Abstract
Enamel matrix proteins, including the most abundant amelogenin and lesser amounts of enamelin, ameloblastin, and proteinases, play vital roles in controlling crystal nucleation and growth during enamel formation. The cooperative action between amelogenin and the 32-kDa enamelin is critical to regulating the growth morphology of octacalcium phosphate crystals. Using biophysical methods, we investigated the interaction between the 32-kDa enamelin and recombinant pig amelogenin 148 (rP148) at pH 6.5 in phosphate-buffered saline (PBS). Dynamic light scattering results showed a trend of increasing particle size in the mixture with the addition of enamelin to amelogenin. Upon addition of the 32-kDa enamelin, the shift and intensity decrease in the ellipticity minima of rP148 in the circular dichroism spectra of rP148 illustrated a direct interaction between the 2 proteins. In the fluorescence spectra, the maximum emission of rP148 was blue shifted from 335 to 333 nm in the presence of enamelin as a result of complexation of the 2 proteins. Our results demonstrate that the 32-kDa enamelin has a close association with amelogenin at pH 6.5 in PBS buffer. Our present study provides novel insights into the possible cooperation between enamelin and amelogenin in macromolecular coassembly and in controlling enamel mineral formation
Key Words: Enamel, 32-kDa enamelin, Amelogenin self-assembly, Circular dichroism, Dynamic light scattering, Fluorescence spectroscopy, Octacalcium phosphate
Introduction
The extracellular matrix proteins secreted by ameloblasts play critical roles in controlling the nucleation, growth, and organization of apatite crystals during enamel development. The dominant proteins in the matrix are amelogenins (>90%) which self-assemble to form nanospheres and higher order structures in vitro and in vivo [Moradian-Oldak and Goldberg, 2005; Margolis et al., 2006]. The functional roles of amelogenin in mineral formation are critical in controlling the morphology, organization, and directionality of apatite crystals, as demonstrated by recent in vitro and in vivo studies [Gibson et al., 2001; Kwak et al., 2009; Yang et al., 2010].
Enamelin is a minor constituent (<5%) of the extracellular matrix but plays a critical role in normal enamel formation [Hu et al., 2008]. Mutations in the ENAM gene result in the formation of abnormal enamel, especially the hypoplastic enamel seen in autosomal-dominant amelogenesis imperfecta [Kida et al., 2002; Ozdemir et al., 2005]. Interestingly some mutations in the ENAM gene causing amelogenesis imperfecta have been reported to be within the 32-kDa enamelin fragment [Gutierrez et al., 2007]. The 32-kDa enamelin fragment is the most stable cleavage product of 186-kDa porcine enamelin and it is a hydrophilic and acidic glycoprotein [Hu and Yamakoshi, 2003].
Our previous in vitro studies revealed that the 32-kDa enamelin in cooperation with amelogenin promotes the kinetics of the nucleation of apatite crystals in a dose-dependent manner [Bouropoulos and Moradian-Oldak, 2004]. We further showed that enamelin undergoes a conformational change with a structural preference for the β-sheet with addition of its potential target, i.e. calcium ions [Fan et al., 2008]. Direct interaction between the full-length amelogenin and enamelin has been demonstrated by in vitro biophysical studies which showed a potential regulating role of the 32-kDa enamelin in the stabilization of amelogenin oligomers, resulting in partial dissociation of the nanospheres formed at pH 8.0 [Fan et al., 2009]. In our most recent studies using a cation-selective membrane system, we demonstrated the cooperative regulatory action of the 32-kDa enamelin and a recombinant pig amelogenin 148 (rP148) on the growth morphology of octacalcium phosphate (OCP) crystals [Iijima et al., 2010]. Remarkably, the presence of enamelin in the amelogenin ‘gel-like matrix’ not only increased the length-to-width ratio (aspect ratio) of OCP but also enhanced the stability of the transient amorphous calcium phosphate (ACP) phase.
In the study presented here, in order to understand the mechanism of this cooperative effect, we investigated the coassembly between amelogenin and the 32-kDa enamelin. We used dynamic light scattering (DLS), circular dichroism (CD), and fluorescence spectroscopy to evaluate the effect of enamelin on amelogenin self-assembly under pH conditions where OCP crystal growth occurs (i.e. pH 6.5) [Iijima et al., 2010].
Materials and Methods
Preparation of the 32-kDa Enamelin and Amelogenin (rP148)
The 32-kDa enamelin was extracted, purified, and characterized following the method reported previously [Fan et al., 2008]. The recombinant porcine amelogenin (rP148) which is amino acids 2–149 of the full-length recombinant porcine amelogenin rP172 and an analog to the major amelogenin proteolytic product (‘20k’) was expressed in Escherichia coli, purified using RP-HPLC, and characterized as previously described [Sun et al., 2006].
Dynamic Light Scattering
The association between the 32-kDa enamelin and amelogenin (rP148, 0.2–0.3 mg/ml) in 20 mM phosphate-buffered saline (PBS) (pH 6.5, 0.15 M NaCl) was investigated at room temperature via DLS using a DynaPro NanoStar (Wyatt Technologies, Santa Barbara, Calif., USA). The mixtures were incubated for 20–30 min before measurements and data were analyzed as previously described [Moradian-Oldak et al., 2000; Fan et al., 2008, Laksminarayanan et al., 2010].
CD Spectroscopy
Measurements were conducted on a JASCO J-810 spectropolarimeter calibrated using a 0.06% (+)-10-camphorsulfonic acid solution. Both the 32-kDa enamelin and the rP148 amelogenin were dissolved in 20 mM PBS buffer (pH 6.5, 0.15 M NaCl). The CD spectra were measured at room temperature in a 1-mm path length quartz cell (300 μl) using a scanning speed of 50 nm/min, a time response of 1 s, a bandwidth of 1 nm, and an average of 4 scans. The CD spectra were taken after a 20-min incubation at room temperature. CD spectra were expressed as the mean residue ellipticity, [Θ]mrw (in degrees cm2 dmol–1), and the [Θ]mrw was calculated as previously described [Lakshminarayanan et al., 2010].
Fluorescence Spectroscopy
Fluorescence spectra were recorded on a QuantaMaster 4 spectrofluorometer (Photon Technology International, Inc., Pemberton, N.J., USA). The acquisition interval and the integration time were maintained at 0.5 nm and 0.5 s, respectively. Fluorescence spectra were obtained by measuring the emission spectra in the range of 310–390 nm at an excitation wavelength 295 nm spaced at 0.5-nm intervals in the excitation domain. Fully corrected spectra were then concatenated into an excitation-emission matrix. Fluorescence intensities were plotted as a function of the excitation wavelength.
Simulated Models for Drawing OCP Crystals
Using the average values of width and thickness of OCP from table 1 [Iijima et al., 2010], we employed 3-D graphic application software Blender (v2.6.2) to model the OCP crystals. In brief, we drew boxes with the correct dimensions for each crystal, placed the ‘camera’ and ‘lighting’ in appropriate places, and rendered the image. We then compiled the 5 rendered images using Adobe Photoshop CS4.
Table 1.
RH, MW, and mass distribution of particles in the enam-elin-rP148 solutions (rP148, 0.25 mg/ml, 20 mM PBS, and 0.15 M NaCl)
| Enamelin:rP148 | RH, nm | MW, kDa | Mass, % |
|---|---|---|---|
| No enamelin | 9.38±1.0 | 619 | 98.1 |
| 82.08±8.9 | 101,230 | 0.1 | |
| 417.38±20.0 | 4,554,670 | 1.8 | |
| 1:100 | 10.28±1.2 | 766 | 93.1 |
| 156.88±19 | 461,123 | 0.4 | |
| 1,2688±0 | 61,316,100 | 6.5 | |
| 1:50 | 10.98±1.0 | 898 | 83.1 |
| 223.58±7.6 | 1,056,590 | 4.1 | |
| 1,6908±0 | 120,200,000 | 12.8 | |
| 1:10 | 12.38±1.4 | 1,189 | 62.0 |
| 276.48±36 | 1,736,840 | 12.8 | |
| 2,5358±0 | 310,402,000 | 25.2 | |
Results
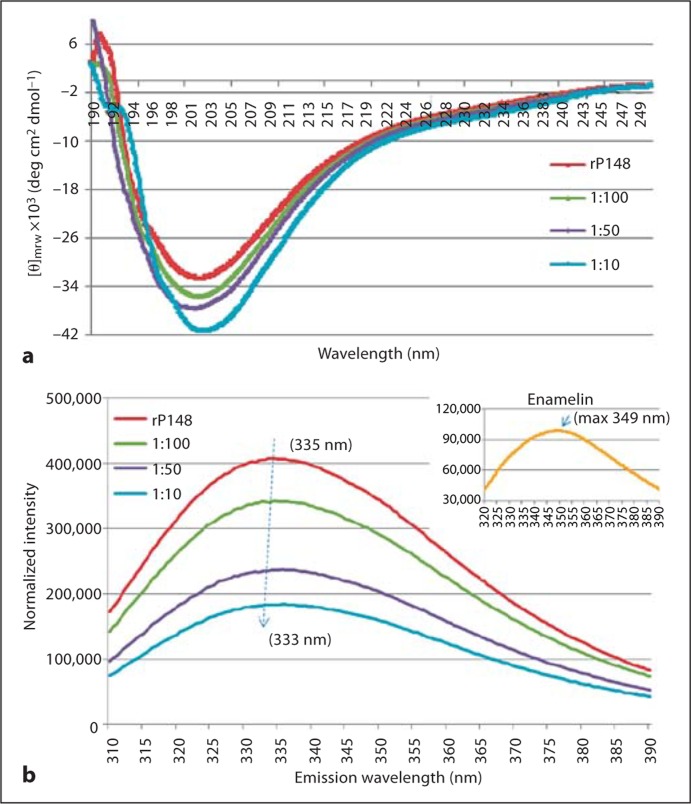
Amelogenin-enamelin interaction and their coassembly were studied in PBS solution with pH 6.5 using CD, DLS, and fluorescence spectroscopy. The CD spectrum of rP148 amelogenin showed a strong negative ellipticity with a minimum [Θ]mrw of –32.5 × 103 degrees cm2 dmol–1 at 201 nm, a characteristic of an unordered polyproline type II structure of amelogenin (Lakshminarayanan et al., 2007). Upon the addition of enamelin to rP148 at molar ratios of 1:100, 1:50, and 1:10, respectively, the intensity of the minima gradually increased in a dose-dependent manner. We subtracted enamelin CD spectra with concentrations equivalent to those in the above mentioned ratios from that of the mixed solution and found that the minimum [Θ]mrw increased to –41.3 × 103 degrees cm2 dmol–1 in the 1:10 (enamelin:rP148) solution (fig. 1a). At the same time, the trough slightly shifted from 201 to 203 nm with the increased ratio of enamelin, indicating a possible change in the conformation of amelogenin. These changes in the CD spectra clearly illustrate that there is a direct interaction between rP148 amelogenin and enamelin.
Fig. 1.
Biophysical studies on the interaction between rP148 and the 32-kDa enamelin in PBS (pH 6.5). a CD spectra of rP148 in association with the 32-kDa enamelin. b Fluorescence spectra of the rP148/enamelin solutions. Inset Spectrum for the 32-kDa enamelin.
The emission maxima in the fluorescence spectra of the 32-kDa enamelin and the rP148 amelogenin are 349 and 335 nm, respectively. Upon the addition of enamelin to the rP148 solution at increasing ratios (1:100, 1:50, and 1:10 of enamelin to rP148), the emission maximum of the solution blue shifted progressively from 335 to 333 nm (fig. 1b). It was also noticeable that the intensity of the maxima decreased significantly corresponding to the amount of enamelin added. The shift of emission maxima and the decrease in intensity illustrate the close association between these 2 enamel proteins, leading to their coassembly. As a result, the tryptophans on the N-terminal of rP148 could be buried inside the oligomers and these consequently give rise to the decreasing intensity and the blue shift of maxima in the fluorescence properties of the rP148/enamelin solution.
The association between the 32-kDa enamelin and rP148 was further analyzed by DLS measurements which provided the estimated hydrodynamic radii (RH), molecular weight (MW), and mass distribution of particles in the rP148-enamelin solutions at pH 6.5 (table 1). The rP148 amelogenin particles had a predominant population with a monodisperse size distribution of a mean RH of 9.3 ± 1 nm and 2 very minor populations with bigger particle sizes. When the 32-kDa enamelin was added to rP148 at a molar ratio of 1:100, the predominant particle size in the rP148-enamelin solution increased slightly to 10.2 ± 1.2 nm while its population decreased. Meanwhile, the mass percentage of the minor populations increased and the particle sizes became larger. At a higher ratio of 1:50, the average RH of the nanoparticles in the major population increased to 10.9 ± 1.0 nm but had a reduced mass percentage, while both the RH and the mass percentage of the 2 minor populations increased. At the highest ratio of enamelin to rP148 (1:10), the average RH of the nanoparticles in the major group increased further to 12.3 ± 1.4 nm while the mass percentage decreased to 62%. The particle sizes and the mass in the minor populations were both increased. This increase in average particle size in the solution was presumably due to the formation of complexes between enamelin and rP148 molecules, indicating a close association between these 2 enamel proteins (table 1).
Discussion
The 32-kDa enamelin and rP148 amelogenin have been shown to have a synergic effect on the regulation of the morphology of OCP crystals grown at pH 6.5 in a cation-selective membrane system [Iijima et al., 2010]. Using the Blender v2.6.2 modeling program which clearly presented the shape and morphology of OCP crystals grown in different amount proteins, we modeled the overall effect of enamelin alone, amelogenin alone, and the combination of the 2 on the shape and habit of the OCP crystals (fig. 2). In the sole presence of 10% rP148, crystals grew in the form of ribbon-like shapes, which is in line with the previous observations of crystal growth regulated by 10% m/v of native bovine amelogenin [Iijima et al., 2001]. On the other hand, in 40En solution (40 μg/ml enamelin alone), the OCP crystals had a slightly bigger width and thickness but also had a substantial decrease in length. The shape and size of OCP crystals were changed remarkably by the coregulatory effect of rP148 and the 32-kDa enamelin. In the rP148 + 4En solution (10% rP148 + 4 μg/ml enamelin), both the width and the thickness of crystals were reduced slightly when compared with those in pure rP148 solution. When more enamelin was added, i.e. in rP148 + 10En (10% rP148 + 10 μg/ml enamelin), the width and thickness of crystals decreased dramatically and the shape of the OCP changed from a ribbon-like plate to a thin needle-like morphology (fig. 2). With the largest amount of enamelin used (10% rP148 + 40 μg/ml enamelin), the width of crystals decreased further but the thickness increased when compared to crystals grown in rP148 + 10En, giving rise to a rod-shaped habit of OCP. It is important to note that the presence of enamelin was critical for the control of OCP crystal arrangement/orientation. This observation led us to investigate the cooperative assembly between these 2 enamel proteins under the conditions of OCP crystal growth (i.e. pH 6.5 and phosphate buffer) and to understand their synergistic action on OCP growth.
Fig. 2.

Computer simulation models for OCP crystals grown in the presence of different amounts of enamelin in 10% rP148. 4En = 4 μg/ml enamelin; 10En = 10 μg/ml enamelin; 40En = 40 μg/ml enamelin. Width and thickness values are based on the average data in the report by Iijima et al., [2010].
rP148 is an analog to the 20-kDa amelogenin fragment, i.e. the dominant cleavage product, and it pos- sesses the unordered polyproline type II structure with a negative ellipticity around 201 nm in the CD spectra [Lakshminarayanan et al., 2007]. The minima increase in intensity following the addition of enamelin and the slight shift of the trough indicates an ellipticity change as the result of a direct interaction between these 2 enamel proteins. Additional support for this notion is provided by the observed increase in the particle sizes of rP148 (from 9.3 ± 1.0 nm to 12.3 ± 1.4 nm) upon addition of the 32-kDa enamelin in a 1:10 ratio. This is an increase in MW from 619 kDa (rP148 alone) to 1,189 kDa (rP148 + enamelin). At pH 6.5, enamelin alone has an RH of 3.3 ± 0.3 nm (data not shown). In the fluorescence spectra as a result of the addition of the 32-kDa enamelin to the rP148 solution, the emission maximum of rP148 shifted from 335 to 333 nm and the emission intensity also decreased substantially. It is important to note that the 32-kDa enamelin has a maximum emission at 349 nm (fig. 1b inset), which corresponds to the 1 tryptophan in its N-terminal region [Fan et al., 2008]. Given the fact that the 2 tryptophans in rP148 are located in the N-terminal region, the emission change in amelogenin upon the addition of enamelin is likely due to the interaction between the N-acetylglucosamine in enamelin and the tyrosyl motif at the N-terminal region of amelogenin [Ravindranath et al., 1999]. Therefore, the complexations of the 2 proteins cause the 2 tryptophans in rP148 to be buried inside the oligomers, and the emission intensities of tryptophans are weakened in the fluorescence spectra. Although we could not unambiguously define the mechanism of the interaction between these 2 enamel proteins, our biophysical data clearly demonstrate a direct interaction and coassembly between rP148 and the 32-kDa enamelin at pH 6.5. Such coassembly may enhance the hydrophilicity of the amelogenin oligomers, resulting in their higher affinity to the OCP crystal faces. We propose that during the postsecretory stage of enamel formation amelogenin and enamelin cooperate synchronically to control crystal growth.
Acknowledgements
This study was supported by NIH-NIDCR R01 grants DE-13414, DE-02009 and DE-15644 to J.M.O. We thank the Nanobiophysics Core Facility at the University of Southern California for use of the CD and fluorescence spectrometers.
Glossary
Abbreviations used in this paper
| ACP | amorphous calcium phosphate |
| CD | circular dichroism |
| DLS | dynamic light scattering |
| MW | molecular weight |
| OCP | octacalcium phosphate |
| PBS | phospate-buffered saline |
| rP148 | recombinant pig amelogenin 148 |
References
- Bouropoulos N., Moradian-Oldak J. Induction of apatite by the cooperative effect of amelogenin and the 32-kDa enamelin. J Dent Res. 2004;83:278–282. doi: 10.1177/154405910408300402. [DOI] [PubMed] [Google Scholar]
- Fan D., Du C., Sun Z., Lakshminarayanan R., Moradian-Oldak J. In vitro study on the interaction between the 32 kDa enamelin and amelogenin. J Struct Biol. 2009;166:88–94. doi: 10.1016/j.jsb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D., Lakshminarayanan R., Moradian-Oldak J. The 32 kDa enamelin undergoes conformational transitions upon calcium binding. J Struct Biol. 2008;163:109–115. doi: 10.1016/j.jsb.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson C.W., Yuan Z.A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J.T., Decker S., Piddington R., Harrison G., Kulkarni A.B. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Gutierrez S.J., Chaves M., Torres D.M., Briceno I. Identification of a novel mutation in the enamelin gene in a family with autosomal-dominant amelogenesis imperfecta. Arch Oral Biol. 2007;52:503–506. doi: 10.1016/j.archoralbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Hu C.C., Hu Y., Smith C.E., McKee M.D., Wright J.T., Yamakoshi Y., Papagerakis P., Hunter G.K., Feng J.Q., Yamakoshi F., Simmer J.P. Enamel defects and ameloblast-specific expression in Enam knock-out/lacZ knock-in mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.C., Yamakoshi Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 2003;14:387–398. doi: 10.1177/154411130301400602. [DOI] [PubMed] [Google Scholar]
- Iijima, M., D. Fan, K.M. Bromley, Z. Sun, J. Moradian-Oldak (2010) Tooth enamel proteins enamelin and amelogenin cooperate to regulate the growth morphology of octacacium phosphate crystals. Crystal Growth Des, in press. [DOI] [PMC free article] [PubMed]
- Iijima M., Moriwaki Y., Takagi T., Moradian-Oldak J. Effects of bovine amelogenins on the crystal morphology of octacalcium phosphate in a model system of tooth enamel formation. J Cryst Growth. 2001;222:615–626. [Google Scholar]
- Kida M., Ariga T., Shirakawa T., Oguchi H., Sakiyama Y. Autosomal-dominant hypoplastic form of amelogenesis imperfecta caused by an enamelin gene mutation at the exon-intron boundary. J Dent Res. 2002;81:738–742. doi: 10.1177/0810738. [DOI] [PubMed] [Google Scholar]
- Kwak S.Y., Wiedemann-Bidlack F.B., Beniash E., Yamakoshi Y., Simmer J.P., Litman A., Margolis H.C. Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro. J Biol Chem. 2009;284:18972–18979. doi: 10.1074/jbc.M109.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laksminarayanan, R., K.M. Bromley, Y.P. Lei, M.L. Snead, J. Moradian-Oldak (2010) Perturbed amelogenin secondary structure leads to uncontrolled aggregation in amelogenesis imperfecta mutant proteins. J Biol Chem, in press. [DOI] [PMC free article] [PubMed]
- Lakshminarayanan R., Fan D., Du C., Moradian-Oldak J. The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys J. 2007;93:3664–3674. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis H.C., Beniash E., Fowler C.E. Role of Macromolecular Assembly of Enamel Matrix Proteins in Enamel Formation. J Dent Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Goldberg M. Amelogenin supra-molecular assembly in vitro compared to the architecture of the forming enamel matrix. Cells Tissue Organs. 2005;181:202–218. doi: 10.1159/000091382. [DOI] [PubMed] [Google Scholar]
- Ozdemir D., Hart P.S., Firatli E., Aren G., Ryu O.H., Hart T.C. Phenotype of ENAM mutations is dosage-dependent. J Dent Res. 2005;84:1036–1041. doi: 10.1177/154405910508401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranath R.H., Moradian-Oldak J., Fincham A.G. Tyrosyl motif in amelogenins binds N-acetyl-D-glucosamine. J Biol Chem. 1999;274:2464–2471. doi: 10.1074/jbc.274.4.2464. [DOI] [PubMed] [Google Scholar]
- Sun Z., Ahsan M.M., Wang H., Du C., Abbott C., Moradian-Oldak J. Assembly and processing of an engineered amelogenin proteolytic product (rP148) Eur J Oral Sci. 2006;114(suppl 1):59–63. doi: 10.1111/j.1600-0722.2006.00296.x. (discussion 93–95, 379–380). [DOI] [PubMed] [Google Scholar]
- Yang X., Wang L., Qin Y., Sun Z., Henneman Z.J., Moradian-Oldak J., Nancollas G.H. How amelogenin orchestrates the organization of hierarchical elongated microstructures of apatite. J Phys Chem B. 2010;114:2293–2300. doi: 10.1021/jp910219s. [DOI] [PMC free article] [PubMed] [Google Scholar]