Abstract
Since first being proposed as a tandem gene family in 2001, the relatedness of the 5 SIBLING proteins (BSP, DMP1, DSPP, MEPE, and SPP1/OPN) has predominantly depended on arguments involving shared intron/exon properties as well as conserved protein biochemical properties (e.g. unstructured and acidic) and specific peptide motifs (e.g. phosphorylation and integrin-binding RGD). This report discusses the evidence that an ancient DMP1 gene underwent a simple duplication in the common ancestor of mammals and reptiles and then separately evolved into DSPP-like paralogs in the 2 classes. Genomic sequence analyses show that different copies of the original DMP1 duplication process were selected by mammalian and reptilian (anole lizard) classes to acquire genetically different but biochemically similar phosphoserine-rich repeat domains by convergent evolution. Mammals, for example, expanded phosphoserine motifs encoded exclusively using motifs containing AGC/T serine codons while the reptile line's repeats also used TCN-encoding serine codons. A similar analysis of the origins of the other 4 SIBLINGs will require even more detailed analysis as genome sequences of various fish and amphibia become available.
Key Words: SIBLINGs, DSPP, DMP1
Introduction
The discovery more than 40 years ago that >90% of the organic matrix of both mineralizing (bone and dentin) and soft connective tissues (skin, tendons, and ligaments) consists of the same collagen proteins (type I) launched the search for other, noncollagenous proteins responsible for initiating and controlling the precisely oriented hydroxyapatite (HA) crystals in calcifying matrices. The working hypothesis was that these would likely be acidic proteins probably posttranslationally modified with phosphate groups that both mimic the phosphate groups in HA and bind readily to calcium ions, the other component of HA. These proteins were presumed to be very abundant to account for the large number of oriented crystals and would remain entrapped within the mineralized matrices. Several laboratories around the world set out to purify and identify the most abundant proteins recovered from demineralized matrices. After eliminating HA-binding serum proteins (albumin and α2HS glycoprotein/fetuin, among others), 3 general categories of proteins were identified: proteoglycans (predominantly biglycan and decorin), γ-carboxylglutamic acid-containing proteins [osteocalcin (OCN)/bone Gla protein (BGP) and matrix Gla protein (MGP)], and the largest group, phosphoproteins [osteonectin/secreted protein, acidic, rich-in-cysteine (SPARC), bone sialoprotein (BSP), osteopontin (OPN)/secreted phosphoprotein-1 (SPP1), and dentin phosphoprotein/phosphophoryn, among others]. Curiously, many years later we still do not have a consensus of which protein(s), if any, definitively control the initiation and/or growth of HA crystals in bones and dentin. The genes encoding the abundant noncollagenous proteins described over the years have been individually deleted in experimental mice and, while many of these knockout mice do have observable if often subtle bone and/or tooth phenotypes, in no case is mineral completely missing from the target tissues (indeed, with the possible exception of OCN, all of the previously hypothesized mineralized tissue-specific proteins have also been found to be expressed in a variety of normal soft tissues). Because each null mouse did have at least a mild bone/tooth phenotype, however, it is also clear that these acid proteins do play at least indirect (if currently undefined) roles in hard tissue biogenesis and/or maintenance.
Over the years, the relatedness of these acidic, HA-binding proteins has engendered lively discussions. OCN (or BGP) and MGP share motifs with each other and proteins in the clotting cascade with respect to the vitamin K-dependent γ-carboxylation of specific glutamate residues [Price and Williamson, 1985]. The 2 small proteoglycans, decorin and biglycan, are easily seen at the amino acid level to be closely related to each other and more distantly related to other small leucine-rich repeat proteins [Fisher et al., 1989]. At first, all of the phosphoproteins appeared to be unrelated to each other at the amino acid level. By the late 1990s the genes encoding 4 of them, i.e. integrin-binding sialoprotein (IBSP) that encodes BSP, SPP1 (or OPN), dentin matrix protein-1 (DMP1), and dentin sialophosphoprotein (DSPP), were mapped to a shared region of human chromosome 4 (mouse chromosome 5) suggesting that their frequent coexpression may be controlled, in part, by relaxation of the local chromosomal regions [Crosby et al., 1996; Feng et al., 1998; Aplin et al., 1999]. Around the millennium, the first drafts of the human and mouse genome sequences enabled us for the first time to report that 4 of the bone/tooth phosphoprotein genes (IBSP, SPP1, DMP1, and DSPP) were tandem genes interrupted only by 1 additional gene, i.e. matrix extracellular phosphoprotein (MEPE), between IBSP and SPP1 (fig. 1a) [Fisher et al., 2001]. Showing that the phosphoprotein genes were clustered together was not, of course, proof that they are genetically related to each other.
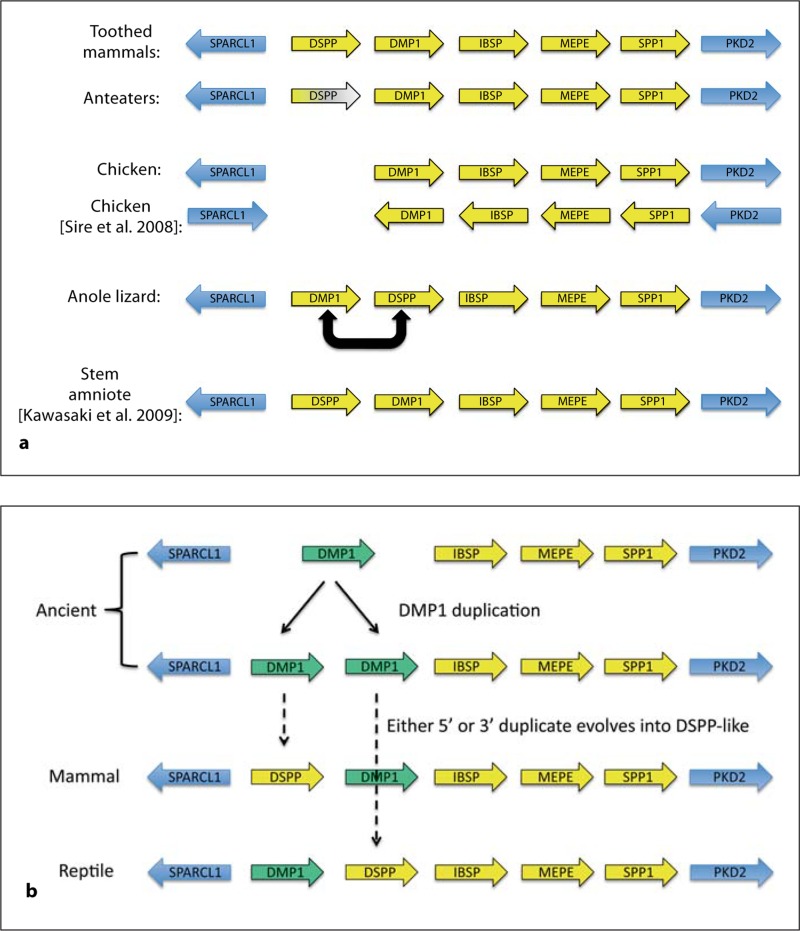
Fig. 1.
Location and transcription direction (arrows) of SIBLING (yellow) and highly conserved flanking (blue) genes along the corresponding chromosomes of mammals, chickens, anoles (reptiles), and the previously proposed stem amniote (a). The color fading of the anteater DSPP gene denotes loss of the translated DPP domain due to frameshift mutations in these toothless mammals. Note the complete loss of DSPP in chickens as well as the inverted order of DSPP and DMP1 genes in anoles compared to both mammals and the proposed stem amniote (a). Our analysis of the transcription directions of chicken genes differs from that proposed by Sire et al. [2008]. Kawasaki [2009] proposed that stem amniote SIBLING genes would be in the same order as in mammals. b Proposed ancient gene duplication of the DMP1 gene (green) and the subsequent independent convergent evolution of DSPP-like proteins by reptile and mammal classes.
When we looked more closely at the properties of the exons for each of the 4 genes, however, we were able to discern a variety of interesting features/motifs that were consistently retained and suggested that they were all the result of ancient gene duplication and subsequent divergent evolution. For example: (1) exon 1 was always noncoding; (2) exon 2 encoded similar secretion signal peptides plus the first 2 amino acids of the mature protein; (3) exons 3 and 5 usually encoded short peptides that included a conserved phosphorylation motif, Ser-Ser-Glu-Glu (SSEE); (4) no intramolecular disulfide bonds were possible due to the presence of, at most, a single Cys (DMP1, DSPP, and MEPE) within the sequence; (5) all genes encoded the integrin-binding tripeptide Arg-Gly-Asp (RGD) within their 3′ one or two large exons, and (6) in every case, all of the introns were type 0 (i.e. interrupting the coding sequences only between codons and thereby permitting all possible splice variants to remain in the frame). Because the functions of the 5 proposed family members were unknown, we chose a function-neutral but biochemically descriptive name: small integrin-binding ligand, N-linked glycoprotein (SIBLING). Analysis of the intervening MEPE gene in many ways bolstered the relatedness hypothesis because this newly described gene had exons 1 and 2 similar to the other 4 genes, the presence of only type 0 introns, and a lack of any intramolecular disulfide bonds, and it encoded the integrin-binding RGD tripeptide as well as several SSEE type phosphorylation motifs. Furthermore, we were able to find evidence in a human brain cDNA library of a previously undisclosed exon similar in size to splice-variant expressed exons observed in the other SIBLINGs [Fisher and Fedarko, 2003]. At that time we proposed that enamelin (ENAM) may also be a SIBLING member, but the RGD is not retained in many other species and ENAM may contain up to 3 intramolecular disulfide bonds. Therefore, we currently limit the SIBLING family to the original 5 tandem gene members.
Recently, we proposed that the SIBLING members share 2 additional properties [Bellahcène et al., 2008]. A variety of reports from many laboratories have shown that all SIBLINGs either are sometimes proteoglycans or at least have conserved motifs that may permit the posttranslational addition of glycosaminoglycan chains under some conditions. Similarly, all SIBLINGs are reported to be cleaved by specific proteases [e.g. bone morphogenetic protein-1 (BMP1) cleaves DMP1 and DSPP and thrombin cleaves SPP1/OPN], although it is not always clear if these processes activate or inactivate the penultimate function(s) of the 5 glycophosphoproteins.
Materials and Methods
Sequences mined from published DNA databases and original sequencing of various mammalian dentin phosphoprotein (DPP) domains of DSPP were previously described [McKnight and Fisher, 2009].
Results
In general, the conservation of a protein's amino acid sequence is driven by the preservation of both its own tertiary structure and surface structures/motifs needed to interact with other molecules. The SIBLINGs, however, appear to belong to the growing class of proteins that remain unstructured in solution (both BSP and OPN have been shown by nuclear magnetic resonance analysis to be flexible, unstructured proteins in solution) [Fisher et al., 2001] and, except for specific motifs (e.g. integrin-binding RGD and SSEE phosphorylation sites), most of its amino acids can change as long as the protein remains unstructured and maintains certain biophysical properties (e.g. remaining acid in nature). The consequences of this unrelenting genetic drift have complicated the search for the homologs of each SIBLING throughout evolution as simple BLAST/BLAT type comparison programs often fail to identify the related proteins in more distant species. Fortunately, the SIBLING gene cluster is usually flanked by 2 highly conserved genes, i.e. SPARC-like-1 (SPARCL1) and pyruvate dehydrogenase kinase isozyme-2 (PDK2), so an investigator can locate one or both of these genes in a genome database and then look for adjacent open reading frames. Using this approach, we searched the available genome databases looking for the homologs for each SIBLING in 34 mammals as well as the chicken and anole lizard [McKnight and Fisher, 2009]. Briefly, the SIBLING genes of all of the toothed mammals investigated had the same gene orientation and transcription direction (fig. 1a). Interestingly, the repeat-containing DPP domain of DSPP of 2 species of toothless anteaters independently acquired frameshift mutations suggesting that this highly phosphorylated protein may provide an evolutionarily required function only in dentin even though DSPP is also expressed at lower levels in many metabolically active ductal epithelial cells. Other mammals such as the baleen whale and platypus, which are commonly thought of as being toothless, actually produce recognizable teeth during development but lose them before adulthood. We found that both bowhead whales and platypuses retain intact DPP domains. Our analysis of the SIBLINGs in the chicken genome (fig. 1a) agreed with 2 previous publications [Kawasaki and Weiss, 2008; Sire et al., 2008] in that this toothless animal has 4 of the SIBLING genes but lacks a DSPP gene. Furthermore, like Kawasaki and Weiss [2008], we found that the order and relative transcriptional directions of the SIBLING genes and SPARCL1 (plus we note PDK2) had the same orientation as in mammals. Our results are in contrast to those of Sire et al. [Sire et al., 2008] who reported that each gene was individually inverted in transcription direction compared to mammals.
The lack of the DSPP gene in the chicken genome raised the question of whether this gene first appeared within the evolutionary branch of mammals or it was also present in earlier animals and, like in the toothless anteaters, was only subsequently lost in birds. In early 2009, Kawasaki [2009] noted that amphibia lacked the DSPP gene (while retaining the more ancient DMP1) and he proposed that stem amniotes (a common ancestor of mammals and reptiles) had all 5 SIBLING genes in the same order as mammals (fig. 1a). Our analysis of the anole reptile genome database, however, shows (fig. 1a) that the order of the SIBLING genes (plus the flanking PDK2) in the anole lizard had the DMP1- and DSPP-like genes reversed without changing their transcription directions vis-à-vis the other genes [McKnight and Fisher, 2009]. One explanation for this observation is that the amphibian-like DMP1 gene may have been duplicated during the evolution of the stem amniote ancestors, and then mammalian and reptilian evolutionary lines independently evolved DSPP-like, highly acidic proteins containing many phosphoserine repeats using different copies of the duplicated DMP1 ancestral genes (fig. 1b).
To test this duplication/convergent evolution hypothesis, both the biochemical properties and the patterns of codon usage of the repeat domain (DPP) of the reptile and mammalian DSPP were studied. DPP (always entirely encoded within the last exon) is the carboxyterminal fragment of DSPP after cleavage by BMP1 at its highly conserved MQXDDP motif, the same motif used by BMP1 to cleave DMP1 [von Marschall and Fisher, 2010] (the other BMP1 fragments, dentin sialoprotein (DSP) for DSPP and the 37-kDa fragment of DMP1, can both be chondroitin sulfate proteoglycans). Convergent evolution processes should result in similar patterns of motifs/biochemical properties of the protein, but both the specific amino acids used and the codons encoding them to generate these domains need not be the same for the 2 independent processes. We mined the available mammalian genome databases as well as directly sequenced the DPP domain of many species whose complete DPP domain was missing or incomplete [McKnight and Fisher, 2009]. The mammals were found to have a wide range in the number of phosphorylation motif (Ser-Ser-Asp, SSD) repeats (from ∼75 in elephants to >230 in humans) interspersed with either few (e.g. primates and some rodents) or several (most mammals) positively charged lysine amino acids. Even the most ancient of mammals, i.e. the opossum and the egg-laying platypus, had the same patterns of primordial repeats. By studying the many homologous changes to the repeat sequences across the mammalian class, we concluded that the number of phosphoserine repeats, like all DNA repeats, is inherently unstable and can easily change in length over evolutionarily short periods of time due to unequal crossover and/or slip-replication events during meiosis. Interestingly, we could find no obvious correlations between simple anatomical aspects of the various species’ teeth/dentin and either the repeat length or the relative abundance of repeat-interrupting lysine amino acids. [We encourage readers to look for correlations between repeat length and/or textural aspects (e.g. the number of positively charged amino acids or dipeptide motifs interrupting the nominal SSD repeats) with various anatomical or biophysical aspects of dentin biology that we have may overlooked in our studies.]
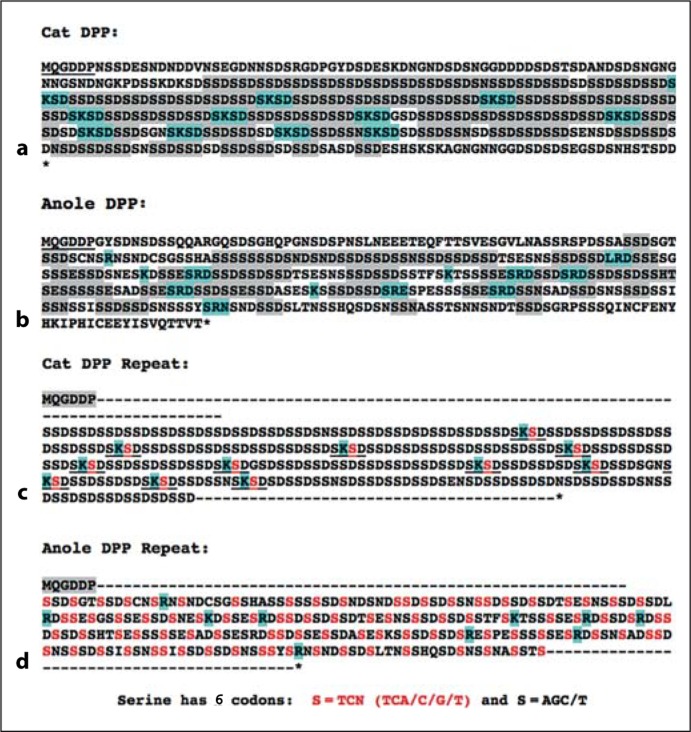
For the sake of simplicity, figure 2 shows the DPP protein sequence of the anole lizard compared to the common cat whose repeat domain is similar in size to that of the reptile. Although it is not composed of as perfect a repeat structure as that in mammals, the repeat domain of the reptile DPP is rich in serines and aspartic acids that likely result in the addition of many phosphate groups. In contrast to the mammals’ lysines (K) interrupting the phosphoserine-rich domains, the anole's DPP sequence is interrupted by arginine (R)-based motifs suggesting an alternative, convergent pathway to spacing single positive charges among the long chain of negative charges (aspartic acids and phosphoserines). Perhaps more convincing is the divergence of codon usage between the 2 classes. There are 6 codons that encode serine, i.e. AGC/T and TC with any 4 bases in the third position (TCN). Note that mammals have expanded various elements of the primordial SSD repeat using exclusively AGC and AGT codons, with the only exception in the cat (and all other mammals) being the second serine (bold red S; fig. 2) in the SKSD tetrapeptide motif that also brings the sole positive charge into its protein sequence. In comparison, the reptile DPP domain expanded a different primordial SSD-rich repeat that uses both AGC/T and TCN type serine codons.
Fig. 2.
Cat (a) and anole (b) DPP domains both contain many phosphorylation repeats (grey) interrupted by motifs (blue) containing single positively charged lysine (K) or arginine (R) amino acids, respectively. The cat DPP repeat domain (c) uses only AGC/T serine-encoding codons (black S), while many of the phosphorylation motifs in anoles (d) also use TCN type serines (red S). The only TCN-encoding repeat serines in the cat (and all other mammals) are those within the lysine-containing tripeptide SKSD. These results support the hypothesis that reptiles and mammals independently expanded different DMP1-derived phosphorylation motifs to generate their DPP-like repeat domains and interspersed them with different positively charged amino acid motifs.
Conclusions
The reptile shows evidence of having evolved a DPP-like, phosphoserine repeat domain interrupted by single positive charges and a carboxyterminal to a highly conserved BMP1 cleavage site. This DPP-like domain is biochemically similar to the mammalian DPP but is encoded by dramatically different DNA repeats that are highly unlikely to ever have been interchanged or interconverted after the initial expansion of their portion of the primordial, DMP1-derived phosphoserine motif(s). This analysis supports the hypothesis that an ancient amniote duplicated the amphibian-like DMP1 gene to form a simple tandem repeat on one end of the SIBLING gene cluster. Reptiles used the DMP1 repeat proximal to the highly conserved SIBLING-flanking gene, SPARCL1, while the mammalian lineage used the copy adjacent to IBSP to separately evolve a DSPP-like protein. Both classes of animals retained their second copy of the DMP1 gene presumably to continue the original functions of this more ancient SIBLING gene product.
Acknowledgment
This research was supported by the Intramural Research Program of the NIH/NIDCR.
Glossary
Abbreviations used in this paper
| BGP | bone Gla protein |
| BMP1 | bone morphogenetic protein-1 |
| BSP | bone sialoprotein |
| DMP1 | dentin matrix protein-1 |
| DPP | dentin phosphoprotein (or phosphophoryn) |
| DSPP | dentin sialophosphoprotein |
| ENAM | enamelin |
| HA | hydroxyapatite |
| IBSP | integrin-binding sialoprotein |
| MEPE | matrix extracellular phosphoprotein (gene for BSP) |
| MGP | matrix Gla protein |
| OCN | osteocalcin |
| OPN | osteopontin |
| PDK2 | pyruvate dehydrogenase kinase isozyme-2 |
| SIBLING | small integrin-binding ligand, N-linked glycoprotein |
| SPARC | secreted protein, acidic, rich-in-cysteine |
| SPARCL1 | SPARC-like-1 |
| SPP1 | secreted phosphoprotein-1 (gene for OPN) |
References
- Aplin H.M., Hirst K.L., Dixon M.J. Refinement of the dentinogenesis imperfecta type II locus to an interval of less than 2 centiMorgans at chromosome 4q21 and the creation of a yeast artificial chromosome contig of the critical region. J Dent Res. 1999;78:1270–1276. doi: 10.1177/00220345990780061201. [DOI] [PubMed] [Google Scholar]
- Bellahcène A., Castronovo V., Ogbureke K.U., Fisher L.W., Fedarko N.S. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): multifunctional proteins in cancer. Nat Rev Cancer. 2008;8:212–226. doi: 10.1038/nrc2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby A.H., Lyu M.S., Lin K., McBride O.W., Kerr O.W., Aplin H.M., Fisher L.W., Young M.F., Kozak C.A., Dixon M.J. Mapping of the human and mouse bone sialoprotein and osteopontin loci. Mamm Genome. 1996;7:149–151. doi: 10.1007/s003359900037. [DOI] [PubMed] [Google Scholar]
- Feng J.Q., Luan X., Wallace J., Jing D., Ohshima T., Kulkarni A.B., D'souza R.N., Kozak C.A., MacDougal l.M. Genomic organization, chromosomal mapping, and promoter analysis of the mouse dentin sialophosphoprotein (Dspp) gene, which codes for both dentin sialoprotein and dentin phosphoprotein. J Biol Chem. 1998;273:9457–9464. doi: 10.1074/jbc.273.16.9457. [DOI] [PubMed] [Google Scholar]
- Fisher L.W., Fedarko N.S. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res. 2003;44:33–40. [PubMed] [Google Scholar]
- Fisher L.W., Termine J.D., Young M.F. Deduced protein sequence of bone small proteoglycan I (biglycan) shows homology with proteoglycan II (decorin) and several nonconnective tissue proteins in a variety of species. J Biol Chem. 1989;264:4571–4576. [PubMed] [Google Scholar]
- Fisher L.W., Torchia D.A., Fohr B., Young M.F., Fedarko N.S. Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem Biophys Res Commun. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- Kawasaki K. The SCPP gene repertoire in bony vertebrates and graded differences in mineralized tissues. Dev Genes Evol. 2009;219:147–157. doi: 10.1007/s00427-009-0276-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K., Weiss K.M. SCPP gene evolution and the dental mineralization continuum. J Dent Res. 2008;87:520–531. doi: 10.1177/154405910808700608. [DOI] [PubMed] [Google Scholar]
- McKnight D.A., Fisher L.W. Molecular evolution of dentin phosphoprotein among toothed and toothless animals. BMC Evol Biol. 2009;9:299. doi: 10.1186/1471-2148-9-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price P.A., Williamson M.K. Primary structure of bovine matrix Gla protein, a new vitamin K-dependent bone protein. J Biol Chem. 1985;260:14971–14975. [PubMed] [Google Scholar]
- Sire J.Y., Delgado S.C., Girondot M. Hen's teeth with enamel cap: from dream to impossibility. BMC Evol Biol. 2008;8:246. doi: 10.1186/1471-2148-8-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Marschall Z., Fisher L.W. Dentin sialophosphoprotein (DSPP) is cleaved into its two natural dentin matrix products by three isoforms of bone morphogenetic protein-1 (BMP1) Matrix Biol. 2010;29:295–303. doi: 10.1016/j.matbio.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]