.van Hijum SA, Zomer AL, Kuipers OP, et al. Projector 2: contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acids Res. 2005;33:W560–6. doi: 10.1093/nar/gki356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubbard TJ, Aken BL, Ayling S, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkatesh B, Kirkness EF, Loh YH, et al. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesh B, Tay A, Dandona N, et al. A compact cartilaginous fish model genome. Curr Biol. 2005;15:R82–3. doi: 10.1016/j.cub.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 12.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–22. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Cole NJ, Currie PD. Insights from sharks: evolutionary and developmental models of fin development. Dev Dyn. 2007;236:2421–31. doi: 10.1002/dvdy.21268. [DOI] [PubMed] [Google Scholar]
- 14.Coolen M, Menuet A, Chassoux D, et al. New York: Cold Spring Harbor Protocols; 2008. The Dogfish Scyliorhinus canicula: a Reference in Jawed Vertebrates. doi:10.1101/pdb.emo111. [DOI] [PubMed] [Google Scholar]
- 15.Kawauchi H, Sower SA. The dawn and evolution of hormones in the adenohypophysis. Gen Comp Endocrinol. 2006;148:3–14. doi: 10.1016/j.ygcen.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Inoue JG, Miya M, Lam K, et al. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol. 2010;27:2576–86. doi: 10.1093/molbev/msq147. [DOI] [PubMed] [Google Scholar]
- 17.Heinicke M, Naylor G, Hedges S. Cartilaginous fishes (Chondrichthyes) In: Kumar S, Hedges S, editors. The Timetree of Life. New York: Oxford University Press; 2009. pp. 320–7. [Google Scholar]
- 18.Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- 19.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–25. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 20.Benson DA, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2009;37:D26–31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 23.Cantarel BL, Korf I, Robb SM, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–96. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mott R. EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci. 1997;13:477–8. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- 26.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–7. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 27.Altenhoff AM, Schneider A, Gonnet GH, et al. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–94. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth AC, Gonnet GH, Dessimoz C. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics. 2008;9:518. doi: 10.1186/1471-2105-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonnet GH, Cohen MA, Benner SA. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–5. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 31.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 32.Parra G, Bradnam K, Ning Z, et al. Assessing the gene space in draft genomes. Nucleic Acids Res. 2009;37:289–97. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dessimoz C, Boeckmann B, Roth AC, et al. Detecting non-orthology in the COGs database and other approaches grouping orthologs using genome-specific best hits. Nucleic Acids Res. 2006;34:3309–16. doi: 10.1093/nar/gkl433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Heijden RT, Snel B, van Noort V, et al. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flicek P, Amode MR, Barrell D, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–6. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gwee PC, Tay BH, Brenner S, et al. Characterization of the neurohypophysial hormone gene loci in elephant shark and the Japanese lamprey: origin of the vertebrate neurohypophysial hormone genes. BMC Evol Biol. 2009;9:47. doi: 10.1186/1471-2148-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yandell M, Mungall CJ, Smith C, et al. Large-scale trends in the evolution of gene structures within 11 animal genomes. PLoS Comput Biol. 2006;2:e15. doi: 10.1371/journal.pcbi.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picardi E, Pesole G. Computational methods for ab Initio and comparative gene finding. In: Carugo O, Eisenhaber F, editors. Data Mining Techniques for the Life Sciences. New York: Springer Verlag; 2010. [Google Scholar]
- 39.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–45. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- 40.Mortazavi A, Schwarz EM, Williams B, et al. Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Res. 2010;20:1740–7. doi: 10.1101/gr.111021.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surget-Groba Y, Montoya-Burgos JI. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 2010;20:1432–40. doi: 10.1101/gr.103846.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.