Abstract
Homeodomain-containing (HOX) factors such as the abdominal class homeodomain protein HOXA10 and the TALE-family protein PBX1 form coregulatory complexes and are potent transcriptional and epigenetic regulators of tissue morphogenesis. We have identified that HOXA10 and PBX1 are expressed in osteoprogenitors; however, their role in osteogenesis has not been established. To determine the mechanism of HOXA10-PBX-mediated regulation of osteoblast commitment and the related gene expression, PBX1 or HOX10 were depleted (shRNA or genetic deletion, respectively) or exogenously expressed in C3H10T1/2, bone marrow stromal progenitors, and MC3T3-E1 (preosteoblast) cells. Overexpression of HOXA10 increased the expression of osteoblast-related genes, osteoblast differentiation and mineralization; expression of PBX1 impaired osteogenic commitment of pluripotent cells and the differentiation of osteoblasts. In contrast, the targeted depletion of PBX1 by shRNA increased the expression of bone marker genes (osterix, alkaline phosphatase, BSP, and osteocalcin). Chromatin-associated PBX1 and HOXA10 were present at osteoblast-related gene promoters preceding gene expression, but PBX1 was absent from promoters during the transcription of bone-related genes, including osterix (Osx). Further, PBX1 complexes were associated with histone deacetylases normally linked with chromatin inactivation. Loss of PBX1 but not of HOXA10 from the Osx promoter was associated with increases in the recruitment of histone acetylases (p300), as well as decreased H3K9 methylation, reflecting transcriptional activation. We propose PBX1 plays a central role in attenuating the activity of HOXA10 as an activator of osteoblast-related genes and functions to establish the proper timing of gene expression during osteogenesis, resulting in proper matrix maturation and mineral deposition in differentiated osteoblasts.
Key Words: Chromatin remodeling during osteogenesis, Osterix histone acetylation, Pbx inhibition of osteoblastogenesis
Introduction
HOX proteins are part of a diverse but closely related family of homeodomain-containing transcription factors that play a fundamental role during development in establishing the body's axis and defining the shape and form of developing limbs. As most of these proteins interact with DNA through a well conserved homeodomain, the binding specificity underlying their specific but diverse biological functions remains largely enigmatic. One primary mechanism by which HOX protein exerts transcriptional specificity is interaction with protein partners. In this regard, several studies have demonstrated that the PBC family protein PBX1 (pre-B-cell leukemia transcription factor 1) interacts with HOX proteins and as such is considered an essential HOX cofactor for developmental gene regulation [Moens and Selleri, 2006].
PBX1 is a TALE (3-amino-acid loop extension) class homeodomain protein [Moens and Selleri, 2006]. Germline deletion of Pbx1 results in death at embryonic day 15.5–16.5 with severe hypoplasia or aplasia of multiple organs and widespread patterning defects of the axial and appendicular skeleton [Selleri et al., 2001]. In addition, Pbx1–/– rib and proximal limb cartilages showed extensive calcium deposition which is indicative of precocious mineralization and accelerated ossification of the mutant cartilage.
While PBX1-mediated gene regulation involves DNA binding activity, PBX1 can also function through epigenetic coactivators such as histone deacetylases (HDACs) and histone acetyltransferases [Choe et al., 2009]. In this study we addressed the role of PBX1 in osteogenesis by reducing PBX1 expression in bone marrow stromal cells (BMSCs) using a targeted shRNA and examined the effect on BMSC differentiation. The mechanism of PBX activity through chromatin remodeling was identified, focusing on the Osterix (Osx) promoter. Osterix is a transcription factor which is essential for the final mineralization stage of osteoblast differentiation [Nakashima et al., 2002] and serves as a model for understanding HOXA10-PBX1 regulatory mechanisms in the commitment and progression of osteoblast maturation.
Materials and Methods
Cell Culture
Human embryonic kidney cells (HEK293T) for generating viruses were obtained from ATCC (Manassas, Va., USA) and maintained as recommended. Primary BMSC osteoprogenitors from 6-week-old mouse femurs were maintained as described [Gordon et al., 2010] and further supplemented with BMP2 (100 ng/ml) (generously provided by John Wozney of the Tissue Repair Research Unit, Pfizer Inc., Cambridge, Mass., USA) for differentiation and/or 280 μM ascorbic acid and 5 mM β-glycerophosphate (Sigma Aldrich, St. Louis, Mo., USA).
Expression Constructs and shRNA Virus Generation
The Xpress-Pbx1 plasmid used in this study was previously described [Gordon et al., 2010]. For knockdown of PBX1, Pbx1-shRNA pGIPZ and control (NS) plasmids were obtained from Open Biosystems (Thermo Fisher Scientific, Huntsville, Ala., USA).
For viral packing, lentiviral vectors containing Pbx1 or Pbx1-shRNA sequences were cotransfected along with pMD2.G and pCMVΔR-8.91 (generously provided by Didier Trono, Addgene plasmids 12259 and 12263) viral plasmids into HEK293T cells. Cells were propagated for approximately 48 h posttransfection, and the supernatant was collected, analyzed for titer in HEK293T cells, and subsequently used for infections.
Antibodies and RT-qPCR
The following antibodies were used for Western blotting: anti-PBX1 (P-20, sc-889; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif., USA), anti-HOXA10 (N-20, sc-17158; Santa Cruz), and anti-RUNX2 (monoclonal, Clone 8G5; MBL International, Woburn, Mass., USA). The following antibodies were used for immunoprecipitations and/or chromatin immunoprecipitations (ChIP) and immunohistochemistry: anti-RNA Pol II (Clone 8WG16; Covance), anti-RUNX2 (M-70, sc-10758; Santa Cruz), anti-HOXA10 (N-20X, sc-17158X; Santa Cruz), anti-PBX1 (4342; Cell Signaling Technology, Danvers, Mass., USA), CBP (C-20, sc-10758; Santa Cruz), anti-p300 (N-20, sc-584; Santa Cruz), anti-penta-acetylated Histone H4 (06-946; Upstate), anti-Histone H3 acetylated lysine 9 (07-352; Upstate), anti-Histone H4 acetylated lysine 16 (ab1762; Abcam, Cambridge, Mass., USA), anti-HDAC2 (C-19, sc-6296; Santa Cruz), anti-HDAC6 (L-18, sc-5258; Santa Cruz), and anti-Histone H3 dimethyl lysine 9 (ab9050; Abcam).
Real-time qPCR was preformed with gene-specific primers as previously described [Gordon et al., 2010].
ChIP Assays
ChIP assays were performed as previously described [Hassan et al., 2007]. Sequential ChIP studies were performed using the primary pull-down from one antibody which was divided into equal aliquots for the second pull-down with antibodies specific for coregulatory molecules. Instead of the elution step (1% SDS and 100 mM Na2HCO3) after cross-linking and washing, immunocomplexes were eluted in 10 mM DTT. The eluate was further diluted 1:40 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM Tris-HCl pH 8.1, and 167 mM NaCl) and used for the secondary immunoprecipitations. Aliquots of each recovered DNA sample were assayed by quantitative PCR to detect the Osx distal (P1) promoter region spanning –1,077 to –988 bp and –849 to –777 bp upstream of the OSX2 transcriptional start site. As a control for nonspecific binding/proper DNA fragmentation, a region of the distal promoter from –1,502 to –1,420 was also analyzed. The primer sequences used for the ChIP experiments are available upon request (Jonathan.Gordon@umassmed.edu).
Results
PBX1 Negatively Regulates the Expression of Osteogenic Genes: Depletion of PBX1 by shRNA Increases Osteoblast Differentiation
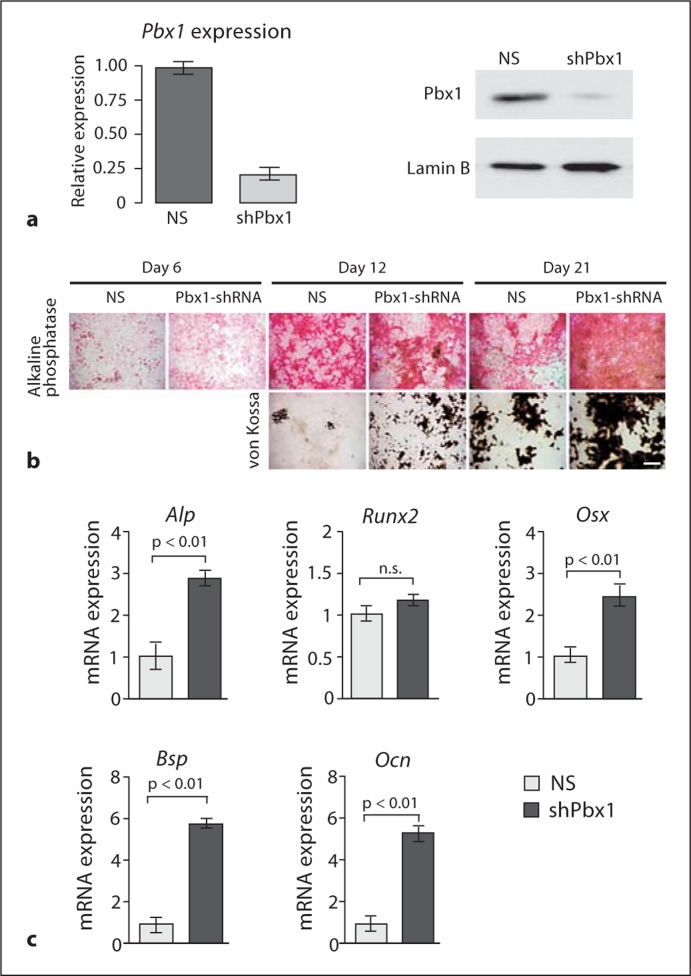
We have previously demonstrated that PBX1 plays a role in the commitment of osteogenic progenitor cells by binding to gene promoters preceding differentiation [Gordon et al., 2010]. In the present study, recombinant lentiviruses encoding either shRNA specific to Pbx1 (shPbx1) or cDNA for full-length Pbx1 (Pbx1) were generated and primary BMSCs were infected with the viruses. The shPbx1 virus successfully decreased PBX1 protein levels (fig. 1a) and this decrease in PBX1 levels was maintained for the period of the experiments (>20 days) (data not shown). Virally infected BMSCs displaying reduced PBX1 expression exhibited increased alkaline phosphatase (ALP) activity and mineralization deposition (von Kossa staining) compared to cells infected with the nonsilencing (NS) control (fig. 1b). Additionally, in Pbx1-shRNA-infected cells the early osteoblastic marker Osx was significantly increased and Ocn and Bsp mRNA levels, markers of the mature osteoblast, were upregulated 5- to 6-fold compared to NS control cells (fig. 1c). However, Runx2 mRNA levels did not change significantly. This result is consistent with our earlier study in which exogenous expression of PBX1 in osteogenic progenitors had the opposite effect. Alkaline phosphatase activity, mineralization, and osteoblastic gene expression were reduced [Gordon et al., 2010]. These findings establish that PBX1 is a potent negative regulator of osteoblastic genes expressed during early and late stages of osteoblast differentiation.
Fig. 1.
Depletion of PBX1 by shRNA results in increased expression of osteoblast-related genes in mesenchymal cells. a–c BMSC cells were infected with ∼100 plaque-forming units/cell of shRNA-encoding recombinant lentivirus (NS or Pbx1-specific target). a Relative mRNA and protein levels of PBX1 in treated BMSCs. b BMSCs treated with Pbx1-shRNA demonstrate increased alkaline phosphatase activity and mineral deposition. Scale bar = 500 μm. c Relative expression of osteoblast-related genes, monitored by RT-qPCR, was significantly increased in Pbx1-shRNA-infected BMSCs. Statistical significance was determined by 1-way ANOVA followed by a Bonferroni post hoc test. Data are presented as the means of 3 experiments ± SEM.
PBX1 Negatively Regulates the Osx Gene through Epigenetic Modification
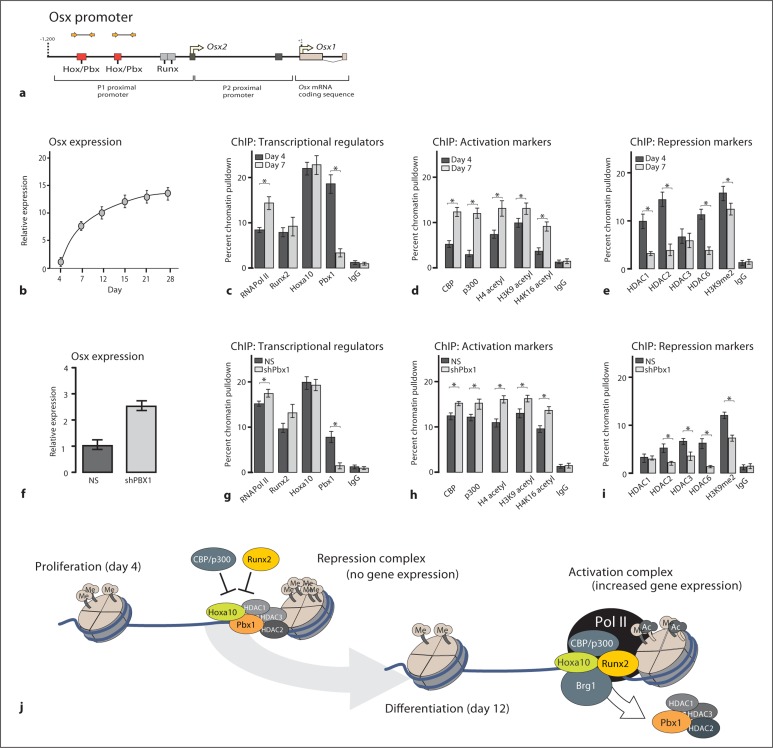
These results suggest that PBX1 may have a biological role in attenuating a wide range of osteoblast-specific genes during differentiation. In silicoanalysis of osteogenic promoters determined that the Osx promoter contains 2 Pbx1 consensus sites (tTGATa) [LaRonde-LeBlanc and Wolberger, 2003] in the distal (P1) promoter region of the gene, suggesting that Osx may be directly regulated by PBX1. The Osx gene promoter (fig. 2a) is tightly regulated and can be induced by numerous transcriptional regulators including MAPK effectors and RUNX2 [Lu et al., 2006]. By ChIP analysis, PBX1 was strongly associated with the Osx promoter in proliferating cells (day 4 prior to gene expression) but not during upregulated gene expression (day 7) (fig. 2c). Several mediators and covalent modifications of histone proteins associated with chromatin remodeling, including CBP/p300, acetylated histone H4, H3 lysine 9 (H3K9), and acetylated H4 lysine 16 (H4K16), were found to increase with osteoblast differentiation (fig. 2d). In contrast, repressive marks on chromatin, including the HDAC1, HDAC2, and HDAC3, as well as the methylation of histone H3K9, demonstrated a decrease upon osteoblast differentiation (fig. 2e). These results strongly suggest that PBX1 is associated with a negative regulation of chromatin remodeling at the osteocalcin promoter.
Fig. 2.
PBX1 displays a pattern of gene-repressive functionality on the osterix promoter. a Diagram of the osterix promoter displaying the relative binding sites and primer sites used for ChIP. BMSCs were isolated from the long bones of 6-week-old mice and collected during the proliferative stage (day 4) or cultured in differentiating conditions and collected during the increase in Osx expression (day 12). b BMSCs were analyzed at the indicated differentiation stages by RT-qPCR to determine the levels of Osx gene expression. c–e ChIP analysis was performed on cleared lysates from BMSCs using ∼5 μg of the indicated antibody. The recovered DNA was then quantified by qPCR using primers specific for the proximal promoter region of the osterix gene to determine the relative occupancy of transcriptional activators (c), activation markers (d), or repressive markers (e). BMSCs (as above) were infected with Pbx1-shRNA or NS-shRNA (NS)-lentiviral constructs (f) and the relative expression of Osx was determined by RT-qPCR. g–i ChIP analysis was performed on cleared lysates from treated BMSCs (as above) to determine the relative occupancy of chromatin modifiers. Statistical significance was determined by Student's t test (∗ p < 0.05 vs. matched control). ChIP experiments were repeated at least 2 times with similar results, and 1 representative experiment is presented ± SD.
In BMSCs with depleted PBX1 expression (shPbx1) (fig. 2f), PBX1 binding to Osx chromatin was decreased compared to NS-infected cells (fig. 2g). While there was no effect on HOXA10 binding, there was an increase in both RUNX2 and p300 occupancy at the Osx promoter in cells deficient in PBX1 as well as a reduction in occupancy of HDAC1 and HDAC2 (fig. 2h, i). These results indicate that PBX1 may be important for recruiting these HDAC factors to the Osx promoter to mark the gene for expression while maintaining a repressive state during proliferation. The data suggest that PBX1 contributes to the switching of chromatin remodeling factors from repression to open chromatin for activation as illustrated in figure 2j.
Discussion
We have provided several lines of evidence that PBX1 is a critical negative regulator of osteoblast differentiation by demonstrating that the depletion of PBX1 by targeted shRNA increases osteoblastogenesis and the expression of osteoblast-related genes. We have also demonstrated that PBX1 acts directly on the Osx promoter as well as Ocn and Bsp [Gordon et al., 2010] to repress the HOXA10-mediated expression of osteoblast genes by a mechanism in which PBX1 recruits negative regulators of chromatin remodeling, thereby decreasing the acetylation of histones adjacent to translational start sites of these genes. In addition, we have demonstrated that PBX1 has the ability to act as a molecular switch functioning with HOXA10 in proliferating cells to maintain osteoblast-specific genes in a repressed state. These key findings highlight a novel regulatory role for PBX1 in repressing and marking osteoblast genes for expression and limiting HOXA10-mediated gene transcription for regulating the temporal activation of postproliferative gene expression.
In conclusion, in this study we have demonstrated interactions between PBX1 and HOXA10 in BMSC control of the temporal expression of bone-specific gene expression at multiple levels, including transcriptional regulation at the promoter level and initiation of transcriptional activity through chromatin remodeling. This characterization of the PBX1 and HOXA10 interacting sites on the Osx promoter has provided a clearer understanding of the chromatin remodeling mechanisms that lead to the expression of tissue-specific genes to establish an osteogenic lineage preceding RUNX2 recruitment in stem cells/mesenchymal progenitors during commitment to the osteoblast lineage. This concept has implications for bone repair and molecular intervention to correct bone disorders.
Acknowledgements
The studies reported were supported in part by grants from the National Institutes of Health (5R37DE012528, 3R37DE012528-S1, 5R01AR039588, and 5P01AR048818). Core resources supported by Diabetes Endocrinology Research Center (DERC) grant DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases were used; J. Lian and G. Stein are members of the UMass DERC (DK32520). The contents of this work are solely our responsibility and do not necessarily represent the official views of the National Institutes of Health.
Glossary
Abbreviations used in this paper
| ALP | alkaline phosphatase |
| BMSC | bone marrow stromal cells |
| BSP | bone sialoprotein |
| ChIP | chromatin immunoprecipitation |
| HDAC | histone deacetylase |
| HOXA10 | homeobox protein A10 |
| NS | nonsilencing |
| OSX | osterix; Sp7 transcription factor |
| OCN | osteocalcin BGLAP2 |
| PBX1 | pre-B-cell leukemia transcription factor 1 |
References
- Choe S.K., Lu P., Nakamura M., Lee J., Sagerstrom C.G. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell. 2009;17:561–567. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J.A., Hassan M.Q., Saini S., Montecino M., van Wijnen A.J., Stein G.S., Stein J.L., Lian J.B. Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol Cell Biol. 2010;30:3531–3541. doi: 10.1128/MCB.00889-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M.Q., Tare R., Lee S.H., Mandeville M., Weiner B., Montecino M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N.A., Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17:2060–2072. doi: 10.1101/gad.1103303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Gilbert L., He X., Rubin J., Nanes M.S. Transcriptional regulation of the osterix (Osx, Sp7) promoter by tumor necrosis factor identifies disparate effects of mitogen-activated protein kinase and NF kappa B pathways. J Biol Chem. 2006;281:6297–6306. doi: 10.1074/jbc.M507804200. [DOI] [PubMed] [Google Scholar]
- Moens C.B., Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Selleri L., Depew M.J., Jacobs Y., Chanda S.K., Tsang K.Y., Cheah K.S., Rubenstein J.L., O’Gorman S., Cleary M.L. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]