Abstract
Cryogenic transmission electron microscopy (cryo-EM) was used to explore the self-assembly of recombinant murine amelogenin (rM179) in vitro. Our cryo-EM data showed that amelogenin self-assembly is a strongly pH-dependent process. At pH 4.4 the main fraction of the protein exists in a monomeric form, although some peculiar structures consisting of chains of monomers were also observed. At pH 5.8 large nanospheres comprising ring-like structures ∼50 nm in diameter were the most abundant particle class. Similarly, at pH 8.0 amelogenins self-assembled into ring-like oligomers of different sizes, which subsequently assembled into nanospheres 15–20 nm in diameter. Furthermore, at pH 7.2, which is close to a physiological pH, branched chains of nanospheres were observed. Our results show that amelogenin assembly is a multistep hierarchical process and provides new insight into the control of enamel mineralization.
Key Words: Cryogenic transmission electron microscopy, Amelogenin, Self-assembly, Nanospheres, pH
Introduction
Mature dental enamel is the most highly mineralized mammalian tissue, and it possesses outstanding mechanical properties which are to a large extent determined by its structural organization. Amelogenin, comprising more than 90% of the extracellular organic matrix of forming enamel, plays an essential role in regulating the growth and organization of enamel crystal during enamel formation [Gibson et al., 2001; Margolis et al., 2006; Pugach et al., 2010]. In vitro, amelogenin has been shown to self-assemble, forming nanospheres of 15–20 nm in diameter similar to those formed in vivo [Fincham et al., 1994; Moradian-Oldak et al., 1994; Fincham et al., 1995]. These nanospheres have been intensively characterized, for example, by dynamic light scattering (DLS) [Uskokovic et al., 2010; Fincham et al., 1998; Wiedemann-Bidlack et al., 2007], atomic force microscopy [Moradian-Oldak et al., 2000], transmission electron microscope (TEM) [Du et al., 2005; Wiedemann-Bidlack et al., 2007], small angle X-ray scattering [Aichmayer et al., 2005], and nuclear magnetic resonance [Buchko et al., 2010; Delak et al., 2009].
It is well established that pH is the major factor regulating amelogenin assembly [Moradian-Oldak et al., 1998; Wiedemann-Bidlack et al., 2007]. Specifically, it has been shown that amelogenin is soluble at low pH values (below 4) and it assembles into nanospheres and chains of nanospheres at pH values above 5.8. In vitro mineralization experiments indicate that amelogenin controls mineral formation and structural organization [Beniash et al., 2005; Kwak et al., 2009; Deshpande et al., 2010]. Despite numerous studies, the underlying molecular mechanisms of amelogenin assembly are not well understood [Margolis et al., 2006]. Understanding these mechanisms is crucial for the development of new therapies and materials for hard tissue repair and regeneration.
One of the major challenges in studies of protein self-assembly is that, although these processes occur in an aqueous environment, their characterization often requires dehydration which can lead to prominent structural changes. To overcome this problem, we used cryogenic transmission electron microscopy (cryo-EM) of vitrified hydrated samples to study recombinant murine amelogenin (rM179) at different pH values. Cryo-EM is ideal for this application since it allows us to perform studies of assembling proteins in their native hydrated state and minimizes dehydration artifacts.
Materials and Methods
Full-length recombinant mouse amelogenin, i.e. rM179, was produced in bacteria Escherichia coli and purified as previously described [Simmer et al., 1994]. Protein stock solution was prepared by dissolution of lyophilized rM179 in distilled water at a concentration of 10 mg/ml. The pH was adjusted to 3.5 using HCl and the solution was kept for at least 24 h at 4°C prior to the experiments to ensure complete dissolution of the protein. rM179 assembly was assessed at 4 pH values, i.e. 4.4, 5.8, 7.2, and 8.0. For experiments at pH values of 4.4 and 5.8, protein stock solution was mixed with distilled deionized water to yield a final concentration of 0.125 mg/ml, whereas for pH values of 7.2 and 8.0 protein stock solution was added to 4 mM PBS on ice to the final protein concentration of 0.1 mg/ml. Although the difference in ionic strength between PBS and DDW can influence the self-assembly process, our recently published study on the effects of ionic strength on amelogenin assembly indicated that it has only a minor affect on amelogenin self-assembly [Wiedemann-Bidlack et al., 2011]. TEM grids coated with lacey carbon (400 mesh; EMS) were placed on top of small, 20-μl droplets of the protein solutions and incubated for 1, 5, 10, or 30 min at room temperature in a humidity chamber. The grids were then blotted and immediately plunge frozen into ethane slush cooled by liquid nitrogen using a Vitrobot automated plunge freezer (Gatan). The frozen grids were cold transferred to an FEI Tecnai F20 TEM with a field emission gun operating at a 200-kV accelerating voltage. The areas of the vitrified ice in the holes of carbon film with an average thickness of 1,000 Å were chosen for the analysis. Data were recorded using a Gatan 4,000 × 4,000 charge-coupled device camera. Images were taken in low-dose mode at 20 e/Å2 to minimize radiation damage to the samples at a nominal magnification of 50,000. To enhance the image contrast, under-focuses in the range of 1–3 μm were used to record the images. The calibrated magnification was 70,093 resulting in a pixel size of 2.14 Å.
Results and Discussion
After 1 min in the reaction at pH 8, a number of different structures were observed (fig. 1a). There were slightly elongated particles less than 3 nm across which we interpreted as amelogenin monomers (fig. 1d). Several dimers were also observed at this stage (fig. 1e). In addition, there were larger ring-like structures of different diameters ranging from 3 to 10 nm which were interpreted as amelogenin oligomers with a different number of protein molecules (fig. 1f-i). Since the TEM micrographs are 2-D projections of 3-D objects, these observations provide only limited structural information, and additional studies are needed to determine the exact structures of monomers and oligomers. After 5 min of incubation, larger oligomers comprised the major fraction, while smaller particles were still quite numerous (fig. 1b). At this time larger aggregates of a roughly spherical shape with an average diameter of 15–20 nm began to appear. We identified these large particles as amelogenin nanospheres, based on their shape and size, consistent with published data. At 10 min of incubation, the majority of the protein particles were nanospheres (fig. 1c). Our studies further reveal that these nanospheres are not homogeneous but possess an internal structure. It appears that these particles are made of a number of oligomeric rings (fig. 1j-l).
Fig. 1.
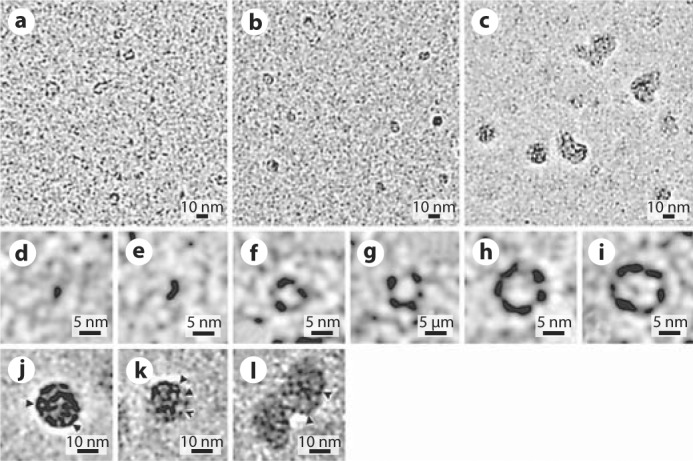
Cryo-EM micrographs of rM179 self-assembly at pH 8.0 at 1 min (a), 5 min (b), and 10 min (c) and close-up images of the particle classes observed: monomer (d), oligomer (e–i), nanosphere (j, k, l). Arrowheads point to individual oligomers.
In contrast, when amelogenin was incubated at pH 7.2, an interconnected network made of nanosphere chains was observed after only 5 min of incubation (fig. 2). As shown in figure 2b and c, nanospheres forming this network have an internal structure similar to that observed at pH 8.0, with apparent ring-like oligomers inside the nanospheres.
Fig. 2.
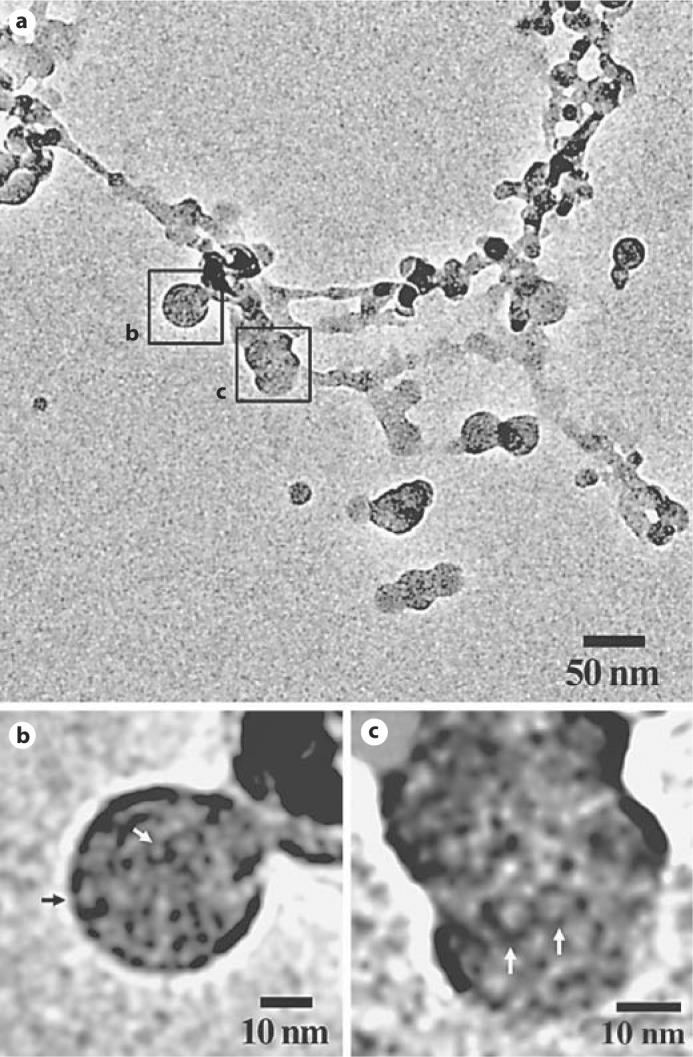
a Full-length rM179 assembly at pH 7.2 for 5 min showing an interconnected network of nanosphere chains. b, c Close-up images of nanospheres in the network. Note that the nanospheres are comprised of ring-like structures (arrows).
We also conducted self-assembly experiments at a low pH of 4.4 (fig. 3). To our surprise in addition to monomers of amelogenin (fig. 3b), partially aligned pearl necklace-like chains of monomers were observed (fig. 3c-e). No intermediate particles were present at this pH. At pH 5.8, however, rM179 assembled into large aggregates with diameters in the range of 20–70 nm (fig. 4a). Interestingly, at this pH oligomeric rings appear similar to those observed in the self-assembly experiments conducted at pH 8.0 as shown in figure 4b and c.
Fig. 3.
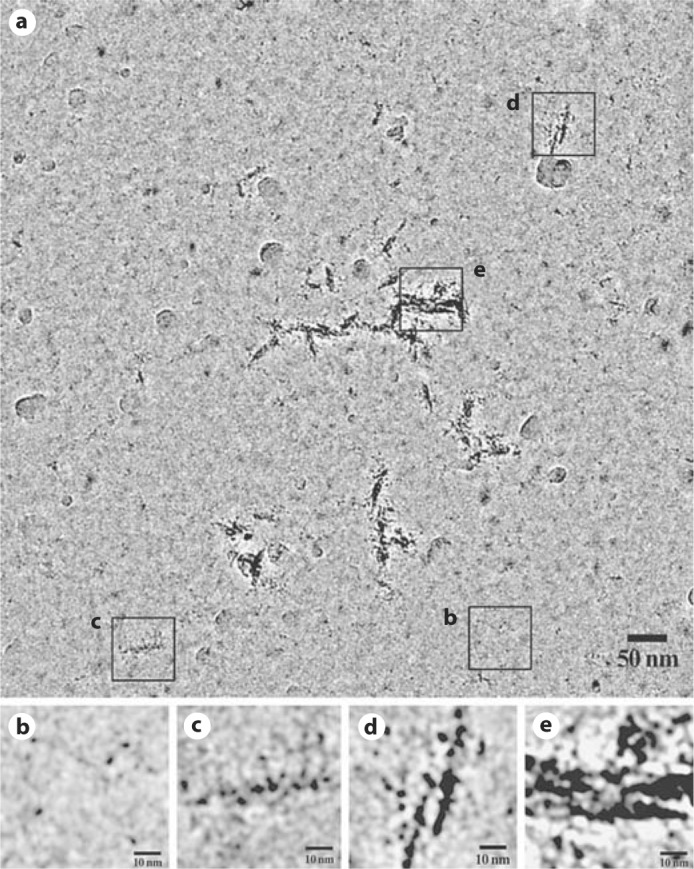
Full-length rM179 assembly at pH 4.4 for 5 min (a) and close-up images of monomers or aggregated monomers (b–e).
Fig. 4.
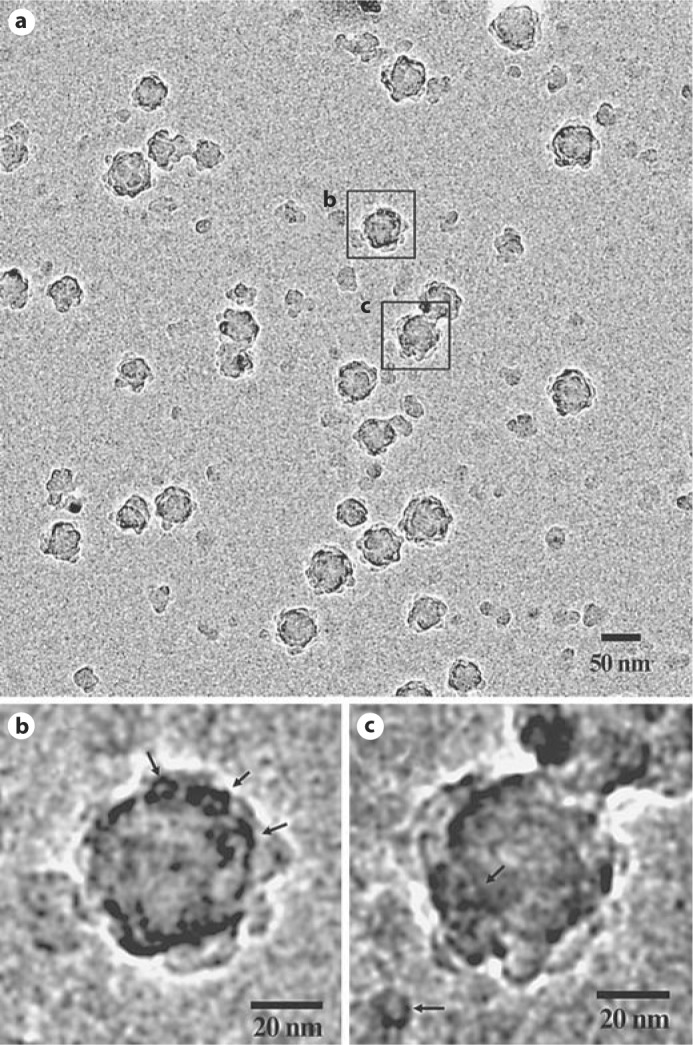
Full-length rM179 assembly at pH 5.8 for 5 min (a) and close-up images of large aggregates of nanospheres comprising oligomers (b, c); arrows point to the oligomeric structures in the nanospheres and one individual oligomer in c.
The use of cryo-EM allowed us to make a number of important observations which further our understanding of amelogenin assembly as well as basic mechanisms of enamel formation. In particular, we provide new insights into the earliest stages of the amelogenin assembly process. The results of our experiments at pH 8.0 demonstrate that amelogenin assembly is a multistep process which occurs via the formation of ring-like oligomers that further assemble to form nanospheres. The resulting nanospheres therefore have an evident internal structure consisting of the oligomers. The presence of intermediate structures in amelogenin assembly has been previously proposed by several researchers [Fincham et al., 1998; Du et al., 2005; Brookes et al., 2006], and our results provide a direct conformation of these hypotheses. Recent studies of amelogenin adsorption onto mineral surfaces and self-assembled monlolayers also indicate that the nanospheres break down into smaller oligomeric particles which interact with the surfaces [Tarasevich et al., 2009], suggesting that these oligomeric subunits might play important roles in the protein-mineral interactions during enamel formation. Interestingly, we observed a number of different classes of ring-like oligomers with diameters ranging from 3 to 10 nm, suggesting that the rings include different numbers of amelogenin molecules. At this point we can only surmise that these variations are related to the fact that amelogenin in solution is an intrinsically disordered protein [Delak et al., 2009] and is likely to be less conformationally rigid during assembly than proteins with well-defined structures. Additional studies will be needed to clarify the question of variability in amelogenin oligomeric structures.
The results of our studies of amelogenin assemblies at pH 7.2 revealed that amelogenin nanospheres form an extended network of branched chains of nanospheres. These results are in agreement with earlier studies [Du et al., 2005; Wiedemann-Bidlack et al., 2007]. At the same time our data indicates that the nanospheres in the chains are structurally similar to those observed at pH 8.0 as they are also composed of ring-like oligomeric structures. These observations suggest that the process of self-assembly at pH 7.2 is similar to the stepwise assembly process observed at pH 8.0. This is an important observation since pH 7.2 is in the physiological range at the sites of initial enamel deposition [Lacruz et al., 2010], and hence such structural organization of amelogenin assemblies can closely resemble the situation in forming enamel in vivo.
The self-assembly experiments at pH 5.8 revealed that the major particle class at this pH is large spherical aggregates. Particles of similar size were reported in earlier studies of amelogenin assembly by DLS at similar pH and temperature [Petta et al., 2006]. Importantly, these nanospheres had an internal structure similar to that observed at pH values of 8.0 and 7.2; namely, they consisted of ring-like substructures. Overall, our results suggest that the major mode of amelogenin nanosphere assembly in a wide range of pH (5.8–8.0) is the stepwise hierarchical assembly via intermediate oligomeric structures. This observation has a number of important implications for our understanding of the function of amelogenin in forming enamel. For example, the modular organization of amelogenin nanospheres might facilitate structural changes in the enamel organic matrix and amelogenin interactions with forming mineral.
Interestingly, at pH 4.4 in addition to monomers we observed elongated protein aggregates up to 100 nm long consisting of aligned chains of monomeric particles. This result was quite unexpected since the current paradigm suggests that at this pH amelogenin is highly soluble and exists in a monomeric form or forms aggregates consisting of only a handful of molecules [Moradian-Oldak et al., 1994, 1998]. However, recently a DLS study from the group of Moradian-Oldak [2000] revealed the presence of larger-size aggregates, with Rh in the range of 50–80 nm, when analyzed using a spherical particle model [Petta et al., 2006]. Our ability to conduct direct structural observation in cryo-EM allowed us to determine that these aggregates have an entirely different organization compared to amelogenin nanospheres. This observation suggests that the mechanisms of amelogenin assembly at low pH might differ from those at high pH. One possibility is that at pH 4.4, which is far removed from the amelogenin isoelectric point, the nature of protein-protein interactions involved in protein assembly may differ. In particular, the side chains of acidic amino acids can be largely protonated. The C-terminal telopeptide of amelogenin has a high concentration of acidic amino acids. Lack of this telopeptide causes major changes as has been shown in studies of knockout and transgenic animal models [Paine et al., 2000; Pugach et al., 2010]. It has been also shown that the C-terminal telopeptide plays an essential role in amelogenin self-assembly and its regulation of mineral formation and organization in vitro [Wiedemann-Bidlack et al., 2007]. Furthermore, it has been proposed that C-terminal telopeptides bind each other via electrostatic interactions between negatively and positively charged amino acids [Fukae et al., 2007]. It is therefore feasible that protonation of acidic amino acids of the C-terminal telopeptide will influence the mode of protein-protein interactions and amelogenin self-assembly.
In conclusion, our cryo-EM data indicate that both the kinetics and the mode of amelogenin self-assembly are strongly pH dependant. Specifically, our data suggest that in a wide range of pH (5.8–8.0) amelogenin assembly is a stepwise process which occurs through the formation of oligomeric ring-like particles that assemble into nanospheres. Our study revealed that at pH 4.4 amelogenin assembles into chains of monomers organized into elongated bundles, suggesting that the mechanism of the assembly at this pH is different from the self-assembly at higher pH values. These results provide new insights into the structural organization of amelogenin supramolecular assemblies and the mechanisms of their formation, and they have important implications for our understanding of the basic mechanisms of enamel formation.
Acknowledgements
Support for this work was provided by NIH grants R01DE016376 (H.C.M.) and R01DE016703 (E.B.), and by the Commonwealth of Pennsylvania grant SAP 4100031302 (J.F.C.).
Glossary
Abbreviations used in this paper
| Cryo-EM | cryogenic transmission electron microscopy |
| DLS | dynamic light scattering |
| rM179 | recombinant murine amelogenin |
References
- Aichmayer B., Margolis H.C., Sigel R., Yamakoshi Y., Simmer J.P., Fratzl P. The onset of amelogenin nanosphere aggregation studied by small-angle X-ray scattering and dynamic light scattering. J Struct Biol. 2005;151:239–249. doi: 10.1016/j.jsb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Beniash E., Simmer J.P., Margolis H.C. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol. 2005;149:182–190. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Brookes S.J., Lyngstadaas S.P., Robinson C., Shore R.C., Kirkham J. Intracellular nanosphere subunit assembly as revealed by amelogenin molecular cross-linking studies. Eur J Oral Sci. 2006;114:280–284. doi: 10.1111/j.1600-0722.2006.00311.x. [DOI] [PubMed] [Google Scholar]
- Buchko G.W., Tarasevich B.J., Roberts J., Snead M.L., Shaw W.J. A solution NMR investigation into the murine amelogenin splice-variant LRAP (Leucine-Rich Amelogenin Protein) Biochim Biophys Acta. 2010;1804:1768–1774. doi: 10.1016/j.bbapap.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delak K., Harcup C., Lakshminarayanan R., Sun Z., Fan Y., Moradian-Oldak J., Evans J.S. The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48:2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A.S., Fang P.A., Simmer J.P., Margolis H.C., Beniash E. Amelogenin-collagen interactions regulate calcium phosphate mineralization in vitro. J Biol Chem. 2010;285:19277–19287. doi: 10.1074/jbc.M109.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Falini G., Fermani S., Abbott C., Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Leung W., Tan J., Moradian-Oldak J. Does amelogenin nanosphere assembly proceed through intermediary-sized structures? Connect Tissue Res. 1998;38:237–240. doi: 10.3109/03008209809017042. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Moradian-Oldak J., Diekwisch T.G.H., Lyaruu D.M., Wright J.T., Bringas P., Slavkin H.C. Evidence for amelogenin nanospheres as functional components of secretory-stage enamel matrix. J Struct Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Moradian-Oldak J., Simmer J.P., Sarte P., Lau E.C., Diekwisch T., Slavkin H.C. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol. 1994;112:103–109. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- Fukae M., Yamamoto R., Karakida T., Shimoda S., Tanabe T. Micelle structure of amelogenin in porcine secretory enamel. J Dent Res. 2007;86:758–763. doi: 10.1177/154405910708600814. [DOI] [PubMed] [Google Scholar]
- Gibson C.W., Yuan Z.A., Hall B., Longenecker G., Chen E.H., Thyagarajan T., Sreenath T., Wright J.T., Decker S., Piddington R., Harrison G., Kulkarni A.B. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Kwak S.Y., Wiedemann-Bidlack F.B., Beniash E., Yamakoshi Y., Simmer J.P., Litman A., Margolis H.C. Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro. J Biol Chem. 2009;284:18972–18979. doi: 10.1074/jbc.M109.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz R.S., Nanci A., White S.N., Wen X., Wang H.J., Zalzal S.F., Luong V.Q., Schuetter V.L., Conti P.S., Kurtz I., Paine M.L. The sodium bicarbonate cotransporter (NBCe1) is essential for normal development of mouse dentition. J Biol Chem. 2010;285:24432–24438. doi: 10.1074/jbc.M110.115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis H.C., Beniash E., Fowler C.E. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Leung W., Fincham A.G. Temperature and pH-dependent supramolecular self-assembly of amelogenin molecules: a dynamic light-scattering analysis. J Struct Biol. 1998;122:320–327. doi: 10.1006/jsbi.1998.4008. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Paine M.L., Lei Y.P., Fincham A.G., Snead M.L. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J Struct Biol. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Simmer J.P., Lau E.C., Sarte P.E., Slavkin H.C., Fincham A.G. Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light-scattering. Biopolymers. 1994;34:1339–1347. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- Paine M.L., Zhu D.H., Luo W., Bringas P., Goldberg M., White S.N., Lei Y.P., Sarikaya M., Fong H.K., Snead M.L. Enamel biomineralization defects result from alterations to amelogenin self-assembly. J Struct Biol. 2000;132:191–200. doi: 10.1006/jsbi.2000.4324. [DOI] [PubMed] [Google Scholar]
- Petta V., Moradian-Oldak J., Yannopoulos S.N., Bouropoulos N. Dynamic light scattering study of an amelogenin gel-like matrix in vitro. Eur J Oral Sci. 2006;114:308–314. doi: 10.1111/j.1600-0722.2006.00325.x. [DOI] [PubMed] [Google Scholar]
- Pugach M.K., Li Y., Suggs C., Wright J.T., Aragon M.A., Yuan Z.A., Simmons D., Kulkarni A.B., Gibson C.W. The amelogenin C-terminus is required for enamel development. J Dent Res. 2010;89:165–169. doi: 10.1177/0022034509358392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer J.P., Lau E.C., Hu C.C., Aoba T., Lacey M., Nelson D., Zeichnerdavid M., Snead M.L., Slavkin H.C., Fincham A.G. Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif Tissue Int. 1994;54:312–319. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- Tarasevich B.J., Lea S., Bernt W., Engelhard M., Shaw W.J. Adsorption of amelogenin onto self-assembled and fluoroapatite surfaces. Journal Phys Chem B. 2009;113:1833–1842. doi: 10.1021/jp804548x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskokovic V., Castiglione Z., Cubas P., Zhu L., Li W., Habelitz S. Zeta-potential and particle size analysis of human amelogenins. J Dent Res. 2010;89:149–153. doi: 10.1177/0022034509354455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann-Bidlack F.B., Beniash E., Yamakoshi Y., Simmer J.P., Margolis H.C. pH triggered self-assembly of native and recombinant amelogenins under physiological pH and temperature in vitro. J Struct Biol. 2007;160:57–69. doi: 10.1016/j.jsb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann-Bidlack F.B., Kwak S.-Y., Beniash E., Yamakoshi Y., Simmer J.P., Margolis H.C. Effects of phosphorylation on the self-assembly of native full-length porcine amelogenin and its regulation of calcium phosphate formation in vitro. J Struct Biol. 2011;173:250–260. doi: 10.1016/j.jsb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


