Abstract
Tooth development is regulated by epithelial-mesenchymal interactions and their reciprocal molecular signaling. Bone morphogenetic protein 2 (Bmp2) is essential for tooth formation. However, the role of Bmp2 during enamel formation remains unknown in vivo. In this study, the role of Bmp2 in the regulation of postnatal enamel formation was investigated via the conditional ablation of Bmp2 in enamel using the (Osx-Cre) mouse. Bmp2 gene ablation was confirmed by PCR analysis in Osx-Cre, Bmp2flox/flox mice. Bmp2-null mice displayed a severe and profound tooth phenotype with asymmetric and open forked incisors. Microradiographs revealed broken incisor tips and dental pulp chamber exposure. The enamel layer of incisors and molars was thin with hypomineralization. Scanning electron microscopy analysis showed that the enamel surface was rough with chipping and the enamel lacked a typical prismatic architecture. These results demonstrate that Bmp2 is essential for enamel formation.
Key Words: Bone morphogenetic protein 2, Conditional knockout, Enamel formation, Amelogenesis imperfecta
Introduction
Tooth development is a highly organized process involving sequential and reciprocal interactions between epithelial and mesenchymal cells. Enamel formation originates from ectodermal cells and proceeds through a series of cytodifferentiations in a spatial-temporal pattern [Linde and Goldberg, 1993]. Amelogenesis is a complex process in which multiple signaling pathways converge to induce enamel formation, and it is controlled by many growth factors and transcriptional factors [Thesleff, 2003]. The balance of stimulatory and inhibitory signals plays an important role in enamel formation.
Members of the bone morphogenetic protein (Bmp) family have diverse biological functions during osteogenesis and embryonic development [Hogan, 1996; Rosen, 2009]. Among the Bmp family members, Bmp2 has been extensively studied for its various biological functions during chondrogenic and osteogenic differentiation [Reddi, 1997; Ducy and Karsenty, 2000]. Bmp2 expression has been detected in epithelial and mesenchymal cells during tooth development [Aberg et al., 1997; Chen et al., 2008]. Bmp2 is able to induce ameloblast differentiation and enamel-related gene expression in vitro [Miyoshi et al., 2008]. These results indicate that Bmp2 is important for ameloblast differentiation and enamel formation. However, the role of Bmp2 in enamel development in vivo, in particular during postnatal enamel formation, has not been described as homozygous mutant embryos for Bmp2 show developmental abnormalities and die at embryonic day 9.5 [Zhang and Bradley, 1996]. Recently, conditional Bmp2 knockout (Bmp2-cKO) mice were generated, and they revealed a critical role of Bmp2 in later stages of osteogenesis [Bandyopadhyay et al., 2006] and bone fracture healing [Tsuji et al., 2006] as well as other forms of organ development [Ma et al., 2005; Lee et al., 2007; Castranio and Mishina, 2009].
In this study, we used an Osterix (Osx)-Cre model that activates Cre as Osx is actively expressed in ameloblasts and other dental tissues [Chen et al., 2009; Hirata et al., 2009]. We characterized the effect of Bmp2 ablation on mice and showed that Bmp2 ablation leads to abnormal enamel formation during postnatal stages.
Animals and Methods
Animals
All animal protocols were reviewed and approved by the Institute of Animal Care at the University of Texas Health Science Center at San Antonio. All animals were fed a soft diet (ClearH2O, Portland, Me., USA), and both Bmp2-cKO mice and Bmp2 control mice (wild type and heterozygote Bmp2) were used for these studies. A conditional allele of the mouse Bmp2 gene was created by introducing Cre recombinase recognition sites (loxP) flanking upstream and downstream of exon 3 of the Bmp2 gene [Ma et al., 2005]. The floxed Bmp2 mice were crossed with Osx-Cre mice (Jackson Laboratory, Bar Harbor, Me., USA) to generate Bmp2-cKO mice. Bmp2-cKO mice with a deletion of exon 3 were identified by PCR analysis using floxed Bmp2-specific primers. Genomic DNA was isolated from mouse tails using a DNA purification kit (Promega, Madison, Wisc., USA). The primers used for detecting the wild and mutant alleles were: forward, 5′-CGGGAACAGATACAGGAAGC-3′; reverse, 5′-GCTGTTTGTGTTTGGCTTGA-3′ (wt); forward, 5′-AGGGTTTCAGGTCAGTTTCCG-3′; reverse, 5′-GATGATGAGGTTCTTGGCGG-3′ (flox); forward, 5′-AGCATGAACCCTCATGTGTTG-3′, and reverse, 5′-GCTGTTTGTGTTTGGCTTGA-3′ (recombinant).
Microscopic Analysis of Teeth
Animals were anesthetized using ketamine (Sigma-Aldrich, St. Louis, Mo., USA). Tooth morphology was recorded under light microscopy. For histological examination, mandibles were fixed in 4% paraformaldehyde overnight. The samples were decalcified, embedded in paraffin, sectioned, and stained with hematoxylin/eosin. Tooth surfaces from control and Bmp-cKO mice were recorded by scanning electron microscopy (SEM) at 20 kV (JEOL JSM 6610 LV; JEOL, Inc., Peabody, Mass., USA).
X-Ray Analysis
Radiography was used to measure changes in teeth and bones. Maxillae and mandibles were radiographed using a Faxitron radiograph inspection unit (Faxitron X-Ray Corporation, Lincolnshire, Ill., USA). Digitized images were analyzed using AnalySIS software to measure the size and width of selected components in the incisors and molars. Tooth mineral density was measured using ImageJ (NIH, Bethesda, Md., USA).
Micro-Computed Tomography
Mandibles from Bmp2-cKO mice plus control mice were stored in 10% neutral buffered formalin at 4°C until processing. Samples were scanned at Numira, Inc. (Salt Lake City, Utah, USA) using a high-resolution scanner (Scanco Medical AG, Basserdorf, Switzerland), and the image data were acquired. The micro-computed tomography (micro-CT)-generated DICOM files were used to analyze the samples and to create volume and density renderings of the enamel of molars and incisors. The raw data files were viewed using Microview (GE Healthcare, Milwaukee, Wisc., USA).
Statistical Analysis
All values are presented as means ± standard deviation (SD). Data were analyzed using an unpaired Student t test. p < 0.05 was considered statistically significant.
Results
Gross Tooth Analysis of Bmp2-cKO Mice
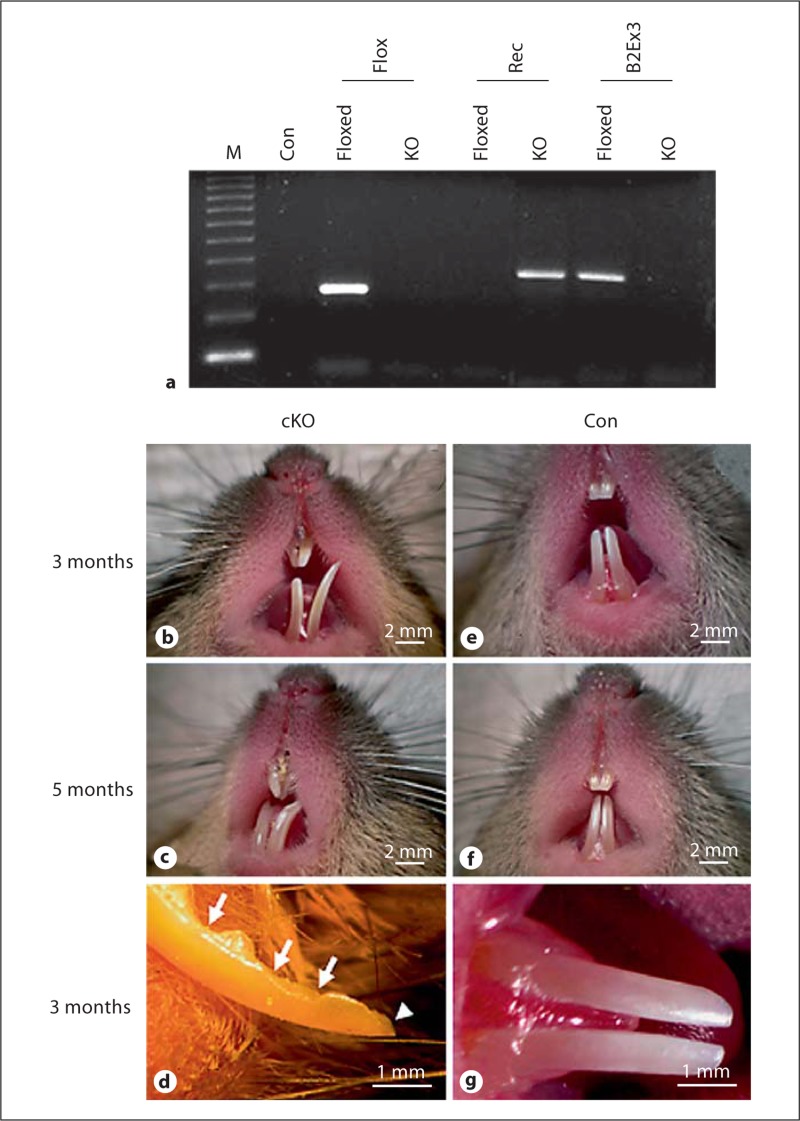
The Bmp2-cKO mice were identified during a large-scale screening for genotypes and phenotypes (fig. 1). The Bmp2-cKO mice were fertile, and newborns appeared normal. However, as early as 4 weeks of age, these mice were small in size compared to the control mice (data not shown). Gross morphological analysis of the teeth of Bmp2-cKO mice revealed small, asymmetric, open forked, and uneven incisors (fig. 1b, c). Noticeably, the incisal edge of the lower incisors was jagged and showed severe wearing, and incisor tips were broken (fig. 1d). In contrast, the incisors of the control mice were symmetric and presented as smooth and opalescent (fig. 1e-g).
Fig. 1.
Gross morphology of teeth. a Genotyping. Tail genomic DNA from floxed Bmp2 and Bmp2-cKO mice was isolated and amplified by PCR using Bmp2-specific primers. The PCR products were run on 1.5% agarose gels and stained with ethidium bromide. M = DNA ladder; Con = negative control. Flox, Rec, and B2Ex3 were flox, recombinant, and Bmp2 exon 3 primers used for PCR, respectively. b–g In the mutant mice, incisors were asymmetric and open forked (b, c). The incisor edge was jagged with chipping (arrows) and the incisor tip was broken (arrowhead) (d). In normal mice, the enamel appeared smooth and opalescent, and incisors were symmetric (e–g). Con = Control mice; cKO = Bmp2 conditional knockout mice.
Enamel Mineral Density and Layer Thickness in Bmp2-cKO Mice
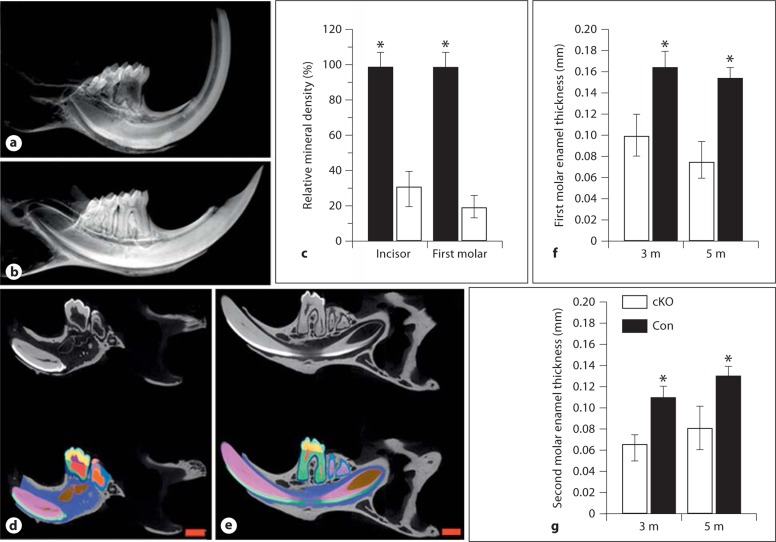
Radiography analysis revealed that the enamel of the Bmp2-cKO mice was thin and the enamel radiopacity decreased compared to the control mice (fig. 2a, b). The mineral density of teeth from Bmp2-cKO mice was reduced compared to that of the control mice (fig. 2c). By Faxitron analysis, incisor tips were broken and dental pulp chambers exposed in these mutant mice. The X-ray data were correlated with the micro-CT study. Quantitative micro-CT analysis demonstrated that the enamel layer was thin and there were significant differences between the control and Bmp2-cKO mice (fig. 2d, e). Mineralized enamel sections through the mandibular first and second molars from the null mice displayed an enamel thickness of 58 and 60% of normal enamel in 3-month-old mice and of 50 and 62% of normal enamel in 5-month-old mice, respectively (fig. 2f-g). Furthermore, micro-CT analysis revealed that the enamel density of incisors and molars was reduced (fig. 2d, e).
Fig. 2.
X-ray and micro-CT of teeth. X-ray analysis of the enamel in the Bmp2-cKO (a) and control (b) mice. The mineral density of incisors and molars from Bmp2-cKO mice was decreased compared to control mice (c). The mineral density of teeth from the control mice acts as 100%, and asterisks (∗) show significant differences between the control and Bmp2-cKO mice. The enamel layer was thin and the mineral density decreased in the 3-month-old Bmp2-null mice. Micro-CT analysis of teeth of the Bmp2-cKO (d) and control (e) mice. Light blue = Enamel thickness; yellow = mantle dentin; green = root dentin; red = dental pulp chamber; blue = alveolar bone. The mandibles from 3-month-old mice were subjected to micro-CT analysis by Numira, Inc. as described in Materials and Methods using parameters that were developed to measure the enamel width of the first and second molars. Significance was different between the control and Bmp2-cKO groups (f, g) (p < 0.05). The mineral density of the enamel from the null mice was reduced as compared to the control groups. m = Months. Scale bar = 1 mm.
Characterization of Null Mouse Enamel by SEM
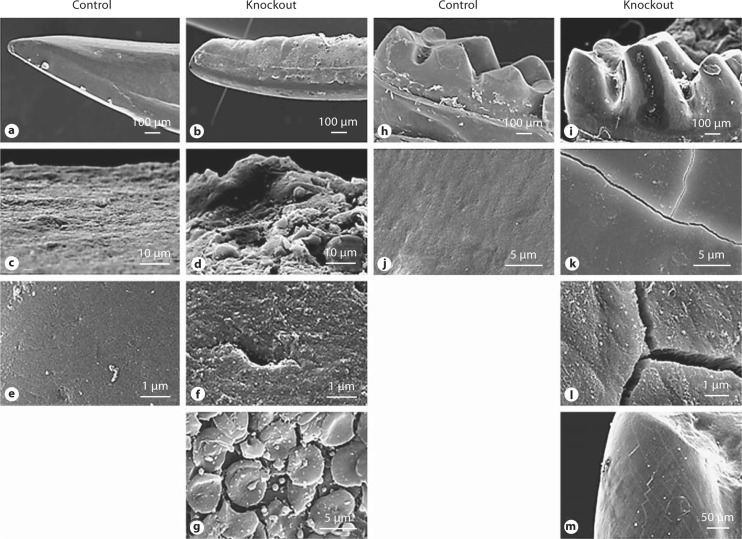
Mandibles from 3- and 5-month-old control and mutant mice were removed and incisors and molars were prepared for SEM. These results showed that incisor surfaces were rough, edges were jagged, and incisor tips were broken (fig. 3b). A higher magnification showed that incisor surfaces appeared cobbled, and the enamel had lost its typical prismatic architecture (fig. 3d, f, g) compared with that of the control group (fig. 3a, c, e). In the Bmp2-null mice, molar cusps appeared rough with abrasion (fig. 3i) and higher magnification photographs showed that the molar surface had fissures (fig. 3k-m) compared to that of the control mice (fig. 3h, j).
Fig. 3.
SEM analysis of teeth. a–g Incisors from 3-month-old control and Bmp2-null mice. The incisor surface was smooth in the control mice (a), whereas the incisor surface was rough and its edge was jagged in the mutant mice (b). Higher-resolution images showed that the incisor appeared cobbled and had an irregular prismatic pattern and ‘hole’ in Bmp2-cKO mice (d, f, g) compared to control mice (c, e). h–m Molars from 3-month-old control and mutant mice. The molar cusps in the mutant mice were rough with shallow grooves (i), whereas the molar surface was smooth in the control mice (h). The molar surface in the mutant mice had fissures (k–m) as compared to the control mice (j).
Discussion
In the present study, we investigated the role of Bmp2 in murine postnatal enamel formation by conditional mutant Bmp2 under the control of Osx-Cre. We found that Bmp2 is required for the specification of normal amelogenesis. Mice with a null Bmp2 gene lead to failure of normal enamel structure. This phenotype of teeth within the mutant mice shows a similar phenotype of amelogenin gene mutations in mice and humans [Lagerström et al., 1991; Gibson et al., 2001].
Bmp2 is expressed in dental epithelium [Aberg et al., 1997; Chen et al., 2008] and its signaling is involved in ameloblast differentiation and enamel marker gene expression in vitro [Miyoshi et al., 2008]. However, the role of Bmp2 in enamel formation in vivo has not been reported because mice with a traditionally targeted deletion of Bmp2 have embryonic lethality due to a failure of proamniotic canal closure or abnormal cardiac development, and they die at embryonic day 9.5 [Zhang and Bradley, 1996].
In order to investigate the role of Bmp2 in postnatal enamel in vivo, we crossed a line of mice in which the Cre recombinase gene was under the control of the Osx gene promoter with a mouse line containing the floxed Bmp2 allele [Ma et al., 2005]. This provides a tissue-specific Bmp2 knockout in dental epithelial cells because Osx is expressed in dental epithelial cells during tooth development [Chen et al., 2009; Hirata et al., 2009]. The phenotype we observed in the enamel of Bmp2-cKO, Osx-Cre mice exhibited severe defects in ameloblast differentiation and enamel mineralization as well as loss of normal enamel structure.
While many Bmp molecules besides Bmp2, including Bmp4 and Bmp7, are expressed during amelogenesis [Aberg et al., 1997], our data suggest that other Bmp molecules cannot compensate for loss of Bmp2 in enamel formation. Mechanisms for the role of Bmp2 in enamel formation are not completely understood. Numerous factors have been shown to be important in enamel development including Dlx2, NF-Y, C/EBP, LEF-1 transcriptional factors, and amelogenin [Xu et al., 2006; Lézot et al., 2008; Tian et al., 2010]. Amelogenin is a highly conserved protein that accounts for 90% of the enamel organic matrix. Mutations of amelogenin in humans and mice are associated with amelogenesis imperfecta, the most common enamel genetic disease [Hart et al., 2002]. Each of these transcriptional factors is essential for amelogenin gene transcription [Xu et al., 2006; Lézot et al., 2008; Tian et al., 2010]. Importantly, Bmp2 upregulates those gene expressions [Harris et al., 2003; Fan et al., 2009; Hoeppner et al., 2009]. In addition, Bmp2 controls the expression of the second most abundant extracellular matrix protein, i.e. ameloblastin, mediated by the Runx2 signaling pathway [Dhamija and Krebsbach, 2001]. Mutations of the ameloblastin gene in humans and mice also cause amelogenesis imperfecta [Mårdh et al., 2001; Hu et al., 2008]. This indicates that Bmp2 has diverse biological functions in controlling enamel gene expression and enamel formation.
In summary, our findings reveal that Bmp2 is required for normal enamel development and mineralization. In future studies, we will elucidate molecular mechanisms of Bmp2 signaling in controlling enamel gene expression in vivo.
Acknowledgements
This work was supported by NIH grant DE019892 (S.C.) and by the San Antonio Area Foundation (S.C.).
Glossary
Abbreviations used in this paper
| Bmp | bone morphogenic protein |
| Bmp2-cKO | conditional Bmp2 knockout |
| micro-CT | micro-computed tomography |
| Osx | osterix |
| SEM | scanning electron microscopy |
References
- Aberg T., Wozney J., Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev Dyn. 1997;210:383–396. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., Tsuji K., Cox K., Harfe B.D., Rosen V., Tabin C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castranio T., Mishina Y. Bmp2 is required for cephalic neural tube closure in the mouse. Dev Dyn. 2009;238:110–122. doi: 10.1002/dvdy.21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Gluhak-Heinrich J., Martinez M., Li T., Wu Y., Chuang H.H., Chen L., Dong J., Gay I., MacDougall M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J Biol Chem. 2008;283:19359–19370. doi: 10.1074/jbc.M709492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Gluhak-Heinrich J., Wang Y.H., Wu Y.M., Chuang H.H., Chen L., Yuan G.H., Dong J., Gay I., MacDougall M. Runx2, osx, and dspp in tooth development. J Dent Res. 2009;88:904–909. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamija S., Krebsbach P.H. Role of Cbfa1 in ameloblastin gene transcription. J Biol Chem. 2001;276:35159–35164. doi: 10.1074/jbc.M010719200. [DOI] [PubMed] [Google Scholar]
- Ducy P., Karsenty G. The family of bone morphogenetic proteins. Kidney Int. 2000;57:2207–2214. doi: 10.1046/j.1523-1755.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Fan Q., Tang T., Zhang X., Dai K. The role of CCAAT/enhancer binding protein (C/EBP)-alpha in osteogenesis of C3H10T1/2 cells induced by BMP-2. J Cell Mol Med. 2009;13:2489–2505. doi: 10.1111/j.1582-4934.2008.00606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson C.W., Yuan Z.A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J.T., Decker S., Piddington R., Harrison G., Kulkarni A.B. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Harris S.E., Guo D., Harris M.A., Krishnaswamy A., Lichtler A. Transcriptional regulation of BMP-2 activated genes in osteoblasts using gene expression microarray analysis: role of Dlx2 and Dlx5 transcription factors. Front Biosci. 2003;8:s1249–s1265. doi: 10.2741/1170. [DOI] [PubMed] [Google Scholar]
- Hart P.S., Hart T.C., Simmer J.P., Wright J.T. A nomenclature for X-linked amelogenesis imperfecta. Arch Oral Biol. 2002;47:255–260. doi: 10.1016/s0003-9969(02)00005-5. [DOI] [PubMed] [Google Scholar]
- Hirata A., Sugahara T., Nakamura H. Localization of runx2, osterix, and osteopontin in tooth root formation in rat molars. J Histochem Cytochem. 2009;57:397–403. doi: 10.1369/jhc.2008.952192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner L.H., Secreto F., Jensen E.D., Li X., Kahler R.A., Westendorf J.J. Runx2 and bone morphogenic protein 2 regulate the expression of an alternative Lef1 transcript during osteoblast maturation. J Cell Physiol. 2009;221:480–489. doi: 10.1002/jcp.21879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B.L. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Hu J.C.C., Hu Y., Smith C.E., McKee M.D., Wright J.T., Yamakoshi Y., Papagerakis P., Hunter G.K., Feng J.Q., Yamakoshi F., Simmer J.P. Enamel defects and ameloblast-specific expression in Enam knock-out/lacZ knock-in mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerström M., Dahl N., Nakahori Y., Nakagome Y., Bäckman B., Landegren U., Pettersson U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- Lee K.Y., Jeong J.W., Wang J., Ma L., Martin J.F., Tsai S.Y., Lydon J.P., DeMayo F.J. Bmp2 is critical for the murine uterine decidual response. Mol Cell Biol. 2007;27:5468–5478. doi: 10.1128/MCB.00342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lézot F., Thomas B., Greene S.R., Hotton D., Yuan Z.A., Castaneda B., Bolaños A., Depew M., Sharpe P., Gibson C.W., Berdal A. Physiological implications of DLX homeoproteins in enamel formation. J Cell Physiol. 2008;216:688–697. doi: 10.1002/jcp.21448. [DOI] [PubMed] [Google Scholar]
- Linde A., Goldberg M. Dentinogenesis. Crit Rev Oral Biol Med. 1993;4:679–728. doi: 10.1177/10454411930040050301. [DOI] [PubMed] [Google Scholar]
- Ma L., Lu M.F., Schwartz R.J., Martin J.F. Bmp2 is essential for cardiac cushion epithelial mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Mårdh C.K., Bäckman B., Simmons D., Golovleva I., Gu T.T., Holmgren G., MacDougall M., Forsman-Semb K. Human ameloblastin gene: genomic organization and mutation analysis in amelogenesis imperfecta patients. Eur J Oral Sci. 2001;109:8–13. doi: 10.1034/j.1600-0722.2001.00979.x. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Nagata H., Horiguchi T., Abe K., Arie Wahyudi I., Baba Y., Harada H., Noma T. BMP2-induced gene profiling in dental epithelial cell line. J Med Invest. 2008;55:216–226. doi: 10.2152/jmi.55.216. [DOI] [PubMed] [Google Scholar]
- Reddi A.H. Bone morphogenetic proteins: an unconventional approach to isolation of first mammalian morphogens. Cytokine Growth Factor Rev. 1997;8:11–20. doi: 10.1016/s1359-6101(96)00049-4. [DOI] [PubMed] [Google Scholar]
- Rosen V. BMP2 signaling in bone development and repair. Cytokine Growth Factor Rev. 2009;20:475–480. doi: 10.1016/j.cytogfr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci. 2003;116:1647–1648. doi: 10.1242/jcs.00410. [DOI] [PubMed] [Google Scholar]
- Tian H., Lv P., Ma K., Zhou C., Gao X. Beta-catenin/LEF1 activated enamelin expression in ameloblast-like cells. Biochem Biophys Res Commun. 2010;398:519–524. doi: 10.1016/j.bbrc.2010.06.111. [DOI] [PubMed] [Google Scholar]
- Tsuji K., Bandyopadhyay A., Harfe B.D., Cox K., Kakar S., Gerstenfeld L., Einhorn T., Tabin C.J., Rosen V. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38:1424–1429. doi: 10.1038/ng1916. [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhou Y.L., Luo W., Zhu Q.S., Levy D., MacDougald O.A., Snead M.L. NF-Y and CCAAT/enhancer-binding protein alpha synergistically activate the mouse amelogenin gene. J Biol Chem. 2006;281:16090–16098. doi: 10.1074/jbc.M510514200. [DOI] [PubMed] [Google Scholar]
- Zhang H., Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977–2986. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]