Abstract
Orthology is a powerful refinement of homology that allows us to describe more precisely the evolution of genomes and understand the function of the genes they contain. However, because orthology is not concerned with genomic position, it is limited in its ability to describe genes that are likely to have equivalent roles in different genomes. Because of this limitation, the concept of ‘positional orthology’ has emerged, which describes the relation between orthologous genes that retain their ancestral genomic positions. In this review, we formally define this concept, for which we introduce the shorter term ‘toporthology’, with respect to the evolutionary events experienced by a gene’s ancestors. Through a discussion of recent studies on the role of genomic context in gene evolution, we show that the distinction between orthology and toporthology is biologically significant. We then review a number of orthology prediction methods that take genomic context into account and thus that may be used to infer the important relation of toporthology.
Keywords: positional orthology, toporthology, homology, synteny, genome alignment
THE CONCEPT OF POSITIONAL ORTHOLOGY
The strengths and weaknesses of the orthology concept
In a seminal article published more than four decades ago, Fitch introduced the terms orthology and paralogy in order to distinguish between different classes of homologous genes [1]. These terms have seen heavy use in the last two decades, primarily due to the abundance of data produced by whole-genome sequencing projects [2]. The relation of orthology, which describes genes that have diverged from a common ancestor due to a speciation event, has been particularly important. Orthologs are the best choices for estimating a species phylogeny because their evolutionary history reflects the species history [3]. In addition, a pair of genes in two genomes are more likely to share a common function if they are orthologs, even though orthology is purely an evolutionary relationship that is independent of function [2, 3]. Thus, the establishment of orthology is critical in the transfer of functional annotations between genomes.
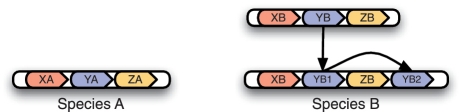
Despite the success of the term orthology, it is apparent from the literature that there has been some friction with its definition. This definitional tension arises because people often use the term ‘orthologs’ to express the notion of ‘equivalent genes’ between two or more genomes, i.e. genes that are the most comparable in terms of their evolutionary histories, irrespective of function. Unfortunately, the relation of orthology is not precise enough for this notion, as not all orthologous relationships appear to be ‘equivalent’. The evolutionary scenario in Figure 1 illustrates this issue. In this scenario, a segmental duplication event occurs in species B creating two copies of gene YB (YB1 and YB2), both of which are orthologous to the single copy of gene Y (YA) in species A. Orthology does not distinguish between the two copies of Y in species B, even though they differ in their genomic context. We would like to express that YA and YB1 are more comparable than YA and YB2, but the terms orthology and paralogy do not allow us to do so. The underlying issue here is that as far as the concepts of homology, orthology and paralogy are concerned, a genome is simply a ‘bag of genes’, i.e. the relative positions of genes within a genome are irrelevant for these concepts.
Figure 1:
An example of the limitations of the orthology concept. A segmental duplication creates two copies of gene Y in species B. Gene YA is orthologous to both YB1 and YB2. However, we would like to distinguish the (YA, YB1) relationship from the (YA, YB2) relationship because YA and YB1 are most representative of the ancestral copy of Y.
To address this issue, many groups have invented terminology to describe a special subset of orthologous relationships. The new terms include ‘true exemplar’ [4], ‘true ortholog’ [5], ‘main ortholog’ [6, 7] and ‘positional ortholog’ [8]. The term ‘positional ortholog’ (or ‘positional homolog’) is currently the most popular, having been used to describe relationships between genes in prokaryotes [8–12], yeast [13] and vertebrates [14]. In general, all of these terms are used to describe orthologous genes whose genomic positions best reflect the genomic position of their most recent common ancestor. For example, Swidan et al. [10] define two genes to be positional orthologs ‘if they are orthologs and have preserved their relative positioning or genomic contexts in the genomes’.
Unfortunately, despite the best intentions of the inventors of these refinements of orthology, the new terms suffer from the fact that they are operationally defined. That is, the terms are defined with respect to some analysis of the relative positions of genes in extant genomes. The definitions depend critically on the exact computations and thresholds used to determine whether a pair of genes are positional orthologs. For example, to assess the positional conservation of a pair of related genes, [9] examined the two genes immediately adjacent to the each of the members of the pair, while [15] used a neighborhood of six genes.
In this review, we argue that we would be better served by a theoretical definition that describes the evolutionary history of genes. The distinction between these two types of definitions is the same as that between the concepts of protein sequence similarity and protein homology [3]. While high sequence similarity is a good predictor of homology, we do not define protein homology in terms of similarity. Instead, we say that two proteins are homologous if they are derived from a common ancestor. Similarly, the evolutionary history of a gene family, rather than the current genomic positions of its members should be used to define positional orthology.
An evolutionary definition for positional orthology
To unify the various genomic-context-dependent orthology concepts described by others, we offer a theoretical definition of the term positional orthology or toporthology (the prefix ‘top’ for the Greek ‘place’). We briefly introduced this concept for understanding the goals of whole-genome alignment [16], and will now provide a more complete description.
To precisely define positional orthology, we must first introduce a series of definitions for genomic duplication events. In general, a duplication event involves the copying of a genomic segment, which may contain any number (including zero) of genes. We begin by classifying genomic duplication events as either ‘symmetric’ or ‘asymmetric’ (or using our previous terminology [16], ‘undirected’ or ‘directed’, respectively). A duplication event is ‘symmetric’ if deleting either copy of the duplicated sequence results in a genome that is identical to the original (pre-duplication) genome. Examples of symmetric duplications are tandem duplications and whole-chromosomal or whole-genome duplications. The general characteristic of such duplications is that one cannot distinguish between the two copies of the duplicated genetic material.
The alternative to a ‘symmetric’ duplication is an ‘asymmetric’ duplication, after the occurrence of which only one of the two copies of the duplicated material may be removed to undo the duplication. The copy that can be removed to return the genome to its original state is called the ‘target’ and the other copy is called the ‘source’ (equivalent terms used previously are ‘daughter’ and ‘parent’, respectively [17]). Examples of events resulting in asymmetric duplications are segmental duplications and retrotranspositions. Generally speaking, the source element remains in the ancestral position while the target element is placed elsewhere in the genome. In the scenario shown in Figure 1, an asymmetric duplication event has occurred in genome B, with gene YB1 as the source and gene YB2 as the target.
With these distinctions made between duplication events, we say that two genes are positionally homologous (topohomologous), if they are homologous and neither gene is derived from the target of an asymmetric duplication since the time of their common ancestor. Two genes are positionally orthologous (toporthologous) if they are both topohomologous and orthologous. Similarly, two genes are positionally paralogous (topoparalogous), if they are topohomologous and paralogous. We use the term atopohomologous for genes that are homologous but not topohomologous. Two genes are atoporthologous if they are both atopohomologous and orthologous. And two genes are atopoparalogous if they are both atopohomologous and paralogous.
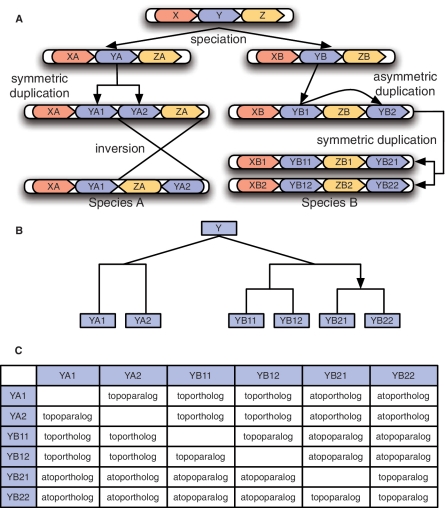
Figure 2 illustrates an evolutionary scenario that gives rise to genes with these relationships. With a gene tree drawn in the form of Figure 2B, one can determine if two genes are topohomologous by tracing up the tree to their common ancestor and noting if the traced path crosses the head of an arrow (which indicates the target of an asymmetric duplication). If the head of an arrow is encountered, then the genes are atopohomologous. Otherwise, they are topohomologous. Note that both whole chromosome and tandem duplications give rise to topohomology, as they are both symmetric duplication processes. Also, although the genomic units (e.g. YA1 and YA2) are described as single genes, they could also represent multi-gene or non-genic genomic segments. Toporthology and the other context-based relationships we have defined may also be applied to arbitrary genomic segments (including single nucleotides).
Figure 2:
A hypothetical evolutionary scenario in which we distinguish between classes of orthologs. (A) A speciation event occurs, creating species A and B. The genome of species A undergoes an undirected duplication (a tandem duplication of gene YA) followed by an inversion. Meanwhile, a directed duplication (a segmental duplication of gene YB) followed by a whole genome duplication event occurs within species B. (B) The evolutionary tree for the descendants of gene Y. The top V-shaped split represents a speciation event, while the rectangular splits represent duplications. For the one directed duplication in the tree, the arrow points towards the target copy of the duplication event. (C) The evolutionary terms used to describe the relationship between each pair of extant genes. Note that the inversion event in species A does not impact the evolutionary relationships.
Unlike the previous operational definitions for positional orthology which rely on measurements between extant genome sequences, these theoretical definitions only depend on the nature (symmetric or asymmetric) of historical duplication events. It follows from our definitions that in the absence of genomic rearrangement events, positional orthologs retain the genomic position of their most recent common ancestor. Thus, orthologs that have similar genomic contexts are more likely to be toporthologs. However, rearrangement events (e.g. inversions and transpositions) and large segmental duplications can complicate matters, and it is possible for two orthologous genes with similar contexts not to be toporthologs. In addition, like orthology, toporthology is not a one-to-one relation. For example, in the scenario depicted in Figure 2, YA1 is toporthologous to both YB11 and YB12. The motivation for concept of toporthology is not to have one-to-one relationships, but rather to distinguish those orthologs that are most comparable in terms of their evolutionary history. However, there will generally be more one-to-one toporthologs than one-to-one orthologs between a pair of genomes.
One ambiguity in the definitions we have presented above is the classification of xenologous relationships, which arise due to horizontal gene transfer (HGT) events. Are xenologous genes topohomologous or atopohomologous? One interpretation would be to say that xenologous genes can be either, depending on the duplication history of their ancestors. That is, we might choose to define xenology and topohomology independently. On the other hand, we could consider HGT to be duplication event, albeit a complicated one, where the system under consideration consists of the genomes of two species. Viewed in this light, HGT is an asymmetric duplication because only the removal of the transferred sequence in the receiving species (the target copy) returns the system to its original state. Thus, with this second interpretation, all xenologs are atopohomologs. Although both interpretations are valid, we prefer the second, as it unifies the relationships through the concepts of symmetric and asymmetric duplications. In fact, the asymmetric nature of HGT has already been used to distinguish between toporthologs and xenologs [8].
Concepts related to positional orthology
The concepts of topohomology, toporthology and topoparalogy are related to some previously described evolutionary terms. The terms of positional homology, positional orthology and positional paralogy have been used in the context of morphological characters, with similar meanings [18]. Ohnologs, defined as paralogs that originated from a whole genome duplication event, are a subset of topoparalogs [19].
Positional orthology is also closely related to what many have referred to as ‘synteny’. The term ‘synteny block’ is often used to refer to orthologous genome segments with conserved gene order. Thus, syntenic blocks are likely to contain pairs of toporthologs. Unfortunately, the term ‘synteny’ is often used incorrectly or ambiguously [20]. By itself, the word ‘synteny’ neither implies homology nor conserved order of related elements. Therefore, we advocate discontinuing the use of such widespread phrases as ‘synteny map’ and ‘synteny block’ in favor of more precise evolutionary terms. Although of similar etymology to ‘synteny’, we prefer the use of the word ‘colinear,’ as famously used by [21], in combination with evolutionary terms to describe relationships between genomic characters.
THE BIOLOGICAL SIGNIFICANCE OF GENOMIC CONTEXT
The concept of positional orthology is important because of the functional consequences of genomic context. Genes that are near each other are more likely to interact [22] and gene expression is significantly affected by genomic position (the ‘position effect’) [23]. In fact, it is hypothesized that in eukaryotes, gene order is non-random because genomic organization is important for coordinated expression [24].
Histone modifications, which play a role in the gene regulatory systems that allow for coordinated expression, have been found to be significantly different between segmental duplications and their source copies [25]. After determining the source and target of 1646 human segmental duplications using macaque as an outgroup, [25] found that the source copies had higher levels of histone modifications, H3K27me1 and H3K9me1 in particular, than their respective target copies. This asymmetry suggested that there are different evolutionary constraints on the source and targets of such duplications, with the source copy more constrained to retain its original function.
Supporting this hypothesis, a study of Caenorhabditis elegans inparalogs (with respect to Saccharomyces cerevisiae) provided evidence that after duplication, there is a tendency for only one gene of each inparalog pair to retain co-regulation with other genes [26]. In this study, the coexpression patterns of 130 Caenorhabditis elegans inparalog pairs with a single Saccharomyces cerevisiae ortholog were analyzed. It was found that the number of pairs for which exactly one member had a conserved coexpression pattern was significantly higher than would be expected if both members were under independent evolutionary constraints. Although genomic context was not analyzed for these inparalogs, we might hypothesize that the genes with conserved co-regulation were more likely to be toporthologs.
The genomic context of a gene is also associated with the gene’s rate and mode of evolution. In general, the sequences of orthologs with conserved genomic positions have been found to be under greater evolutionary constraint [8, 9, 11, 15, 27–29] than those in non-ancestral positions. Duplicated genes in non-ancestral positions due to retrotransposition or DNA-mediated transposition not only evolve more rapidly, but also are more likely to undergo positive selection [30]. The accelerated rate of sequence evolution in relocated duplicated regions occurs in both coding and non-coding sequence [31]. Although duplicate genes resulting from asymmetric duplication events generally evolve faster than their source (parent) genes, the opposite phenomenon has also been observed. A study of prokaryotic genomes found that source genes had lower sequence similarity with their orthologs in 29–38% of cases, indicating that using sequence similarity alone does not reliably predict positional orthologs and that genomic context must be taken into account [15].
Although toporthologs may be more likely to retain the function of their ancestors than atoporthologs, there are certainly exceptions. First, it is not necessarily the case that after an asymmetric gene duplication, one copy retains the ancestral function while the other diverges. One possible outcome is that both copies undergo sub-functionalization, in which each copy retains a complementary subset of the functions of the ancestral gene (see e.g. [32]). Second, even if one copy of an asymmetric duplication acquires a new function (neo-functionalization) or degenerates (non-functionalization), it is not necessarily the case that this copy will have been the target of the duplication. For example, in a study of epistatic interactions in A. thaliana, [33] discovered a recent asymmetric gene duplication for which the target retained functionality in one strain while the source was the functional copy in another strain. For large-scale asymmetric duplications that retain much of the cis-regulatory elements around the duplicated genes, we might expect this type of outcome to be more frequent, as there is less of a distinction between the source and target copies in these scenarios. Thus, toporthology inferences should not be used alone for the purpose of gene function prediction. Rather, toporthology should be used in combination with other data, such as regulatory module predictions and structural similarity, for assigning functions to genes.
THE USE OF GENOMIC CONTEXT FOR ASSESSING ORTHOLOGY
The problem of predicting orthology between whole genomes has been addressed by numerous groups [34]. None of the methods that have been developed explicitly predict positional orthology as it is defined in this article. However, there are a variety of methods that go beyond the ‘bag of genes’ model of genomes and use genomic context to predict or refine orthologous relationships. Based on the fact that gene neighborhood conservation has been used to benchmark orthology predictions [35–37] and visualized to aid in the manual inspection of orthology predictions [38–40], it is evident that genomic context provides valuable information for this problem.
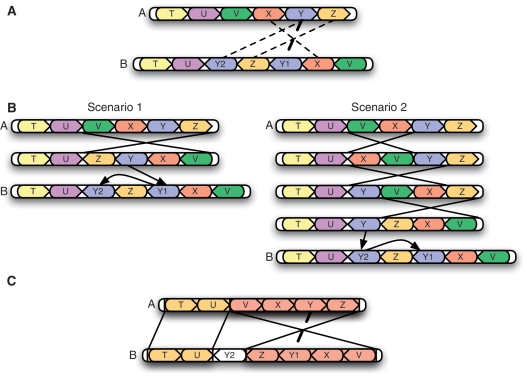
Orthology prediction methods that take genomic context into account can be classified into three categories. The methods of the first category use conserved gene order or conserved gene neighborhoods in addition to gene similarity scores to predict gene-level orthology. A second class includes methods that also predict gene-level orthology, but do so using explicit models of gene order evolution and a parsimony objective. The third class of methods includes ‘synteny block’ generators and whole-genome aligners, which instead aim to predict orthology at the nucleotide level and rely on the large regions of colinearity found between closely related genomes. Related to these methods are others that aim to classify recent duplication events based on genomic context. Such methods do not predict orthology but can be used to distinguish toporthologs from atoporthologs. We review these various classes of methods in the following sections. Figure 3 provides an example of how each class of methods uses genomic context.
Figure 3:
The three classes of orthology prediction methods that take genomic context into account. (A) Methods that consider the conservation of local gene order or gene neighborhood into account would predict Y and Y1 to be toporthologs because they have two neighbors in common, whereas Y and Y2 only have one common neighbor (where we only consider immediately adjacent neighbors). (B) Methods based on gene order evolution models infer toporthology by finding an assignment between the gene sets that minimizes the number of evolutionary events required to explain the genomes. Assignment of Y and Y2 as toporthologs (scenario 2) requires four events (three inversions and one duplication), whereas assignment of Y and Y1 as toporthologs (scenario 1) only requires two events (one inversion and one duplication). Thus, Y and Y1 would be predicted as toporthologs by these methods. (C) Whole-genome alignment methods disregard gene boundaries and find colinear orthologous segments between genomes (the shaded blocks). From this alignment, we would infer that Y and Y1 are toporthologs because they fall within in the same block.
Orthology prediction with conserved gene order or neighborhood
Because toporthologs tend to retain the genomic positions of their ancestors, the genomic contexts of a pair of similar genes can provide valuable information for assessing the likelihood that the genes are orthologous or toporthologous. However, genomic context is usually only informative for orthology prediction between closely related species, as gene order often evolves faster than amino acid sequences [41]. This is particularly true for prokaryotic genomes, in which gene order changes rapidly, even disrupting operons [42]. Even if gene neighborhoods are rarely conserved between a pair of genomes, when they can be found, they are significant evidence for orthology.
The relationship between conserved gene context and positional orthology has been used to help predict positional orthologs in a variety of ad hoc ways for genomic studies. One of the simplest approaches has been to first compute best reciprocal hit (BRH) pairs between two gene sets and then predict as orthologous the BRH pairs and any pair of similar genes adjacent (in both genomes) to a BRH pair [43]. Similarly, in [9], ‘positional homologs’ were operationally defined as pairs of genes that were both inferred to be homologous and were adjacent to at least one other homologous pair. Other studies have examined a larger neighborhood of genes surrounding a gene pair to evaluate genomic context conservation. In both [15] and [44], the number of orthologous pairs within six neighboring genes (three upstream and downstream) of a given gene pair was used to determine if its genomic context was conserved. Others have used external ‘synteny information’ to infer positional orthologs [45, 14]. In a testament to the information gained from genomic context, [44] found that mammalian orthologs predicted from gene neighborhood conservation alone were 93% concordant with those predicted by INPARANOID [6], a method that relies exclusively on protein sequence similarity.
A first class of general methods that incorporate genomic context information into orthology prediction formulate the problem as finding an optimal matching between the gene sets of a pair of genomes, with an objective function that takes into account gene neighborhood conservation. One of the earliest context-aware methods falls into this class, as it finds a maximum matching in a weighted bipartite graph [46]. The vertices of the graph correspond to the genes of two genomes and the edges represent significant gene sequence similarities. The weight of an edge is initially set to the sequence similarity of its connected genes and then boosted based on the amount of gene neighborhood conservation surrounding the gene pair. Orthologs are predicted by finding a maximum matching in the graph, which can be solved efficiently with the Hungarian method [47]. A recent method, EGM [48], uses a remarkably similar approach, with some differences in how the edge weights are determined from gene neighborhood conservation. The method of [49] also seeks to find a matching, but instead uses the notion of a ‘common interval’, a pair of genomic intervals with the same gene family content, to formulate its objective. These methods produce a one-to-one orthology mapping between a pair of genomes, which likely makes them specific to toporthologs, at the expense of missing one-to-many or many-to-many toporthologs.
Another method that uses idea of combining gene similarity scores with gene neighborhood conservation is SYNERGY [50]. However, unlike the matching-based methods, SYNERGY does not seek a one-to-one map between a pair of genomes. Instead, it combines the hit-clustering and tree-building strategies for orthology prediction to predict orthologs and paralogs across multiple genomes. Genomic context is taken into account by using gene pair distances that are a combination of a protein-level evolutionary distance estimate and a gene neighborhood similarity score. These distances are used both for clustering orthologs and building phylogenetic trees. For their yeast data set, the authors of SYNERGY found that taking genomic context into account played a minor role in predicting orthologs, but significantly improved the estimated gene trees.
A second class of general methods use gene order conservation to disambiguate the orthology relationships for genes that do not have a clear ‘best hit’ in another genome, in terms of sequence similarity [51–55]. These methods first identify blocks of conserved gene order from gene pairs that are predicted to be one-to-one orthologs with high confidence. Genes for which an orthology assignment is ambiguous based on sequence similarity and that fall within a conserved gene order block have their candidate orthologs filtered according to the block. This filtering often results in the discovery of more unambiguous orthologous gene pairs. While this general technique most often results in one-to-one orthology assignments, a number of methods also handle many-to-many orthology relations, particularly those resulting from tandem duplication [52, 53]. A related method, OrthoParaMap [56], uses conserved gene order to distinguish between orthologs and paralogs, but only does so for gene families that are provided to it as input. The HomoloGene database [57] is also said to use conserved gene order as part of its build procedure, although the exact details of this procedure have not been described.
Orthology prediction with gene order evolution models
An independent line of methods has emerged that aims to predict orthology by reconstructing parsimonious gene order evolution scenarios with explicit models of genome evolution. This area of research began with Sankoff’s introduction of the ‘true exemplar problem’ [4]. In this problem, we are given an ordered set of genes from two genomes, with each gene labeled by the gene family to which it belongs. Many families may consist of just one gene from each genome, representing high confidence ortholog pairs, while others may contain multiple genes from a single genome. The goal is to select a pair of genes, one per genome, from each gene family such that if you remove all unselected genes, the distance between the resulting gene orders is minimized. The selected genes are called the ‘true exemplars’ and are suggested to be the best estimates of (positional) orthology between the genomes. In the original article, the distance between two gene orders was defined as either the reversal distance (the minimum number of inversion events required to transform one gene order into the other) or the breakpoint distance (the number of breaks in gene order colinearity).
Although this problem was proven to be NP-hard [58], several algorithms have been developed for its solution. Sankoff originally provided a branch-and-bound algorithm. Later, a divide-and-conquer algorithm was developed that was shown to be more efficient than the original branch-and-bound approach [59]. As the primary focus of these algorithms was to calculate evolutionary distance and not ortholog assignments, the SOAR method [60] was developed as a complete approach for predicting orthology between a pair of genomes with the reversal distance minimization objective. SOAR decomposes the problem into a pair of new optimization problems, minimum common partition and minimum cycle decomposition, for which efficient approximate and heuristic algorithms are used.
These initial methods have been extended to model evolutionary events other than inversions. One of the first extensions was to simultaneously model both reversal and duplication events [61]. Later, a theoretical framework for orthology assignment by minimizing the number of inversion, deletion, insertion and duplication events was presented [62]. Following SOAR, MSOAR [7] was developed to additionally model duplications, translocations and chromosomal fusions and fissions. MSOAR 2.0 [63] improved on MSOAR by handling tandem duplication events, and MultiMSOAR [64] and MultiMSOAR 2.0 [65] extended these methods to multiple genomes. Algorithms and challenges of the parsimony approach to orthology prediction were recently reviewed [66].
The advantages of these approaches are that they explicitly model genome evolution and do not rely on the existence of conserved gene order or neighborhoods. Thus, they may be more effective in utilizing genomic context information, even when genomes have significantly diverged. Although they are perhaps more appealing in principle, these approaches are significantly more complex than those that simply use conserved gene order or neighborhoods.
Whole-genome alignment
Whole-genome alignment, the task of predicting nucleotide-level orthology relations [16, 67], relies heavily on genomic context information. This task is generally restricted to genomes that have significant nucleotide-level similarity and some amount of colinearity. While methods for aligning whole genomes are generally focused on nucleotide-level assignments, the output of such methods can easily be used to establish gene-level orthology predictions as well. Whole genome aligners vary in whether they estimate one-to-one, many-to-many or one-to-many (in the case of reference-based alignment) orthologs, most restrict themselves to predicting one-to-one toporthologs [16]. Methods for this task can generally be categorized into three groups: hierarchical, local or hybrid methods [16].
The hierarchical approach is to first construct a high-level colinear orthology map between a set of genomes and then compute a nucleotide-level global alignment (which requires colinearity) on each colinear orthologous block specified by the map. Examples of this approach are the combinations of Mercator and MAVID [68], Enredo and PECAN [69], Shuffle-LAGAN and LAGAN [70], and Nucmer and SeqAn::TCoffee (Mugsy) [71]. The problem of determining a colinear orthology map between a set of genomes is often referred to as the ‘synteny block’ finding problem. Numerous methods have been developed for this task alone [72–77].
In the local approach to whole genome alignment, a high-sensitivity genomic local alignment method is used to find local regions of similarity between pairs of genomes. Longer colinear segments are found by ‘chaining’ the resulting local alignments and possibly performing global alignment in between neighboring local alignments within a chain. These chains can represent both orthologous and paralogous sequences, but are typically filtered with alignment score thresholds or ‘best hit’ criteria to retain primarily orthologous sequences. Alignments of multiple genomes can be obtained by a progressive merging of overlapping pairwise alignments. Examples of this approach are the programs BLASTZ [78], MUMmer [79], MULTIZ/TBA [80] and CHAINNET [81].
A couple of hybrid methods have also been developed that blend aspects of the local and hierarchical approaches. In general, these methods perform several rounds of finding local alignments, identifying a set of one-to-one colinear segments and filtering of segments that are small or likely to be paralogs. The primary examples of this approach are progressiveMauve [82] and MAGIC [10].
Classification of duplications with genomic context
Related to orthology prediction is the problem of classifying recent genomic duplications. Recent duplications result in inparalogs and orthologous relationships that are not one-to-one. By determining the type and directionality of these duplications, we may be able to distinguish which copies are positional orthologs. A number of methods have been developed for classifying duplication events [83]. These methods use evolutionary divergence (temporal) information, genomic context (spatial) information or both to analyze duplicated genomic regions [83]. Here we describe some more recent methods that have used genomic context information for this problem. In a study of human segmental duplications, the ‘ancestral copy’ (positional ortholog) for a segment with multiple copies was determined using BRH nucleotide-level alignments [84]. To assess the evolutionary rates of retrotranspositions, DNA-mediated interspersed repeats and tandem duplications, another group used local gene neighborhood conservation (looking at 10 genes surrounding the duplicate) and intron conservation to distinguish between duplicated copies [28]. Recently, a general method, PRIUS, for classifying duplicated gene copies as either ‘parents’ (toporthologs) or ‘daughters’ has been published [17]. PRIUS uses a probabilistic model of the length of conserved gene neighborhoods to distinguish between parent and daughter copies.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
The concept of a distinguished subclass of orthologs that retain their ancestral positions has been used in evolutionary studies for over a decade. In this review, we have formalized this concept with an evolutionary definition of positional orthology or toporthology. An important distinction between the definition presented here and the operational definitions used previously, is that the former is concerned with past events, while the latter are generally concerned with present positions. As such, toporthology is defined in the same spirit as orthology, paralogy and homology, and is a relation that cannot be inferred with absolute certainty from present-day data. Despite the uncertainty that we must accept, the inference of toporthologs is an important task, as genes that remain in their ancestral positions are more likely to retain the functions of their ancestors.
A number of methods are already predicting what are likely to be toporthologs by using genomic context information. Many of these work at the gene level and either use local gene neighborhood conservation or explicit models of gene order evolution. Methods for whole-genome alignment, which predict orthology at the nucleotide level, are often restricted to predicting toporthologous sequence. Critical to the inference of toporthology is the classification of duplication events as either asymmetric or symmetric, for which there has also been some methodological work.
A future challenge for this field is to develop methods that predict all homologous relationships between a set of genomes and that distinguish both between orthologs, paralogs and xenologs as well as between toporthologs and atoporthologs (or topoparalogs and atopoparalogs). Accompanying this challenge is the open question of how distantly related two species can be before we can no longer reliably distinguish between topohomologs and atopohomologs. Although the limits to the detection of homology from sequence have been well-studied (e.g. [85, 86]), the limits to the detection of topohomology using both sequence and context data are not well understood. While it is likely that homology is more easily detected than topohomology, accurate inference of the context-based relations we have defined will allow us to better understand the evolution of genomes and the functions of the genes encoded within them.
Key Points.
Positional orthology or toporthology is an important subrelation of orthology.
Toporthologs generally evolve more slowly than atoporthologs and are more likely to retain the function of their ancestors.
A variety of computational methods have been developed for taking genomic context into account during orthology prediction.
FUNDING
The National Science Foundation (DEB 0936214).
Acknowledgements
We thank Eugene Koonin and Lior Pachter for valuable discussions regarding the concept and naming of the term toporthology. We also thank three anonymous reviewers for their constructive comments on an earlier version of this article.
Biography
Colin N. Dewey is an assistant professor in the Department of Biostatistics and Medical Informatics and the Department of Computer Sciences at the University of Wisconsin-Madison.
References
- 1.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 2.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–38. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 3.Fitch W. Homology: a personal view on some of the problems. Trends Genet. 2000;16:227–31. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- 4.Sankoff D. Genome rearrangement with gene families. Bioinformatics. 1999;15:909–17. doi: 10.1093/bioinformatics/15.11.909. [DOI] [PubMed] [Google Scholar]
- 5.Bandyopadhyay S, Sharan R, Ideker T. Systematic identification of functional orthologs based on protein network comparison. Genome Res. 2006;16:428–35. doi: 10.1101/gr.4526006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–52. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 7.Fu Z, Chen X, Vacic V, et al. MSOAR: a high-throughput ortholog assignment system based on genome rearrangement. J Comput Biol. 2007;14:1160–75. doi: 10.1089/cmb.2007.0048. [DOI] [PubMed] [Google Scholar]
- 8.Koski LB, Morton RA, Golding GB. Codon bias and base composition are poor indicators of horizontally transferred genes. Mol Biol Evol. 2001;18:404–12. doi: 10.1093/oxfordjournals.molbev.a003816. [DOI] [PubMed] [Google Scholar]
- 9.Burgetz IJ, Shariff S, Pang A, et al. Positional homology in bacterial genomes. Evolu Bioinform Online. 2006;2:77–90. [PMC free article] [PubMed] [Google Scholar]
- 10.Swidan F, Rocha EPC, Shmoish M, et al. An integrative method for accurate comparative genome mapping. PLoS Comput Biol. 2006;2:e75. doi: 10.1371/journal.pcbi.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemoine F, Lespinet O, Labedan B. Assessing the evolutionary rate of positional orthologous genes in prokaryotes using synteny data. BMC Evol Biol. 2007;7:237. doi: 10.1186/1471-2148-7-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penn K, Jenkins C, Nett M, et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009;3:1193–203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson AP, Gamble JA, Yeomans T, et al. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 2009;19:2231–44. doi: 10.1101/gr.097501.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeppner MP, White S, Jeffares DC, et al. Evolutionarily stable association of intronic snoRNAs and microRNAs with their host genes. Genome Biol Evol. 2009;1:420–8. doi: 10.1093/gbe/evp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Notebaart RA, Huynen MA, Teusink B, et al. Correlation between sequence conservation and the genomic context after gene duplication. Nucleic Acids Res. 2005;33:6164–71. doi: 10.1093/nar/gki913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewey CN, Pachter L. Evolution at the nucleotide level: the problem of multiple whole-genome alignment. Hum Mol Genet. 2006;15:R51–6. doi: 10.1093/hmg/ddl056. [DOI] [PubMed] [Google Scholar]
- 17.Han MV, Hahn MW. In: Biocomputing 2009 - Proceedings of the Pacific Symposium. Identifying parent-daughter relationships among duplicated genes. Volume 125, p. 114–25. World Scientific Publishing Co. Pte. Ltd., Singapore. [PubMed] [Google Scholar]
- 18.Albert V, Gustafsson M, Di Laurenzio L. Ontogenetic systematics, molecular developmental genetics, and the angiosperm petal, Springer. In: Soltis D, Soltis P, Doyle J, editors. Molecular Systematics of Plants II. Boston: Kluwer Academic Publishers; 1998. pp. 349–74. [Google Scholar]
- 19.Wolfe K. Robustness–it’s not where you think it is. Nat Genet. 2000;25:3–4. doi: 10.1038/75560. [DOI] [PubMed] [Google Scholar]
- 20.Passarge E, Horsthemke B, Farber RA. Incorrect use of the term synteny. Nat Genet. 1999;23:387. doi: 10.1038/70486. [DOI] [PubMed] [Google Scholar]
- 21.Yanofsky C, Carlton BC, Guest JR, et al. On the colinearity of gene structure and protein structure. Proc Natl Acad Sci USA. 1964;51:266–72. doi: 10.1073/pnas.51.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynen M, Snel B, Lathe W, et al. Exploitation of gene context. Curr Opin Struct Biol. 2000;10:366–70. doi: 10.1016/s0959-440x(00)00098-1. [DOI] [PubMed] [Google Scholar]
- 23.Kleinjan D, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet. 1998;7:1611–8. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 24.Hurst LD, Pál C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 25.Zheng D. Asymmetric histone modifications between the original and derived loci of human segmental duplications. Genome Biol. 2008;9:R105. doi: 10.1186/gb-2008-9-7-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snel B, van Noort V, Huynen MA. Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes. Nucleic Acids Res. 2004;32:4725–31. doi: 10.1093/nar/gkh815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cusack BP, Wolfe KH. Not born equal: increased rate asymmetry in relocated and retrotransposed rodent gene duplicates. Mol Biol Evol. 2007;24:679–86. doi: 10.1093/molbev/msl199. [DOI] [PubMed] [Google Scholar]
- 28.Jun J, Ryvkin P, Hemphill E, et al. Duplication mechanism and disruptions in flanking regions determine the fate of Mammalian gene duplicates. J Comput Biol. 2009;16:1253–66. doi: 10.1089/cmb.2009.0074. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Dong X, Ding G, et al. Comparing the retention mechanisms of tandem duplicates and retrogenes in human and mouse genomes. Genet Select Evol. 2010;42:24. doi: 10.1186/1297-9686-42-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han MV, Demuth JP, McGrath CL, et al. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–67. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostka D, Hahn MW, Pollard KS. Noncoding sequences near duplicated genes evolve rapidly. Genome Biol Evol. 2010;2:518–33. doi: 10.1093/gbe/evq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–37. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 33.Bikard D, Patel D, Le Metté C, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–6. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- 34.Kuzniar A, van Ham RCHJ, Pongor S, et al. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–51. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Hulsen T, Huynen M, de Vlieg J, et al. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Heijden RTJM, Snel B, van Noort V, et al. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–61. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehmann J, Stadler PF, Prohaska SJ. SynBlast: assisting the analysis of conserved synteny information. BMC Bioinformatics. 2008;9:351. doi: 10.1186/1471-2105-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemoine F, Labedan B, Lespinet O. SynteBase/SynteView: a tool to visualize gene order conservation in prokaryotic genomes. BMC Bioinformatics. 2008;9:536. doi: 10.1186/1471-2105-9-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huynen MA, Bork P. Measuring genome evolution. Proc Natl Acad Sci. 1998;95:5849–56. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf YI, Rogozin IB, Kondrashov AS, et al. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–72. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 43.Stein LD, Bao Z, Blasiar D, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jun J, Mandoiu II, Nelson CE. Identification of mammalian orthologs using local synteny. BMC Genomics. 2009;10:630. doi: 10.1186/1471-2164-10-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Touchon M, Hoede C, Tenaillon O, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bansal A, Bork P, Stuckey P. Automated pair-wise comparisons of microbial genomes. Math Model Sci Comput. 1998;9:1–23. [Google Scholar]
- 47.Kuhn H. The Hungarian method for the assignment problem. Nav Res Logist Q. 1955;2:83–97. [Google Scholar]
- 48.Mahmood K, Konagurthu AS, Song J, et al. EGM: encapsulated gene-by-gene matching to identify gene orthologs and homologous segments in genomes. Bioinformatics. 2010;26:2076–84. doi: 10.1093/bioinformatics/btq339. [DOI] [PubMed] [Google Scholar]
- 49.Blin G, Chateau A, Chauve C, et al. Inferring positional homologs with common intervals of sequences. In: Comparative Genomics. Vol. 4205 Lecture Notes in Computer Science. Berlin/Heidelberg: Springer, 2006, 24–38. [Google Scholar]
- 50.Wapinski I, Pfeffer A, Friedman N, et al. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:i549–58. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- 51.Clamp M, Andrews D, Barker D, et al. Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res. 2003;31:38–42. doi: 10.1093/nar/gkg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kellis M, Patterson N, Birren B, et al. Methods in comparative genomics: genome correspondence, gene identification and regulatory motif discovery. J Comput Biol. 2004;11:319–55. doi: 10.1089/1066527041410319. [DOI] [PubMed] [Google Scholar]
- 53.Zheng XH, Lu F, Wang ZY, et al. Using shared genomic synteny and shared protein functions to enhance the identification of orthologous gene pairs. Bioinformatics. 2005;21:703–10. doi: 10.1093/bioinformatics/bti045. [DOI] [PubMed] [Google Scholar]
- 54.Lam WWM, Chan KCC, Chiu DKY, et al. In: 2007 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2007). IEEE. Fremont, CA: IEEE Computer Society Press; 2007. MAGMA: an algorithm for mining multi-level patterns in genomic data; pp. 89–94. [Google Scholar]
- 55.Yang K, Setubal JC. In: Proceedings of the First ACM International Conference on Bioinformatics and Computational Biology - BCB 10. New York, USA: ACM Press; 2010. Homology prediction refinement and reconstruction of gene content and order of ancestral bacterial genomes; p. 230. 56. [Google Scholar]
- 56.Cannon SB, Young ND. OrthoParaMap: distinguishing orthologs from paralogs by integrating comparative genome data and gene phylogenies. BMC Bioinformatics. 2003;4:35. doi: 10.1186/1471-2105-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wheeler DL, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bryant D. The complexity of calculating exemplar distances, Springer. In: Sankoff D, Nadeau J, editors. Comparative Genomics: Empirical and Analytical Approaches to Gene Order Dynamics, Map Alignment, and the Evolution of Gene Families. Boston: Kluwer Academic Publishers; 2000. pp. 207–12. [Google Scholar]
- 59.Nguyen CT, Tay YC, Zhang L. Divide-and-conquer approach for the exemplar breakpoint distance. Bioinformatics. 2005;21:2171–6. doi: 10.1093/bioinformatics/bti327. [DOI] [PubMed] [Google Scholar]
- 60.Chen X, Zheng J, Fu Z, et al. Assignment of orthologous genes via genome rearrangement. IEEE/ACM Trans Comput BiolBioinform. 2005;2:302–15. doi: 10.1109/TCBB.2005.48. [DOI] [PubMed] [Google Scholar]
- 61.El-Mabrouk N. Reconstructing an ancestral genome using minimum segments duplications and reversals. J Comput System Sci. 2002;65:442–64. [Google Scholar]
- 62.Swenson K, Pattengale N, Moret BME. A Framework for Orthology Assignment from Gene Rearrangement Data. 2005. Vol. 3678, Lecture Notes in Computer Science. Berlin/Heidelberg: Springer, 2005, 153–66. [Google Scholar]
- 63.Shi G, Zhang L, Jiang T. MSOAR 2.0: incorporating tandem duplications into ortholog assignment based on genome rearrangement. BMC Bioinformatics. 2010;11:10. doi: 10.1186/1471-2105-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu Z, Jiang T. Clustering of main orthologs for multiple genomes. J Bioinform Comput Biol. 2008;6:573–84. doi: 10.1142/s0219720008003540. [DOI] [PubMed] [Google Scholar]
- 65.Shi G, Peng M, Jiang T. In: Proceeding LSS Comput Syst Bioinform Conference Stanford, CA. Vol. 2. Stanford, CA: Life Sciences Society; 2010. Accurate identification of ortholog groups among multiple genomes; pp. 166–179. [Google Scholar]
- 66.Jiang T. Some algorithmic challenges in genome-wide ortholog assignment. J Comput Sci Technol. 2010;25:42–52. [Google Scholar]
- 67.Blanchette M. Computation and analysis of genomic multi-sequence alignments. Annu Rev Genomics Hum Genet. 2007;8:198–213. doi: 10.1146/annurev.genom.8.080706.092300. [DOI] [PubMed] [Google Scholar]
- 68.Dewey CN. Aligning multiple whole genomes with Mercator and MAVID. Methods Mol Biol. 2007;395:221–36. doi: 10.1007/978-1-59745-514-5_14. [DOI] [PubMed] [Google Scholar]
- 69.Paten B, Herrero J, Beal K, et al. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–28. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubchak I, Poliakov A, Kislyuk A, et al. Multiple whole-genome alignments without a reference organism. Genome Res. 2009;19:682–9. doi: 10.1101/gr.081778.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Angiuoli SV, Salzberg SL. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2010;27:334–42. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pevzner P, Tesler G. Genome rearrangements in mammalian evolution: lessons from human and mouse genomes. Genome Res. 2003;13:37–45. doi: 10.1101/gr.757503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rödelsperger C, Dieterich C. Syntenator: multiple gene order alignments with a gene-specific scoring function. Algorithms Mol Biol. 2008;3:14. doi: 10.1186/1748-7188-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lemaitre C, Tannier E, Gautier C, et al. Precise detection of rearrangement breakpoints in mammalian chromosomes. BMC Bioinformatics. 2008;9:286. doi: 10.1186/1471-2105-9-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng Q, Alekseyev M, Tesler G, et al. In: Proceedings of the Ninth International Conference on Algorithms in Bioinformatics. Berlin, Heidelberg: Springer-Verlag; 2009. Decoding synteny blocks and large-scale duplications in mammalian and plant genomes; pp. 220–32. [Google Scholar]
- 76.Hachiya T, Osana Y, Popendorf K, et al. Accurate identification of orthologous segments among multiple genomes. Bioinformatics. 2009;25:853–60. doi: 10.1093/bioinformatics/btp070. [DOI] [PubMed] [Google Scholar]
- 77.Pham SK, Pevzner PA. DRIMM-Synteny: decomposing genomes into evolutionary conserved segments. Bioinformatics. 2010;26:2509–16. doi: 10.1093/bioinformatics/btq465. [DOI] [PubMed] [Google Scholar]
- 78.Schwartz S, Kent WJ, Smit A, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–7. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blanchette M, Kent WJ, Riemer C, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–15. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kent WJ, Baertsch R, Hinrichs A, et al. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci. 2003;100:11484–9. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Durand D, Hoberman R. Diagnosing duplications–can it be done? Trends Genet. 2006;22:156–64. doi: 10.1016/j.tig.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 84.Jiang Z, Tang H, Ventura M, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–8. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 85.Rost B. Twilight zone of protein sequence alignments. Protein Engineering. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 86.Spang R, Vingron M. Limits of homology detection by pairwise sequence comparison. Bioinformatics. 2001;17:338–42. doi: 10.1093/bioinformatics/17.4.338. [DOI] [PubMed] [Google Scholar]